Abstract
Analysis of gene function in apicomplexan parasites is limited by the absence of reverse genetic tools that allow easy and rapid modulation of protein levels. The fusion of a ligand-controlled destabilization domain (ddFKBP) to a protein of interest enables rapid and reversible protein stabilization in T. gondii. This allows an efficient functional analysis of proteins that have a dual role during host cell invasion and/or intracellular growth of the parasite.
Although conditional mutagenesis to study the function of vital genes has been achieved in apicomplexan parasites, the current major limitation is the slow kinetics of the expression switch, which requires several hours to days of downregulation until the phenotypic consequences can be analyzed1. Rapid regulation of protein abundance has recently been demonstrated for mammalian cells. Selective stabilization is achieved by reversible binding of a synthetic ligand, termed Shield-1 (Shld1), to an otherwise short-lived fusion of a protein of interest and a destabilization domain (ddFKBP) based on the “rapamycin binding protein” FK-506 (FKBP12)2.
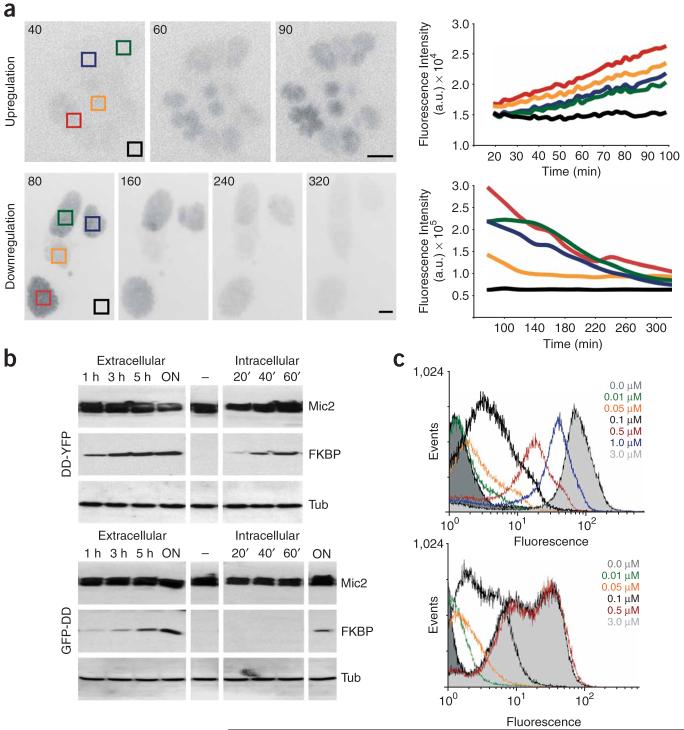
To test whether this system can be applied in apicomplexan parasites, we used T. gondii as a model system. We generated genetic fusions, linking the ddFKBP N-terminally to yellow fluorescent protein (ddFKBP-YFP) and C-terminally to green fluorescent protein (GFP-ddFKBP; Supplementary Methods online). In clonal parasite strains we were able to specifically modulate the levels of the respective fusion protein in a Shld1-dependent manner. In the presence of 1 μM Shld1 we detected high levels of the respective fusion protein, whereas in absence of Shld1 the fusion proteins were not detectable (Fig. 1a,b). To analyze the kinetics of up- and downregulation, we inoculated HFF cells with clonal parasites expressing ddFKBP-YFP in presence or absence of Shld1. After addition or removal of Shld1 from the medium, we imaged YFP fluorescence by time-lapse microscopy (Fig. 1a, Supplementary Figs. 1 and 2 and Supplementary Movies 1 and 2 online). Although we used a clonal population for infection, we detected different levels of fluorescence in independently established para-sitophorous vacuoles (Fig. 1a and Supplementary Figs. 1 and 2). Upregulation of ddFKBP-YFP to full induction was achieved within 90 min, whereas downregulation of ddFKBP-YFP to background levels took 320 min (Fig. 1a). We obtained similar results with a clonal parasite strain expressing GFP-ddFKBP (data not shown). Although tight regulation of GFP-ddFKBP is possible, we observed slower kinetics of up- and downregulation compared to that of ddFKBP-YFP (Fig. 1b). Slower degradation kinetics of C-terminal ddFKBP fusions also has been demonstrated in mammalian cells2 and Plasmodium falciparum3. We detected ddFKBP-YFP as soon as 20 min and less then 1 h after induction in intra- and extracellular parasites, respectively (Fig. 1b). In case of GFP-ddFKBP, more than 5 h were necessary to reach full induction (Fig. 1b). Notably, we did not detect any change in endogenous protein levels owing to the presence or absence of Shld1 (Fig. 1b).
Figure 1.
Rapid regulation of proteins fused to ddFKBP. (a) To examine upregulation, fluorescence micrographs were acquired after induction of intracellular parasites expressing ddFKBP-YFP with 1 μm Shld1 (top). To examine downregulation, infected cells treated with Shld1 were placed in medium without Shld1. Acquisition of fluoresence micrographs was started 80 min after removal of Shld1 (bottom). Scale bars, 10 μm. Fluorescence intensities for the indicated vacuoles (colored squares) were measured for the entire sequence and plotted for every time point (lines correspond to respectively colored squares; right). Black, background fluorescence. (b) Immunological analysis of intra- or extracellular parasites expressing either ddFKBP-YFP or GFP-ddFKBP induced with 1 μM Shld1 for the indicated amount of time before collection. The respective blot was simultaneously probed with polyclonal anti-FKBP12 (ABCAM) and monoclonal anti-MIC2 (6D10; ref. 9). The same lysates were probed with anti-TUB1 (ref. 10) in an independent blot. (c) Flow cytometry analysis of parasites expressing either ddFKBP-YFP (top) or GFP-ddFKBP (bottom) grown in the indicated concentration of Shld1. After egress parasites were subjected to flow cytometry analysis. We collected 100,000 data points for each sample.
The possibility to specifically regulate a protein of interest in extra- versus intracellular parasites is of importance when proteins with dual functions (for example during host cell invasion and intracellular development of the parasite) need to be analyzed. Next we inoculated parasites expressing either ddFKBP-YFP or GFP-ddFKBP in presence of different concentrations of Shld1 (0.01 to 3 μM). We found maximum regulation of protein levels over two orders of magnitude. Whereas stabilization of GFP-ddFKBP was saturated at 0.5 μM of Shld1, in the case of ddFKBP-YFP, higher concentrations of up to 3 μM Shld1 were required to reach full stabilization of the fusion protein (Fig. 1c). This demonstrates that the efficiency of this system allows a fine tuning of protein levels by simply adjusting Shld1 concentrations.
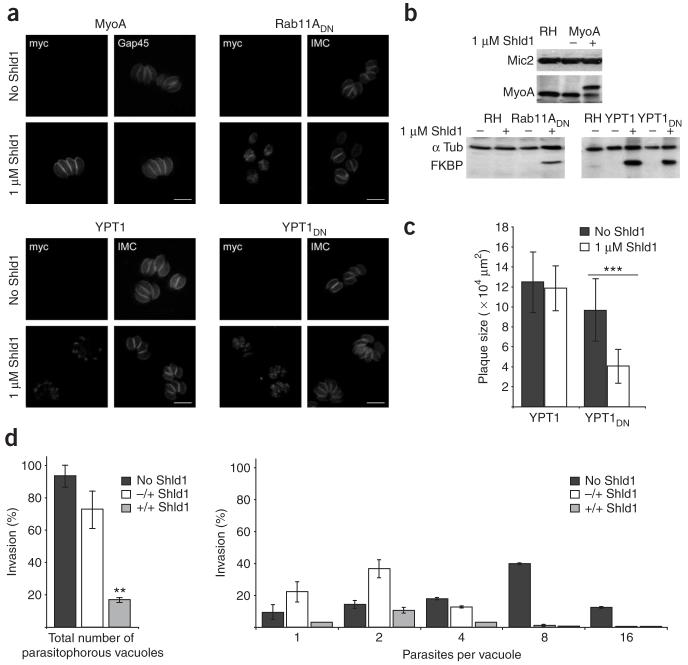
Next we tested whether essential parasite proteins could be regulated and functionally characterized. Previously, a conditional mutant for the motor protein TgMyoA had been established and functionally described in detail using a tetracycline-inducible system4. Hence we chose TgMyoA as a candidate protein to evaluate the potential of the ddFKBP system. In a first attempt we generated stable parasites expressing TgMyoA, N-terminally fused to ddFKBP and a myc tag (ddFKBP-MyoA) under control of the strong constitutive promoter p5RT70 (ref. 5). We generated clonal transgenic parasites expressing ddFKBP-MyoA that could be regulated in a Shld1-dependent manner (Fig. 2a,b). Stabilization of transgenic ddFKBP-MyoA resulted in substantial downregulation of endogenous TgMyoA (Fig. 2b). A similar observation has been reported upon regulated overexpression of TgMyoA using a tetracycline-inducible repression system, and it has been speculated that the observed downregulation of endogenous TgMyoA occurs post-transcriptionally6. Using the ddFKBP system we demonstrated that endogenous TgMyoA is downregulated post-translationally. It thus appears that ectopic overexpression of TgMyoA results in competition for interaction partners for endogenous TgMyoA like the myosin light chain (TgMLC) that stabilizes the motor complex7. Next we attempted to remove endogenous myoA using homologous recombination, as described before4. To do so, we maintained transgenic ddFKBP-MyoA-expressing parasites in presence of Shld1 to increase protein stability. In several independent attempts we did not obtain any conditional knockouts for TgMyoA, suggesting that N-terminal tagging of TgMyoA with ddFKBP results in a nonfunctional motor. In support of these data we were unable to complement the previously established conditional knockout for MyoA with ddFKBP-MyoA (data not shown).
Figure 2.
Functional analysis of essential proteins using the ddFKBP system. (a) Immunological fluorescence analysis of T. gondii parasite strains expressing ddFKBP-MyoA, ddFKBP-YPT1wt, ddFKBP-YPT1DN or ddFKBP-Rab11ADN for 4 h after induction using monoclonal anti-myc (9E10, Sigma) and polyclonal anti-IMC1 (ref. 11) or anti-GAP45 (ref. 12). Scale bar, 10 μm. (b) Immunological analysis on transgenic parasites shown in a. Parasites were grown in presence and absence of Shld1 for 48 h (ddFKBP-MyoA), for 8 h (ddFKBP-YPT1 and ddFKBP-YPT1DN) or 2 h (ddFKBP-Rab11ADN) before immunological analysis using the indicated antibodies. (c) Determination of plaque size after 5 d by parasite strain ddFKBP-YPT1 and ddFKBP-YPT1DN (see Supplementary Fig. 3). Data represent mean values of 20 independent plaques ± s.d. Asterisks indicate significant difference in plaque sizes between ddFKBP-YPT1DN grown in presence or absence of Shld1 (P < 0.001, two-tailed Student’s t-test). (d) Quantification of invasion and replication of parasite strain ddFKBP-Rab11ADN (-/+, parasites treated with Shld1 after invasion; +/+, parasites treated with Shld1 before and after invasion). Total number of parasitophorous vacuoles and number of parasites per parasitophorous vacuole were determined. Mean values of 3 independent experiments ± s.d. are shown. Asterisks indicate significant difference in total invasion between parasite strain Rab11ADN not treated with Shld1 and those treated with Shld1 before and after invasion (P < 0.01, two tailed Student’s t-test).
An alternative approach often used to examine the functions of essential genes is to determine the phenotypic consequences of overexpression of dominant-negative alleles. We used the ddFKBP-system to characterize the function of two small G proteins, Rab1 and Rab11A, by Shld1-dependent stabilization of dominant negative versions. For the initial analysis of Rab1 we fused the highly conserved wild-type (YPT1) and dominant negative (YPT1DN) versions of Candida albicans8 proteins to the ddFKBP. Both versions could be efficiently regulated in a Shld1-dependent way, and we detected the respective fusions as soon as 4 h after induction (Fig. 2a,b). We assayed parasite growth in presence or absence of Shld1 for 5 d. Whereas overexpression of Ypt1 had no effect on the propagation of the parasites, as judged from the size of plaques formed in a HFF monolayer, stabilization of YPT1DN resulted in substantially reduced parasite propagation (Fig. 2c and Supplementary Fig. 3 online), indicating that expression of YPT1DN, but not YPT1 is deleterious for parasite growth.
For analysis of Rab11A we generated a dominant negative version of TgRab11A (N126I), which inactivates the GTPase domain. Upon stable transfection of ddFKBP-Rab11ADN, we isolated clonal parasites in which ddFKBP-Rab11ADN was detectable 2 h after addition of 1 μM Shld1 (Fig. 2a,b). In growth assays we detected normal plaque formation, when compared to RH wild-type (RHwt) parasites in the absence of Shld1. However, in the presence of inducer we did not detect growth of the parasite, demonstrating that expression of this dominant negative G protein is deleterious for parasite growth (Supplementary Fig. 3).
To assess whether Rab11A might have different roles in intra- and extracellular parasites, we took advantage of the rapid induction kinetics of the ddFKBP system. We pre-incubated extracellular parasites for 20 min in the presence or absence of Shld1, allowed them to invade host cells for 3 h and then removed remaining parasites. We incubated intracellular parasites in presence or absence of Shld1 for 16 h, and then analyzed invasion and replication. Whereas RHwt parasites do not show any invasion or replication defect upon treatment with Shld1 (Supplementary Fig. 4 online), stabilization of ddFKBP-Rab11ADN in extracellular parasites reduced the ability of the parasites to invade the host cell by ∼85% when compared to parasites not treated with Shld1 (Fig. 2d). Additionally, there was a substantial reduction in the number of parasites per vacuole when parasites were treated with Shld1 after invasion of the host cell. This indicates a role of Rab11A in invasion of the host cell and in the intracellular propagation (replication or maturation) of the parasite (Fig. 2d). This suggests that Rab11A has an essential function for both extra- and intra-cellular parasites and shows that the ddFKBP system can be used to dissect the role of a protein in extra- and intracellular parasites.
Although the ddFKBP system can be applied in T. gondii, some proteins, as we demonstrated for MyoA, might not be functional when fused to ddFKBP. Another limitation might be that not all fusion proteins can be efficiently targeted to the proteasome. However, the rapid induction kinetics of this system allows specific modulation of protein levels in extra- and intracellular parasites, and will allow a detailed, functional dissection of specific processes during the life cycle of the parasite with high precision. Analysis of protein functions is often performed with the help of dominant negative mutants. Using the ddFKBP system, it is now possible to apply this strategy in T. gondii and possibly P. falciparum3, which may find broad application in research on apicomplexans.
Supplementary Material
ACKNOWLEDGMENTS
We thank B. Striepen (University of Georgia), D.L. Sibley (Washington University), V.B. Carruthers (Johns Hopkins School of Public Health), C.J. Beckers (The University of North Carolina) and D. Soldati (University of Geneva) for sharing reagents. We thank K. Matuschewski, G. Langsley and D. Goldberg for critically reading the manuscript, M. Rauch for technical assistance, and M. Kudryashev for help with image analysis. This work was funded by the BioFuture-Programm (grant 0311897) of the German ministry of science and education (BMBF).
References
- 1.Meissner M, Agop-Nersesian C, Sullivan WJ., Jr. Appl. Microbiol. Biotechnol. 2007;75:963–975. doi: 10.1007/s00253-007-0946-4. [DOI] [PubMed] [Google Scholar]
- 2.Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong CM, Goldberg DE. Nat. Methods. 2007 November 11; advance online publication. doi:10.1038/nmeth1132. [Google Scholar]
- 4.Meissner M, Schluter D, Soldati D. Science. 2002;298:837–840. doi: 10.1126/science.1074553. [DOI] [PubMed] [Google Scholar]
- 5.Soldati D, Boothroyd JC. Mol. Cell. Biol. 1995;15:87–93. doi: 10.1128/mcb.15.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meissner M, Brecht S, Bujard H, Soldati D. Nucleic Acids Res. 2001;29:e115. doi: 10.1093/nar/29.22.e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herm-Gotz A, et al. EMBO J. 2002;21:2149–2158. doi: 10.1093/emboj/21.9.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee SA, Mao Y, Zhang Z, Wong B. Microbiology. 2001;147:1961–1970. doi: 10.1099/00221287-147-7-1961. [DOI] [PubMed] [Google Scholar]
- 9.Wan KL, Carruthers VB, Sibley LD, Ajioka JW. Mol. Biochem. Parasitol. 1997;84:203–214. doi: 10.1016/s0166-6851(96)02796-x. [DOI] [PubMed] [Google Scholar]
- 10.Morrissette NS, Sibley LD. J. Cell Sci. 2002;115:1017–1025. doi: 10.1242/jcs.115.5.1017. [DOI] [PubMed] [Google Scholar]
- 11.Mann T, Gaskins E, Beckers CJ. J. Biol. Chem. 2002;12:41240–41246. doi: 10.1074/jbc.M205056200. [DOI] [PubMed] [Google Scholar]
- 12.Gaskins E, et al. J. Cell Biol. 2004;165:383–393. doi: 10.1083/jcb.200311137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.