Summary
γδ T cells contribute uniquely to host immune competence, but how they do so remain unclear. Here, by analyzing T10/T22-specific γδ T cells in mice with different T10/T22 expression patterns, we find that encountering antigen in the thymus is neither required nor inhibitory for the development of these cells. Instead, ligand recognition determines which of two distinct functional subsets γδ T cells will become. When triggered through the TCR, lymphoid-γδ T cells that encounter ligand during development produce IFNγ, while those that develop in the absence of ligand make IL-17, a major inducer of granulopoiesis during inflammation. Indeed, we find large fractions of IL-17+ γδ T cells from the draining lymph nodes immediately after peptide/CFA immunization and days before the appearance of antigen specific IL-17+ αβ T cells. This suggests a critical role for γδ T cells as ‘initial providers’ of IL-17 in an inflammatory response to novel antigens.
Introduction
γδ T cells and αβ T cells are present together in all but the most primitive vertebrates. In most adult animals, αβ T cells are the predominant T-cell population and also perform many of the well-defined functions attributed to T cells. Nevertheless, in experimental systems where αβ T cell and/or γδ T cell deficient mice are infected with pathogens, the absence of both T-lymphocyte populations generally results in a more severe infection. In particular, γδ T cell-deficient mice usually fare worse in neutrophil-dominated inflammatory responses, heat-, ozone- or chlorine-induced injuries and in bacterial infections (Nocardia asteroids, Klebsiella pneumonia) (King et al., 1999; Koohsari et al., 2007; Moore et al., 2000; Toth et al., 2004). In these cases, fewer infiltrating neutrophils, increased bacterial load, early dissemination and higher mortality rates are noted. Furthermore, patients with bacterial, parasitic and viral infections often have increased numbers of γδ T cells in the peripheral blood (from <5% in healthy individuals to >45% in patients) (De Paoli et al., 1990; Ho et al., 1990; Jason et al., 2000). It was also reported that γδ T cells are over-represented among infiltrating T cells in the early but not in the late lesions of MS patients (up to 20-30% of the total number of T cells) (Wucherpfennig et al., 1992). These observations suggest that γδ T cells play a unique role in the initial host response to tissue damage and infection. However, it is unclear why and how γδ T cells are preferentially suited for this task.
γδ T cells, like αβ T cells, develop in the thymus before entering the periphery. In the case of αβ T cells, thymic development entails ligand driven positive and negative selection, which determine what αβ T cell can recognize (Huseby et al., 2005; Van Laethem et al., 2007); and whether these T cells will develop into CD4+ helper or CD8+ cytolytic T cells. Thus, understanding γδ T cell selection in the thymus could provide valuable clues as to their likely targets and function.
Previous analysis of the role of thymic selection in the establishment of a functional γδ T cell repertoire has focused mostly on the studies of KN6 and G8 γδ TCR transgenic mice. KN6 and G8 are two independently derived γδ T cell clones that recognize the same closely related, β2m-associated non-classical MHC class I molecules, T10 and T22 (Ito et al., 1990; Schild et al., 1994; Weintraub et al., 1994). In both systems, transgenic mice were crossed to the C57BL/6 (B6) background, which express both the inducible T10 and constitutively expressed T22; BALB/c mice which only express T10, or to the β2m-/- background which do not have cell surface T10/T22 expression. It was reported that in C57BL/6 mice, there were significantly lower numbers of or no transgenic T cells in the spleens of BALB/c mice. There were also fewer G8 γδ thymocytes (Dent et al., 1990). While KN6 thymocytes were present, their ability to secrete IL-2/IL-4 and to proliferate was much reduced (Bonneville et al., 1990). When G8 and KN6 transgenic T cells were expressed in β2m-/- mice (B6 background), there were fewer transgenic cells in the periphery and transgenic thymocytes showed a reduced ability to secrete cytokine and proliferate when stimulated in vitro (Pereira et al., 1992; Wells et al., 1991). Based on these observations, it was concluded γδ T cells, similar to αβ T cells, undergo ligand driven positive and negative selection in the thymus. However, analyzing the same G8 transgenic mice, Schweighoffer and Fowlkes found that G8 T cells were able to mature in β2m-/- mice, contradicting the conclusion that positive selection is required (Schweighoffer and Fowlkes, 1996).
In addition to the KN6 and G8 transgenic systems, the role of ligand recognition in the development of murine skin-dendritic epidermal T cells (DETCs) has also been analyzed. These γδ T cells express the same TCR and are the first to appear during fetal thymic development (Havran and Allison, 1988). While the ligand of these cells has yet to be identified, DETCs are reactive to keratinocytes in a TCR dependent manner (Havran et al., 1991). Here, all experimental results suggest that encountering thymic ligand is necessary for DETCs to migrate to the skin and to acquire their ability to react to keratinocytes (Lewis et al., 2006; Mallick-Wood et al., 1998; Xiong et al., 2004).
Previously, we found that a sizable population (0.1-1%) of γδ T cells in normal un-immunized mice recognize T10/T22 (Crowley et al., 2000). Surprisingly, a comparable frequency of T10/T22-specific γδ T cells was also found in β2m-/- mice (Crowley, 1998). Moreover, in analyzing the antigen recognition determinant of T10/T22-specific γδ T cells, we found that the T10/T22 specificity is largely encoded by amino acid residues on Vδ and Dδ gene elements which are brought together by rearrangement; and that 0.85% of the non-selected TCRδ sequences (from CD3ε-deficient murine thymocytes and from out-of-frame VDJ recombination events) contain the T10/T22 recognition motif (Shin et al., 2005). This is within the observed range of T10/T22-specific γδ T cells in normal mice. Therefore, this repertoire seems to be determined largely by gene rearrangement instead of ligand dependent selection-observations, which present a significant departure from the analysis of T10/T22-specific γδ TCR transgenic mice.
If γδ T cells require no ligand driven positive or negative selection to develop, then the repertoire of γδ T cell antigens will be significantly enlarged to include pathogens, which do no cross-react to host thymic molecules, as well as infection- or stress-induced antigens which express in the thymus, such as T10/T22. Furthermore, although γδ T cells and αβ T cells secrete similar cytokines and mount cytolytic responses, there is very little information on how γδ T cell effector functions develop. Since we have developed a T22 tetrameric staining reagent (Crowley et al., 2000), which allows us to follow and analyze this substantial population of T10/T22-specific γδ T cells in normal non-transgenic mice, we decided to re-evaluate these issues.
Here, we find that (1) encountering antigen in the thymus is neither required nor inhibitory for the development of T10/T22-specific γδ T cells, (2) self-dimerization of γδ TCRs may be sufficient to drive γδ thymocytes development, (3) a sizable number of the γδ T cells in normal mice are phenotypically and functionally similar to β2m-/- T10/T22-specific cells, suggesting that most γδ T cells in the periphery have yet to encounter antigen, and (4) when activated through the TCR, cells with prior antigen exposure produce IFNγ, while cells that develop in the absence of ligand make IL-17, a major initiator of inflammation and that is elicited without prior antigen. Indeed, we find that γδ T cells are major IL-17 producers in the draining lymph nodes after peptide/Complete Freund’s Adjuvant (CFA) immunization. These results suggest that a functional γδ T repertoire can be divided into two subsets, influenced by ligand recognition, and uniquely equipped to initiate and regulate the inflammatory response.
Results
The presence of T10/T22-specific γδ T cells is neither inhibited nor enhanced by host T10/T22 expression
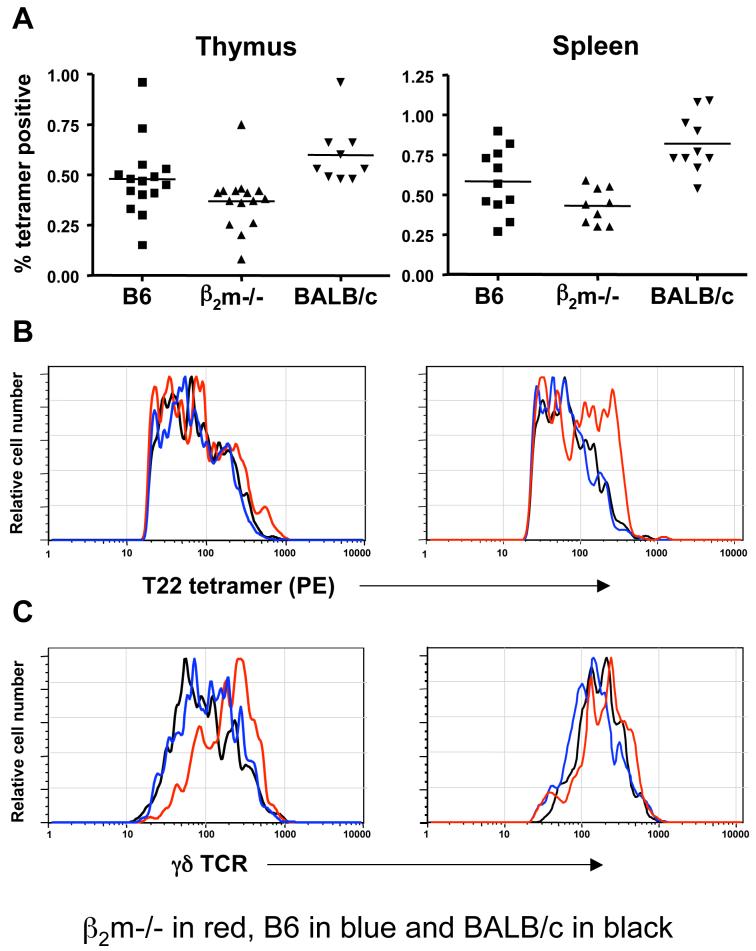
To determine the role of ligand recognition in the development of a functional γδ T cell repertoire, we first analyzed the frequency of T10/T22-specific γδ T cells in C57BL/6, BALB/c, and β2m-/- mice using a fluorescently labeled T22 tetramer. T10/T22-specific γδ T cells were found within a similar range (0.1-1% of the total γδ T cell population) in the thymuses, spleens and the inter-epithelial lymphocyte (IEL) compartments of all three strains of mice (Fig. 1A, and data not shown). Regardless of the genetic background of the mice, T10/T22-specific γδ T cells also show a spectrum of affinity to T22 as evaluated by tetramer staining intensity (Fig. 1B). They express TCRs that are diverse in V gene usage and CDR3 junctional sequences but with the same (W-SEGYEL) recognition motif in the TCRδ CDR3 (data not shown) as previously described for T10/T22-specific γδ T cells from B10.BR mice (Shin et al., 2005). These results show that endogenous ligand expression does little to influence the number, TCR usage and the recognition motif of T10/T22-specific γδ T cells.
Figure 1. Frequency and surface phenotypes of T10/T22-specific γδ T cells from mice with and without T10/T22 expression.
(A) Frequency; (B) tetramer staining intensity; and (C) TCR levels of T10/T22 specific γδ thymocytes (left column) and splenocytes (right column) from C57BL/6 (B6) (T10+T22+) (blue), C57BL/6 β2m-/- (T10-T22-) (red) and BALB/c (T10+T22-) (black) mice. Each symbol or histogram represents the result of one mouse.
(D) Representative FACS analysis of tetramer positive (red) and tetramer negative (blue) γδ thymocytes and splenocytes for the expression of CD122 (IL-2/IL-15R β chain) from B6, BALB/c and β2m-/- mice.
(E) γδ hymocytes, (F) γδ splenocytes and (E) lymph node γδ T cells from B6 and β2m-/- mice for the expression of surface markers commonly associated with antigen recognition by T cells.
γδ T cells were first enriched with the pan-γδ antibody GL-3, followed by staining with PE labelled T22 tetramer, antibodies to cell surface markers and Cy5PE labelled anti- CD19, anti-TCRβ, Armenian Hamster isotype IgG2κ, streptavidin and propidium iodide (PI) as described in Experimental Procedures. Cy5PE and PI positive cells were excluded from analysis. Representative histogram plots are shown (n≥3).
A similar frequency of T10/T22-specific cells was observed in the thymuses and spleens of mice lacking both β2m and class II MHC molecules, whether or not they were treated with cyclosporin A (to inhibit αβ T cell positive selection) (data not shown). These results show that the development of the T10/T22-specific γδ repertoire is neither inhibited by nor dependent on the expression of T10/T22, of class II MHC or any other β2m-associated molecules, nor does it require signaling pathways utilized for αβ T cell positive selection. Thus, the development of T10/T22-specific γδ T cells does not require either positive or negative selection, as described for the development of αβ T cells.
Encountering T10/T22 is not necessary for T10/T22-specific γδ thymocytes to signal through the TCR or to exit from the thymus
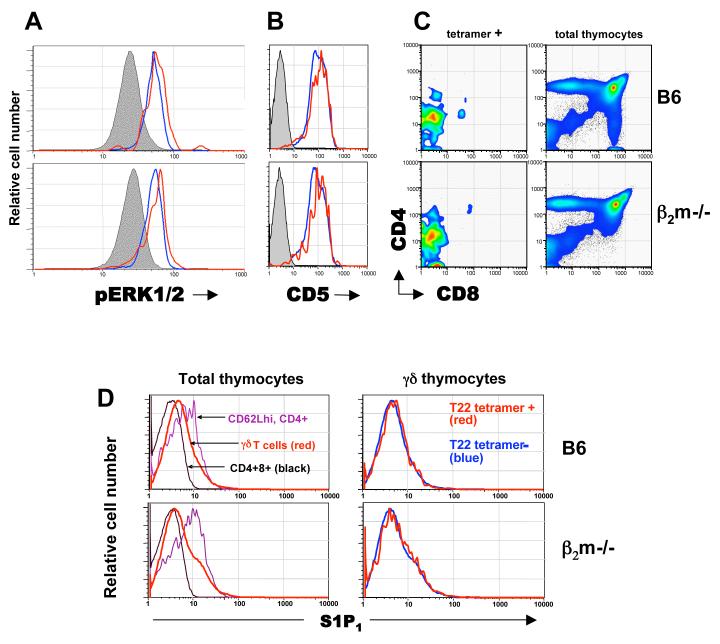
The maturation of γδ thymocytes requires signalling through the TCR (Haks et al., 2005; Hayes et al., 2005). T10/T22-specific γδ thymocytes from different ligand expressing environments show similarly high levels of phosphorylated ERK1/2 (pERK1/2) (Fig. 2A), a TCR signaling intermediate in the MAP kinase pathway, and of CD5 (Fig. 2B), a stable indicator of signaling strength (Azzam et al., 1998; Tarakhovsky et al., 1995). In fact, the pERK1/2 levels in B6 and β2m-/- γδ thymocytes, regardless of their T10/T22 specificities are higher than those from CD4+CD8+ and CD4+CD8- αβ thymocytes (Fig. 2A, S1). Although it was reported that in the absence of ligand, KN6 TCR transgenic γδ T cells adopt an αβ T cell fate as defined by CD4/CD8 expression and lowered γδ TCR levels (Haks et al., 2005), we find that tetramer positive γδ T cells in β2m-/- mice remain CD4-CD8-(Fig. 2C) and show no TCR down-regulation (Fig. 1C). In this regard, although a >20-fold increase in CD4+8+ KN6+ T cells was reported in the thymuses of KN6 β2m-/- RAG-/- mice, there was also a two-fold increase in the number of CD4-8-transgenic T cells (Haks et al., 2005). Thus, the expansion of the double positive cells may not have been at the expense of the γδ lineage cells in that setting.
Figure 2. Host T10/T22 expression does not affect signalling, lineage commitment and S1P1 levels of T10/T22-specific γδ thymocytes.
Representative FACS analysis of direct ex vivo γδ thymocytes (n=3), (A) Intracellular expression of phosphorylated ERK1/2 in tetramer+ (red), total γδ thymocytes (blue) and CD4+8+ thymocytes (shaded).
(B) CD5 expression on tetramer+ (red) and tetramer- (blue) γδ hymocytes, isotype control on GL-3+ γδ thymocytes (shaded).
(C) CD4 and CD8 expression on tetramer positive GL-3+ γδ thymocytes (left column), or total thymocytes (right column) from B6 and β2m-/- mice.
(D) Sphingosine-1 Phosphate receptor-1 (S1P1) surface expression on CD4+CD8+ (black), mature CD4+CD62Lhi αβ T cells that have gone through positive and negative selection and ready for thymic exit (purple), GL-3+ γδ thymocytes (red) (left column); or tetramer positive (red) and negative (blue) GL-3+ γδ thymocytes (right column) from B6 and β2m-/- mice. S1P1 expression was identified with a polyclonal rabbit antiserum to S1P1 (Jason Cyster, UCSF) and detected with donkey anti-rabbit IgG FITC.
Up-regulation of sphingosine-1-phosphate receptor 1 (S1P1) is necessary for mature thymocytes to exit the thymus (Matloubian et al., 2004), and we find no observable difference in the high S1P1 expression between tetramer-positive and tetramer-negative thymocytes from B6 and β2m-/- mice (Fig. 2D). Taken together, these results indicate that endogenous thymic ligand expression does very little to constrain γδ thymocyte development, thymic exit and maturation. These aspects represent a significant departure from what has been known for αβ T cell development and what has been thought previously about γδ T cell selection.
TCR dimerization provides a possible ligand independent mechanism for signaling through the TCR
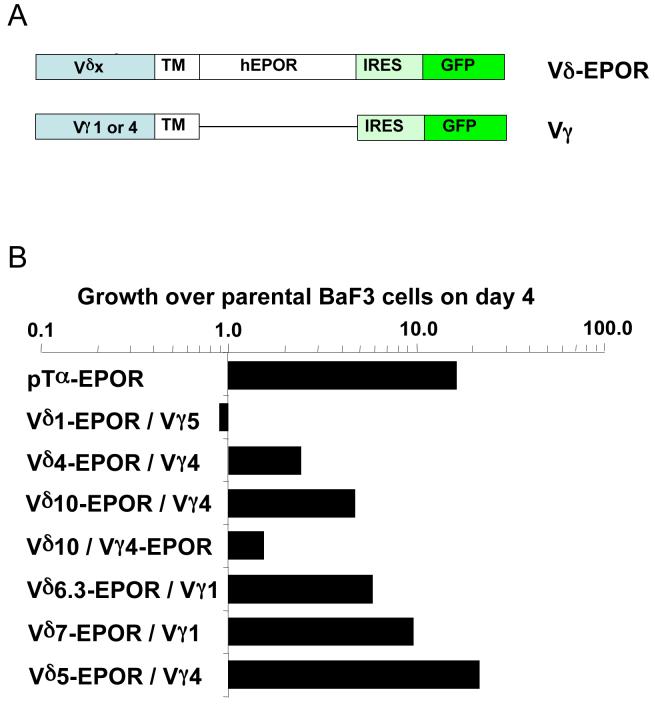
In αβ thymocyte development, autonomous signaling mediated by the dimerization of the pre-Tα has been suggested for pre- T cell activation (Yamasaki et al., 2006). Interestingly, the crystal structure of the G8 γδ TCR with its T22 ligand showed a dimerized complex between the TCR Vδ domains (Adams et al., 2005). We therefore tested whether or not γδ TCRs can dimerize to induce signaling without cross-linking by ligand. The extracellular domain of the human erythropoietin receptor (EPOR) gene was replaced with the TCRδ chain extracellular domain and expressed together with the extracellular domain of the TCRγ chain in BaF3 cells (Fig. 3A). EPOR signaling mediated through dimerization of the extracellular domain allows these cells to grow in the absence of IL-3 (Yoshimura et al., 1990). We found that all of the Vγ/Vδ-EPOR fusion protein pairs, except for Vγ5/Vδ1-EPOR, were able to promote the growth of BaF3 cells after IL-3 withdrawal (Fig. 3B, and Fig. S2). The Vδ1/Vγ5 TCR we tested here is from the DETCs, which constitute the first wave of T cells that appear during fetal thymic development and later populate the murine skin. Various reports suggest that these cells require antigen-driven positive selection for development (Lewis et al., 2006; Mallick-Wood et al., 1998; Xiong et al., 2004). Thus, our results offer a possible ligand-independent mechanism for signaling through the γδ TCR (with the exception of the DETC TCR), which may be sufficient to drive the development of γδ thymocytes, which have high pERK1/2 levels (Fig. S1). In this context, Prinz et al. analyzed the development of γδ thymocytes and suggested that surface expression of γδ TCR on thymocytes is the only “check point” in the development which involves a phase of proliferation with magnitude similar to that induced by Pre-Ta (Prinz et al., 2006).
Figure 3. TCRδ-δ interactions as assayed by the induction of BaF3 cell autonomous growth.
(A) Schematic representation of TCR-EPOR chimeric genes. The TCRγ chain is fused to the transmembrane region of the human EPOR but lacking the cytoplasmic hEPOR domain, whereas TCRδ is fused to the transmembrane and cytoplasmic domains of the hEPOR (also see Experimental Procedures).
(B) γδ TCR-EPOR mediated signaling allows BaF3 cell growth in the absence of IL-3. The relative growth of TCR-EPOR expressing BaF3 cells to parental BaF3 cells day 4 after IL-3 withdraw. Ratios defined as: [number of chimeric EPOR transfectants at day 4 after IL-3 removal / number of transfectants at day 0] / [number of parental BaF3 cells at day 4 after IL-3 removal / number of parental BaF3 cells at day 4]. The survival curves of the TCR expressing BaF3 cells are shown in supplementary Figure S2. pTα-EPOR as previously reported induces BaF3 cell growth (Yamasaki et al., 2006) and serves as a positive control. The Vδ10/Vγ4 G8 TCR was assayed two different ways: Vδ10-EPOR/Vγ4 and Vγ4-EPOR/Vδ10. Consistent with the crystal structure of G8 (Adams et al., 2005), the Vδ chain mediates dimerization, Vδ10-EPOR/Vγ4 signals better than Vγ4-EPOR/Vδ10.
A large fraction of lymphoid γδ T cells exhibit an ‘antigen-naïve’ phenotype
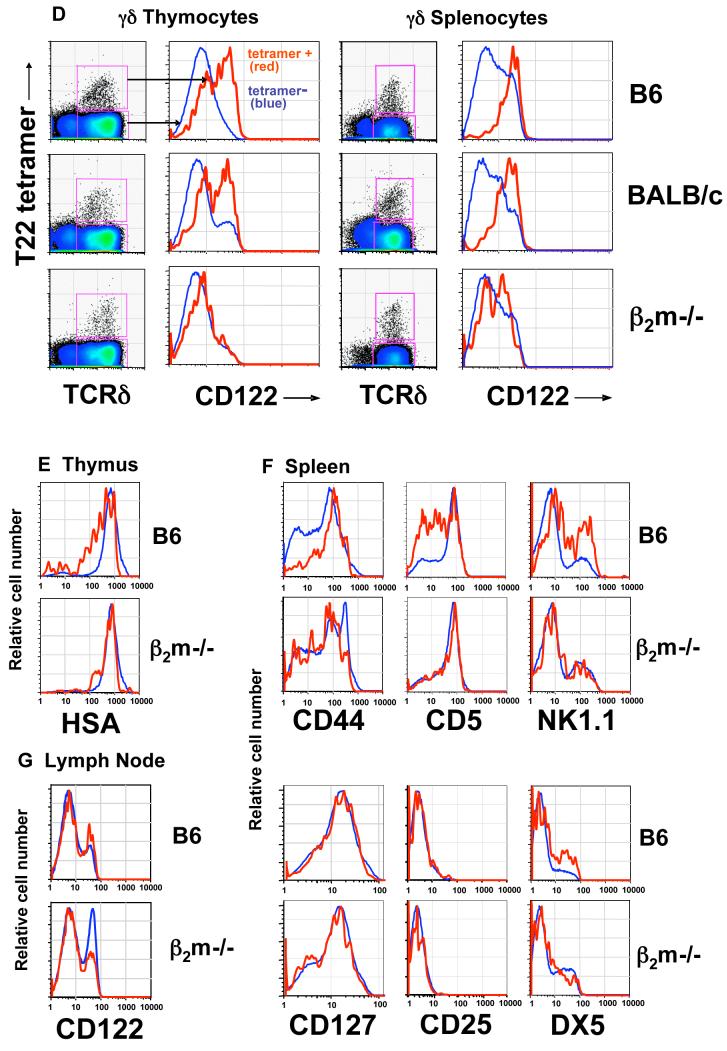
While T10/T22-specific thymocytes in B6, BALB/c, and β2m-/- mice are similar in number and have comparable tetramer staining intensities, their phenotypes suggest that majority from B6 and BALB/c mice encounter antigen and those from β2m-/- mice do not. In particular, T10/T22-specific cells from B6 and BALB/c mice express lower levels of TCR (Fig. 1C) and heat stable antigen (HSA, or J11D), but higher levels of the IL-2/IL-15 receptor common β chain (CD122) than those from β2m-/- mice (Fig. 1D, E). αβ Thymocytes that have encountered ligand express lower levels of HSA, and a HSAlo profile has also been reported for G8 γδ TCR transgenic thymocytes that develop in the presence of ligand (Schweighoffer and Fowlkes, 1996; Wells et al., 1991). Furthermore, the upregulation of CD122 has been used as an indicator of self-ligand recognition in αβ thymocytes (Hanke et al., 1994) and during murine skin γδ dendritic epidermal cells (DETCs) development (Xiong et al., 2004).
Keeping with the idea that exposure to ligand is not necessary for γδ T cells to exit from the thymus, T10/T22-specific γδ splenocytes from β2m-/- mice exhibit the phenotype of T cells which have not encountered antigen. Specifically, the majority of these cells are CD44lo,int and CD122lo,int while splenocytes from B6 mice are CD44hi, CD122hi, and a greater fraction of these cells are DX5+, NK1.1+, and CD5lo (Fig. 1F). In addition, there are consistently lower numbers of γδ splenocytes with very high tetramer staining in B6 mice than in β2m-/- mice (Fig. 1B) suggesting that cells with TCRs possessing very high affinity to T10/T22 have been deleted in B6 mice. Since the G8 TCR has the highest affinity to T10/T22 among all the T10/T22 specific receptor pairs that we have analyzed (Shin et al., 2005) (and data not shown), it is not surprising that G8 transgenic T cells were not found in the spleen of B6 mice (Dent et al., 1990).
Significantly, the surface marker expression pattern of tetramer-negative γδ thymocytes and splenocytes (i.e. >99% of the total γδ T cell population) in all strains of mice is more similar to that of T10/T22-specific γδ T cells from β2m-/- mice, than to those from B6 and BALB/c mice (Fig. 1D-F), suggesting that a significant fraction of γδ cells may not have encountered ligand during their development or in the periphery.
It has been reported that peripheral CD44hi γδ T cells have a higher turnover rate than their CD44lo-int counterparts (Tough and Sprent, 1998), and IL-15 has been observed to enhance homeostatic proliferation of γδ T cells in lymphopenic animals (Baccala et al., 2005; French et al., 2005). Since T10/T22-specific γδ T cells that develop in B6 and BALB/c mice generally express higher levels of CD122 and CD44 than the rest of γδ T cells, we asked whether these cells have enhanced turnover rates in vivo and if this would lead to a ligand driven bias in the repertoire. To test this, we measured the amount of BrdU incorporation in tetramer-positive and tetramer-negative γδ splenocytes from different strains of mice.
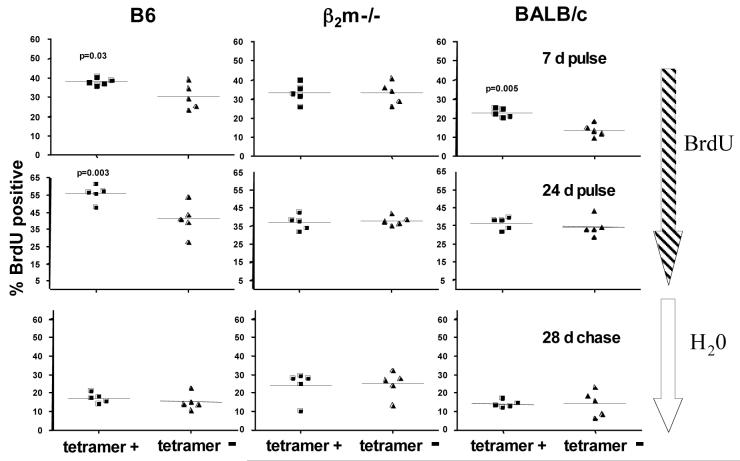
As shown in Fig. 4, T10/T22-specific γδ T cells from B6 and BALB/c mice incorporated significantly more BrdU after 7-day labeling than the vast majority of γδ T cells. However, this difference is no longer apparent after a 28-day chase preceded by 24-day labeling. Thus, encountering antigen increases the turnover of T10/T22-specific γδ T cells, but does not ‘fix’ this specificity in the repertoire. Indeed, we found no significant differences in the frequency of tetramer-positive cells from any strain of mice between two, seven, twelve, fourteen and twenty weeks of age (data not shown).
Figure 4. Host T10/T22 expression enhances T10/T22-specific γδ T cell turnover but does not fix this specificity in the repertoire.
Turnover rates of tetramer positive and negative γδ splenocytes from B6, β2m-/- and BALB/c mice. The percentage of BrdU+ cells among tetramer positive or negative γδ plenocytes were analyzed by intracellular BrdU staining as described in Experimental Procedures after mice were fed with 0.8 mg/ml BrdU in their drinking water for 7 days (upper panels), 24 days (middle panels) or chased with normal drinking water for 28 days after the 24-day labeling phase (lower panels). Each dot represents the analysis of one mouse. Data was analyzed by a paired, two-tailed student t test.
Importantly, tetramer-positive and tetramer-negative γδ T cells from β2m-/- mice had similar levels of BrdU incorporation across all experimental conditions, indicating that, lack of ligand recognition during development and in the periphery does not compromise the turn-over ability of antigen specific γδ T cells. In addition, the turnover rate of the majority of the splenic γδ T cells are similar to that of T10/T22-specific γδ T cells which develop in β2m-/- mice, further supporting the notion that a significant number of peripheral γδ T cells have yet to encounter antigen.
Antigen recognition during development defines γδ T cell functional subsets
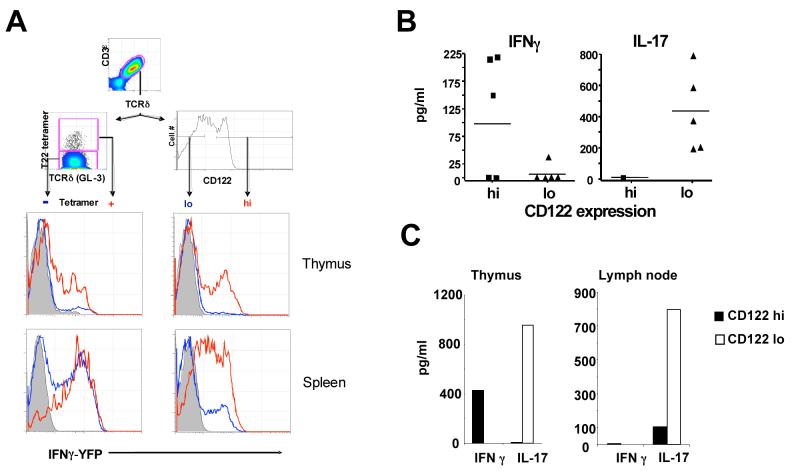
To test whether encountering ligand during development affects the ability of γδ T cells to make cytokines, we first analyzed YETI mice (B6 background), which express an IFNγ-YFP bicistronic reporter (Stetson et al., 2003). This allowed us to evaluate IFNγ expression using FACS analysis for YFP, as γδ T cells are known to make this cytokine (Ferrick et al., 1995). We found that T10/T22-specific γδ splenocytes in YETI mice are mostly YFP+, indicating that developing in the presence of antigen, does not compromise their ability to make IFNγ. Consistent with this supposition, most of the CD122hi YETI splenocytes are also YFP positive (Fig. 5A). In addition, CD122hi γδ splenocytes from B6 mice stimulated with plate-bound TCRδ cross-linking antibody GL-4 secrete high levels of IFNγ (Fig. 5B). In contrast, the majority of the CD122lo γδ splenocytes from YETI mice do not express YFP (Fig. 5A), and CD122lo B6 γδ splenocytes make very little or no IFNγ upon stimulation (Fig. 5B). Instead, we found that freshly isolated CD122lo γδ splenocytes respond to GL-4 stimulation by secreting high levels of IL-17. Thus, naïve γδ cells respond to TCR triggering readily. This is in stark contrast to the activation requirements of αβ T cells, which require an initial antigen specific priming event by professional antigen presenting cells before developing into effectors cells with the capability to secrete cytokines.
Figure 5. γδ T cell functional subsets.
(A) γδ hymocytes (upper row) and splenocytes (lower row) from YETI (Yellow Enhanced Transcript for IFNγ) mice (H-2b, T10+T22+) were analyzed for YFP (IFNγ) expression in tetramer+ (red) and tetramer- (blue) cells (left column); or CD122hi (red) and CD122lo (blue) cells right column (n=3); total γδ thymocytes or splenocytes from YFP negative littermate controls were also analyzed (shaded). FACS analysis was performed as described in Fig. 1.
(B) IFNγ and IL-17 production by CD122hi and CD122lo γδ plenocytes, (C) γδ thymocytes and γδ lymph node cells were FACS sorted into CD122hi and CD122lo populations according to the indicated gate in (A), and stimulated with plate bound anti-TCRδ (GL-4) for 40 hours. Supernatants were assayed for the production of IFNγ and IL-17 by ELISA. In (B) each symbol represents the result of one mouse, in (C) a representative graph.
To test whether the ability of γδ T cells to respond is acquired in the thymus, we first analyzed γδ thymocytes in YETI mice. As shown in Fig. 5, ∼20% of CD122hi cells and a similar percentage of the T10/T22-specific thymocytes in YETI mice are YFP+. In contrast, most of the CD122lo γδ thymocytes in YETI mice are YFP-. As in the spleen, CD122hi γδ thymocytes make IFNγ and only CD122lo cells make high levels of IL-17 after GL-4 stimulation (Fig. 5C). Taken together, these results indicate that γδ T cells can acquire their effector functions in the thymus.
In B6 and BALB/c mice, T10/T22-specific γδ hymocytes show a range of CD122 expression (Fig. 1D), which may reflect the heterogeneity in TCR-ligand binding affinities, Vδ gene usage (which have different dimerization potentials), developmental stages of these cells and/or IL-2, IL-15 signalling. However, most of the T10/T22-specific γδ T cells in the spleen of B6 mice are CD122hi (Fig. 1D), and in the lymph nodes, CD122lo (Fig. 1G). In fact, most of the γδ splenocytes express higher levels of CD122 than lymph node γδ T cells do (Fig. 1G, 1F). Although the mechanism leading to this biased distribution is unclear, it appears that αβ T cells also show similar differential ‘sorting’, where memory αβ T cells (that have encountered antigens previously), are preferentially found in the spleen than in the lymph nodes (Forster et al., 1999). Similar to γδ hymocytes and splenocytes, CD122lo lymph node γδ T cells respond to TCR cross-linking by preferentially making IL-17 (Fig. 5C). Our results here indicate that T10/T22-specific γδ T cells in the spleen and the thymus of B6 and β2m-/- mice will mount different cytokine responses. This difference may explain in part the perceived requirement for ligand-driven positive selection during the development of G8 and KN6 transgenic γδ T cells, where cytokines IL-2/4 was used as a readout for thymocyte maturation (Pereira et al., 1992; Wells et al., 1993).
While lymphoid γδ T cells are CD122lo and make IL-17, this is not the case for γδ IELs, which are noted for being cytolytic directly ex vivo in re-directed lysis assays (Lefrancois and Goodman, 1989). Regardless of antigen specificity and the host’s genetic background, these cells are uniformly CD122lo but constitutively express high levels of Eomes, Granzyme A, B (Fahrer et al., 2001; Shires et al., 2001)(data not shown). In this context, it should be noted that γδ IELs are not only very different than lymphoid γδ T cells in gene expression but also in their development, maturation, and survival requirement (Hayes and Love, 2007).
IL-17 producing γδ T cells appear early after peptide/CFA immunization
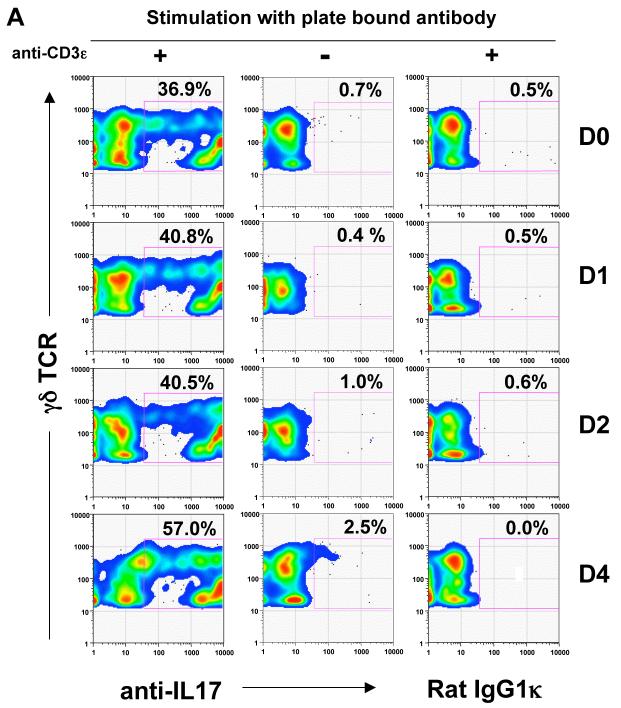
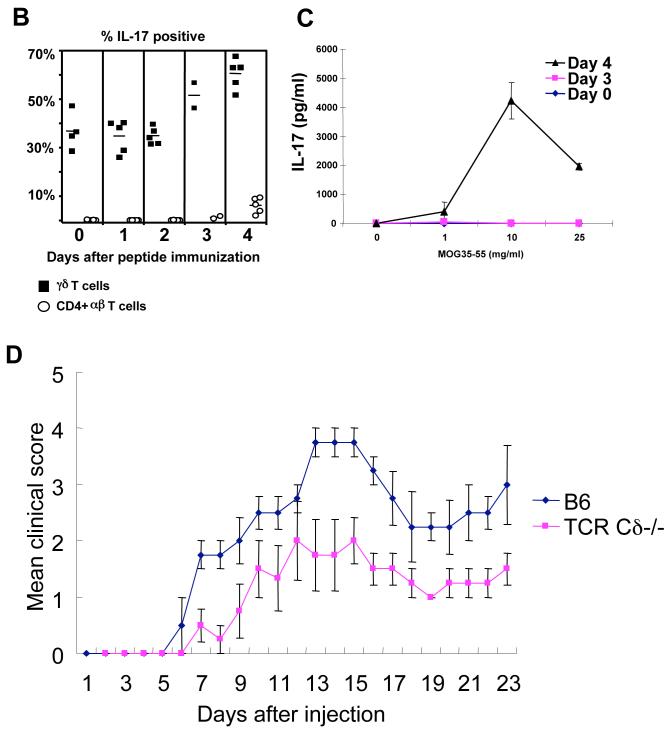
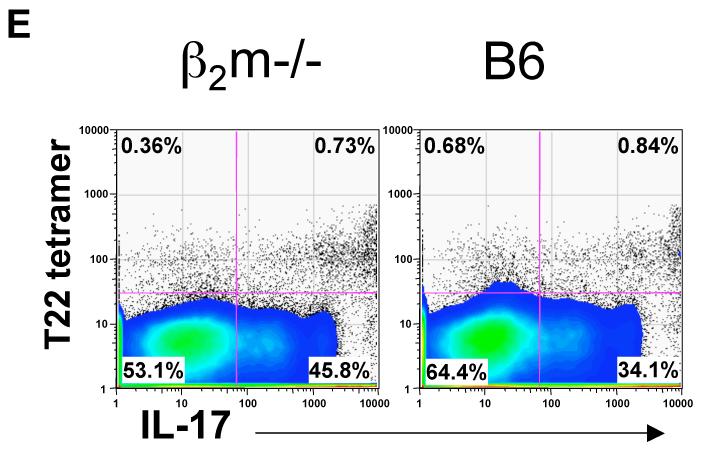
IL-17 regulates the expansion and recruitment of neutrophils and monocytes to initiate the inflammatory response (Ley et al., 2006; Stark et al., 2005). In an acute inflammatory response, a swift IL-17 response must be elicited without prior antigen exposure, suggesting that γδ T cells may be uniquely suited to produce IL-17 at the onset of the inflammatory response. To test this directly, we immunized B6 mice with myelin oligodendrocyte glycoprotein (MOG) peptide (35-55), in Complete Freund’s Adjuvant (CFA), as a surrogate to induce acute inflammation, and to prime antigen specific αβ T cells. T cells were isolated from the draining lymph nodes and stimulated with anti-CD3 for 24 hours in vitro before intracellular IL-17 staining. As shown in Figure 6, although the MOG-specific, as well as a significant CD4+ αβ T cell IL-17 response is not observed until day 3 or 4, a large fraction of lymph node γδ T cells from B6 mice produce IL-17 before and immediate after immunization. These include T10/T22-specific γδ T cells from B6 as well as from β2m-/- mice (Fig. 6E), demonstrating that encountering ligand during development is neither required nor inhibitory for γδ T cell to respond by producing cytokine.
Figure 6. IL-17+ T cells from draining lymph nodes after peptide/CFA immunization.
(A) Representative intracellular IL-17 staining of γδ T cells (GL-3+CD3ε+) and; (B) Percentage of IL-17+ cells among total γδ T cells (■) or CD4+CD8- αβ T cells (CD3ε+) (○) from the draining lymph nodes at the indicated days after CFA/MOG35-55 immunization. Each symbol represents the result of one mouse. Lymph node cells were isolated and stimulated with (+) or without (-) plate bound anti-CD3ε for 24 hours followed by staining with antibodies against CD4, CD8α, CD3ε, TCRδ, F4/80, CD11b, Gr-1 and CD19 and for intracellular IL-17 as described in Experimental Procedures. F4/80, CD11b, Gr-1 and CD19 positive cells were excluded from analysis.
(C) Representative dose response curves of the MOG33-55 specific IL-17 response at days 0, 3, and 4 after immunization. Lymph node cells were isolated from individual mice and incubated with various concentrations of MOG33-55 peptide for 48 hours and assayed for IL-17 in the supernatant.
(D) Progression of EAE disease in TCR Cδ-/- and B6 mice after immunization with MOG33-55 and pertussis toxin. The daily mean clinical score of each group is plotted (see Experimental Procedures).
(F) IL-17 response of T10/T22-specific γδ T cells from β2m-/- or B6 mice 4 days after MOG33-55/CFA immunization, draining lymph node cells from 12 β2m-/-, or B6 mice were pooled and stimulated as in 6A. γδ T cells were enriched, stained with T22 tetramer and assayed for intracellular IL-17 or stained with Rat IgG1κ isotype control (data not shown). Dot plots display gated GL-3+ H57- CD11b- F4/80- Gr1- CD19- γδ T cells.
In vertebrates, antigen specific αβ T cell responses are initiated by the inflammatory response. In this context, we have confirmed an earlier report by Weiner and colleagues (Spahn et al., 1999) that that experimental autoimmune encephalomyelitis (EAE) can be induced readily by MOG/CFA immunization in C57BL/6 mice, but with a delayed onset and much-reduced severity in TCRδ-/- mice on the same genetic background (Fig. 6D). This disease model has been used extensively in the evaluation of the development of Th17 αβ T cells (Bettelli et al., 2007).
Discussion
In the data presented here we show that a population of antigen specific γδ T cells is neither positively nor negatively selected in the thymus. This is in contrast to some (but not all (Schweighoffer and Fowlkes, 1996)) of the previous findings using mice expressing transgenic TCRs with the same specificity. These differences may also be due to the tendency of endogenous pre-rearranged TCR genes to skew T lymphocyte development (Serwold et al., 2007). While the development of dendritic epidermal T cells (DETCs) seem to require ligand driven positive selection, these cells are derived from the very first wave of γδ T cells during fetal thymic development and are unusual in that they have a largely monomorphic TCR and reside only in the skin of mice. Thus, it is arguable that the T10/T22-specific γδ T cells studied here are more typical of γδ T cells in adult mice.
Our results also showed that Vδ’s can self-dimerize as assayed with chimeric EPOR molecules in BAF3 cells. We suggest that this mechanism may allow γδ T cells to signal and mature in the thymus in the absence of a TCR ligand. TCR dimerization may also enhance antigen specific activation of peripheral γδ T cells, which are known to signal more efficiently than αβ T cells (Hayes et al., 2003) despite lacking CD4/CD8 co-receptor expression and having low levels of pERK1/2 similar to those of naïve αβ T cells (Fig. S2B). These are testable hypothesis and we are actively pursuing them.
Surprisingly, while ligand expression does little to constrain antigen specificities of the γδ T cell repertoire, it does play a role in endowing lymphoid γδ T cells with different functional programs. We find very clear evidence for two distinct functional subsets of lymphoid γδ T cells, namely ligand naïve cells which can make IL-17, designated as Tγδ-17s, and ligand experienced cells which secrete Interferon γ, designated Tγδ-IFNγ. While a significant population of splenic γδ T cells are Tγδ-IFNγ, the majority of the lymph node γδ cells, are Tγδ-17s. This is particularly significant as lymph nodes serves largely as the initial collection point of foreign antigens and a site for propagating inflammation. We are currently investigating whether the functional programs determined by ligand recognition can be ‘overwritten’ by tissue specific environments, and whether there is functional plasticity in these types of cells.
It is of particular importance that regardless of ligand experience, γδ T cells are able to make cytokines immediately upon TCR engagement. In higher vertebrates, IL-17 is made early in the host response to tissue damage or infection, leading to neutrophil maturation and their recruitment from the bone marrow (Stark et al., 2005). The major source of IL-17 is from T lymphocytes. Yet, it has been unclear how the early stages of the IL-17 response could be elicited from the classic antigen-specific αβ Th17 cells, which can take 4 to 7 days to develop, and requires both antigen-specific priming, co-stimulation provided by professional antigen presenting cells and a particular cytokine environment (Weaver et al., 2006). Thus, the new subset of γδ T cells that we describe here is ideally suited to provide IL-17 in the very earliest stages of an inflammatory response, before antigen specific αβ T cells are able to. This may explain reports showing that γδ T cells are particularly important in neutrophil dominated infectious diseases such as- Mycobacterium tuberculosis (Lockhart et al., 2006), Nocardia asteroides (Umemura et al., 2007), Escherichia coli (Shibata et al., 2007) and pulmonary aspergillosis in chronic granulomatous disease (CGD) (Romani et al., 2008), and in a mouse model of autoimmunity (Roark et al., 2007).
The influx of neutrophils sets in motion the recruitment of other leukocytes at the site of inflammation, which in turn affects the local cytokine milieu and subsequent antigen specific B and T cell responses. Thus, γδ-17s could play a crucial role in B and αβ T cell differentiation. Consistent with this supposition, various reports have observed deviated αβ T cell and B cell responses in γδ T cell deficient mice (Zuany-Amorim et al., 1998), including the lack of EAE induction as first shown by Weiner and colleagues (Spahn et al., 1999) and reproduced by us in this work.
Cumulatively, the data points to a γδ T cell repertoire largely determined by VDJ rearrangement and not restricted to recognize self antigens encountered during development. In addition, we have defined two distinct subsets of lymphoid γδ T cells in mice, Tγδ-17s and Tγδ-IFNγ, which are heavily influenced by ligand recognition. The ability of γδ T cells to produce IL-17 rapidly without prior antigen exposure suggests that they are uniquely suited to initiate and regulate the inflammatory response. This may be the key to understanding how they contribute to host immune competence and why they have been maintained throughout vertebrate evolution.
Experimental procedures
Mice
6-8 week age and sex matched C57BL/6, BALB/c, β2m-/- (C57BL/6), TCR Cδ-/- (B6.129P2-Tcrdtm1Mom/J) and β2m-/- MHC II -/- (B6.129-H2-Ab1tm1Gru B2mtm1Jae N17) mice were purchased from either Jackson or Taconic laboratories and housed in the Stanford Animal Facility according to guidelines set by Stanford. YETI (Yellow Enhanced Transcript for IFNγ) mice (Stetson et al., 2003)(C57BL/6, H-2b N6) were housed at UCSF according to guidelines set by UCSF.
Antibodies
All antibodies were purchased from eBioscience or BD Pharmingen unless otherwise stated.
FACS analysis of γδ T cells
To enrich γδ T cells, total thymocytes and splenocytes (108 cells/ml) were first incubated with normal hamster serum, normal mouse serum (Jackson ImmunoResearch), 5 μg/ml anti-CD16/32 FcBlock, followed by fluorescently labeled GL-3 (anti-TCRδ) antibody and enriched with anti-fluorescein isothiocyanate (FITC) or allophycocyanin (APC) MicroBeads (Miltenyi).
To stain GL-3 enriched γδ T cells with T22 tetramer, cells were further blocked with 4 μg/mL CD8αCT (CALTAG) to prevent CD8-tetramer binding, followed by staining with 5-7 μg/ml phycoerythrin (PE) labeled T22 tetramer, together with anti-CD19 Cy5PE (MB 19-1), anti-TCRβ Cy5PE (H57), anti-KLH Armenian Hamster IgG2κ Cy5PE (GL-3 isotype) and streptavidin Cy5PE (BD Pharmingen) for 1.5 hours on ice. After washing, cells were suspended in FACS buffer (2% FCS PBS) with 1 μg/ml propidium iodide (PI). Six-color FACS was performed on Vantage or LSR and data was collected using Cell Quest software. Cells positive for PI and Cy5PE were excluded from analysis. FACS data was compensated and analyzed using FlowJo software (Treestar).
To stain tetramer positive γδ T cells for the expression of cell surface markers, enriched and tetramer stained γδ T cells were concurrently stained with one of the following antibodies labeled with FITC (unless otherwise stated): anti-CD122 (TM-β1), anti-HSA (M1/69), anti-CD127 (A7R34), anti-CD5 (53-7.3), anti-CD25 Alexa488 (3C7), anti-NK1.1 APC (NKR-P1C), anti-CD49b (DX5), anti-CD44 (IM7), anti-CD4 APC-Cy7 (15-8-A2), anti-CD8 (CD8αCT, CALTAG), Rat IgG2aκ FITC (53-7.3 isotype), or anti-S1P1 polyclonal (UCSF) detected with goat anti-rabbit secondary (Jackson ImmunoResearch). Tetramer positive and negative, GL-3 positive cells were analyzed for maker expression and plotted by a histogram. For analyzing YETI mice, TCRδ (GL-3) and CD3ε Cy7APC (145-2C11) double positive cells were analyzed for tetramer or anti-CD122 PE staining, and YFP (IFNγ) expression.
Intracellular staining of pERK1/2 or BrdU in tetramer positive γδ T cells
To detect intracellular levels of pERK1/2 or BrdU in T22 tetramer positive cells, cell suspensions from thymus or spleen, were blocked with serums, FcBlock and CD8αCT as described above; stained with 15-20 μg/ml PE labeled tetramer and enriched with anti-PE MicroBeads (Miltenyi). After enrichment, approximately 50-70% of the TCRδ+CD3ε+ cells were tetramer positive. Tetramer enriched cells were stained with relevant antibodies, then fixed with Cytofix/Cytoperm™ solution on ice and permeabilized with Cytoperm™/Wash buffer (BD Pharmingen). For the detection of phosphorylated ERK1/2, cells were stained with rabbit 197G2 (Cell Signaling) for 45 min. at room temperature. After washing, cells were stained with anti-rabbit FITC (Jackson ImmunoResearch) for 30 min. at room temp, followed by washing with Cytoperm™/Wash and FACS buffers. For the detection of BrdU incorporation, samples were further permeabilized with Cytofix/Cytoperm™ Plus buffer (BD Pharmingen) then treated with 30 μg DNase (Sigma-Aldrich) for 60 min at 37°C to expose BrdU epitopes. After washing, cells were stained with anti-BrdU APC (3D4), for 45 min. at room temperature followed by washing.
In vitro stimulations and the detection of IFNγ and IL-17
CD122hi and CD122lo γδ thymocytes, splenocytes and lymph node cells were FACS sorted using a Vantage cell sorter and 105 cells/well were plated on a 96 well plate (Costar) coated with 10 μg/ml anti-TCRδ (GL-4). Cells were cultured in RPMI 1640+GlutaMAX™-1 supplemented with 10% fetal calf serum (Hyclone), 0.1 mM non-essential amino acids (Cellgro), 1 mM sodium pyruvate (Irvine Scientific), 100 U/ml penicillin, 100 μg/ml streptomycin (Gibco) and 3.5 μl/L 2-mercaptoethanol (Sigma). 40 hours later supernatant was analyzed for IFNγand IL-17 by ELISA (eBioscience).
Construction, expression of TCR-EPOR hybrid genes in BaF3 cells and the proliferation assays
The TCRδextracellular region (defined by an IMGT multiple sequence alignment, http://imgt.cines.fr) was linked to the transmembrane (TM) and intracellular sequence of the human EPOR; and the TCRγ extracellular region (defined as above) was linked to and terminated at the end of the EPOR TM region by PCR. PCR products were TA cloned (Invitrogen), sequenced and inserted into a pMX IRES-GFP vector(Yamasaki et al., 2006), and transfected into ecotrophic Phoenix cells with FuGene 6 Transfection reagent (Roche) according to manufacture’s suggestions. Viral supernatant was obtained as described (http://www.stanford.edu/group/nolan/protocols/pro_helper_dep.html) and Vδ-EPOR/Vγ chain pairs were used to infect BaF3 cells grown in 1 ng/ml mouse IL-3 (R & D systems). Transfectants were FACS sorted for high (upper 10%) of GFP expression. For the proliferation assays, sorted transfectants were washed 3 times and mixed with equal numbers of washed parental BaF3 (GFP negative) cells, such that 105 cells/400 μl of each cell type were plated in a 48 well plate. Various days after IL-3 removal, PI was added directly to the medium, cells were resuspended and enumerated by FACS. The absolute number of GFP+ or GFP- (parental BaF3) cells various days after IL-3 removal divided by the absolute number obtained on day 0 was plotted for Supplemental Figure S2. For Figure 3B, the fold growth over parental BaF3 cells on day 4 was plotted and calculated as follows: [number of chimeric EPOR transfectants at day 4 after IL-3 removal /number of transfectants at day 0] / [number of parental BaF3 cells at day 4 after IL-3 removal / number of parental BaF3 cells at day 4].
MOG35-55 / CFA immunization, intracellular IL-17 detection and the MOG35-55 specific IL-17 lymph node response
Mice were immunized subcutaneously with 100 μg of HPLC purified MOG35-55 peptide (MEVGWYRSPFSRVVHLYRNGK), 200μg of heat killed mycobacterium (DIFCO) emulsified in 1 part mineral oil and 1 part H20 (Complete Freund’s Adjuvant). Axillary, brachial and inguinal lymph nodes were harvested at various days post immunization. Total cell suspensions were plated at 5×106 cells per well in 48 well plates (Falcon) pre-coated with 10 μg/ml anti-CD3ε (145-2C11, no azide, low endotoxin) or Armenian Hamster IgG1κ (BD). After incubating 24 hours at 37°C, cells were harvested, blocked with serums and FcBlock as described above, and stained with antibodies against CD4 (GK1.5), CD8 (CD8αCT), CD3ε (145-2C11), TCRδ (GL-3), F4/80 (BM8), CD11b (M1/70), Gr-1 (RB6-8C5), and CD19 (MB19-1). For tetramer analysis, cells were enriched with GL-3 and anti-FITC microbeads before staining with APC labeled T22 tetramer and dump antibodies as described above. Cells were then washed with FACS buffer, fixed and permeabilized with Cytofix™ and Cytoperm™/Wash buffers respectively. Cells were stained with 2 μg/ml PE labeled anti-IL-17 (TC11-18H10, BD Pharmingen) or IgG1,κ isotype (R3-34, BD Pharmingen) for 30 minutes on ice in Cytoperm™/Wash buffer. Cells were washed as described before. CD11b, Gr-1, F4/80, CD19 positive cells were excluded from analysis. Isotype staining was used to define cytokine positive cells.
To detect the MOG 33-55 specific IL-17 lymph node response, lymph node cells from individual mice were isolated various days before and after peptide/CFA immunization. 106 cells were plated in a 96 well plate and incubated with various concentrations of MOG33-55 peptide in RPMI culture medium. 48 hours later supernatant was harvested and monitored for IL-17 using anti-IL-17 beads (Bender MedSystems®) according to the manufacturer’s protocol.
EAE was induced in C57BL/6 and TCR Cδ-/- (B6) (N=4) mice by subcutaneous immunization with MOG peptide as described above. On the day of immunization and 2 days later, mice were injected intravenously with 50 ng of Bordetella pertussis toxin in PBS. Mice were examined each day for clinical signs of EAE and scored as follows: 0, no paralysis; 1, loss of tail tone; 2, hindlimb weakness; 3, hindlimb paralysis; 4, hindlimb and forelimb paralysis; 5, moribund or dead.
Supplementary Material
Acknowledgements
We thank Dr. J. Cyster (UCSF) for the anti-S1P1 polyclonal rabbit serum, E. Gallo and J. Crabtree for the use of CSA treated β2m-/-, MHC II null mice, the 197G2 antibody and for discussions regarding its use, C. Franco for assistance with cloning, Dr. L. Reinhardt for facilitating experiments with YETI mice, Dr. Y. Konigshofer for comments. Author contributions: K.J., X.S. S.S and Y.C. designed experiments; K.J., X.S., S.S., L.L., Sa.Y. performed experiments; Sho.Y. and T.S. provided reagents and advice on the EPOR-BAF3 experiments; L.S. provided advice on the MOG/CFA inflammatory response model; N.B. performed qPCR analysis on IELs; R.L. provided YETI mice and advice; K.J., X.S., S.S. and Y.C. analyzed the results; K.J., L.L., M.M.D. and Y.C. wrote the paper. K.J. was supported by the Stanford Graduate Fellowship and the NIH Cell and Molecular Biology (CMB) training grant. This work was supported by NIH grants (Y.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams EJ, Chien YH, Garcia KC. Structure of a gammadelta T cell receptor in complex with the nonclassical MHC T22. Science. 2005;308:227–231. doi: 10.1126/science.1106885. [DOI] [PubMed] [Google Scholar]
- Azzam HS, Grinberg A, Lui K, Shen H, Shores EW, Love PE. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J Exp Med. 1998;188:2301–2311. doi: 10.1084/jem.188.12.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccala R, Witherden D, Gonzalez-Quintial R, Dummer W, Surh CD, Havran WL, Theofilopoulos AN. Gamma delta T cell homeostasis is controlled by IL-7 and IL-15 together with subset-specific factors. J Immunol. 2005;174:4606–4612. doi: 10.4049/jimmunol.174.8.4606. [DOI] [PubMed] [Google Scholar]
- Bettelli E, Oukka M, Kuchroo VK. T(H)-17 cells in the circle of immunity and autoimmunity. Nat Immunol. 2007;8:345–350. doi: 10.1038/ni0407-345. [DOI] [PubMed] [Google Scholar]
- Bonneville M, Ishida I, Itohara S, Verbeek S, Berns A, Kanagawa O, Haas W, Tonegawa S. Self-tolerance to transgenic gamma delta T cells by intrathymic inactivation. Nature. 1990;344:163–165. doi: 10.1038/344163a0. [DOI] [PubMed] [Google Scholar]
- Crowley MP. Characterization of the Ligand Recognition Properties and Function of a Gamma/Delta T cell Receptor and Ligand Pair. Stanford University; Stanford: 1998. [Google Scholar]
- Crowley MP, Fahrer AM, Baumgarth N, Hampl J, Gutgemann I, Teyton L, Chien Y. A population of murine gammadelta T cells that recognize an inducible MHC class Ib molecule. Science. 2000;287:314–316. doi: 10.1126/science.287.5451.314. [DOI] [PubMed] [Google Scholar]
- De Paoli P, Gennari D, Martelli P, Cavarzerani V, Comoretto R, Santini G. Gamma delta T cell receptor-bearing lymphocytes during Epstein-Barr virus infection. J Infect Dis. 1990;161:1013–1016. doi: 10.1093/infdis/161.5.1013. [DOI] [PubMed] [Google Scholar]
- Dent AL, Matis LA, Hooshmand F, Widacki SM, Bluestone JA, Hedrick SM. Self-reactive gamma delta T cells are eliminated in the thymus. Nature. 1990;343:714–719. doi: 10.1038/343714a0. [DOI] [PubMed] [Google Scholar]
- Fahrer AM, Konigshofer Y, Kerr EM, Ghandour G, Mack DH, Davis MM, Chien YH. Attributes of gammadelta intraepithelial lymphocytes as suggested by their transcriptional profile. Proc Natl Acad Sci U S A. 2001;98:10261–10266. doi: 10.1073/pnas.171320798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrick DA, Schrenzel MD, Mulvania T, Hsieh B, Ferlin WG, Lepper H. Differential production of interferon-gamma and interleukin-4 in response to Th1- and Th2-stimulating pathogens by gamma delta T cells in vivo. Nature. 1995;373:255–257. doi: 10.1038/373255a0. [DOI] [PubMed] [Google Scholar]
- Forster R, Schubel A, Breitfeld D, Kremmer E, Renner-Muller I, Wolf E, Lipp M. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell. 1999;99:23–33. doi: 10.1016/s0092-8674(00)80059-8. [DOI] [PubMed] [Google Scholar]
- French JD, Roark CL, Born WK, O’Brien R,L. {gamma}{delta} T cell homeostasis is established in competition with {alpha}{beta} T cells and NK cells. Proc Natl Acad Sci U S A. 2005;102:14741–14746. doi: 10.1073/pnas.0507520102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haks MC, Lefebvre JM, Lauritsen JP, Carleton M, Rhodes M, Miyazaki T, Kappes DJ, Wiest DL. Attenuation of gammadeltaTCR signaling efficiently diverts thymocytes to the alphabeta lineage. Immunity. 2005;22:595–606. doi: 10.1016/j.immuni.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Hanke T, Mitnacht R, Boyd R, Hunig T. Induction of interleukin 2 receptor beta chain expression by self-recognition in the thymus. J Exp Med. 1994;180:1629–1636. doi: 10.1084/jem.180.5.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havran WL, Allison JP. Developmentally ordered appearance of thymocytes expressing different T-cell antigen receptors. Nature. 1988;335:443–445. doi: 10.1038/335443a0. [DOI] [PubMed] [Google Scholar]
- Havran WL, Chien YH, Allison JP. Recognition of self antigens by skin-derived T cells with invariant gamma delta antigen receptors. Science. 1991;252:1430–1432. doi: 10.1126/science.1828619. [DOI] [PubMed] [Google Scholar]
- Hayes SM, Li L, Love PE. TCR signal strength influences alphabeta/gammadelta lineage fate. Immunity. 2005;22:583–593. doi: 10.1016/j.immuni.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Hayes SM, Love PE. A retrospective on the requirements for gammadelta T-cell development. Immunol Rev. 2007;215:8–14. doi: 10.1111/j.1600-065X.2006.00476.x. [DOI] [PubMed] [Google Scholar]
- Hayes SM, Shores EW, Love PE. An architectural perspective on signaling by the pre-, alphabeta and gammadelta T cell receptors. Immunol Rev. 2003;191:28–37. doi: 10.1034/j.1600-065x.2003.00011.x. [DOI] [PubMed] [Google Scholar]
- Ho M, Webster HK, Tongtawe P, Pattanapanyasat K, Weidanz WP. Increased gamma delta T cells in acute Plasmodium falciparum malaria. Immunol Lett. 1990;25:139–141. doi: 10.1016/0165-2478(90)90105-y. [DOI] [PubMed] [Google Scholar]
- Huseby ES, White J, Crawford F, Vass T, Becker D, Pinilla C, Marrack P, Kappler JW. How the T cell repertoire becomes peptide and MHC specific. Cell. 2005;122:247–260. doi: 10.1016/j.cell.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Ito K, Van Kaer L, Bonneville M, Hsu S, Murphy DB, Tonegawa S. Recognition of the product of a novel MHC TL region gene (27b) by a mouse gamma delta T cell receptor. Cell. 1990;62:549–561. doi: 10.1016/0092-8674(90)90019-b. [DOI] [PubMed] [Google Scholar]
- Jason J, Buchanan I, Archibald LK, Nwanyanwu OC, Bell M, Green TA, Eick A, Han A, Razsi D, Kazembe PN, et al. Natural T, gammadelta, and NK cells in mycobacterial, Salmonella, and human immunodeficiency virus infections. J Infect Dis. 2000;182:474–481. doi: 10.1086/315740. [DOI] [PubMed] [Google Scholar]
- King DP, Hyde DM, Jackson KA, Novosad DM, Ellis TN, Putney L, Stovall MY, Van Winkle LS, Beaman BL, Ferrick DA. Cutting edge: protective response to pulmonary injury requires gamma delta T lymphocytes. J Immunol. 1999;162:5033–5036. [PubMed] [Google Scholar]
- Koohsari H, Tamaoka M, Campbell HR, Martin JG. The role of gamma delta T cells in airway epithelial injury and bronchial responsiveness after chlorine gas exposure in mice. Respir Res. 2007;8:21. doi: 10.1186/1465-9921-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrancois L, Goodman T. In vivo modulation of cytolytic activity and Thy-1 expression in TCR-gamma delta+ intraepithelial lymphocytes. Science. 1989;243:1716–1718. doi: 10.1126/science.2564701. [DOI] [PubMed] [Google Scholar]
- Lewis JM, Girardi M, Roberts SJ, Barbee SD, Hayday AC, Tigelaar RE. Selection of the cutaneous intraepithelial gammadelta+ T cell repertoire by a thymic stromal determinant. Nat Immunol. 2006;7:843–850. doi: 10.1038/ni1363. [DOI] [PubMed] [Google Scholar]
- Ley K, Smith E, Stark MA. IL-17A-producing neutrophil-regulatory Tn lymphocytes. Immunol Res. 2006;34:229–242. doi: 10.1385/IR:34:3:229. [DOI] [PubMed] [Google Scholar]
- Mallick-Wood CA, Lewis JM, Richie LI, Owen MJ, Tigelaar RE, Hayday AC. Conservation of T cell receptor conformation in epidermal gammadelta cells with disrupted primary Vgamma gene usage. Science. 1998;279:1729–1733. doi: 10.1126/science.279.5357.1729. [DOI] [PubMed] [Google Scholar]
- Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, Brinkmann V, Allende ML, Proia RL, Cyster JG. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427:355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- Moore TA, Moore BB, Newstead MW, Standiford TJ. Gamma delta-T cells are critical for survival and early proinflammatory cytokine gene expression during murine Klebsiella pneumonia. J Immunol. 2000;165:2643–2650. doi: 10.4049/jimmunol.165.5.2643. [DOI] [PubMed] [Google Scholar]
- Pereira P, Zijlstra M, McMaster J, Loring JM, Jaenisch R, Tonegawa S. Blockade of transgenic gamma delta T cell development in beta 2-microglobulin deficient mice. Embo J. 1992;11:25–31. doi: 10.1002/j.1460-2075.1992.tb05023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz I, Sansoni A, Kissenpfennig A, Ardouin L, Malissen M, Malissen B. Visualization of the earliest steps of gammadelta T cell development in the adult thymus. Nat Immunol. 2006;7:995–1003. doi: 10.1038/ni1371. [DOI] [PubMed] [Google Scholar]
- Roark CL, French JD, Taylor MA, Bendele AM, Born WK, O’Brien RL. Exacerbation of collagen-induced arthritis by oligoclonal, IL-17-producing gamma delta T cells. J Immunol. 2007;179:5576–5583. doi: 10.4049/jimmunol.179.8.5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild H, Mavaddat N, Litzenberger C, Ehrich EW, Davis MM, Bluestone JA, Matis L, Draper RK, Chien YH. The nature of major histocompatibility complex recognition by gamma delta T cells. Cell. 1994;76:29–37. doi: 10.1016/0092-8674(94)90170-8. [DOI] [PubMed] [Google Scholar]
- Schweighoffer E, Fowlkes BJ. Positive selection is not required for thymic maturation of transgenic gamma delta T cells. J Exp Med. 1996;183:2033–2041. doi: 10.1084/jem.183.5.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwold T, Hochedlinger K, Inlay MA, Jaenisch R, Weissman IL. Early TCR expression and aberrant T cell development in mice with endogenous prerearranged T cell receptor genes. J Immunol. 2007;179:928–938. doi: 10.4049/jimmunol.179.2.928. [DOI] [PubMed] [Google Scholar]
- Shin S, El-Diwany R, Schaffert S, Adams EJ, Garcia KC, Pereira P, Chien YH. Antigen recognition determinants of gammadelta T cell receptors. Science. 2005;308:252–255. doi: 10.1126/science.1106480. [DOI] [PubMed] [Google Scholar]
- Shires J, Theodoridis E, Hayday AC. Biological insights into TCRgammadelta+ and TCRalphabeta+ intraepithelial lymphocytes provided by serial analysis of gene expression (SAGE) Immunity. 2001;15:419–434. doi: 10.1016/s1074-7613(01)00192-3. [DOI] [PubMed] [Google Scholar]
- Spahn TW, Issazadah S, Salvin AJ, Weiner HL. Decreased severity of myelin oligodendrocyte glycoprotein peptide 33 - 35-induced experimental autoimmune encephalomyelitis in mice with a disrupted TCR delta chain gene. Eur J Immunol. 1999;29:4060–4071. doi: 10.1002/(SICI)1521-4141(199912)29:12<4060::AID-IMMU4060>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Stark MA, Huo Y, Burcin TL, Morris MA, Olson TS, Ley K. Phagocytosis of apoptotic neutrophils regulates granulopoiesis via IL-23 and IL-17. Immunity. 2005;22:285–294. doi: 10.1016/j.immuni.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Mohrs M, Reinhardt RL, Baron JL, Wang ZE, Gapin L, Kronenberg M, Locksley RM. Constitutive cytokine mRNAs mark natural killer (NK) and NK T cells poised for rapid effector function. J Exp Med. 2003;198:1069–1076. doi: 10.1084/jem.20030630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarakhovsky A, Kanner SB, Hombach J, Ledbetter JA, Muller W, Killeen N, Rajewsky K. A role for CD5 in TCR-mediated signal transduction and thymocyte selection. Science. 1995;269:535–537. doi: 10.1126/science.7542801. [DOI] [PubMed] [Google Scholar]
- Toth B, Alexander M, Daniel T, Chaudry IH, Hubbard WJ, Schwacha MG. The role of gammadelta T cells in the regulation of neutrophil-mediated tissue damage after thermal injury. J Leukoc Biol. 2004;76:545–552. doi: 10.1189/jlb.0404219. [DOI] [PubMed] [Google Scholar]
- Tough DF, Sprent J. Lifespan of gamma/delta T cells. J Exp Med. 1998;187:357–365. doi: 10.1084/jem.187.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Laethem F, Sarafova SD, Park JH, Tai X, Pobezinsky L, Guinter TI, Adoro S, Adams A, Sharrow SO, Feigenbaum L, Singer A. Deletion of CD4 and CD8 Coreceptors Permits Generation of alphabetaT Cells that Recognize Antigens Independently of the MHC. Immunity. 2007;27:735–750. doi: 10.1016/j.immuni.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Weaver CT, Harrington LE, Mangan PR, Gavrieli M, Murphy KM. Th17: an effector CD4 T cell lineage with regulatory T cell ties. Immunity. 2006;24:677–688. doi: 10.1016/j.immuni.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Weintraub BC, Jackson MR, Hedrick SM. Gamma delta T cells can recognize nonclassical MHC in the absence of conventional antigenic peptides. J Immunol. 1994;153:3051–3058. [PubMed] [Google Scholar]
- Wells FB, Gahm SJ, Hedrick SM, Bluestone JA, Dent A, Matis LA. Requirement for positive selection of gamma delta receptor-bearing T cells. Science. 1991;253:903–905. doi: 10.1126/science.1831565. [DOI] [PubMed] [Google Scholar]
- Wells FB, Tatsumi Y, Bluestone JA, Hedrick SM, Allison JP, Matis LA. Phenotypic and functional analysis of positive selection in the gamma/delta T cell lineage. J Exp Med. 1993;177:1061–1070. doi: 10.1084/jem.177.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wucherpfennig KW, Newcombe J, Li H, Keddy C, Cuzner ML, Hafler DA. Gamma delta T-cell receptor repertoire in acute multiple sclerosis lesions. Proc Natl Acad Sci U S A. 1992;89:4588–4592. doi: 10.1073/pnas.89.10.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong N, Kang C, Raulet DH. Positive selection of dendritic epidermal gammadelta T cell precursors in the fetal thymus determines expression of skin-homing receptors. Immunity. 2004;21:121–131. doi: 10.1016/j.immuni.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Yamasaki S, Ishikawa E, Sakuma M, Ogata K, Sakata-Sogawa K, Hiroshima M, Wiest DL, Tokunaga M, Saito T. Mechanistic basis of pre-T cell receptor-mediated autonomous signaling critical for thymocyte development. Nat Immunol. 2006;7:67–75. doi: 10.1038/ni1290. [DOI] [PubMed] [Google Scholar]
- Yoshimura A, Longmore G, Lodish HF. Point mutation in the exoplasmic domain of the erythropoietin receptor resulting in hormone-independent activation and tumorigenicity. Nature. 1990;348:647–649. doi: 10.1038/348647a0. [DOI] [PubMed] [Google Scholar]
- Zuany-Amorim C, Ruffie C, Haile S, Vargaftig BB, Pereira P, Pretolani M. Requirement for gammadelta T cells in allergic airway inflammation. Science. 1998;280:1265–1267. doi: 10.1126/science.280.5367.1265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.