Abstract
The Gli code hypothesis postulates that the three vertebrate Gli transcription factors act together in responding cells to integrate intercellular Hedgehog (Hh) and other signaling inputs, resulting in the regulation of tissue pattern, size and shape. Hh and other inputs are then just ways to modify the Gli code. Recent data confirm this idea and suggest that the Gli code regulates stemness and also tumor progression and metastatic growth, opening exciting possibilities for both regenerative medicine and novel anticancer therapies.
Introduction
How organs develop to attain specific adult sizes and shapes and then maintain them throughout life is not fully understood. Similarly, how closely related species can vary in size is not clear. These evolutionarily plastic mechanisms are thought to have a basis in the control of cell behavior and much effort has been devoted over the last three decades to elucidating them. Interestingly, pathological changes in cell number, such as in cancer, seem to result from abnormal tissue and organ patterning, indicating that morphogenesis, homeostasis and cancer are intimately interconnected.
A handful of intercellular signaling pathways are known to operate in most instances of animal patterning. They include the Hedgehog (Hh)-Gli, Notch, Wnt, epidermal growth factor (EGF), fibroblast growth factor (FGF), insulin-like growth factor (IGF) and bone morphogenetic protein/transforming growth factor β (BMP/TGFβ) pathways as well as other peptide growth factor-tyrosine kinase receptor intracellular signaling cascades. Of these, Hh-Gli signaling has crucial roles in development, as well as in stemness and cancer [1] and is the focus of this review. Secreted Hh ligands act on responding cells by turning on an intracellular signaling pathway (Figure 1) that ultimately induces activating and inhibits repressive activities of the Gli transcription factors. Hh signaling thus regulates the combinatorial and cooperative function of the Gli proteins, which together form the Gli code [2,3].
Figure 1.
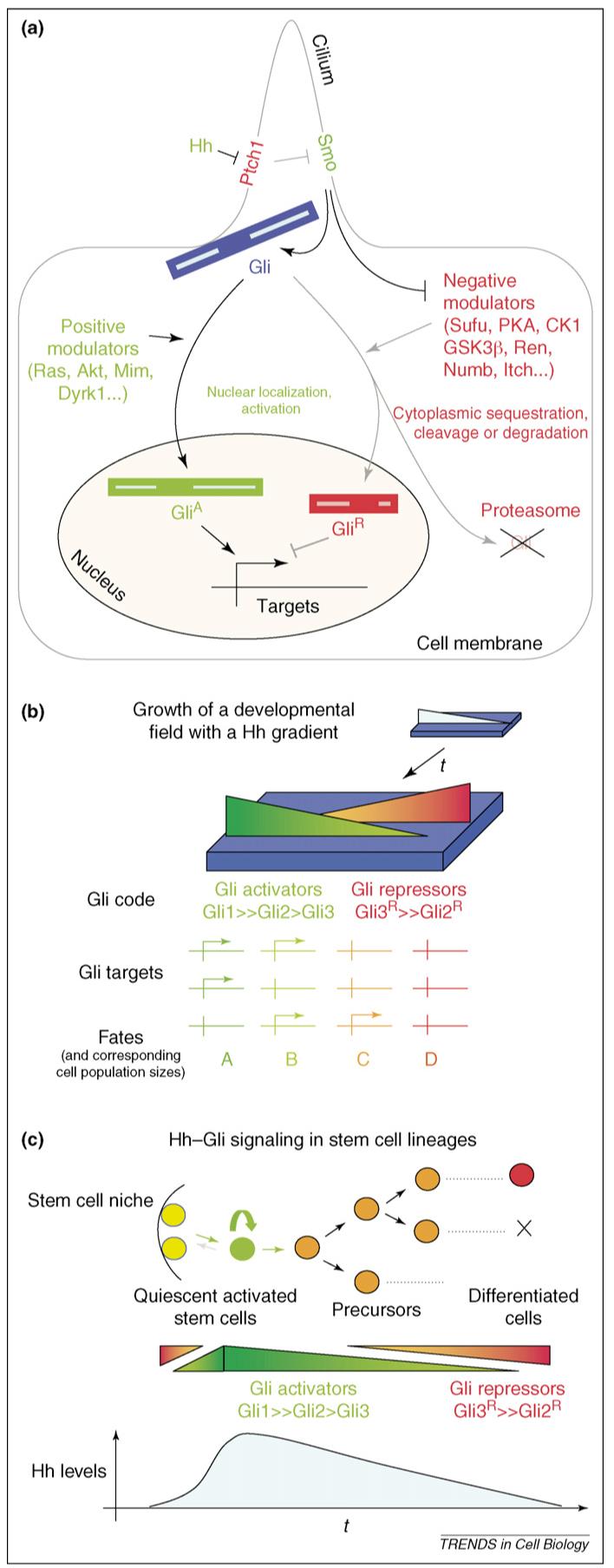
The Hh-Gli pathway and the Gli code in tissue patterning and stem-cell lineages. (a) Representation of the Gli code acting downstream of Hh-Smo signaling. On inhibition of Ptch1 function by the Hh ligands, the repression of Smo by Ptch1 is ended. Smo then orchestrates changes in the Gli code by stabilizing and activating full-length Gli proteins (GliA) and blocking the action of inhibitors and the production of Gli repressors (GliR), mostly Gli3 in this case. Positive modulators enhance the nuclear localization and the activation of full-length Gli proteins. In the absence of Hh ligands, Ptch1 inhibits Smo and negative modulators sequester full-length Gli proteins in the cytoplasm and target them for cleavage to produce C-terminal truncations (which act as constitutive nuclear repressors) or for complete degradation by the proteasome. Examples of positive and negative modulators are given here and in the text. Note that reception of the Hh signal and the orchestration of the Gli code are proposed to occur in the primary cilium [100,102]. (b) Model of a Hh gradient in a developmental field (top) that grows and becomes patterned through the action of inverse gradients of Gli activators (green) and repressors (red). Different combinations of the Gli code are proposed to activate overlapping sets of targets that result in distinct fates and proliferation/survival rates and thus sizes of tissues and organs. (c) Model of a stem-cell lineage with the activation of stem cells from quiescent cells in the niche by an upsurge of Hh signals. This leads to self-renewal and the production of derived precursors and differentiated cells. The temporal gradient of Hh is interpreted by varying levels of activators and repressors of Gli proteins, leading to a changing Gli code.
Gli proteins: context-dependent transcriptional regulators acting in a combinatorial and cooperative fashion
The three Gli proteins are zinc-finger transcription factors of >1000 amino acids that encode both activator and repressor functions. In frogs, fish, mice and humans, Gli1 is a strong transcriptional activator; Gli2 has both activator and repressor functions; and Gli3 is mostly a repressor, although it can also have positive effects [4-10]. The fly Gli protein, Cubitus interruptus (Ci), is also both an activator and a repressor [11,12]. Full-length Gli proteins, possibly modified, encode strong activator function, whereas repressor activity is found mostly in C-terminally deleted forms of Gli2 and Gli3 processed by proteasome-mediated cleavage (see, for example, Refs [11-15]; Figure 1).
In the absence of Hh signaling, Gli1 is transcriptionally silent but Gli2 and Gli3 can be expressed [4,7-9]. In this case, Gli2 is not an activator and Gli3 is present mostly as a cleaved repressor, silencing Hh-Gli targets (e.g. [5,7,9]). In the presence of Hh ligands and activation of the transmembrane protein Smoothened (Smo), the Gli code is changed: Gli1 is activated transcriptionally, possibly by preexisting Gli2 or Gli3; Gli2 becomes an activator; and Gli3 is no longer cleaved: Gli repressors are lost and Gli activators are made (Figure 1a). Gli1 has a positive autoregulatory role, extending the duration and possibly strength of signaling, although this is competed against by higher levels of feedback inhibitors also induced by Gli1, such as Patched1 (Ptch1) and Hip1. Thus, the timing and strength of signaling is regulated precisely by the timing of ligand action as well as by positive feed-forward and negative feedback mechanisms.
Gli activator and repressor targets are only defined partially (see, for example, Refs [10,16-18]) and can respond to combinatorial and cooperative Gli activity [4,5,10,12,19-21]. For example, in the frog embryo neural plate, distinct Hh-Gli targets display differential requirements for each individual Gli factor and these requirements are dynamic, as shown by differences in the regulation of the same target in embryos aged a few hours apart [10]. The way the Gli proteins act is thus likely to be specific to tissue, age, state and species, possibly owing to their ability to form Gli-Gli complexes [10]. In addition, the Gli code is also modified by the related Zic zinc-finger transcription factors. The Gli proteins interact physically with Zic1 and Zic2 [10,22,23]. Known targets that can partly explain the orchestrated effect of the Gli code on cell behavior include, for example, CyclinD1, N-Myc, Bcl2, Bmi1 and Snail, because these participate in the regulation of cell-cycle entry, proliferation, survival, self-renewal and epithelial-mesenchymal transitions, respectively (see, for example, Refs [20,24-26]).
The Gli code in vertebrate tissue patterning acts through gradients of repressor and activator functions in time and space
During normal development, Hh ligands have been proposed to act as morphogens in developmental fields, although they also act as mitogens (Figure 1b). In the vertebrate neural plate and neural tube, a ventral gradient of Hh contributes to the patterning of this tissue. It induces a double Gli gradient with opposite polarities of activators (high to low from ventral or medial to dorsal or lateral) and repressors (high to low from dorsal or lateral to ventral or medial) [2,4,7,9,27,28]. A similar situation seems to take place in other tissues and species (e.g. [29-32]).
Graded Hh-Gli signaling and the Gli code are not the sole effectors of neural-tube patterning, although they are crucial components of the elaboration of final pattern. In the neural tube of mouse embryos lacking one of the Hh genes, Sonic Hh (Shh), ventral pattern is largely lost [33]. In this case, no activating Gli proteins are found ventrally and there is predominant Gli3 repressor function. However, Shh Gli3 double null embryos display a partial restoration of the dorsoventral pattern lost in single Shh mutants, although the number of cells in different cell-type pools is abnormal [34]. In the neural tube, Hh signaling thus acts to inhibit Gli3 repressor function but its induction of activating forms of Gli1 and Gli2 appear to be required to orchestrate size and shape (including the correct number of cells in each pool), in cooperation with prepattern mechanisms (reviewed in Ref. [3]).
The Gli code is thus thought to specify position within a developmental field by responding to Hh signaling but also by integrating information from other sources (see later). The duration of overall signaling and other inputs, as well as their strength, would then determine the ratio of activator to repressor Glis and the extent of the morphogenetic and/or proliferative effects (Figure Ia in Box 1). For instance, in the developing cerebellum, Hh-Gli signaling is crucial to induce the proliferative expansion of granule neuron precursors in the external germinal layer [35-37] and the timing and strength of such signaling is proposed to regulate the growth and patterning of the cerebellum [35].
Box 1. The Gli code in development and cancer.
During development (Figure Ia), Gli activator (GliA) function is dependent on signaling by Hedgehog (Hh) ligands that leads to Smo activation. The kinetics of GliA activation follows that of Hh-Smo. Hh-pathway activity thus promotes the formation of labile GliA, enabling the precise control of the response to Hh signaling. Before and after the pulse of Hh signaling, dominant Gli repressors (GliR) inhibit all Hh-Gli responses. These changes are seen by following the ratio of GliA:GliR in time. See the main text for the specific role of each of the three Gli proteins.
In cancers dependent on sustained Hh-Gli pathway activity (Figure Ib), tumor initiation could take place in cells with an already active Hh-Gli pathway, either through ligand expression or activation of Smo, for instance, or in cells that acquire active Hh-Gli signaling. These tumor initiating cells, possibly stem/precursor cells, then accumulate oncogenic hits that drive tumor progression, in part, by locking the Gli code in progressively higher activating states, which increases the GliA:GliR ratio. Oncogenic RAS or AKT do not seem to affect Gli3R nuclear localization or function [48].

The Gli code is targeted by oncogenes: implications for senescence and tumor progression
With this developmental background, how are we to view cancer? We think cancer is a disease of patterning [38] and not of single cells gone amok as a result of the disregulation of the cell cycle, for instance. In this context Hh-Gli signaling is taking center stage in the understanding and possible treatment of many kinds of human sporadic cancers, including those of skin, brain, lung, prostate, pancreas and stomach ([39-48]; reviewed in Ref. [1]).
GLI1 was identified originally as an amplified gene in a human glioma cell line [49]. However, the suggestion that amplification was important was denied shortly after [50]. Later, the HH-GLI pathway was implicated independently in familial basal-cell carcinomas (through loss of PTCH1; reviewed in Ref. [1]), and GLI1 function was implicated in the sporadic forms of this tumor type [40,51]. The functional role of GLI1 in human gliomas has been defined only recently [39,47].
Cerebellar tumors or medulloblastomas also have an active HH-GLI pathway and require its activity for sustained growth [39,41,52,53]. The biological context of Hh-Gli signaling in the cerebellum [35-37] suggests that the normal patterning of granule-cell precursors in the external germinal layer is disrupted in tumors so that precursors persist in a proliferative state through inappropriate sustained activation of HH-GLI signaling [39], explaining the development of medulloblastomas in Ptch1+/- mice [54,55].
Similarly, inappropriate activation of Hh-Gli signaling in GLI1+ neural stem cells (see later) or precursors might provide a developmental context to understand gliomas [39,47]. The final example is melanomas, which also express and depend on sustained Hh-Gli activity [48]. Here, the context is the proliferating melanocyte precursors or committed stem cells in the hair-follicle matrix [48]. In all of these cases, normal development or homeostasis is disrupted and abnormal patterning and proliferative programs ensue.
All brain, prostate and skin tumors that we have tested, through a variety of methods (using the SMO antagonist cyclopamine; siRNAs against each of the three GLI mRNAs; or with a lentivector expressing a shRNA against SMO [39,40,44,47,48]), require HH-GLI signaling at one level or another, although there is a clear predilection for activation at the level of SMO or above. SMO could be a good target because it efficiently regulates the GLI code. The ubiquitous requirement that we have documented also implies that HH-GLI-dependent cancers must harbor the common mutations in oncogenes and tumor suppressors described for each of these tumor classes. For example, there is frequent activation of EGF receptor (EGFR)-RAS signaling in gliomas through mutation of EGFR to a dominant-active state, and melanomas display a high incidence of N-RAS and B-RAF mutations. It is therefore possible to suggest a funnel hypothesis in which oncogenic inputs converge on GLI activity to promote tumor progression [38] (Figure Ib in Box 1).
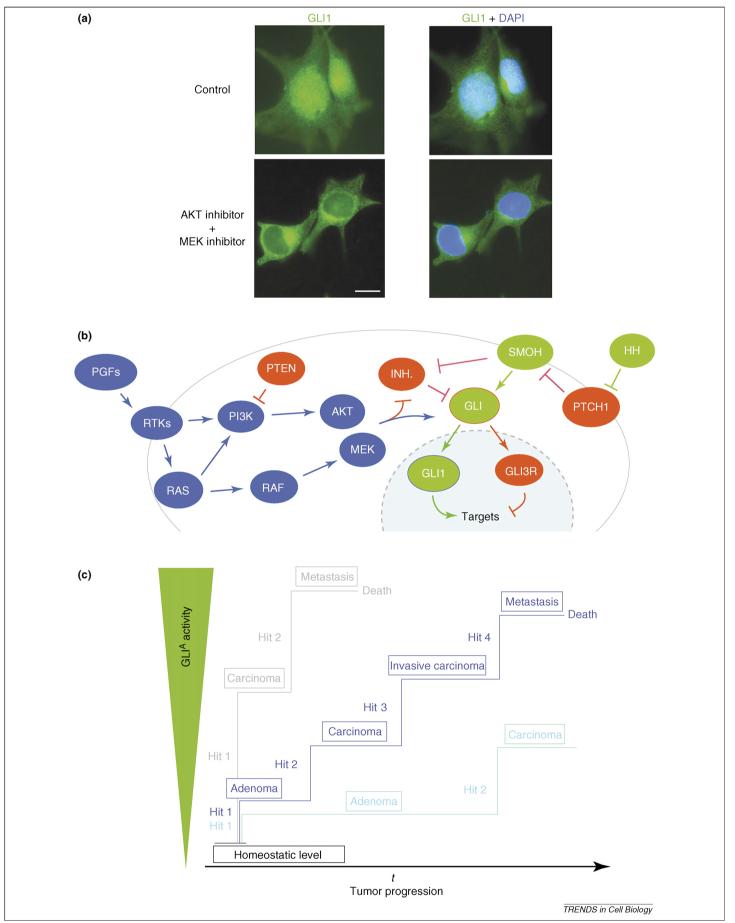
EGF cooperates and synergizes with Hh-Gli signaling in normal stem cells [56,57]. This finding further supports the funnel hypothesis, which predicts that oncogenic EGFR-RAS-AKT signaling could affect the Gli code. Indeed, in melanomas, gliomas and prostate-cancer cells, GLI1 activity requires both endogenous AKT and MEK function, whereas oncogenic H- or N-RAS, AKT1 or MEK1 potentiate GLI1 function [48] (Figure 2a,b; Box 2). Moreover, the colony-forming activity of GLI1 and RAS in SK-Mel2 human melanoma cells require endogenous AKT and MEK function, and SMO and GLI1 activity, respectively [48]. Similarly, K-RAS up-regulates GLI1 activity in pancreatic-cancer cells [58]. RAS and AKT signaling enhances the nuclear localization of GLI1 [48], counteracting its suppression by other modifiers that retain it in the cytoplasm, such as Suppressor of fused (Sufu) (Box 2) [48]. It remains unclear what are the primary targets of the oncogenic pathways because they might not only modify the GLI proteins themselves but also (or rather) modify crucial regulators of GLI function, such as SUFU. Thus, signaling by peptide growth factors (PGFs), receptor tyrosine kinases (RTKs), RAS, RAF, MEK, phosphoinositide-3 kinase (PI3K) and AKT is an essential regulator of the GLI code: oncogenic RAS or AKT highjack and lock the GLI code in a hyperactivating state (Box 1). Our findings are consistent with the requirement of both oncogenic AKT and RAS functions to induce gliomas from neural precursors [59], which express Gli1 [39]. Moreover, suppressors of RAS-AKT function, such as the tumor-suppressor PTEN, and the attenuation of RAS signaling involved in senescence [60], could be thus viewed as modulators of the GLI code. Finally, melanomas induced by expression of oncogenic RAS in mice lacking the INK4a tumor suppressor express Gli1 and depend on sustained Hh-Gli signaling for growth and survival [48].
Figure 2.
Integration of HH-GLI and oncogenic RAS-AKT signaling and a model for the role of positive activating GLI function in tumor progression. (a) Immunolocalization of endogenous GLI1 protein in untreated SK-Mel2 human melanoma cells or in cells treated with AKT and MEK inhibitors [48]. GLI1 protein (green) is mostly nuclear but relocalizes to the cytoplasm in the absence of endogenous RAS-MEK/AKT signaling. The position of the nucleus is shown by DAPI staining (blue) in the right-hand panel. Scale bar = 20 μm. (b) Representation of the peptide-growth factor (PGF)-receptor tyrosine kinase (RTK)-RAS-RAF-MEK and PI3K-AKT pathways (blue ovals), which lead to enhanced nuclear localization of GLI1, downstream of the classical HH pathway (green ovals). Inhibitors are in red. The effect could be direct on the GLI proteins or indirect by repressing GLI inhibitors. In both cases, there would be an enhancement of GLI nuclear levels [48]. (c) Model for the role of GLI activators (GLIA) in tumor progression. Increasing levels of positive GLI activity (mostly GLI1) induce distinct sets of targets, produce increases in tumor-cell number and induce tumor progression. Increases in GLI activity are brought about by sequential oncogenic hits, such as those activating EGFR, RAS or RAF, those amplifying AKT or those inhibiting the tumor suppressor PTEN. The hypothetical model depicts three scenarios leading to distinct states with different kinetics.
Box 2. The Gli code is regulated exquisitely by multiple mechanisms.
There is a wealth of negative regulators that finely tune Gli activity triggered by Hh ligands. Within the classical Hh-Gli pathway, protein kinase A (PKA), glycogen synthase kinase 3β (GSK3β) and casein kinase 1 (CK1), phosphorylate and tag Glis for cleavage or complete proteasome degradation through multiple E3-ligases (see, for example, Ref. [15]). Interestingly, Numb, which is often associated with asymmetric cell divisions and is an inhibitor of Notch signaling, targets Gli1 for proteasome destruction through the E3 ligase Itch [83]. Even if the Gli proteins survive these attacks, other proteins prevent them from entering the nucleus: Sufu and Ren, two unrelated proteins, then sequester them in the cytoplasm and inhibit their nuclear function [84-86]. All of these elements must be orchestrated to enable Hh signaling to produce a positive Gli code to induce a specific genetic program (Figure 1a in the main text). How this takes place is not clear, although initial hints suggest, for example, that Smo activation inhibits Sufu function [85].
Positive regulators of Gli function have also been found, including the kinase Dyrk1 [87] and the actin-binding protein Missing in metastasis (MIM, also called BEG4) [88]. Others include peptide-growth factor (PGF)-activating receptor tyrosine kinases, such as EGF, PDGF, FGF and IGF, which synergize with or affect Hh-Gli function [21,48,56,57,89]. Consistently, PI3K-AKT, PKCδ and RAS-RAF-MEK pathways potentiate positive Gli function [21,48,58,90-92]. Moreover, Hh-Gli signaling can feed back positively on PGF-RAS/AKT function by inducing expression of ligands and receptors (e.g. IGF2 and PDGFRA [39,47,93]). These interactions are also context dependent because oncogenic RAS inhibited GLI1-mediated 10T1/2-cell differentiation ([48] and data not shown).
Hh-Gli signaling interacts with other signaling pathways in bidirectional and context-specific ways. For example, Gli1 and Gli2 induce batteries of Wnt genes in ectoderm [94] and Gli3-repressor function inhibits Wnt-β-catenin signaling [95], however, Wnt-β-catenin signaling can also induce Hh-Gli pathway activity [96]. Other forms of interactions also take place, such as Smad-Gli binding [97]. Understanding the cross-regulation and integration of multiple signaling pathways to yield reproducible species- and context-specific effects is one of the main goals of current research [98]. Finally, it is probable that the Gli proteins are regulated by additional mechanisms that are yet to be discovered and fully detailed. For instance, talpid3 gene function affects the Gli code but how it does so is unclear [99]; also, the primary cilium seems to be a site where the Gli proteins are modified [100,102] (Figure 1a in the main text), suggesting a physical site for the transformation of extracellular Hh information into intracellular Gli code function.
Conversely, the well documented progression of tumors owing to consecutive increments on RAS-RAF-MEK and PI3K-AKT signaling might be dependent or derive from sequential increases in the activating strength of the GLI code. We have proposed that the cell-autonomous sequential increases in GLI1 activity are a crucial component of tumor progression [44,48] (Figure 2c) and the findings on the impact of the RAS-AKT oncogenic pathways on GLI1 activity lend support to this hypothesis. Consistent with this, tyr-NRASQ61K;INK4a-/- mouse melanomas show higher levels of Gli1 in the lymph nodes after metastasis than in primary skin sites [48]. Analyses of limited collections of primary and metastatic human tumors are so far inconclusive but there is a suggestion of higher expression of GLI1 in metastases [46].
The finding that EGF signaling alters the kinds of targets induced by GLI1 [21] raises the possibility that oncogene-driven increases in GLI1 function result in the differential expression of distinct target sets involved in the different phases of tumor progression. How EGF-RAS affects target selection is unclear but it might relate to differential binding-site affinity and the number of binding sites in the regulatory regions of target genes.
Tumor growth involves interactions between tumor cells and stroma. Whereas there is clear evidence for the action of the GLI code in tumor cells, the evidence for its implication in stroma is unresolved (but see Ref. [101]). For example, the human prostate gland exhibits HH-GLI pathway activity in the epithelium but not in the mesenchyme and GLI1 is not expressed appreciably in the tumor stroma [44]. By contrast, Gli1 is expressed in the stroma of the rodent prostate, where it is proposed to participate in tumor-stroma cross-talk [61].
The Gli code controls the behavior of activated stem cells and cancer stem cells
Hh-Gli signaling regulates the behavior of a variety of precursors in different tissues, including the cerebellum (see earlier), cerebral cortex and colliculi [39]. It also regulates the behavior of stem cells, notably of neural stem cells in neurogenic niches of the subgranular zone of the hippocampus and the subventricular zone of the lateral ventricle of the forebrain [56,57,62-64], as well as other stem-cell types [65]. Quiescent epithelial stem cells seem to have little or no Gli1 expression and Hh-Gli signaling instead regulates their activation to do their sine qua non job: self-renewal and production of daughter cells with restricted potentials (Figure 1c). The finding that Hh signaling expands neural adult brain stem cells, increasing neurosphere size and number [57], raises the possibility of manipulating the Gli code to produce large numbers of stem cells and derived precursors for the treatment of neurodegenerative diseases. One potential problem is that normal stem cells or derived precursors induced to proliferate might acquire tumorigenic properties, possibly through the acquisition of oncogenic hits that further increase GLI1 activity.
Human cancer stem cells are tumor cells that self-renew, give rise to the tumor bulk and appear to be the sole cells able to reinduce tumorigenesis after transplantation in nude mice. Cancer stem cells are often rare and have been described in nonsolid tumors as well as in breast, pancreas, skin, brain and other cancers [66-70].
The clonogenicity and tumorigenicity of human glioma cancer stem cells depends on sustained HH-GLI signaling [47]. These cells display a GLI code that is locked in a hyperactivating state: GLI1 and often GLI2 are required positively (with GLI1 having a more central role and being required for survival), whereas GLI3 has little or no role. However, inhibition of HH-GLI signaling through treatment with cyclopamine (and thus, through SMO inhibition) leads to the reversal of the code so that negative modulators and repressive GLI proteins now have major inhibitory roles. Indeed, GLI3 mediates the repressive effects of SMO shut-down [47,48]. Cancer stem cells in other tumors, such as breast [71] and multiple myeloma [72], might similarly require HH-GLI signaling.
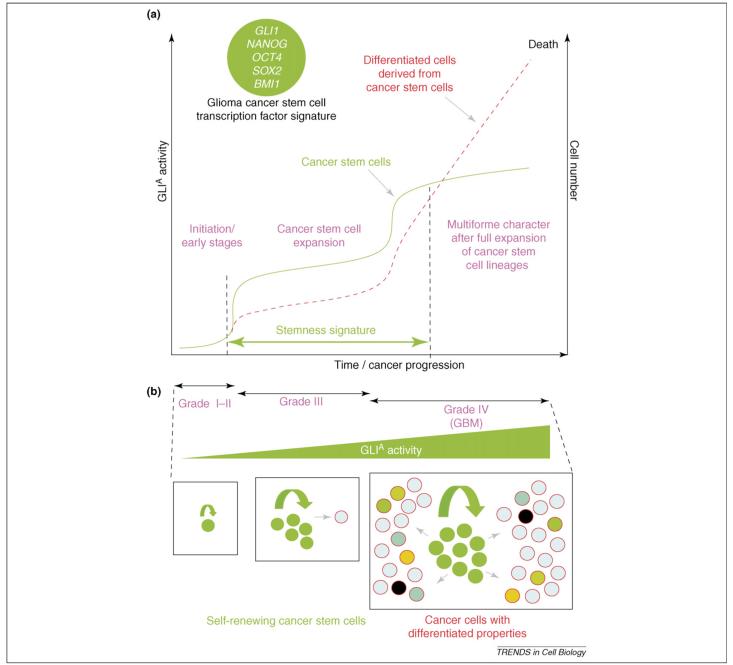
We propose that tumors start as an amplification of abnormally proliferating (cancer) stem cells, inducing a benign in situ tumor (or an adenoma), which can then give rise to a high number of derived precursors with more differentiated progeny leading to the development of a high grade tumor (or a carcinoma) (Figure 3a,b). Such an increase in cell number, locally or metastatically, kills the individual. In support of this idea, grade III gliomas display a stemness signature that is lost in grade IV gliomas [or glioblastoma multiforme (GBMs)] [47]. This is probably due to the dilution of GBM cancer stem cells, and thus of their stemness signature, in the tumor pool by derived precursors and the increased participation of host cells, for instance, through enhanced angiogenesis [47]. How cancer stem cells progress to activate their lineage potential fully while also preserving self-renewal is not clear. Changes in the putative perivascular cancer stemcell niche could have a role in this change.
Figure 3.
Activating GLI function, tumor progression and cancer stem cells in human gliomas. (a) Model for the progression of human tumors, in which there is an initial expansion of cancer stem cells (green curve), enabling the detection of a stemness signature (doubled arrowed green line) as the cancer stem cells predominate in the tumor population [47]. High-grade tumors (here grade IV tumors) display an expansion of stem cell-derived lineages (red broken line), which dilutes the stem cells and their stemness signature, leading to the killing of the individual. The transcription-factor signature of glioma cancer stem cells [47] is illustrated in the inset. (b) The model in (a) proposes an increase of positive-activating GLI function (mostly GLI1) through the different tumor grades, leading to maximal stem-cell self-renewal and full expansion of derived lineages that can acquire differentiated phenotypes, giving a multiforme character in the case of high-grade gliomas, which are also known as glioblastoma multiforme.
The origin of cancer stem cells is also uncertain. Glioma cancer stem cells might be derived from stem cells in postnatal neurogenic niches or from dedifferentiated astrocytes [73]. We suggest that cancer stem cells either derive from normal stem cells or derived precursors that already express GLI1 and have an active HH-GLI pathway, or from cells that necessarily acquire active HH-GLI signaling. It is still unproven whether or not classical oncogenic hits affect cancer stem cells, but it is probable. This would suggest that classical oncogenic and tumor-suppressor pathways modify the tumor GLI code to pervert the behavior of stem cells and precursors, giving rise to cancer stem cells with a perennially hyperactivating code. Tumor progression might then depend on the sequential increases in overall GLI1 activity, which is driven by accumulating hits in cancer stem cells.
The GLI code and metastasis
Metastatic founder or pioneer cells are likely to be cancer stem cells that undergo selection to perform organ-specific colonization. Recent data involve the HH-GLI pathway and the GLI code in metastasis. Human malignant melanoma, pancreas and prostate cells grow metastatically into the lungs of nude mice and this is prevented by systemic treatment with cyclopamine [45,46,48] (Figure 4). Cyclopamine treatment changes the GLI code from a locked hyperactivating state to a repressive one [48]. Conversely, GLI1 can also enhance the metastatic growth potential of a stably transfected prostate cancer cell line in nude mice [45], although it is unclear if the cells themselves were changed in the process of in vitro selection. The inhibition of metastases by systemic cyclopamine treatment together with the dependence of cyclopamine on GLI3 repressor function indicate that, in these contexts, the GLI code is an essential determinant of metastatic behavior. One key GLI target seems to be Snail [26], which promotes epithelial-mesenchymal transitions [74,75] that are essential for metastasis of epithelial solid tumors. We propose that high levels of cell-autonomous GLI1 activity, including enhancements by integrating oncogenes and loss of tumor suppressors (see earlier), promote metastasis.
Figure 4.
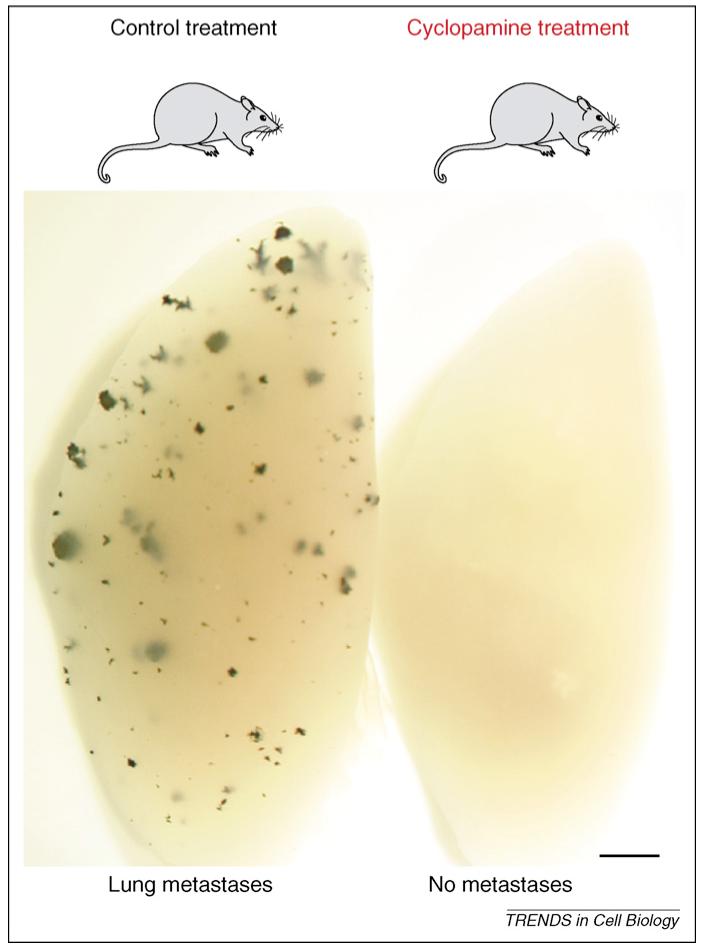
Inhibition of metastatic tumor growth by interference with HH-GLI signaling in vivo. Intravenous injection of lacZ-transduced human melanoma cells into nude mice leads to the metastatic growth of cancer cells in the lungs, here revealed as blue masses after XGal staining. Metastatic growth can be prevented fully by systemic treatment of the injected mice with cyclopamine, which inhibits the HH-GLI pathway and reverts the GLI code to a repressive state (see Ref. [48]). Scale bar = 1 mm.
Combinatorial cancer therapies and the GLI code
The fact that all gliomas, medulloblastomas, prostate cancers and melanomas that we have tested require SMO and/or GLI1 function is striking [39,44,47,48]. It is even more striking that this is the case for many kinds of unrelated human sporadic cancers. Indeed, although some authors have tried to divide tumors based on transcriptome profiles or levels of expression of GLI1 or PTCH1, these evaluations remain artificial at best because expression of GLI1 per se is an indicator of an active pathway [76]. Thus, so far, our functional studies suggest a ubiquitous requirement for HH-GLI by many tumor types and many tumor grades, although this needs further and extensive evaluation. These findings suggest the exciting possibility that manipulation of the GLI code could provide a wide-spectrum anticancer target for therapeutic intervention. The finding that oncogenic RAS-AKT pathways regulate GLI1 and that there is a mutual proliferative dependence in some assays between oncogenic RAS and HH-GLI function [48], further supports this idea and raises the interesting prospect of using combinatorial therapies targeting oncogenic RAS, AKT and HH-GLI.
In addition, there is an additive or synergistic effect of classical chemotherapeutic (nonspecific, universally damaging) drugs with alterations of the GLI code. Temozolomide, the leading (DNA-alkylating) chemotherapeutic drug for GBM, has additive and synergistic effects with cyclopamine in GBM stem-cell cultures [47]. Similarly, there are positive combined effects of gemcitabine (a nucleoside analogue that disrupts DNA replication) and cyclopamine in pancreatic cancer cells [46] and also among cyclopamine, gefitinib (an EGFR inhibitor) and docetaxel (a taxol analogue that inhibits microtubule-dependent cell division) [77]. Although how such synergisms occur is not understood, they have clear implications for therapy.
Cyclopamine treatment has proven good at inhibiting tumor growth and survival of both human cell lines and primary cultures [39,41-48]. It is also effective in inhibiting proliferation and reducing the bulk of human xenografts [41-43,45-48] and of endogenous genetic tumor mouse models (Ptch1+/- p53-/- medulloblastoma [53] and Tyr-RASQ61K INK4a-/- melanoma [48]). Even though the effects of systemic treatment with cyclopamine are limited in large tumors [48], probably owing to inefficient drug delivery into the tumor, this drug can prevent recurrence locally [48] and inhibit metastatic growth systemically [45,46,48]. Thus, whereas developing novel small-molecule antagonists of HH-GLI signaling [52,78-80] with optimal pharmacokinetic properties and efficacy is important, it is still worth considering cyclopamine as a candidate drug for clinical trials. Cyclopamine is a natural plant alkaloid that is available and effective orally in farm and range (see, for example, Ref. [81]) as well as laboratory animals with minimal side effects (diarrhea [47,48,53,57]).
Concluding remarks
Work over the last decade has provided multiple lines of support for the Gli code hypothesis and has crucially extended it to stemness and cancer. Although our focus on the GLI code might seem narrow at first and many other genes and pathways play important roles in stemness, tumorigenesis and tumor progression, the evaluation of current data strongly indicates a central role of the GLI code in human cancer. We envision a unified therapy for many human cancers of different grades, including metastatic cancers, based on drugs that, although acting at different levels (e.g. GLI, SMO, RAS and AKT), have a combined effect of modulating the GLI code, from the locked hyperactivating state in cancer to a repressive mode that is inconsistent with continued cancer-cell proliferation and survival. In this context, GLI-GLI interactions [10] might also provide a therapeutic target. Importantly, such treatments will modify the GLI code in cancer stem cells [47], indicating that they will both debulk the tumors and prevent metastasis and recurrence. Normal stem cells and their lineages would naturally be affected but, because quiescent stem cells do not seem to require positive GLI activity, they will regenerate any damage to stem-cell lineages following cessation of treatment. In mice, approximately 20 days of treatment with cyclopamine are sufficient to abolish the tumors and prevent recurrence and metastasis [48], suggesting that limited treatments will be efficacious. Finally, the finding that oncogenic RAS requires positive GLI activity in several contexts [48] and that HH-GLI is also activated by other cancer-inducing agents, such as tobacco smoke [82], suggests that the GLI code participates in a vast number of human cancers. The dependence of many human tumors on high positive GLI activity thus represents an unexpected common target. Finding the best arrow to cast is now priority.
Acknowledgements
We thank present and previous lab members, collaborators and colleagues for discussion. Work in the authors’ laboratory is supported currently by grants from the Swiss National Foundation, the National Institutes of Health, the Swiss Cancer League, the Louis-Jeantet and Leenaards Foundations and the State of Geneva. We apologize to colleagues whose work could not be referenced owing to space restrictions.
References
- 1.Ruiz i Altaba A, editor. Hedgehog-Gli signaling in human disease. Landes Bioscience; Georgetown: 2006. [Google Scholar]
- 2.Ruiz i Altaba A. Catching a Gli-mpse of Hedgehog. Cell. 1997;90:193–196. doi: 10.1016/s0092-8674(00)80325-6. [DOI] [PubMed] [Google Scholar]
- 3.Ruiz i Altaba A, et al. The emergent design of the neural tube: prepattern, SHH morphogen and GLI code. Curr. Opin. Genet. Dev. 2003;13:513–521. doi: 10.1016/j.gde.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Ruiz i Altaba A. Combinatorial Gli gene function in floor plate and neuronal inductions by Sonic hedgehog. Development. 1998;125:2203–2212. doi: 10.1242/dev.125.12.2203. [DOI] [PubMed] [Google Scholar]
- 5.Ruiz i Altaba A. Gli proteins encode context-dependent positive and negative functions: implications for development and disease. Development. 1999;126:3205–3216. doi: 10.1242/dev.126.14.3205. [DOI] [PubMed] [Google Scholar]
- 6.Aza-Blanc P, et al. Expression of the vertebrate Gli proteins in Drosophila reveals a distribution of activator and repressor activities. Development. 2000;127:4293–4301. doi: 10.1242/dev.127.19.4293. [DOI] [PubMed] [Google Scholar]
- 7.Bai CB, et al. All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev. Cell. 2004;6:103–115. doi: 10.1016/s1534-5807(03)00394-0. [DOI] [PubMed] [Google Scholar]
- 8.Tyurina OV, et al. Zebrafish Gli3 functions as both an activator and a repressor in Hedgehog signaling. Dev. Biol. 2005;277:537–556. doi: 10.1016/j.ydbio.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Stamataki D, et al. A gradient of Gli activity mediates graded Sonic Hedgehog signaling in the neural tube. Genes Dev. 2005;19:626–641. doi: 10.1101/gad.325905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen V, et al. Cooperative requirement of the Gli proteins in neurogenesis. Development. 2005;132:3267–3279. doi: 10.1242/dev.01905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aza-Blanc P, et al. Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell. 1997;89:1043–1053. doi: 10.1016/s0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- 12.Methot N, Basler K. Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell. 1999;96:819–831. doi: 10.1016/s0092-8674(00)80592-9. [DOI] [PubMed] [Google Scholar]
- 13.Huntzicker EG, et al. Dual degradation signals control Gli protein stability and tumor formation. Genes Dev. 2006;20:276–281. doi: 10.1101/gad.1380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan Y, Wang B. A novel protein-processing domain in Gli2 and Gli3 differentially blocks complete protein degradation by the proteasome. J. Biol. Chem. 2007;282:10846–10852. doi: 10.1074/jbc.M608599200. [DOI] [PubMed] [Google Scholar]
- 15.Di Marcotullio L, et al. Multiple ubiquitin-dependent processing pathways regulate Hedgehog/gli signaling: implications for cell development and tumorigenesis. Cell Cycle. 2007;6:390–393. doi: 10.4161/cc.6.4.3809. [DOI] [PubMed] [Google Scholar]
- 16.Muller B, Basler K. The repressor and activator forms of Cubitus interruptus control Hedgehog target genes through common generic gli-binding sites. Development. 2000;127:2999–3007. doi: 10.1242/dev.127.14.2999. [DOI] [PubMed] [Google Scholar]
- 17.Yoon JW, et al. Gene expression profiling leads to identification of GLI1-binding elements in target genes and a role for multiple downstream pathways in GLI1-induced cell transformation. J. Biol. Chem. 2002;277:5548–5555. doi: 10.1074/jbc.M105708200. [DOI] [PubMed] [Google Scholar]
- 18.Vokes SA, et al. Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development. 2007;134:1977–1989. doi: 10.1242/dev.001966. [DOI] [PubMed] [Google Scholar]
- 19.Lei Q, et al. Transduction of graded Hedgehog signaling by a combination of Gli2 and Gli3 activator functions in the developing spinal cord. Development. 2004;131:3593–3604. doi: 10.1242/dev.01230. [DOI] [PubMed] [Google Scholar]
- 20.Regl G, et al. Activation of the BCL2 promoter in response to Hedgehog/GLI signal transduction is predominantly mediated by GLI2. Cancer Res. 2004;64:7724–7731. doi: 10.1158/0008-5472.CAN-04-1085. [DOI] [PubMed] [Google Scholar]
- 21.Kasper M, et al. Selective modulation of Hedgehog/GLI target gene expression by epidermal growth factor signaling in human keratinocytes. Mol. Cell. Biol. 2006;26:6283–6298. doi: 10.1128/MCB.02317-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brewster R, et al. Gli/Zic factors pattern the neural plate by defining domains of cell differentiation. Nature. 1998;393:579–583. doi: 10.1038/31242. [DOI] [PubMed] [Google Scholar]
- 23.Koyabu Y, et al. Physical and functional interactions between Zic and Gli proteins. J. Biol. Chem. 2001;276:6889–6892. doi: 10.1074/jbc.C000773200. [DOI] [PubMed] [Google Scholar]
- 24.Kenney AM, et al. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development. 2003;130:15–28. doi: 10.1242/dev.00182. [DOI] [PubMed] [Google Scholar]
- 25.Leung C, et al. Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature. 2004;428:337–341. doi: 10.1038/nature02385. [DOI] [PubMed] [Google Scholar]
- 26.Li X, et al. Snail induction is an early response to Gli1 that determines the efficiency of epithelial transformation. Oncogene. 2006;25:609–621. doi: 10.1038/sj.onc.1209077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Persson M, et al. Dorsal-ventral patterning of the spinal cord requires Gli3 transcriptional repressor activity. Genes Dev. 2002;16:2865–2878. doi: 10.1101/gad.243402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer NP, Roelink H. The amino-terminal region of Gli3 antagonizes the Shh response and acts in dorsoventral fate specification in the developing spinal cord. Dev. Biol. 2003;257:343–355. doi: 10.1016/s0012-1606(03)00065-4. [DOI] [PubMed] [Google Scholar]
- 29.Mullor JL, et al. Hedgehog activity, independent of decapentaplegic, participates in wing disc patterning. Development. 1997;124:1227–1237. doi: 10.1242/dev.124.6.1227. [DOI] [PubMed] [Google Scholar]
- 30.Litingtung Y, et al. Shh and Gli3 are dispensable for limb skeleton formation but regulate digit number and identity. Nature. 2002;418:979–983. doi: 10.1038/nature01033. [DOI] [PubMed] [Google Scholar]
- 31.Cayuso J, et al. The Sonic hedgehog pathway independently controls the patterning, proliferation and survival of neuroepithelial cells by regulating Gli activity. Development. 2006;133:517–528. doi: 10.1242/dev.02228. [DOI] [PubMed] [Google Scholar]
- 32.Furimsky M, Wallace VA. Complementary Gli activity mediates early patterning of the mouse visual system. Dev. Dyn. 2006;235:594–605. doi: 10.1002/dvdy.20658. [DOI] [PubMed] [Google Scholar]
- 33.Chiang C, et al. Cyclopia and defective axial patterning in mice lacking Sonic Hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 34.Litingtung Y, Chiang C. Specification of ventral neuron types is mediated by an antagonistic interaction between Shh and Gli3. Nat. Neurosci. 2000;3:979–985. doi: 10.1038/79916. [DOI] [PubMed] [Google Scholar]
- 35.Dahmane N, Ruiz i Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development. 1999;126:3089–3100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- 36.Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 1999;22:103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 37.Wallace VA. Purkinje-cell-derived Sonic Hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr. Biol. 1999;9:445–448. doi: 10.1016/s0960-9822(99)80195-x. [DOI] [PubMed] [Google Scholar]
- 38.Ruiz i Altaba A, et al. Hedgehog-Gli signaling in brain tumors: stem cells and paradevelopmental programs in cancer. Cancer Lett. 2004;204:145–157. doi: 10.1016/S0304-3835(03)00451-8. [DOI] [PubMed] [Google Scholar]
- 39.Dahmane N, et al. The Sonic Hedgehog-Gli pathway regulates dorsal brain growth and tumorigenesis. Development. 2001;128:5201–5212. doi: 10.1242/dev.128.24.5201. [DOI] [PubMed] [Google Scholar]
- 40.Dahmane N, et al. Activation of the transcription factor Gli1 and the Sonic hedgehog signalling pathway in skin tumours. Nature. 1997;389:876–881. doi: 10.1038/39918. [DOI] [PubMed] [Google Scholar]
- 41.Berman DM, et al. Medulloblastoma growth inhibition by hedgehog pathway blockade. Science. 2002;297:1559–1561. doi: 10.1126/science.1073733. [DOI] [PubMed] [Google Scholar]
- 42.Thayer SP, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watkins DN, et al. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 2003;422:313–317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 44.Sanchez P, et al. Inhibition of prostate cancer proliferation by interference with SONIC HEDGEHOG-GLI1 signaling. Proc. Natl. Acad. Sci. U. S. A. 2004;101:12561–12566. doi: 10.1073/pnas.0404956101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karhadkar SS, et al. Hedgehog signalling in prostate regeneration, neoplasia and metastasis. Nature. 2004;431:707–712. doi: 10.1038/nature02962. [DOI] [PubMed] [Google Scholar]
- 46.Feldmann G, et al. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res. 2007;67:2187–2196. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clement V, et al. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr. Biol. 2007;17:165–172. doi: 10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stecca B, et al. Melanomas require HEDGEHOG-GLI signaling regulated by interactions between GLI1 and the RAS-MEK/AKT pathways. Proc. Natl. Acad. Sci. U. S. A. 2007;104:5895–5900. doi: 10.1073/pnas.0700776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kinzler KW, et al. Identification of an amplified, highly expressed gene in a human glioma. Science. 1987;236:70–73. doi: 10.1126/science.3563490. [DOI] [PubMed] [Google Scholar]
- 50.Xiao H, et al. A search for gli expression in tumors of the central nervous system. Pediatr. Neurosurg. 1994;20:178–182. doi: 10.1159/000120783. [DOI] [PubMed] [Google Scholar]
- 51.Nilsson M, et al. Induction of basal cell carcinomas and trichoepitheliomas in mice overexpressing GLI-1. Proc. Natl. Acad. Sci. U. S. A. 2000;97:3438–3443. doi: 10.1073/pnas.050467397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Romer JT, et al. Suppression of the Shh pathway using a small molecule inhibitor eliminates medulloblastoma in Ptc1+/-p53-/- mice. Cancer Cell. 2004;6:229–240. doi: 10.1016/j.ccr.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 53.Sanchez P, Ruiz i Altaba A. In vivo inhibition of endogenous brain tumors through systemic interference with Hedgehog signaling in mice. Mech. Dev. 2005;122:223–230. doi: 10.1016/j.mod.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 54.Goodrich LV, et al. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 55.Hahn H, et al. Rhabdomyosarcomas and radiation hypersensitivity in a mouse model of Gorlin syndrome. Nat. Med. 1998;4:619–622. doi: 10.1038/nm0598-619. [DOI] [PubMed] [Google Scholar]
- 56.Palma V, Ruiz i Altaba A. Hedgehog-GLI signaling regulates the behavior of cells with stem cell properties in the developing neocortex. Development. 2004;131:337–345. doi: 10.1242/dev.00930. [DOI] [PubMed] [Google Scholar]
- 57.Palma V, et al. Sonic Hedgehog controls stem cell behavior in the postnatal and adult brain. Development. 2005;132:335–344. doi: 10.1242/dev.01567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ji Z, et al. Oncogenic kras suppresses GLI1 degradation and activates hedgehog signaling pathway in pancreatic cancer cells. J. Biol. Chem. 2007;282:14048–14055. doi: 10.1074/jbc.M611089200. [DOI] [PubMed] [Google Scholar]
- 59.Holland EC, et al. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat. Genet. 2000;25:55–57. doi: 10.1038/75596. [DOI] [PubMed] [Google Scholar]
- 60.Courtois-Cox S, et al. A negative feedback signaling network underlies oncogene-induced senescence. Cancer Cell. 2006;10:459–472. doi: 10.1016/j.ccr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pu Y, et al. Sonic hedgehog-patched Gli signaling in the developing rat prostate gland: lobe-specific suppression by neonatal estrogens reduces ductal growth and branching. Dev. Biol. 2004;273:257–275. doi: 10.1016/j.ydbio.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lai K, et al. Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nat. Neurosci. 2003;6:21–27. doi: 10.1038/nn983. [DOI] [PubMed] [Google Scholar]
- 63.Machold R, et al. Sonic hedgehog is required for progenitor cell maintenance in telencephalic stem cell niches. Neuron. 2003;39:937–950. doi: 10.1016/s0896-6273(03)00561-0. [DOI] [PubMed] [Google Scholar]
- 64.Ahn S, Joyner AL. In vivo analysis of quiescent adult neural stem cells responding to Sonic Hedgehog. Nature. 2005;437:894–897. doi: 10.1038/nature03994. [DOI] [PubMed] [Google Scholar]
- 65.Trowbridge JJ, et al. Hedgehog modulates cell cycle regulators in stem cells to control hematopoietic regeneration. Proc. Natl. Acad. Sci. U. S. A. 2006;103:14134–14139. doi: 10.1073/pnas.0604568103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ignatova TN, et al. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia. 2002;39:193–206. doi: 10.1002/glia.10094. [DOI] [PubMed] [Google Scholar]
- 67.Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 68.Collins AT, et al. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 69.Fang D, et al. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005;65:9328–9337. doi: 10.1158/0008-5472.CAN-05-1343. [DOI] [PubMed] [Google Scholar]
- 70.Li C, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 71.Liu S, et al. Hedgehog signaling and Bmi-1 regulate selfrenewal of normal and malignant human mammary stem cells. Cancer Res. 2006;66:6063–6071. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peacock CD, et al. Hedgehog signaling maintains a tumor stem cell compartment in multiple myeloma. Proc. Natl. Acad. Sci. U. S. A. 2007;104:4048–4053. doi: 10.1073/pnas.0611682104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sanai N, et al. Neural stem cells and the origin of gliomas. N. Engl. J. Med. 2005;353:811–822. doi: 10.1056/NEJMra043666. [DOI] [PubMed] [Google Scholar]
- 74.Batlle E, et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 75.Cano A, et al. The transcription factor snail controls epithelial mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2000;2(2):76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 76.Lee J, et al. Gli1 is a target of Sonic Hedgehog that induces ventral neural tube development. Development. 1997;124:2537–2552. doi: 10.1242/dev.124.13.2537. [DOI] [PubMed] [Google Scholar]
- 77.Mimeault M, et al. Cytotoxic effects induced by a combination of cyclopamine and gefitinib, the selective Hedgehog and epidermal growth factor receptor signaling inhibitors, in prostate cancer cells. Int. J. Cancer. 2006;118:1022–1031. doi: 10.1002/ijc.21440. [DOI] [PubMed] [Google Scholar]
- 78.Frank-Kamenetsky M, et al. Small-molecule modulators of Hedgehog signaling: identification and characterization of Smoothened agonists and antagonists. J. Biol. 2002;1:10. doi: 10.1186/1475-4924-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen JK, et al. Small molecule modulation of Smoothened activity. Proc. Natl. Acad. Sci. U. S. A. 2002;99:14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lauth M, et al. Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc. Natl. Acad. Sci. U. S. A. 2007;104:8455–8460. doi: 10.1073/pnas.0609699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Keeler RF. Teratogenic compounds of Veratrum californicum (Durand) X. Cyclopia in rabbits produced by cyclopamine. Teratology. 1970;3:175–180. doi: 10.1002/tera.1420030210. [DOI] [PubMed] [Google Scholar]
- 82.Lemjabbar-Alaoui H, et al. Wnt and Hedgehog are critical mediators of cigarette smoke-induced lung cancer. PLoS ONE. 2006;1:e93. doi: 10.1371/journal.pone.0000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Di Marcotullio L, et al. Numb is a suppressor of Hedgehog signalling and targets Gli1 for Itch-dependent ubiquitination. Nat. Cell Biol. 2006;8:1415–1423. doi: 10.1038/ncb1510. [DOI] [PubMed] [Google Scholar]
- 84.Kogerman P, et al. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat. Cell Biol. 1999;1:312–319. doi: 10.1038/13031. [DOI] [PubMed] [Google Scholar]
- 85.Svard J, et al. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev. Cell. 2006;10:187–197. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 86.Di Marcotullio L, et al. REN(KCTD11) is a suppressor of Hedgehog signaling and is deleted in human medulloblastoma. Proc. Natl. Acad. Sci. U. S. A. 2004;101:10833–10838. doi: 10.1073/pnas.0400690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mao J, et al. Regulation of Gli1 transcriptional activity in the nucleus by Dyrk1. J. Biol. Chem. 2002;277:35156–35161. doi: 10.1074/jbc.M206743200. [DOI] [PubMed] [Google Scholar]
- 88.Callahan CA, et al. MIM/BEG4, a Sonic hedgehog-responsive gene that potentiates Gli-dependent transcription. Genes Dev. 2004;18:2724–2729. doi: 10.1101/gad.1221804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brewster R, et al. Gli2 functions in FGF signaling during anteroposterior patterning. Development. 2000;127:4395–4405. doi: 10.1242/dev.127.20.4395. [DOI] [PubMed] [Google Scholar]
- 90.Riobo NA, et al. Protein kinase C-delta and mitogen-activated protein/extracellular signal-regulated kinase-1 control GLI activation in hedgehog signaling. Cancer Res. 2006;66:839–845. doi: 10.1158/0008-5472.CAN-05-2539. [DOI] [PubMed] [Google Scholar]
- 91.Riobo NA, et al. Phosphoinositide 3-kinase and Akt are essential for Sonic Hedgehog signaling. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4505–4510. doi: 10.1073/pnas.0504337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lauth M, et al. Phorbol esters inhibit the Hedgehog signalling pathway downstream of Suppressor of Fused, but upstream of Gli. Oncogene. 2007;26:5163–5168. doi: 10.1038/sj.onc.1210321. [DOI] [PubMed] [Google Scholar]
- 93.Hahn H, et al. Patched target Igf2 is indispensable for the formation of medulloblastoma and rhabdomyosarcoma. J. Biol. Chem. 2000;275:28341–28344. doi: 10.1074/jbc.C000352200. [DOI] [PubMed] [Google Scholar]
- 94.Mullor JL, et al. Wnt signals are targets and mediators of Gli function. Curr. Biol. 2001;11:769–773. doi: 10.1016/s0960-9822(01)00229-9. [DOI] [PubMed] [Google Scholar]
- 95.Ulloa F, et al. Inhibitory Gli3 activity negatively regulates Wnt/beta-catenin signaling. Curr. Biol. 2007;17:545–550. doi: 10.1016/j.cub.2007.01.062. [DOI] [PubMed] [Google Scholar]
- 96.Watt FM. Unexpected Hedgehog-Wnt interactions in epithelial differentiation. Trends Mol. Med. 2004;10:577–580. doi: 10.1016/j.molmed.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 97.Liu F, et al. Carboxy-terminally truncated Gli3 proteins associate with Smads. Nat. Genet. 1998;20:325–326. doi: 10.1038/3793. [DOI] [PubMed] [Google Scholar]
- 98.Bertrand N, Dahmane N. Sonic hedgehog signaling in forebrain development and its interactions with pathways that modify its effects. Trends Cell Biol. 2006;16:597–605. doi: 10.1016/j.tcb.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 99.Davey MG, et al. The chicken talpid3 gene encodes a novel protein essential for Hedgehog signaling. Genes Dev. 2006;20:1365–1377. doi: 10.1101/gad.369106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Haycraft CJ, et al. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PloS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dierks, et al. Essential role of stromally induced Hedgehog signaling in B-cell malignancies. Nat. Med. 2007;13:944–951. doi: 10.1038/nm1614. [DOI] [PubMed] [Google Scholar]
- 102.Rohatgi R, et al. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]