Summary
Chromosome movement is prominent during meiosis. Here, using a combination of in vitro and in vivo approaches, we elucidate the basis for dynamic mid-prophase chromosome movement in budding yeast. Diverse finding reveal a process in which, at the pachytene stage, individual telomere/nuclear envelope (NE) ensembles attach passively to, and then move in concert with, nucleus-hugging actin cables that are continuous with the global cytoskeletal actin network. Other chromosomes move in concert with lead chromosome(s). The same process, in modulated form, explains the zygotene "bouquet" configuration in which, immediately preceding pachytene, chromosome ends colocalize dynamically in a restricted region of the NE. Mechanical properties of the system and biological roles of mid-prophase movement for meiosis, including recombination, are discussed.
Introduction
Movement plays fundamental roles in basic chromosomal processes. Organized chromosomes congress and segregate at mitosis. Movement also occurs during DNA replication, DNA repair, gene activation and transcription and also globally (reviewed in Kumaran et al., 2008). Movement is especially significant during meiosis, when homologous maternal and paternal chromosomes ("homologs"), must recognize one another and “pair”, efficiently and without entanglements.
Meiotic homolog juxtaposition culminates in formation of synaptonemal complex (SC) between homolog axes all along their lengths. SC formation defines "zygotene"; presence of complete SC defines "pachytene". Pachytene chromosomes are well-organized, with chromatin loops of homologs emanating outward from either side of the SC. Zygotene and pachytene also include global changes in chromosome disposition. Before zygotene, chromosome ends associate with the NE in meiosis-specific complexes. During zygotene, these ends cluster into a localized area of the NE (the "bouquet ") while, at early pachytene, they redistribute throughout the nuclear periphery (Zickler and Kleckner, 1998; Bass, 2003; Scherthan 2007; Harper et al., 2004). Zygotene and pachytene chromosomes also exhibit dynamic movements over shorter time scales. Such movements have been described thus far in rat spermatocytes (Parvinen and Soderstrom, 1976), S. pombe meiosis (reviewed in Chikashige et al., 2007) and budding yeast (Trelles-Stricken et al., 2005; Scherthan et al., 2007; this work; Conrad et al., 2008; Wanat et al., in press; Kosaka et al., in press; this work) and C. elegans and Drosophila (A. Dernburg and R.S. Hawley, personal communications).
In pombe, telomeres cluster at the spindle pole body (SPB) and are moved dramatically back and forth along microtubules via dynein motors; in budding yeast, both telomeres and actin are implicated (references above). The current study further examines meiotic chromosome movement in budding yeast with respect to mechanism and biological significance.
Results
Active chromosome movement is required in vivo to overcome constraints on thermal motion
Mechanical properties of in vitro pachytene chromosomes
We developed a method to release pachytene chromosomes as singlets and interconnected groups (Figure 1A; Movie S1 and Movie S2). Compared to in vivo counterparts, in vitro DAPI-stained chromosomes present a similar length (1.3 µm versus 1.1 µm; n = 30), and are slightly wider (0.4–0.6 µm vs. ~0.35-0.4 µm), perhaps reflecting slight chromatin decondensation. Singlet chromosomes exhibit a random walk path with step-sizes conforming to a Gaussian distribution (Figure 1Bi). In vitro chromosomes also exhibit thermal bending which, for any filament, reflects its intrinsic stiffness/flexibility as discussed in terms of "persistence length" (Lp). Lp is the distance over which the correlation between the directions of two vectors tangent to the skeleton path of the polymer is lost (Doi and Edwards, 1986). Appropriate analysis of singlet in vitro pachytene chromosomes (n=40) (Ott et al., 1993; Figure 1Bii; Note S1) defines a set of (Gaussian-distributed) bending angles (Figure 1Biii, yellow curve) which in turn define Lp = ~12 µm (Note S1), a value characteristic of a "semi-flexible" object (Discussion).
Figure 1. in vivo and in vitro properties of pachytene chromosomes.
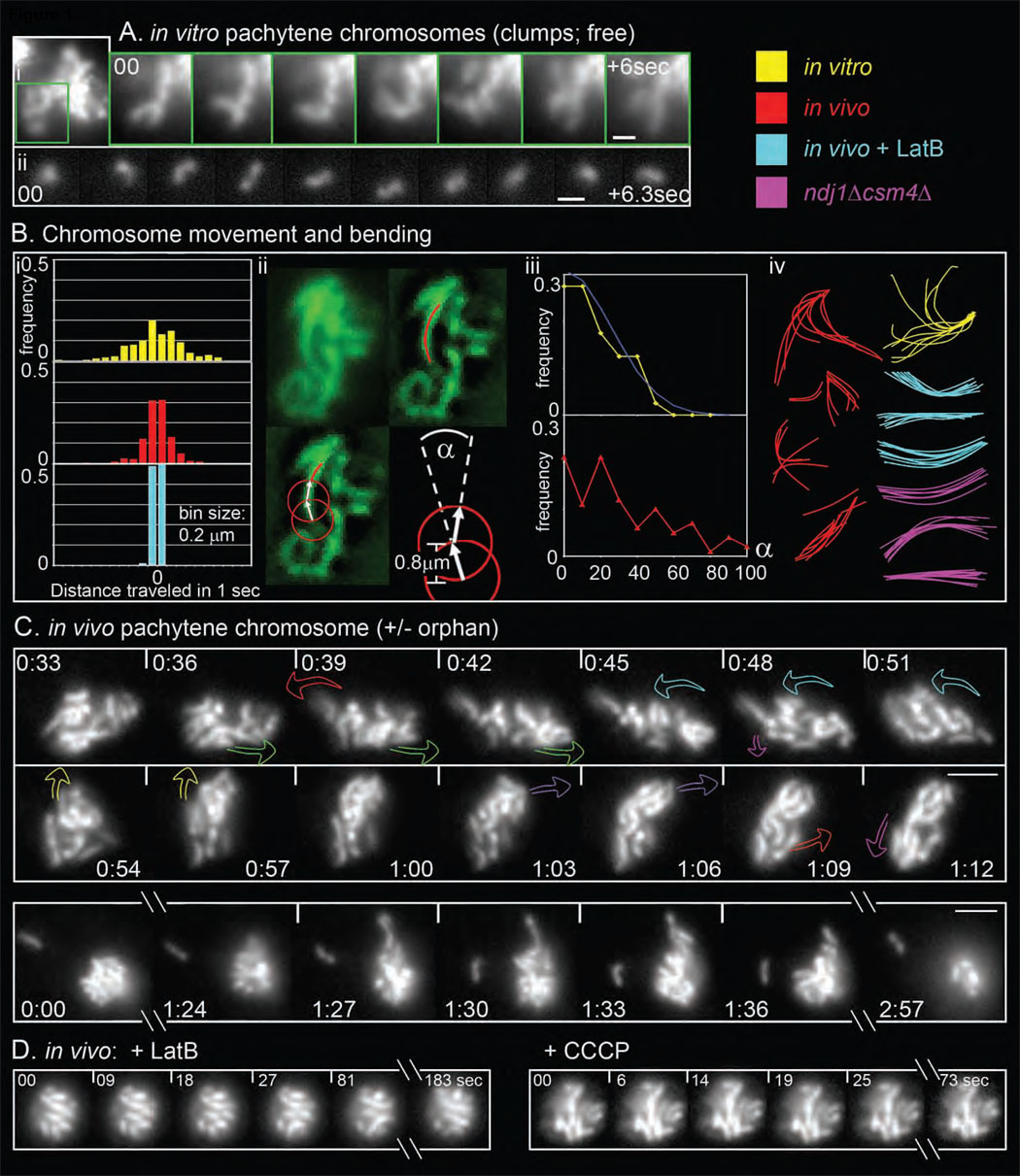
(A) Time series, non-deconvolved, of DAPI-labeled in vitro chromosomes. (i) individual chromosomes emerging from the compact mass of a partially-disrupted nucleus (1 sec intervals; MovieS1). (ii) free chromosome released from disrupted nucleus (0.7 sec intervals; MovieS2). (B) (i) Step-sizes histograms of the displacement of in vitro (yellow), in vivo (red) and in vivo + LatB (turquoise) pachytene chromosome ends, tracked every second for 91, 60 and 60 seconds, respectively. (ii) Calculation of bending angles and persistence lengths. Images of Zip1-GFP-labeled chromosomes (top left) were deconvolved and the skeleton paths of lengthy chromosome segments were outlined manually (top right, red line). Consecutive tangent vectors were then drawn beginning at one end of the chromosome using an arc length of 0.8µm (bottom left; Note S1). The angle α between two vectors is then determined (bottom right). For in vitro chromosomes, these angles are used to calculate persistence length (Lp). (iii) Distributions of (α) (in absolute values within 0–180°) for in vitro (yellow; n = 40 chromosomes; corresponding normal curve in blue) and in vivo chromosomes (red; n=50). (iv) Representative series of chromosome outlines taken every second over 8–10 sec and superimposed. (C) 2D time series of pachytene Zip1-GFP chromosomes (5h–6h after meiosis induction; 3 sec intervals; strain NKY3834). Top: Movie S3; colored arrows indicate abrupt, discrete transition(s) (see Figure 2Ai, ii). Bottom: Movie S4 illustrates long-lived orphan plus dramatic outward protrusion [1:27–1:36]. (D) Same as (A) but with 30 µM LatB (left) or 40 µM CCCP (right). (A) Scale bar = 1 µm. (C, D) Bars = 2 µm.
Actively-promoted dynamic movements in vivo
Scherthan et al (2007) showed that pachytene Zip1-GFP chromosomes in living cells exhibit dynamic, dramatic motion over time intervals of a few seconds. We confirmed this observation (e.g. Figure 1C top, Movie S3) and noticed that sometimes a single chromosome becomes well separated from the main group (5 occurrences in 100 nuclei over 3 min). “Orphans" usually persist ≤ 5 sec. but occasionally "dangle in space" much longer (Figure 1C bottom, Movie S4).
Chromosome movements in vivo are similar to those in vitro, suggesting that in vivo movements might be thermally promoted. This is not the case: Scherthan et al. (2007) find that in vivo motion is inhibited by LatrunculinB (LatB), which binds to actin monomers and prevents their incorporation into filaments. We also see this effect (Figure 1D; e.g. Movie S5, n>100) and, further, have found that inhibition is a specific effect of LatB on actin: a mutant whose actin no longer binds LatB is resistant to its effects (Figure S1). LatB severely reduces both translational step-sizes (Figure 1B, compare red and turquoise) and bending angles (Figure 1Biv, turquoise versus red; data not shown). We also find that movement is inhibited by carbonyl cyanide m-chloro phenyl hydrazone (CCCP), which blocks mitochondrial ATP synthesis (Figure 1D, e.g. Movie S6, n>100; Experimental Procedures). In contrast, neither inhibition of microtubule (MT) polymerization by nocodazole nor deletion of the cytoplasmic dynein-encoding gene DHC1 detectably reduces pachytene motion (Movie S7; data not shown), implying a fundamentally different mechanism from that described for pombe (Introduction).
In vivo constraints
Chromosome movement in vivo in the presence of LatB/CCCP is also much less dynamic than movement in vitro (Figure 1A versus 1D; Figures 1Bi and 1Biv, turquoise versus yellow). This difference suggests that, in vivo, spontaneous thermal motions are severely constrained such that an active mechanism is required to provide significant displacement. Interestingly, in vivo chromosomes often exhibit bending angles greater than any observed during spontaneous movement (Figure 1Biii, red vs. yellow); thus, actin-mediated forces must be significantly greater forces than thermal forces. Also, abrogation of telomere/NE tethering (by the ndj1Δ mutation; below) does not restore vigorous movement to chromosomes in motion-defective conditions (e.g. ndj1Δ csm4Δ, Figure 1Biv pink; or ndj1Δ + LatB, data not shown), implying that constraints on spontaneous movement reflect conditions internal within the nucleus.
Dissection of in vivo chromosome motion
Analysis of individual Zip1-GFP chromosome paths over time reveals five features of pachytene movement.
(i) Abrupt, discrete, directed transitions of chromosome groups
A given nucleus experiences a series of abrupt, discrete transitions. In each, a group of nearby chromosomes (or sometimes a single chromosome) moves coordinately in an outward-directed and/or peripheral and curvilinear path. Eight transitions are indicated in Figure 1C (top; Movie S3); five of these are documented by color-coding of individual chromosomes and sets of colored arrows describing frame-to-frame displacements (Figure 2Ai, ii). This and other movies (below; Figure S2) show that a given nucleus exhibits one transition every ~6–9 sec., with each transition lasting <3–9 sec. A nucleus sometimes experiences two transitions simultaneously. However, such pairs are only partially contemporaneous, involve different sets of chromosomes and occur in different directions, implying that they are independent events (e.g. red versus green arrows and magenta versus pink arrows in Figure 1C and 2Ai, ii). The dramatic nature of transitions is further illustrated by Figure 1D [1:24–1:36] (Movie S4); illumination of recombination-related Zip3-GFP foci (Figure 2D; Movie S8) (Agarwal and Roeder, 2000); and examples below.
Figure 2. Pachytene chromosome motion.
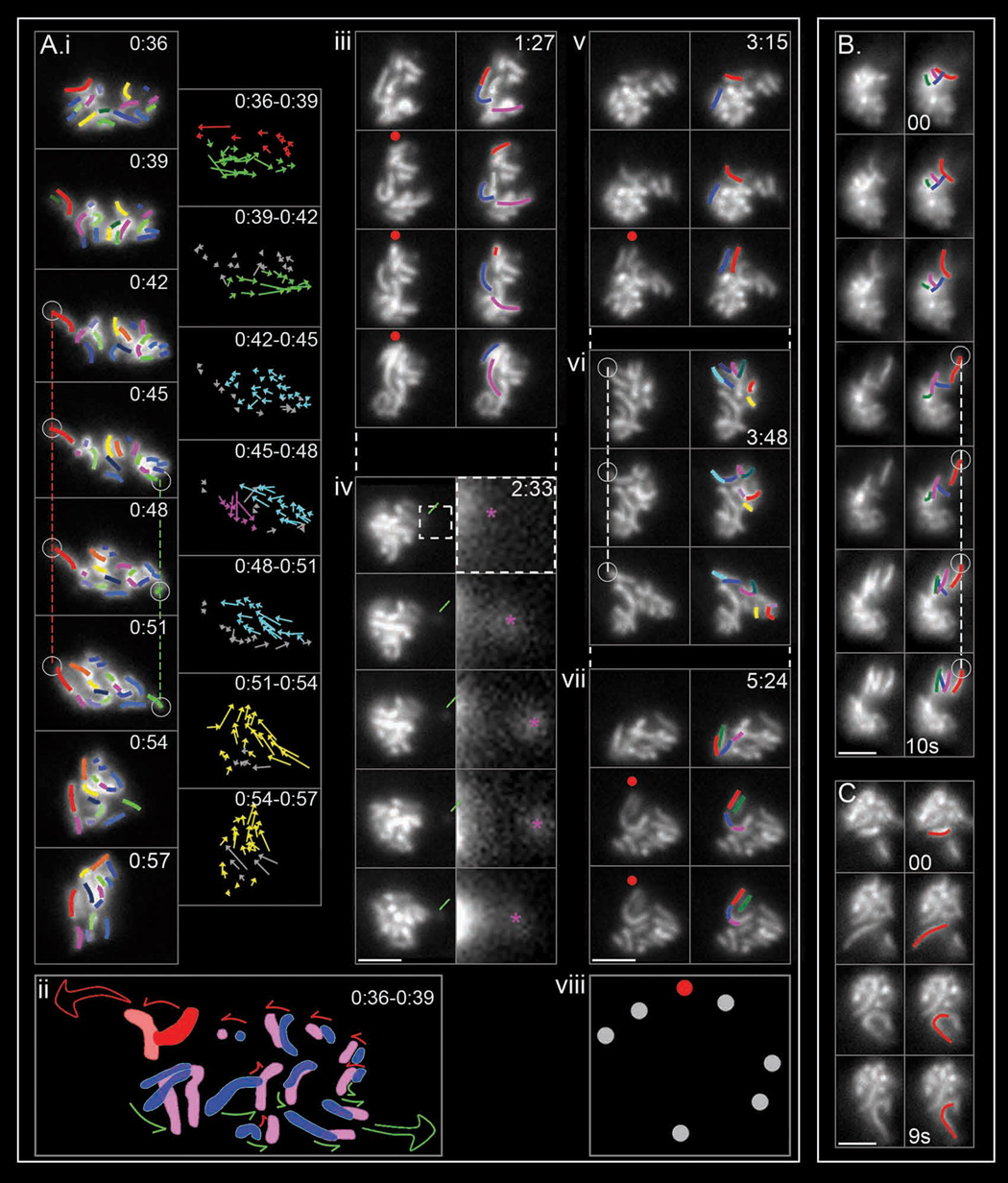
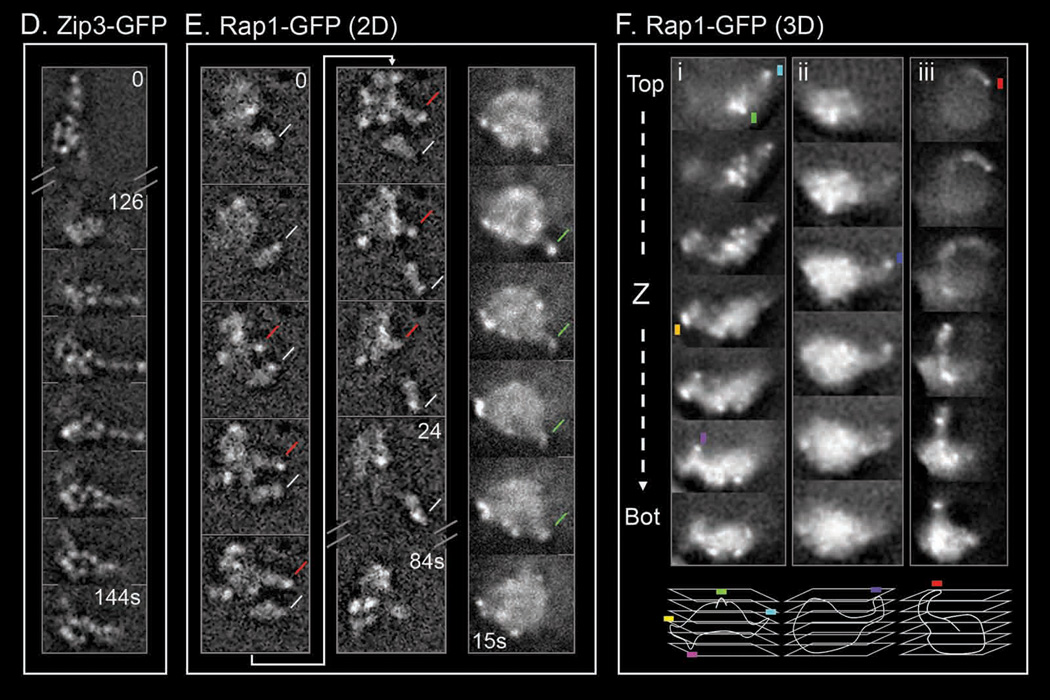
2D time series of Zip1-GFP chromosomes (bars = 2 µm) and analysis. ((Ai, iii–vii): same nucleus as Figure 1C (top); Movie S3; 3sec intervals. (B, C): Movie S9 and Movie S10; 1sec intervals). (A) (i) Left column: all chromosomes in the focal plane are outlined (left column; red color stands for “leaders”). Vertical dashed lines indicate anchoring of "leader" (red) and putative “leader” (green) chromosomes. Right column: for each chromosome, the direction and range of motion is depicted by vectors drawn between apparent telomeric positions in successive frames with different colors denoting different transitions. (ii) Stylized depiction of motion seen in (i; [0:36–0:39]) with positions in blue and pink, respectively (leader in red) and directional effects implied by arrows. (Aiii–vii) and (B, C): Seven additional discrete transitions. Chromosomes undergoing dramatic movement are highlighted, with red color representing the leader (see also Movie S10). Green line indicates an orphan (magnification at right, pink asterisk). Red dots highlight repeated use of a single trajectory in independent transitions. Open circle and dotted line indicate “anchoring”. (viii) Summary of all focal points over 6 minutes for Movie S3 (see also Figure S2) (D) Time series for a Zip3-GFP nucleus (NKY3501). (E) 2D dynamics of Rap1-GFP signals in nuclei of two living cells (NKY4000). Telomeres positions marked by stronger GFP foci; chromatin shape given by general background. 3 sec intervals except at discontinuous points (Movie S11). Left and middle: deconvolved frames showing a long transient nuclear protrusion with a GFP telomere focus at its extremity (red line), and an orphan chromosome (white line). Right: non-deconvolved frames of a short chromatin protrusion with a telomere at the edge (green line). (F) 3D reconstructions of chromatin protrusions revealed by Rap1-GFP. Top: consecutive 0.4 µm optical "Z" sections taken at 600ms intervals, from top to bottom, of three nuclei (i–iii). Chromatin protrusions (and essentially all protrusions in such analyses) have telomeric GFP foci at their outer extremity (colored squares). Bottom: schematic 3D reconstruction of each nucleus.
(ii) Telomere-led movement of a "lead" chromosome
In many transitions, a chromosome located at the leading edge of motion moves dramatically, and in a way which strongly suggests that force is being exerted specifically on its "leading end". Such effects are indicated by "red" chromosomes in Figure 2 (Figure 2Ai [0:36–0:39]; 2Aiii, v, vi, vii; cartoon 2Aii); Figure 2B top four panels (Movie S9); Figure 2C plus additional dramatic examples in the same series (Movie S10). A leading end typically moves 0.5 to 1 µm (and up to 2 µm) at ~0.3 – 0.5 µm per second (up to ~0.8 µm/sec). No particular chromosome(s) occur preferentially as "leaders", as all possible sizes of chromosomes are seen in this role. “Maverick” chromosomes noted by Scherthan et al. (2007), correspond to “leaders” exhibiting especially accentuated displacement.
(iii) Coordinated movement via interconnections among the "lead" chromosome and others
In transitions involving multiple chromosomes, the “followers” (non-red chromosomes in all transitions shown in Figures 2Ai, iii, v–vii, B and C) re-orient and move in the direction defined by the “leader” (arrows in Figures 2Ai, ii; non red chromosomes in other panels). When two transitions occur simultaneously, chromosomes located between the two moving groups respond to both tendencies (middle chromosomes, Figure 2Aii). Some transitions involve movement only of a lead chromosome, occasionally yielding a well-isolated "orphan" (e.g. Figure 2Aiv).
Coordinately moving chromosomes always comprise a spatially associated group (e.g. Figure 2A–C). Often, chromosomes nearer to the lead chromosome exhibit greater movement than those located farther away (e.g. relative magnitudes of arrows in Figure 2Ai; relative movements of non-red chromosomes in other transitions of Figure 2A–C). Nonetheless, spatial proximity to the lead chromosome is not sufficient to determine coordinate movement: motion often involves one group of chromosomes close to the lead chromosome without involving others equally close (Figure 2Avi). Also, affected chromosomes sometimes form a chain that follows the lead chromosome like a string of sausages (Figure 2Aiii, vii). Such phenomena suggest that movement of the lead chromosome provokes movement of a subset of nearby chromosomes which happen to be interconnected.
(iv) Peripheral tracks
Other phenomena point to involvement of stably-positioned extra-nuclear "tracks". (i) A chromosome can move back and forth along a particular trajectory (Figure 2Aiv). (ii) Directed movement of one chromosome can be followed, shortly afterward, by directed movement of another chromosome(s) along the same trajectory (Figure 2B, first four frames versus last three frames). (iii) Independent discrete transitions, separated by many seconds or minutes, occur along the same or closely-adjacent trajectories (Figure 2Aiii, v and vii). The focal points defined for a single nucleus over several minutes seem to define short linear arrays (Figure 2Aviii, Figure S2) implying different focal points along a single "track" and/or drifting of a single focal point.
(v) Lead chromosome movement often culminates in spatial fixation
“Leader” chromosome ends also tend, at the end of the transition, to become fixed in space (dotted vertical lines, Figure 2Ai, vi and 2B). An end that has become fixed sometimes remains so until a major transition occurs in a different direction, whereupon it moves away in the new direction (Figure 2Ai, red and green vertical lines until "yellow" transition at [0:51–0:54]). It appears that spatially fixed chromosome ends are physically anchored to an external structure from which they are eventually released when pulled away by the force of another transition. Such associations could also explain dangling of orphans at fixed positions (Figure 1D).
Telomeres occur at the leading ends of chromosomal protrusions
We also examined dynamic behavior of Rap1-GFP, which localizes prominently to chromosome telomeres and less prominently throughout the chromosomes (Klein et al., 1992; Trelles-Sticken et al., 2005), thus permitting differential visualization of both features. 2D movies reveal dynamic, outward-directed chromatin protrusions, essentially every one of which has a bright (telomeric) focus at its leading point (Figure 2E, Movie S11). Correspondingly, 3D reconstructions of entire nuclei, via images obtained from living cells by rapid collection of optical z-sections (streaming), reveal long protrusions with bright foci at their leading ends (Figure 2F). These results confirm that chromosome movement is telomere-led.
The NE undergoes local shape changes in correlation with chromosome movement
Since pachytene chromosome ends are robustly associated with the NE (Introduction), telomere-led chromosome movement should be accompanied by concomitant local NE deformation. 2D time lapse analysis of nuclear pores illuminated with Nup49-GFP (Belgareh and Doye, 1997) show this to be true.
Cones that give rise to short and long protrusions
In vegetative growth or early meiosis, the NE exhibits a static spherical or oval shape (Heun et al., 2001). In contrast, the pachytene NE exhibits highly dynamic local shape changes. Most common are outward-directed angular deformations (Figure 3Ai), which correspond to “cones” in 3D space. Many of these deformations then evolve into thin protrusions or "NE tubes", which fall into two length categories, "short" (up to ~0.3 µm; Figure 3Ai; Movie S12) and "long" (>>0.3 µm; Figure 3Aii–iv; Movie S13–Movie S15). The two types are similar in diameter (~0.3–0.4 µm, n=20 and n=35 each), extend outward at similar velocities (~0.3–0.5 µm/sec, n=30 NE each) and are usually transient, though long tubes can be quite stable (>100 sec).
Figure 3. Pachytene nuclear envelope dynamics.
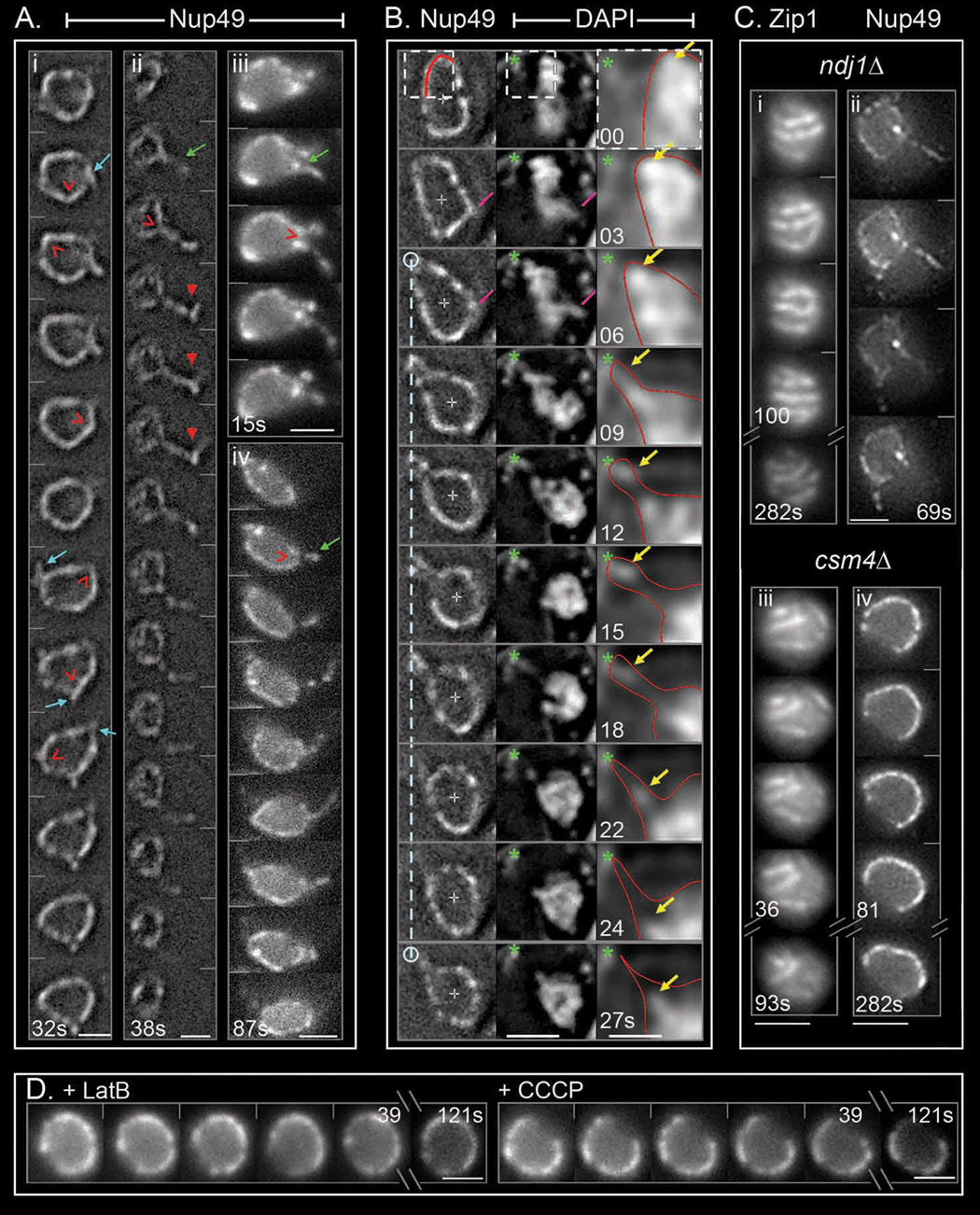
2D time series of pachytene nuclei with Nup49-GFP labeled NE, taken at two- (Ai–ii) or three- (Aiii–iv, B, C) second intervals. Scale bars = 2 µm. (A) Deconvolved frames showing NE angular deformations (red empty arrowheads) eventually evolving into small (i) or long (ii – iv) transient protrusions (turquoise and green arrows indicate the birth of such protrusions, respectively). Long protrusions occasionally exhibit angular path (≥1%, n=200; red arrowhead). (B) Co-visualization of chromosomes and NE (DAPI and Nup49-GFP, respectively; strain NKY3992). Pink lines indicate concomitant NE angular deformation and an outward-directed chromosome (deconvolved images). White dotted vertical line indicates "anchoring" of a NE region. The section corresponding to white dotted squares is magnified in the right column as a less contrasted, deconvolved image (red lines indicate NE outline). Yellow arrows indicate outward movement of a chromosome mass and emergence and movement of an orphan. Green asterisk indicates the continuous presence of a DAPI-stained mitochondrion. (C) Dynamics of Zip1-GFP chromosomes and Nup49-GFP NE in ndj1Δ and csm4Δ pachytene cells (NKY3837, 3993, 4002 and 3994). Due to delayed progression of these mutants through meiosis, cells examined at t=6 hr and 7 hr after meiosis induction, respectively. (D) Reduction of NE movements after addition of either 30 µM LatB (left) or 40 µM CCCP (right).
Correlations with chromosome movement
Dynamic NE shape changes can be correlated with directional, telomere-led chromosome movement by direct co-visualization with DAPI-stained chromosomes. As illustrated in the sequence of Figure 3B (see also Movie S16): (i) At [03–06] sec, an angular deformation, likely with a short protrusion, occurs at the leading end of a single chromosome that protrudes from the main chromosome mass (pink lines). (ii) At a different position in the nucleus, a DAPI signal (yellow arrow) undergoes outward-directed movement, accompanied by elongation of the surrounding NE segment (outlined with a red line inside the box). (iii) This change is followed by movement of the body of the nucleus in the opposite direction via global "drift” (below), leaving behind an "anchored" DAPI signal plus an associated NE segment and an elongated NE connector (Figure 3B; [09–18] sec). (iv) The orphan eventually separates from the NE anchor point and moves to the main chromosome mass while the NE tube collapses (Figure 3B, [18–24] sec).
NE shape changes and chromosome movements are correlated in other ways. Both are eliminated by CCCP or LatB, which cause the NE to assume a regular spherical shape (Figure 3D; Movie S17, Movie S18; below). Chromatin protrusions seen by Rap1-GFP or Zip3-GFP and NE tubes (short and long) have the same width (~0.4±0.1 µm; n=6 protrusions and n=35 tubes) and emanate outward at the same velocity (0.3–0.5 µm/sec; above). Finally, both effects initiate at the onset of zygotene and continue through the end of pachytene (below).
We conclude that directional force is exerted specifically on telomere/NE ensembles, moving the targeted chromosome end and concomitantly deforming the NE at the affected point.
Telomere/NE uncoupling in ndj1Δ
Ndj1 localizes to chromosome ends and mediates their robust meiosis-specific association with the NE, though weaker mitotic-like associations are seen in its absence (Conrad et al., 1997; Trelles-Sticken et al., 2000). Pachytene chromosome motion is strongly reduced in ndj1Δ(Scherthan et al., 2007; Figure 3Ci, Movie S19; n=150 nuclei during 3min). Thus, robust telomere/NE association is required for active chromosome movement. In contrast, dynamic NE deformations still occur in ndj1Δ, similarly to WT (~40% of 100 nuclei over 3min; Figure 3Cii, Movie S20, Movie S21), and are still LatB sensitive (data not shown). Thus: actin-mediated forces are exerted on the NE component of telomere/NE complexes, independent of meiosis-specific telomere association. Correspondingly, ndj1Δ chromosomes still exhibit significant residual movement (Movie S19), which is also eliminated by LatB (not shown), presumably via non-specific chromosome/NE associations.
Aberrant dynamics in csm4Δ
Csm4 is a putative tail-anchored membrane protein which interacts physically and functionally with Ndj1, localizes to the NE in mitotic and meiotic cells, and is strongly induced in meiosis (Conrad et al., 2008; Wanat et al., in press; Kosaka et al., in press). Csm4 is not required for robust telomere/NE association (Wanat et al., in press). Nonetheless, absence of Csm4 dramatically reduces not only pachytene chromosome motion, more than absence of Ndj1 (Figure 3Ciii, Movie S22), but also NE deformations, which are now sporadic and less pronounced than in either WT or ndj1Δ (Figure 3Civ, Movie S23). These phenotypes suggest that Csm4 couples telomere/NE ensembles to the force-generation process.
Identification of actin filaments that hug the NE and extend outward into the global cytoskeletal network
We visualized actin cables illuminated by Abp140-GFP (Yang and Pon, 2002) in combination with DAPI-stained chromosomes, the NE (Nup49-GFP) and/or the SPB (via SPC42-YFP). Meiotic nuclei, unlike their mitotic counterparts, exhibit a complex, highly dynamic, non-polarized cage-like network of cytoplasmic actin cables (Taxis et al., 2006). Two- and three-dimensional movies of living cells at times when most cells are in zygotene or pachytene (t=4 or 5 hours after initiation of meiosis) confirm existence of network at both stages and reveals additional features common to both time points:
- The nucleus is a "black hole" within which no actin cables are visible (Figure 4ABC, Movie S24, Movie S25). Also, irrespective of the highly dynamic nature of the cytoskeletal actin network and ongoing nucleus/chromosome shape changes, the nucleus is usually quite fixed within the cell, with occasional slow drift and abrupt movement (Figure S1).
Figure 4. Actin cables and their relationships to NE and chromosome movement.
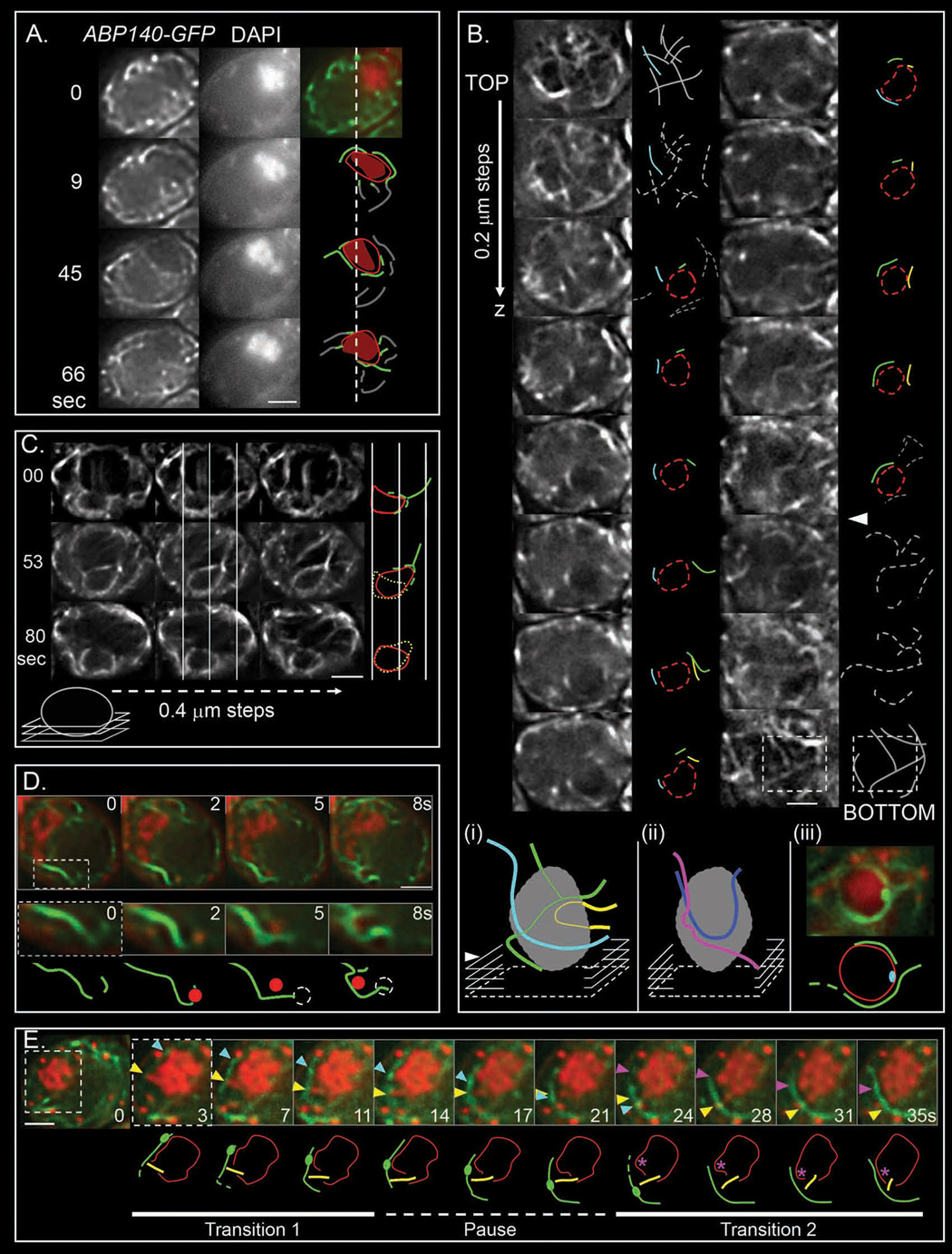
(A) Time series (Movie S24; strain YCT791) of the actin cable network (labeled with Abp140-GFP; left panel) and chromosomal mass (DAPI stained; middle panel). GFP (green) and DAPI (red) signals are merged and interesting features of the actin cable network, “black hole” (text) and DNA mass are schematized with lines, red ellipse and orange surface, respectively (right panel). Cables in contact with the chromosomal mass are outlined in green. Dotted line indicates the original position of the upper left side of the nuclei. (B) Consecutive optical sections (0.2 µm × 600 ms) spanning an entire cell reveal the actin cable network in 3D. Schematic representations show the approximate localization of the NE (red dotted ellipse), as deduced by concomitant DAPI staining, and the actin network (grey lines). Cables in contact with the nucleus are outlined in colors and followed through successive sections. (i) 3D reconstruction of actin cables disposition along the nuclei; planes containing the bottom of the nucleus and of the cell are indicated by white arrowhead and dotted square, respectively. (ii) Schematic 3D reconstruction of a second nucleus. (iii) Position of a zygotene SPB labeled with SPC42-YFP (at 4h; NKY3999) relatively to actin cables, and corresponding schematic representation (Movie S26). (C) Consecutive optical sections were extracted from live-cell 3D frame series at three different time points. NE and actin cables were co-labeled with Nup49-GFP and Abp140-GFP, respectively (NKY3995) and their 3D organization (horizontally) was assessed as a function of time (vertically). Their relative disposition is outlined by white lines, and schematized in red and green, respectively (green signal may eventually but unlikely be a thin NE protrusion). The dotted yellow line tracks the former position of the nucleus. The sections are close to the bottom of the cell, as schematized. (D, E) Time series of the actin cable network (labeled with Abp140-GFP) and chromosomal mass (DAPI stained). (D) Top: merge images of GFP (green) and dapi (red) signals. The section framed by a white dotted box is magnified and schematized with actin in grey and DAPI stained orphan in red. The dotted circles represent the position of the orphan in the former frame. (E) An actin fiduciary mark and an outward directed chromosome are indicated by turquoise and yellow arrowheads, respectively. A pink arrowhead points at the localization of a secondary chromatin deformation. The bottom panel shows the chromatin outline (red circle) as well as the actin cable of interest (green line). The fiduciary mark is schematized as a green patch, the outward directed chromosome as a yellow line, and the position of secondary chromatin movement as a pink asterisk. All cells examined 5 hr – 6 hr after meiosis induction and all scale bars are 2 µm.
- In every cell, a few cables make circumvolutions such that internal segments “hug” the external nuclear periphery, with their ends extending outward beyond the nucleus into the general cytoskeletal network. This pattern appears clearly in 3D reconstructions (Figure 4Bi, Movie S25) and correlates to 2D patterns (Figure 4AC). Actin cable/NE association is very intimate: co-visualization of Nup49-GFP and Abp140-GFP presents only a single nucleus-peripheral signal, even throughout dynamic nucleus shape changes (Figure 4C).
- Nucleus-hugging actin cables are located preferentially at/around point at which the nucleus is juxtaposed to the edge of the cell (Figure 4Bii). The SPB is also nearly always located at the nucleus/cell periphery interface (~84% of cells at t=4 hours, n = 32; Figure 4Biii). It jiggles at a relatively stationary position over the time intervals monitored (1min), with abrupt shifts occasionally observed (data not shown). These features may be due to cytoplasmic astral microtubules holding the SPB in a fixed position, similarly to nuclear positioning during mitosis (Shaw et al., 1997). Correspondingly, nucleus-hugging actin cables often occur in the vicinity of the SPB, e.g. in half of t=4 hours nuclei (n=34) (Figure 4Biii; Movie S26). This correspondence may explain the spatial bias of the "bouquet" (Discussion).
Coordinately dynamic associations between chromosomes and actin cables
Correlated motion
Chromosome movements described above could be explained if, in each transition, a single telomere/NE complex associates with a nucleus-hugging actin cable; moves for a few seconds in conjunction with that cable; and then stops, without necessarily being released. Other chromosomes would move coordinately according to connectedness with this leader. Time lapse co-visualization of Abp140-GFP and DAPI reveals sequences matching this description. In Figure 4E (Movie S27), a chromosome end is closely spatially associated an actin cable (yellow triangle and corresponding yellow line below; t = 3 sec). The end of this chromosome then moves, maintaining its association with the cable, for ~7 sec. (transition 1), pauses, remains in the vicinity of the cable for ~10 sec, and then resumes its cable-associated movement for another ~7 sec. (transition 2). During this sequence, a second chromosome associates with the same cable and then moves in the same way (pink arrow and corresponding pink asterisk below). In all three transitions, ends move at ~ 0.2 µm/sec. These images support the proposed model not only qualitatively but with respect to the frequency, velocity and duration of movement. Such movies also confirm that telomere-led motion results in movement of other chromosomes. In Figure 4E, the overall chromatin pattern changes during the first period of telomere-led movement, does not change during the pause period, and then begins to change again thereafter, concomitant with later movements.
Co-visualization movies also provide examples in which an orphan chromosome is associated with an actin cable through a period of dynamic changes, in correspondence to “dangling of orphans” (above). In Figure 4D, an isolated DAPI signal is associated with a "hook" in an actin cable and then remains associated as the hook changes shape and drifts (from right to left).
A passive attachment mechanism
Co-visualization movies also reveal that lead chromosome end appears to move via passive attachment to a cable which is, independently, being propelled through space. Movement of an actin cable within the nucleus is revealed by brightly-staining "bulges" that remain fixed in position along the cable, thus providing "fiduciary marks" (Yang and Pon, 2002). In Figure 4E, in transition 1, the chromosome end can be seen to move in parallel with the indicated fiduciary mark (Figure 4E; yellow and turquoise triangles at top; yellow line and green bulge, bottom), implying that the end is remaining at a fixed position along the cable, while the cable itself is sliding along the surface of the NE and beyond, thereby mediating curvilinear, outward-directed movement of the associated chromosome end. Cable movement is thought to occur due to addition of subunits at various positions within the network. In further support of a passive attachment mechanism, typical rates of movement for leader chromosome ends correspond to the previously defined speed of actin cable extension (0.3–0.5µm/sec versus ~0.3µm/sec (Yang and Pon, 2002)).
Chromosome and NE movement begin coordinately at zygotene
In yeast cultures undergoing synchronous meiosis, the zygotene and pachytene stages (nuclei with forming and formed SC; Introduction) occur sequentially, as seen e.g. by analysis of Zip1-GFP patterns (Figure 5Ai). The timing with which cells enter and exit each stage is given by cumulative curve analysis (Hunter and Kleckner, 2001; Figure 5Av). Parallel analysis of Nup49-GFP dynamics reveals that NE deformations begin at zygotene and become more pronounced at pachytene, with long protrusions limited to the latter period (Figures 5Aii, iv, v). Chromosome movement exhibits the same progression. Motion was examined independently of the SC via a telomere-localized lacO-LacI-GFP array and via monitoring of Zip-GFP (Experimental Procedures). Step-size distributions of focus displacements reveal that mobility is unchanged from onset of meiosis (t=0) through leptotene (t = 3 hours), increases significantly at zygotene (t= 4 hours) and is even more dramatic at pachytene (t = 5 hours) (Figure 5Aiii, 5B). Interestingly, even though onset of zygotene is tightly linked to a key step of meiotic recombination, motion initiates at approximately the usual time in mid-prophase even in the absence of recombination, as seen by analogous analysis in a spo11Δ mutant (data not shown). In WT meiosis, onset of motion may be part of the basic cellular program which becomes coupled to progression of recombinational progression by coordinating regulatory mechanisms.
Figure 5. Timing, quantification and potential effects of motion.
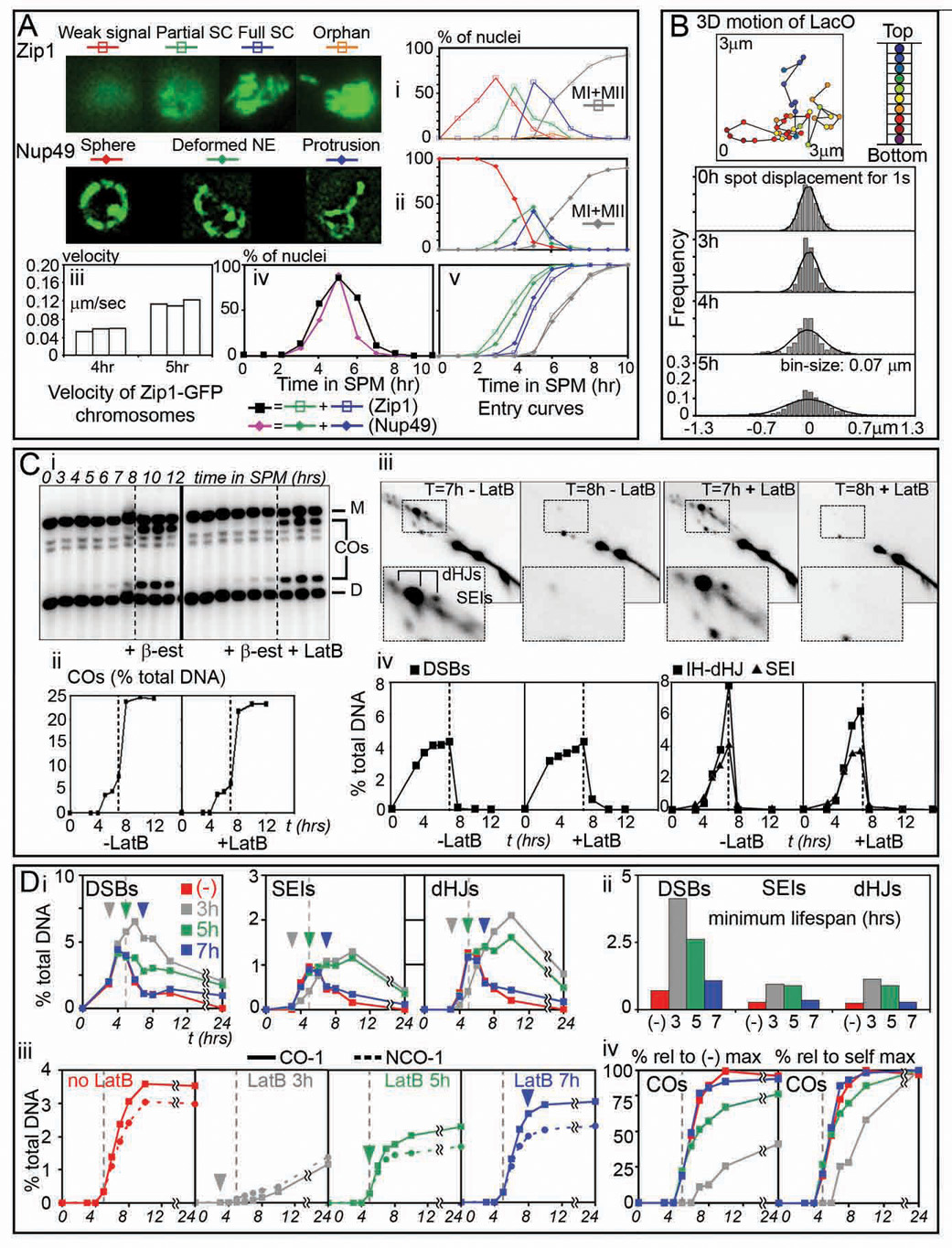
(A) Categories of expanding SC (i) and NE deformations (ii), quantified over time during meiosis in cells expressing either Zip1-GFP or Nup49-GFP (NKY3834 and NKY3992, respectively; n=100). Meiotic divisions (MI+MII) shown in grey. (iii) Mean velocities of random chromosomes (n=30) tracked at one sec intervals in 3 random nucleus during zygotene (4 hr) and pachytene (5 hr). (iv) Proportions of cells showing NE deformations (pink) and full or partial SC chromosomes (black) (same time courses as in i and ii, respectively). (v) Cumulative analysis showing when cells exhibiting NE deformations or onset of SC formation, detailed in (i) and (ii), have entered the corresponding stage. (B) Top: 3D trajectory of a telomeric lacO/lacI-GFP spot (NKY3835). Bottom: step-size distribution histograms of the 2D displacement of projected lacO/lacI-GFP signal, tracked every second for 2 min in 3 random nucleus at 0, 3, 4 and 5 hours in SPM. Predicted Normal distributions are indicated. (C) Meiotic recombination in ndt80-arrested cells, released from arrest at t=7 hr in the presence or absence of LatB (NKY3889). Recombination analyzed at HIS4LEU2 as described (Hunter and Kleckner 2001; Experimental Procedures). (i) Parental homologs, “Dad” (D) and “Mom” (M), DSBs and COs are distinguished on Southern blots via restriction site polymorphism. (ii, iv) DNA species as percent of total hybridizing signal with time after transfer to SPM. (iii) Representative 2D gels of SEIs and dHJs from the timecourses shown in (i). (D) Effect of LatB on recombination progression (NKY3990). Arrowheads indicate the time of LatB addition to aliquots of a same premeiotic culture [color code corresponds to (no addition), addition at t=3, 5, and 7 hours]. Dotted line at 5h helps graph to graph comparison. (i) Quantification of DSBs, SEIs, and dHJs species over time and (ii) corresponding lifespans (i.e. time when 50% of the initiated recombination events have entered or exited the corresponding stage). (iii) CO and NCO formation as percent of total hybridizing signal in LatB time-course experiments. (iv) Left panel: COs level relatively to the maximum level observed in the control culture (−). Right panel: for each culture, relative COs levels when compared to the maximum level observed in the corresponding culture.
LatB disrupts early post-initiation steps of recombination
Programmed DNA recombination, a prominent feature of meiosis (Bishop and Zickler, 2004), initiates at early prophase (leptotene) via programmed double-strand breaks (DSBs). At late leptotene, some DSBs are designated to mature into crossover (CO) recombination products; the remainders are fated for maturation into noncrossovers (NCOs). CO-designation also locally nucleates SC formation (onset of zygotene). For CO recombination, the two DSB ends interact sequentially with a partner DNA segment, giving single-end invasions (SEIs) during zygotene and then, during pachytene, double Holliday junctions (dHJs) which mature into products.
To ask whether chromosome motion might be important for recombination, we first used a system in which a transcription factor required for late stages of CO formation, Ndt80, is expressed from a hormone-inducible promoter (Benjamin et al., 2003). Recombination was assayed at the well-characterized HIS4LEU2 hot spot (Hunter and Kleckner, 2001). Prior to Ndt80 expression, dHJs and some SEIs accumulate while COs are rare. When hormone is added, intermediates disappear, and COs appear, extremely rapidly (Figure 5C). If LatB is added concomitantly with hormone, chromosome movement stops immediately (data not shown) but completion of CO formation is absolutely unaffected (Figure 5C compare +LatB with −LatB for all panels). Thus, motion is not required for these zygotene/pachytene events.
To probe for earlier effects, we added LatB to cells proceeding synchronously through meiosis at various times during the process (see Figure S3). Addition at t=3 hours, when many DSBs have formed but no later species are present (Figure 5Di, (−) LatB data), confers delayed turnover of DSBs, SEIs and dHJs, as shown by increases in the "lifespans" of these species (Hunter and Kleckner, 2001; Figure 5Dii). Also, CO and NCO products form at reduced levels and with delayed timing (Figure 5Diii, iv). Thus, LatB can disrupt recombination if added early enough. Addition of LatB at t=0 has a similar effect to addition at t=3 hours (data not shown), suggesting that the early events of DNA replication and DSB formation are not LatB/motion-sensitive (data not shown). Thus, the period of direct LatB sensitivity is mid-late leptotene through early pachytene, consistent with a role for zygotene/pachytene movement in recombination.
There appear to be three distinct effects of LatB on recombination. (i) Addition of LatB at t=3 hrs differentially affects CO formation versus NCO formation and severely reduces both types of products. This phenotype closely resembles that conferred by absence of Ndt80 (Figure 5C and K.K. unpublished) and thus could reflect an indirect effect of LatB on transcription. (ii) Addition of LatB at t=5 hrs coordinately affects both CO and NCO products, which are modestly reduced in level and modestly delayed in timing (Figure 5Diii, iv). This phenotype resembles that conferred by absence of Ndj1/Csm4 and thus reflects loss of motion per se (Discussion). (iii) LatB affects pachytene steps when added at or before zygotene (t = 4 hours) but not when added directly to pachytene (Ndt80-blocked) cells. Thus, pachytene defects occur only as an indirect consequence of earlier problems.
Discussion
The current study provides new information about the mechanics and movement of budding yeast meiotic chromosomes and points to a possible role for movement in resolution of topological irregularities among pairing chromosomes.
Meiotic pachytene chromosomes are semi-flexible objects
The current study provides the first experimental assessment of the thermal persistence length of a meiotic pachytene chromosome. The obtained value of ~10µm implies that these chromosomes are semi-flexible objects. The stiffness represented by this value could be determined primarily by properties of the (structurally prominent) SC. However, a more intriguing possibility is that the primary determinant is the state of the chromatin. This possibility arises because bending of the SC requires compression of the chromatin along the inner surface of the bend and thus could be limited by the stiffness of that chromatin (Almagro et al., 2004; Marko and Siggia, 1997; Kleckner et al., 2004; below). Indeed, a ~10µm persistence length is close to that predicted by a “bottle-brush” polymer model for meiotic pachytene chromosomes in which axis stiffness makes no contribution (Marko and Siggia, 1997; data not shown). Lp = ~10µm is also interesting because it is similar to that of actin filaments (~17 µm; Ott et al., 1993), consistent with mechanical interplay between the chromosomes and nucleus-hugging actin cables.
How do pachytene chromosomes move?
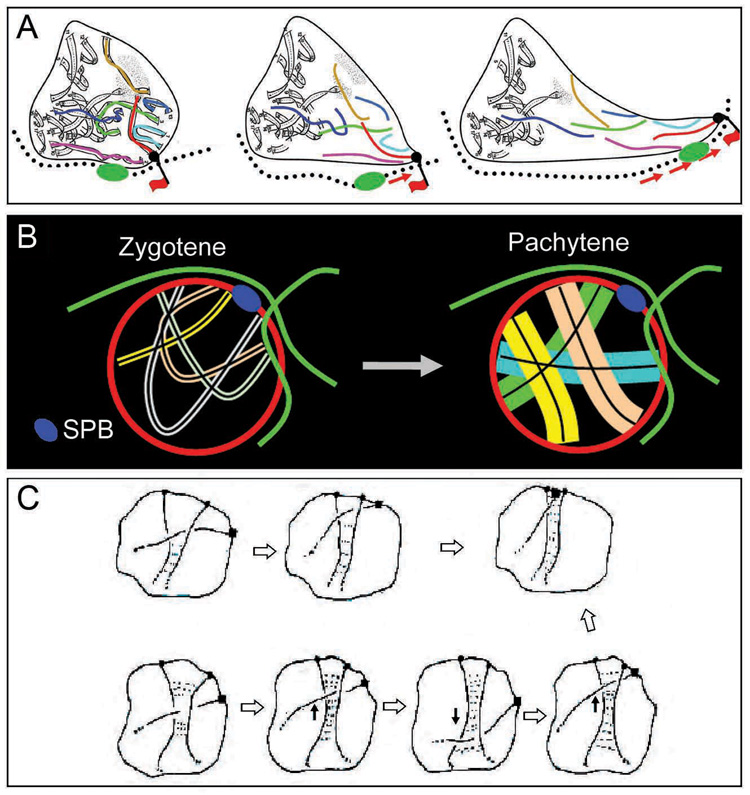
Zygotene/pachytene chromosome motion requires both actin and telomere/NE association (references above; this work). We now find that motion involves cytoskeletal actin cables, rather than intra-nuclear actin as sometimes proposed. We find specifically that segments of the cytoskeletal actin network are intimately associated with the NE and that complexes involving both chromosome ends and their associated NE regions move along these “nucleus-hugging” cables, with concomitant deformation of the NE and nearby chromosomes (Figure 6A).
Figure 6. Zygotene/pachytene chromosome dynamics.
(A) “Piggy-back” model for pachytene movements. A chromosome/NE complex (black ball) binds passively along a nucleus-hugging actin cable (red flag). The cable moves/slides, as indicated by movement of the fiduciary mark (green thickening), dragging the telomere/NE complex in parallel, and concomitantly causing a local deformation of the NE. Intermingled chromosomes move coordinately. (B) Thinner zygotene chromosomes (left) are able to have multiple telomeres simultaneously associated with nucleus-hugging actin cables (thus can colocalize with one another in the vicinity of the SPB) whereas fat, stiff pachytene chromosomes (right) can attach only one at a time and, additionally, can not co-aggregate. (C) Interlocks could be resolved via movement of chromosome ends with accompanying adjustment of synapsis, in response and/or as a driving force (from Kleckner and Weiner, 1993; Copyright holder is Cold Spring Harbor Laboratory Press).
A “piggy-backing” mechanism
We present evidence that motion occurs by passive association of telomere/NE complexes to these cables which are, irrespective of such association, moving dynamically through the cytoplasm. This “piggy-backing” mechanism is analogous to that proposed for movement of mitochondria through the cytoplasm in yeast (Fehrenbacher et al., 2004). Other mechanisms described for actin-mediated movement are: (i) cargo transport mediated by motor proteins and (ii) direct actin polymerization on the object itself. A motor mechanism is unlikely here because it does not predict coordinate movement of chromosome ends with fiduciary marks. A directed polymerization model is unlikely as it would place the moving object at the ends of, rather than along, actin tracks.
An integrated mechanical system
When force is exerted on the NE component of the telomere/NE complex, that motion is constrained by linkages within the system. Those linkage ultimately undergo local mechanical disruption. (i) The lengths of the longest chromosomal protrusions match that of pachytene bivalents [1.1±0.5 µm (n=10) and 1.1±0.35 µm (n=30), respectively], suggesting that outward-directed movement limited by anchoring of the distal end, e.g. to the NE. (ii) Long NE protrusions, in contrast, are usually much longer (2.1±0.7 µm, n=34), suggesting that once chromosome extension is limited, the NE portion of the telomere complex often dissociates and continues to move outward. (iii) Orphans separate from the main chromosome complement at distances matching the maximum protrusion distance (2.3±1.1 µm, n=6 orphans), suggesting that orphans arise when a moving chromosome loses its distal constraints. (iv) NE deformations arise as cones which often evolve into longer or shorter protrusions. This pattern implies initial resistance from the membrane and its substructure (matrix and/or chromatin) followed by dissociation from the substructure and elongation into a (chromosome-containing) membrane tube. (iv) Coordinate movement of spatially-associated chromosomes could involve direct inter-chromosomal linkages (e.g. as in mitotic chromosomes; reviewed in Poirier and Marko, 2003) and/or co-association of ends to a common area of interconnected NE-embedded proteins, e.g. Mps3/Csm4, which exhibit patchy NE staining (Conrad et al., 2007; Conrad et al., 2008; Wanat et al., in press).
Reconciling zygotene and pachytene
As a specific feature of the yeast zygotene stage, chromosome ends tend to colocalize near, but not at the SPB (the “bouquet” configuration”); at pachytene, ends are distributed relatively evenly throughout the nuclear periphery. Movements involved in this progression are presumably provided by actin-mediated telomere-led movement, which occurs specifically at these two stages. What, then, determines the spatial localization at zygotene and the abrogation of that localization at pachytene? The current work provides an important clue: nucleus-hugging actin cables tend to occur preferentially near, but not at, the SPB, and near the edge of the cell, thus explaining how dynamic movements can have the zygotene spatial bias. Moreover, since pachytene chromosome movement involves tracks and preferred focal points, the same spatial bias likely occurs also at that stage but with single chromosome ends involved. Thus, the determinative difference between the two stages is the state of the chromosomes rather than the nature of motion.
We propose that the zygotene/pachytene progression is directly driven by changes in chromatin compaction. In many organisms, including yeast, chromatin is more compact at zygotene and more expanded at pachytene (Kleckner et al., 2004). More compact chromatin will make chromosomes not only more compact, but also floppier (above), thus permitting closer juxtaposition of multiple chromosome ends into the bouquet (Figure 6B, left panel). Expansion of chromatin at early pachytene will then produce fatter, stiffer chromosomes which can only behave independently, like sausages contained within a small sphere (Figure 6B, right panel; see also Figure 3B, 4E; Note S2 and Figure S4).
Role(s) of actin-mediated mid-prophase motion
The bouquet phenomenon has long been discussed in relation to the overall dynamics of homolog pairing and juxtaposition. Classically the bouquet was thought to promote initial homolog contacts by reducing the dimensionality of homology searching. Similarly, dynamic chromosome movements might provide “stirring forces” needed to bring corresponding regions into spatial proximity.
An alternative possibility is that chromosome movement, over short and long time scales, is required to help prevent topological entanglements during the latter stages of homolog juxtaposition and to eliminate such entanglements if they remain into the pachytene stage (for previous discussions see Rasmussen, 1986; Scherthan et al., 1994). We favor this view. We propose specifically that movement permits an entrapped chromosome to be released by sliding out to the end(s) of the entrapping chromosomes (Figure 6C). In support of this idea: (i) The dynamic movements in the three published examples are all of a “back-and-forth” character, “rotationally” in rat (Parvinen and Soderstrom, 1976), along the length of the nucleus in S. pombe (Chikashige et al., 2007), and along actin cables in S. cerevisiae (above). These are not the effects required for “stirring” but are exactly what is needed for resolution of entanglements or other types of non-specific connections. (ii) Primary homolog recognition has already occurred before movement begins (Zickler 2006; Hunter and Kleckner, 2001). Correspondingly, stirring forces needed for juxtaposition can be provided by basal movement known to occur even in mitotic G1 cells (Introduction). (iii) This model can explain how abrogation of motion might affect recombination during early post-DSB stages (above). An irregular topological relationship will preclude the occasional DSB from finding or then mediating close juxtaposition of chromosome axes. Such a defect should be sensed by the cell, not as “damage”, but as “not ready to progress”, resulting in a global temporal delay without (necessarily) any major defect in product formation. The milder effect of LatB on recombination is of this nature, as is that conferred by ndj1 or csm4 mutation(s) (Conrad et al., 2008; Wanat et al., in press; Kosaka et al., in press). Long-term persistence of incomplete interactions might then confer regulatory delays at later stages, thus explaining the indirect effects of LatB on pachytene events. (iv) A maize mutant defective in bouquet co-localization of NE-associated telomeres (perhaps analogously to csm4Δ above) shows an increase in pachytene interlocks (Golubovskaya et al., 2002).
Summary
Taken together the above findings permit a coherent model in which, concomitant with the latter stages of homolog juxtaposition, telomere-led movement in combination with compaction and expansion of chromatin produces an overall dynamic whose primary role is regularization of topological relationships among pairing chromosomes.
Experimental Procedures
Strains and Meiotic Time Courses
Yeast strains are isogenic MATa/MATα derivatives of wild-type SK1 (Table S1). Details on strains constructions are available in (Note S3). Pregrowth and synchronous meiosis as described, with all media equilibrated to 30°C prior to use, and synchrony of DNA replication and divisions monitored (Hunter and Kleckner, 2001).
Analysis of isolated pachytene chromosomes in vitro
Yeast cells of pachytene stage were spheroplasted (Keeney et al., 1997) and resuspended in spheroplast buffer (0.4 M sorbitol, 0.4 M KCl, 40 mM dipotassium phosphate, 0.5 mM MgCl2, Sigma Protease inhibitor cocktail P8215 1:300). One µl was loaded on the slide glass (plain, non-treated) at room temperature, gently mixed with 6 µl of distilled water and covered with a coverslip and incubated at room temperature for ~20 min.
Chemical treatments of pachytene cells
All chemicals are from Sigma and were dissolved in dimethyl-sulfoxide (DMSO). Cell aliquots of interest were incubated in 30 µM Latrunculin A or B 3 or 15 min prior time-lapse, 40 µM CCCP (20 min at 30°C to deplete pre-existing ATP), 15 µg/ml nocodazole (20 min at 30°C) and BDM 200 µm to 100 mM (15 min at 30°C) (Heun et al., 2001). Control aliquots treated with DMSO (up to 1% final concentration) always exhibit motions similar to non-treated cells.
NDT80 arrest and release
A single culture of NKY3889 was synchronized and fractionated into two identical sporulation cultures in 1% potassium acetate and 0.02% raffinose. At t= 7 hours, 1 µm β-estradiol (Sigma) was added to both cultures to release mid-prophase arrest. In one culture, fresh 15 µM LatB (Sigma) was concomitantly added. DNA events were analyzed as in (Hunter and Kleckner, 2001; Probe “A”).
In vivo DAPI staining of chromosomes
DAPI (Sigma) was directly added to cells samples to a final concentration of 10–20 µg/ml. After incubation overnight on ice, ~10% of cells present faintly stained chromosomes, all still showing dynamic movements. Analysis of Zip1-GFP chromosomes confirms that DAPI-treatment does not affect chromosome motion.
Live-Cell imaging
Cell samples were vortexed at full speed for 10 sec and ~3 µl quickly spread onto a glass slide (plain, non-treated). A 22 × 22mm coverslip was added in such a way that several air bubbles remain trapped, with cells located in their vicinity (up to ~30 µm) exhibiting motion for a few minutes. This oxygenation is crucial: motion quickly stops if the air bubble drifts away from an observed cell. Cells are observed at room temperature using an epifluorescence microscope (Zeiss) equipped with GFP, DAPI and TexRed filters, a Cascade 512b CCD camera (Roper Scientific), and a PIFOC® piezo device (Physik Instrumente) to drive a 100X oil immersion objective (NA 1.45) for acquiring Z-stacks. Images were acquired using Metamorph™ software. 2 color movies were made from sequential acquisition of frames alternating with switch of filters, generating a ~300 ms discrepancy between two superimposed frames.
Image analysis
Deconvolution of 2D and 3D acquisitions was performed using AutoDeblur®. For bending and step size studies, outlines of Zip1-GFP chromosomes were drawn by thresholding and centroids automatically determined by Integrated Morphometry Analysis function of Metamorph™. For tracking of LacO-telomeres, Z-stacks time-series were recorded every 3sec during 3min, planes separated by 0.20 µm (21 total). For each plane, X- and Y-coordinates of the LacI-GFP spot centroid were calculated by SpotTracker2D ImageJ Plug-in (Sage et al. 2005). The Z-coordinate of the spot was approximated as coordinate of the plane containing the brightest centroid. Velocities were calculated from relative positions of spots in successive Z-series (three independent nuclei per time point).
Supplementary Material
Acknowledgements
We thank A.Shinohara, E.Alani, J.Wanat and M.Dresser for unpublished data; A.Amon, J.Fung, E.White, S.Gasser, I.Nachman, K.Thorn, A.Murray, K.Nasmyth and D.Knop for plasmids/strains; and R.Padmore and D.Zickler for reproduction of images in Figure S4. Support for M.P. was provided by Harvard University. R.K., S.K., K.K. and this research were supported by grants to N.K. (NIH RO1 GM-044794 and/or NIH RO1 GM-025326).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarwal S, Roeder GS. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell. 2000;102:245–255. doi: 10.1016/s0092-8674(00)00029-5. [DOI] [PubMed] [Google Scholar]
- Almagro S, Riveline D, Hirano T, Houchmandzadeh B, Dimitrov S. The mitotic chromosome is an assembly of rigid elastic axes organized by structural maintenance of chromosomes (SMC) proteins and surrounded by a soft chromatin envelope. J. Biol. Chem. 2004;279:5118–5126. doi: 10.1074/jbc.M307221200. [DOI] [PubMed] [Google Scholar]
- Bass HW. Telomere dynamics unique to meiotic prophase: formation and significance of the bouquet. Cell Mol. Life Sci. 2003;60:2319–2324. doi: 10.1007/s00018-003-3312-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh N, Doye V. Dynamics of nuclear pore distribution in nucleoporin mutant yeast cells. J. Cell Biol. 1997;136:747–759. doi: 10.1083/jcb.136.4.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin KR, Zhang C, Shokat KM, Herskowitz I. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 2003;17:1524–1539. doi: 10.1101/gad.1101503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DK, Zickler D. Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell. 2004;117:9–15. doi: 10.1016/s0092-8674(04)00297-1. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Haraguchi T, Hiraoka Y. Another way to move chromosomes. Chromosoma. 2007;116:497–505. doi: 10.1007/s00412-007-0114-8. [DOI] [PubMed] [Google Scholar]
- Conrad MN, Dominguez AM, Dresser ME. Ndj1p, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science. 1997;276:1252–1255. doi: 10.1126/science.276.5316.1252. [DOI] [PubMed] [Google Scholar]
- Conrad MN, Lee CY, Wilkerson JL, Dresser ME. MPS3 mediates meiotic bouquet formation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2007;104:8863–8868. doi: 10.1073/pnas.0606165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad MN, Lee CY, Chaol G, Shinohara M, Kosaka H, Shinohara A, Conchello JA, Dresser ME. Telomeres Drive Rapid Movements Regulated by Chromosome Status in Meiotic Prophase. (in press) [Google Scholar]
- Doi M, Edwards SF. The Theory of Polymer Dynamics. Oxford, UK: The Clarendon Press; 1986. [Google Scholar]
- Fehrenbacher KL, Yang HC, Gay AC, Huckaba TM, Pon LA. Live cell imaging of mitochondrial movement along actin cables in budding yeast. Curr. Biol. 2004;14:1996–2004. doi: 10.1016/j.cub.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Golubovskaya IN, Harper LC, Pawlowski WP, Schichnes D, Cande WZ. The pam1 gene is required for meiotic bouquet formation and efficient homologous synapsis in maize (Zea mays L.) Genetics. 2002;162:1979–1993. doi: 10.1093/genetics/162.4.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper L, Golubovskaya I, Cande WZ. A bouquet of chromosomes. J Cell Sci. 2004;117:4025–4032. doi: 10.1242/jcs.01363. [DOI] [PubMed] [Google Scholar]
- Heun P, Laroche T, Raghuraman MK, Gasser SM. The positioning and dynamics of origins of replication in the budding yeast nucleus. J. Cell Biol. 2001;152:385–400. doi: 10.1083/jcb.152.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- Kleckner N, Weiner BM. Potential advantages of unstable interactions for pairing of chromosomes in meiotic, somatic, and premeiotic cells. Cold Spring Harbor Symp. Quant. Biol. 1993;58:553–565. doi: 10.1101/sqb.1993.058.01.062. [DOI] [PubMed] [Google Scholar]
- Kleckner N, Zickler D, Jones GH, Dekker J, Padmore R, Henle J, Hutchinson J. A mechanical basis for chromosome function. Proc. Natl. Acad. Sci. USA. 2004;101:12592–12597. doi: 10.1073/pnas.0402724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Laroche T, Cardenas ME, Hofmann JF, Schweizer D, Gasser SM. Localization of RAP1 and topoisomerase II in nuclei and meiotic chromosomes of yeast. J. Cell Biol. 1992;117:935–948. doi: 10.1083/jcb.117.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran RI, Thakar R, Spector DL. Chromatin dynamics and gene positioning. Cell. 2008;132:929–934. doi: 10.1016/j.cell.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka H, Shinohara M, Shinohara A. Csm4-dependent telomere movement on nuclear envelope promotes meiotic recombination. doi: 10.1371/journal.pgen.1000196. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko JF, Siggia ED. Polymer models of meiotic and mitotic chromosomes. Mol. Biol. Cell. 1997;8:221722–221731. doi: 10.1091/mbc.8.11.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott A, Magnasco M, Simon A, Libchaber A. Measurement of the persistence length of polymerized actin using fluorescence microscopy. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics. 1993;48:R1642–R1645. doi: 10.1103/physreve.48.r1642. [DOI] [PubMed] [Google Scholar]
- Parvinen M, Soderstrom KO. Chromosome rotation and formation of synapsis. Nature. 1976;260:534–535. doi: 10.1038/260534a0. [DOI] [PubMed] [Google Scholar]
- Poirier MG, Marko JF. Micromechanical studies of mitotic chromosomes. Curr. Top. Dev. Biol. 2003;55:75–141. doi: 10.1016/s0070-2153(03)01002-0. [DOI] [PubMed] [Google Scholar]
- Rasmussen SW. Initiation of synapsis and interlocking of chromosomes during zygotene in Bombyx spermatocytes. Carlsberg Res. Commun. 1986;51:401–432. [Google Scholar]
- Sage D, Neumann FR, Hediger F, Gasser SM, Unser M. Automatic tracking of individual fluorescence particles: application to the study of chromosome dynamics. IEEE Trans. Image Process. 2005;14:1372–1383. doi: 10.1109/tip.2005.852787. [DOI] [PubMed] [Google Scholar]
- Scherthan H. Telomere attachment and clustering during meiosis. Cell. Mol. Life Sci. 2007;64:117–124. doi: 10.1007/s00018-006-6463-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H, Wang H, Adelfalk C, White EJ, Cowan C, Cande WZ, Kaback DB. Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2007;104:16934–19939. doi: 10.1073/pnas.0704860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw SL, Yeh E, Maddox P, Salmon ED, Bloom K. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J.Cell Biol. 1997;139:985–994. doi: 10.1083/jcb.139.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxis C, Maeder C, Reber S, Rathfelder N, Miura K, Greger K, Stelzer EH, Knop M. Dynamic organization of the actin cytoskeleton during meiosis and spore formation in budding yeast. Traffic. 2006;7:1628–1642. doi: 10.1111/j.1600-0854.2006.00496.x. [DOI] [PubMed] [Google Scholar]
- Tessé S, Storlazzi A, Kleckner N, Gargano S, Zickler D. Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc Natl Acad Sci U S A. 2003;100:12865–12870. doi: 10.1073/pnas.2034282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trelles-Sticken E, Adelfalk C, Loidl J, Scherthan H. Meiotic telomere clustering requires actin for its formation and cohesin for its resolution. J. Cell Biol. 2005;170:213–223. doi: 10.1083/jcb.200501042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trelles-Sticken E, Dresser ME, Scherthan H. Meiotic telomere protein Ndj1p is required for meiosis-specific telomere distribution, bouquet formation and efficient homologue pairing. J. Cell Biol. 2000;151:95–106. doi: 10.1083/jcb.151.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanat J, Kim K, Koszul R, Zanders S, Weiner B, Kleckner N, Alani E. Csm4, in collaboration with Ndj1, mediates telomere-led chromosome dynamics and recombination during yeast meiosis. doi: 10.1371/journal.pgen.1000188. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HY, Burgess SM. Ndj1, a telomere-associated protein, promotes meiotic recombination in budding yeast. Mol Cell Biol. 2006;10:3683–3694. doi: 10.1128/MCB.26.10.3683-3694.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, West RR, McIntosh JR, Hiraoka Y. A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J. Cell Biol. 1999;145:1233–1249. doi: 10.1083/jcb.145.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HC, Pon LA. Actin cable dynamics in budding yeast. Proc. Natl. Acad. Sci. USA. 2002;99:751–756. doi: 10.1073/pnas.022462899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D. From early homologue recognition to synaptonemal complex formation. Chromosoma. 2006;115:158–174. doi: 10.1007/s00412-006-0048-6. [DOI] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 1998;32:619–697. doi: 10.1146/annurev.genet.32.1.619. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.