Abstract
Standard polymerase chain reaction methods often cannot detect drug-resistance mutations in Plasmodium falciparum infections if the mutation is present in ≤ 20% of the parasites. A heteroduplex tracking assay was developed that can detect dihydrofolate reductase 164-L mutations in variants representing 1% of the parasites in an individual host. Using this assay, we confirmed the presence of the mutation in P. falciparum infections in Malawi.
Drug resistance to sulfadoxine-pyrimethamine (SP) is widespread in Malawi and is conferred by mutations in dihydrofolate reductase (DHFR) and dihydropteroate synthase (DHPS).1–4 The combination of five mutations (DHFR-51, -59, and -108, and DHPS-437 and -540) has been strongly associated with SP failure.5,6 Other mutations, such as DHFR-164, DHPS-581, and DHPS-613, develop later and sequentially add to SP resistance.7–9 In addition, mutations in DHFR-164 can confer resistance to pyrimethamine and chlorproguanil.7,10 This mutation is particularly important because its presence could limit the usefulness of the new artemisinin combination therapy chlorproguanil-dapsone-artesunate, which is currently under development.11
Plasmodium falciparum infections are almost always mixtures of genetically distinct variants12 (Juliano J. J. and others, unpublished data). The DHFR-164 mutations are usually measured using a conventional polymerase chain reaction (PCR) that cannot detect minority variants.13–15 In this report, we report a new heteroduplex tracking assay (HTA) for DHFR-164 that can detect minority variants. We use this method to screen patients and characterize previously reported mutant DHFR-164 isolates from Malawi.2
Clinical samples from two cohorts in Malawi were evaluated by HTA. First, we screened the 85 pregnant women in labor who came to the Queen Elizabeth Central Hospital in Blantyre who had originally been tested by real-time PCR.2 Second, we tested 52 samples from an intermittent preventive therapy (IPTp) trial that took place at rural health clinics in Mpemba and Madziabango.16 Among the patients from Mpemba and Madziabango, only samples from the initial visit were used. Therefore, the women had not received any IPTp at the time of collection. Informed consent, as approved by the ethics committees of the University of North Carolina and the Malawi College of Medicine/Ministry of Health, was obtained form all participants in this research study. The clinical characteristics of the patient samples have been described previously.2,13,16,17
DNA was extracted from filter paper blood spots or frozen blood using the Qiamp DNA minikit (Qiagen, Valencia, CA). The method for developing HTAs for the detection of point mutations in malaria has been previously described, and the differences for the DHFR-164 HTA are noted below.13 Briefly, wild-type DHFR is amplified at the region of interest. The PCR was carried out in a 50-μL reaction containing 5 μL of DNA, 1.25 units of HotStar Taq DNA polymerase (Qiagen), 5 μL of 10× PCR buffer, 1 μL of dNTP mixture (catalog no. U1511; Promega, Madison, WI), 300 nm forward primer (5′-ATCATTAACAAAGTTGAAGATCTAATAGTTTTAC-3′), and 500 nM reverse primer (5′-TCGCTAACAGAAATAATTTGATACTCAT-3′).2 This reaction was amplified by preheating to 95°C for 15 minutes, followed by 35 cycles of 95°C for 30 seconds, 50°C for 30 seconds, and 72°C for 1 minute. The reaction was completed with a 10-minute hold at 72°C. The PCR products were cloned into a PCR2.1 TOPO plasmid as described in the protocol of the Topo TA cloning kit (In-vitrogen, Carlsbad, CA), and mutations were randomly introduced into the construct at the −3, −1, +1, and +3 nucleotides relative to the single nucleotide polymorphism using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, LaJolle, CA).13 Screening of probes, probe construction, and labeling of HTA probes were done per the methods previously described.13,18 The successful probe had the following mutations: +1 T→G and +3 G→T.
The HTAs were performed by a modification of the method of Ngrenngarmlert and others.18 An annealing reaction consisting of 8 μL of PCR product (either a control or sample DNA) was mixed with 1 μL of 10× annealing buffer (1 M NaCl, 100 mM Tris-HCl, pH 7.5, 20 mM EDTA), 2 μL of 6× loading dye, 0.25 μL of 100 pM forward primer, 0.25 μL of 100 pM reverse primer, and 0.5 μL of radiolabeled probe in a total volume of 12 μL. All HTA gels included the following controls: water, a non-template control PCR, and PCRs from wild-type (MR4, MRA-102G) and mutant (MR4, MRA-176G) DNA stocks. Gels were analyzed as previously described.13
The sensitivity of the DHFR-164 HTA to detect minority variant drug-resistant parasites was tested using artificial mixtures of wild-type and mutant genomic P. falciparum DNA.14 These mixtures were at a final total concentration of 0.1 ng of DNA per microliter and all tests were done in duplicate. The HTA accurately and reproducibly detected mutant variants comprising as little as 1% of the total parasite population (Table 1).
Table 1.
Sensitivity of the dihydrofolate reductase–164 heteroduplex tracking assay for minority variant drug-resistant parasites
| % Mutant control DNA | % Mutant detected (replicate 1) | % Mutant detected (replicate 2) |
|---|---|---|
| 50 | 47.9 | 42.1 |
| 20 | 22.3 | 16.3 |
| 10 | 9.3 | 11.1 |
| 5 | 5.9 | 6.9 |
| 1 | 0.8 | 2.2 |
| 0.1 | 0 | 0 |
Of the 90 patient samples from Blantyre previously genotyped by Alker and others, 85 samples were available to be re-evaluated.2 The other samples were missing or depleted. Of these, 77 (91%) were successfully genotyped by the DHFR-164 HTA. The DHFR-164 mutant variants were found in all four samples previously found to have the DNFR-164 mutation by real-time PCR.2 No other DHFR-164 mutants were found.
All four samples were found to be mixed infections by HTA. The relative amount of DHFR-164–bearing parasites was determined for each sample by Phosphorimager evaluation of the gels.13 The samples contained the following amounts of DHFR-164 mutant parasites: patient 1249 (32.8%, Figure 1, lanes I and J), patient 1367 (89.4%, lanes K and L), patient 1628 (20.6%, lanes M and N), and patient 1238 (11.4%, lanes O and P). The isolate from patient 1367 was originally identified as pure mutant DHFR-164 by real-time PCR.2 Thus, the DHFR-164 mutation is present only in patients with wild-type parasites.
Figure 1.
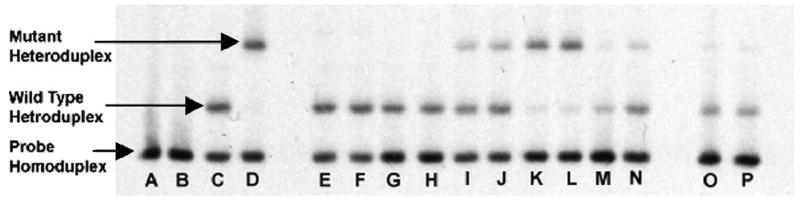
Dihydrolate reductase (DHFR)–164 heteroduplex tracking assay of malaria isolates from clinical samples. Lanes A–D, controls (water, non-template control [NTC], wild-type DHFR-164 DNA, and mutant DHFR-164 DNA, respectively). Each clinical sample is done in duplicate: Lanes E and F, patient 1458 (0% mutant); lanes G and H, patient 1468 (0% mutant); lanes I and J, patient 1249 (32.8% mutant); lanes K and L, patient 1367 (89.4% mutant); lanes M and N, patient 1628 (20.6% mutant); lanes O and P, patient 1238 (11.4% mutant).
Parasite DNA from patients 1249 and 1628 were TA-cloned per protocol (Invitrogen). Twenty-five colonies from each of these samples were screened by colony real-time PCR.2,13 From both samples, a selection of 3 mutant and 2 wild-type plasmids isolated from these colonies was then sequenced at the University of North Carolina-Chapel Hill Automated DNA Sequencing Facility. Sequencing confirmed that the three putative mutant clones were mutant and that the two putative wild-type clones were wild type. The presence of mutant DHFR-164 was previously confirmed by sequencing in patient 1367.2
The results of the HTAs on the other 73 isolates from Queen Elizabeth Central Hospital were all pure wild-type. All 45 of the patient samples from Mpemba and Madziabango that were successfully amplified were also pure wild type at DHFR-164. All of these latter samples were also genotyped by real-time PCR and found to be wild type by this method. Thus, in total between the two cohorts, we detected mutant DHFR-164 in 4 (3.3%) of 122 successfully amplified samples by HTA and real-time PCR.
Mutations at other sites in DHFR (DHFR-51, -59, and -108) are common in Malawi. Among the malaria and human immunodeficiency virus in pregnancy (MHP) samples evaluated, 63 contained parasites that were genotypically pure mutant at all three other DHFR sites by real-time PCR, including samples 1458 and 1468 (Figure 1, lanes E–H).1 In addition, there were 10, 6, and 2 MHP samples containing either pure wild-type or mixed genotype parasites at DHFR-51, -59, and -108, respectively. Because the forward primer of the DHFR-164 HTA lies between the DHFR-108 and DHFR-164 mutation, the assay is highly specific for the detected mutation. None of the samples containing other DHFR mutations migrated differently, or contained additional heteroduplexes compared with the control DNA from 3d7 (genotypically wild type at all three sites).
Thus, this report confirms the presence of the DHFR-164L mutation in Malawi by a second assay method as well as DNA sequencing. We also demonstrate 100% concordance for the presence of mutant DHFR-164 between the HTA and real-time PCR assay.
The DHFR-164 mutation remains rare in Africa, but has been documented in Central African Republic, Kenya, Uganda, the Comoros, Tanzania, and among travelers returning to Sweden from Africa.14,15,19–23 These observations, together with the data presented here, suggest that DHFR-164L is a problem in Africa.24 Thus, monitoring for the DHFR-164 mutation should be continued. The ability of the HTA to detect DHFR-164L–bearing variants representing ≥ 1% of the parasites in an individual host suggest that the method is more sensitive than standard PCR methods and should be used if proof of the mutation’s absence is needed.
The presence of this mutation in only one of the two cohorts studied here could serve as an important lesson. The samples from Mpemba and Madziabango were taken early in pregnancy, before subjects received SP IPTp. The samples from Queen Elizabeth Central Hospital were obtained at labor and delivery, after the women had probably received IPTp. This suggests that the difference might be in part caused by in vivo selection for the DHFR-164L genotype. In vivo selection of drug resistance mutations has been seen for other dhfr/dhps mutations as well as in other genes in malaria parasites.25,26 Second, this study and all previous studies of dhfr/dhps mutations in Malawi used conveniently available samples, not samples that were collected in a manner representative of the general population. An accurate assessment of the prevalence of this mutation can only be done with a true population-based survey.
Acknowledgments
We are grateful to Ella Nkhoma and Alisa Alker for performing the real-time PCR on clinical samples.
Financial support: This study was supported by ASPH/CDC S1935-21/21, NIAID/NIH AI49804, Infectious Disease Pathogenesis Research Training grant (DHHS/NIH/NIAID 5 T32 AI07151-29), and the 2007 IDSA ERF/NFID Merle A. Sande/Pfizer Fellowship in International Infectious Diseases. These agencies had no involvement in the design, collection, analysis, or interpretation of data in this study or in writing this paper or submitting it for publication.
References
- 1.Alker A, Mwapasa V, Meshnick SR. Rapid real-time PCR genotyping of mutations associated with sulfadoxine-pyrimethamine resistance in Plasmodium falciparum. Antimicrob Agents Chemother. 2004;48:2924–2929. doi: 10.1128/AAC.48.8.2924-2929.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alker A, Mwapasa V, Purfield A, Rogerson SJ, Molyneux M, Kamwendo D, Tadesse E, Chaluluka E, Meshnick SR. Mutations Associated with sulfadoxine-pyrimethamine and chlorproguanil resistance in Plasmodium falciparum isolates from Blantyre, Malawi. Antimicrob Agents Chemother. 2005;49:3919–3921. doi: 10.1128/AAC.49.9.3919-3921.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bwijo B, Kaneko A, Takechi M, Zungu IL, Moriyama Y, Lum LK, Tsukahara T, Mita T, Takahashi N, Bergqvist Y, Bjorkman A, Kobayakawa T. High Prevalence of quintuple mutant dhfr/dhps genes in Plasmodium falciparum infections seven years after introduction of sulfadoxine and pyrimethamine as first line treatment in Malawi. Acta Trop. 2003;85:363–373. doi: 10.1016/s0001-706x(02)00264-4. [DOI] [PubMed] [Google Scholar]
- 4.Nkhoma S, Molyneux M, Ward S. Molecular surveillance for drug-resistant Plasmodium falciparum malaria in Malawi. Acta Trop. 2007;102:138–142. doi: 10.1016/j.actatropica.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Kublin JG, Dzinjalamala FK, Kamwendo DD, Malkin EM, Cortese JF, Martino LM, Mukadam RA, Rogerson SJ, Lescano AG, Molyneux ME, Winstanley PA, Chimpeni P, Taylor TE, Plowe CV. Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. J Infect Dis. 2002;185:380–388. doi: 10.1086/338566. [DOI] [PubMed] [Google Scholar]
- 6.Plowe CV. Folate antagonists and mechanisms of resistance. In: Rosenthal PJ, editor. Antimalarial Chemotherapy: Mechanisms of Action, Resistance, and New Directions in Drug Discovery. Totowa, NJ: Humana Press; 2001. pp. 173–190. [Google Scholar]
- 7.Foote SJ, Galatis D, Cowman AF. Amino acids in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum involved in cycloguanil resistance differ from those involved in pyrimethamine resistance. Proc Natl Acad Sci USA. 1990;87:3014–3017. doi: 10.1073/pnas.87.8.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plowe CV, Cortese JF, Djimde A, Nwanyanwu OC, Watkins WM, Winstanley PA, Estrada-Franco JG, Mollinedo RE, Avila JC, Cespedes JL, Carter D, Doumbo OK. Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase and epidemiologic patterns of pyrimethamine-sulfadoxine use and resistance. J Infect Dis. 1997;176:1590–1596. doi: 10.1086/514159. [DOI] [PubMed] [Google Scholar]
- 9.Sirawaraporn W, Sathitkul T, Sirawaraporn R, Yuthavong Y, Santi DV. Antifolate-resistant mutants of Plasmodium falciparum dihydrofolate reductase. Proc Natl Acad Sci USA. 1997;94:1124–1129. doi: 10.1073/pnas.94.4.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krudsood S, Imwong M, Wilairatana P, Purkittayakamee S, Nonprasert A, Snounou G, White NJ, Looaresuwan S. Artesunate-dapsone-proguanil treatment of falciparum malaria: Genotypic determinants of therapeutic response. Trans R Soc Trop Med Hyg. 2005;99:142–149. doi: 10.1016/j.trstmh.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Winstanley P. Chlorproguanil-dapsone (LAPDAP) for uncomplicated falciparum malaria. Trop Med Int Health. 2001;6:952–954. doi: 10.1046/j.1365-3156.2001.00751.x. [DOI] [PubMed] [Google Scholar]
- 12.Kwiek JJ, Alker A, Wenink E, Chaponda M, Kalilani LV, Meshnick SR. Estimating true antimalarial efficacy by hetero-duplex tracking assay in patients with complex Plasmodium falciparum infections. Antimicrob Agents Chemother. 2007;51:521–527. doi: 10.1128/AAC.00902-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Juliano JJ, Kwiek JJ, Cappell K, Mwapasa V, Meshnick SR. Minority-variant pfcrt K76T mutations and chloroquine resistance, Malawi. Emerg Infect Dis. 2007;13:873–877. doi: 10.3201/eid1306.061182. [DOI] [PubMed] [Google Scholar]
- 14.Farnert A, Tengstam K, Palme IB, Bronner U, Lebbad M, Swedberg G, Bjorkman A. Polyclonal Plasmodium falciparum malaria in travelers and selection of antifolate mutations after proguanil prophylaxis. Am J Trop Med Hyg. 2002;66:487–491. doi: 10.4269/ajtmh.2002.66.487. [DOI] [PubMed] [Google Scholar]
- 15.Staedke SG, Sendagire H, Lamola S, Kamya MR, Dorsey G, Rosenthal PJ. Relationship between age, molecular markers, and response to sulphadoxine-pyrimethamine treatment in Kampala, Uganda. Trop Med Int Health. 2004;9:624–629. doi: 10.1111/j.1365-3156.2004.01239.x. [DOI] [PubMed] [Google Scholar]
- 16.Kalilani L, Mofolo I, Chaponda M, Rogerson SJ, Alker AP, Kwiek JJ, Meshnick SR. A randomized controlled pilot trial of azithromycin or artesunate added to sulfadoxine-pyrimethamine as treatment for malaria in pregnant women. PLoS ONE. 2007;2:e1166. doi: 10.1371/journal.pone.0001166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mwapasa V, Rogerson SJ, Molyneux ME, Abrams ET, Kamwendo DD, Lema VM, Tadesse E, Chaluluka E, Wilson PE, Meshnick SR. The effect of Plasmodium falciparum malaria on peripheral and placental HIV-1 RNA concentrations in pregnant Malawian women. AIDS. 2004;18:1051–1059. doi: 10.1097/00002030-200404300-00014. [DOI] [PubMed] [Google Scholar]
- 18.Ngrenngarmlert W, Kwiek JJ, Kamwendo DD, Ritola K, Swanstrom R, Wongsrichanalai C, Miller RS, Ittarat W, Meshnick SR. Measuring allelic heterogeneity in Plasmodium falciparum by heteroduplex tracking assay. Am J Trop Med Hyg. 2005;72:694–701. [PubMed] [Google Scholar]
- 19.Hastings MD, Bates SJ, Blackstone EA, Monks SM, Mutabingwa TK, Sibley CH. Highly pyrimethamine-resistant alleles of dihydrofolate reductase in isolates of Plasmodium falciparum from Tanzania. Trans R Soc Trop Med Hyg. 2002;96:674–676. doi: 10.1016/s0035-9203(02)90349-4. [DOI] [PubMed] [Google Scholar]
- 20.Kamya MR, Gasasira AF, Achan J, Mebrahtu T, Ruel T, Kekitiinwa A, Charlebois ED, Rosenthal PJ, Havlir D, Dorsey G. Effects of trimethoprim-sulfamethoxazole and insecticide treated bednets on malaria among HIV-infected Ugandan children. AIDS. 2007;21:2059–2066. doi: 10.1097/QAD.0b013e3282ef6da1. [DOI] [PubMed] [Google Scholar]
- 21.Maiga O, Djimde AA, Hubert V, Renard E, Aubouy A, Kironde F, Nsimba B, Koram K, Doumbo OK, Le Bras J, Clain J. A shared Asian origin of the triple-mutant dhfr allele in Plasmodium falciparum from sites across Africa. J Infect Dis. 2007;196:165–172. doi: 10.1086/518512. [DOI] [PubMed] [Google Scholar]
- 22.McCollum AM, Poe AC, Hamel M, Huber C, Zhou Z, Shi YP, Ouma P, Vulule J, Bloland P, Slutsker L, Barnwell JW, Udhayakumar V, Escalante AA. Antifolate resistance in Plasmodium falciparum: Multiple origins and identification of novel dhfr alleles. J Infect Dis. 2006;194:189–197. doi: 10.1086/504687. [DOI] [PubMed] [Google Scholar]
- 23.Menard D, Djalle D, Yapou F, Manirakiza A, Talarmin A. Frequency distribution of antimalarial drug-resistant alleles among isolates of Plasmodium falciparum in Bangui, Central African Republic. Am J Trop Med Hyg. 2006;74:205–210. [PubMed] [Google Scholar]
- 24.Nzila A, Ochong E, Nduati E, Gilbert K, Winstanley P, Ward S, Marsh K. Why has the dihydrofolate reductase 164 mutation not consistently been found in Africa yet? Trans R Soc Trop Med Hyg. 2005;99:341–346. doi: 10.1016/j.trstmh.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Jelinek T, Kilian AH, Curtis J, Duraisingh MT, Kabagambe G, von Sonnenburg F, Warhurst DC. Plasmodium falciparum: selection of serine108 of dihydrofolate reductase during treatment of uncomplicated malaria with co-trimoxazole in Ugandan children. Am J Trop Med Hyg. 1999;61:125–130. doi: 10.4269/ajtmh.1999.61.125. [DOI] [PubMed] [Google Scholar]
- 26.Sisowath C, Stromberg J, Martensson A, Msellem M, Obondo C, Bjorkman A, Gil JP. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem) J Infect Dis. 2005;191:1014–1017. doi: 10.1086/427997. [DOI] [PubMed] [Google Scholar]


