Abstract
Autophagy is responsible for nonspecific, bulk degradation of cytoplasmic components. Recent work has revealed also that there is specific, autophagic degradation of polyubiquitinated protein aggregates, whose buildup occurs during neurodegenerative disease. Here, we report that simple mono-ubiquitination of normally long-lived cytoplasmic substrates is sufficient to target these substrates for autophagic degradation in mammalian cells. That is, upon their ubiquitination, both small [i.e., red fluorescent protein (RFP)] and large (i.e., peroxisomes) substrates are efficiently targeted to autophagosomes and then degraded within lysosomes upon autophagosome-lysosome fusion. This targeting requires the ubiquitin-binding protein, p62, and is blocked by the Class III phosphatidylinositol 3-kinase (PI3K) inhibitor, 3-methyladenine (3-MA), or by depletion of the autophagy-related-12 (Atg12) protein homolog. Mammalian cells thus use a common pathway involving ubiquitin and p62 for targeting diverse types of substrates for autophagy.
Keywords: autophagy, p62, pexophagy
To destroy soluble proteins, large aggregates and organelles in the cytoplasm, cells use macroautophagy, a highly conserved bulk degradation pathway in eukaryotes (1). In the initial step of this pathway, herein referred to as autophagy, an isolation membrane forms in the cytoplasm through the activity of specific autophagy effectors, including LC3 (microtubule-associated protein 1 light chain 3) (1). The nascent membrane then wraps around a portion of cytoplasm (including the soluble proteins, aggregates, or organelle) to eventually form a double membrane-bounded structure called the autophagosome. When this structure fuses with the lysosome, the sequestered materials are degraded by lysosomal hydrolytic enzymes and recycled as free amino acids, lipids, and carbohydrates for macromolecular synthesis and/or energy production. Emerging evidence indicates that autophagy plays a critical role not only as a supplier of amino acids for cell survival under stress conditions, but as a cytoprotector in removing long-lived proteins, aggregated protein complexes, and excess or damaged organelles (1). Defects in autophagy, therefore, underlie various pathological conditions within organisms, including tumorigenesis, defects in developmental programs and the buildup of toxic, protein aggregates involved in neurodegeneration (2–4).
An important unanswered question regarding autophagy is how its diverse array of substrates are selectively sequestered by autophagosomes. Although a major avenue of substrate degradation is mediated via nonspecific, bulk engulfment of these substrates by the autophagosome, recent evidence suggests that some type of specific autophagic targeting mechanism also plays a role. Mitochondria, peroxisomes and ribosomes, for example, are sequestered into autophagosomes in a selective manner upon specific metabolic changes within the cell (5–9). Likewise, many viral and bacterial pathogens are efficiently enveloped into autophagic membranes after being taken up into cells (10, 11). Finally, aggregates of polyubiquitinated, misfolded proteins formed in the cytoplasm are selectively removed from cells by autophagy (12).
Recent data have shown also that the autophagic turnover of protein aggregates (called inclusion bodies) is facilitated by the signaling adaptor scaffold protein p62 (also referred to as SQSTM1), which binds both to polyubiquitinated proteins in aggregates and to LC3 (13, 14). It is now well established that p62 regulates inclusion body formation and degradation by autophagy (13, 14). However, although p62-mediated cross-talk between polyubiquitinated aggregate proteins and LC3 helps explain how inclusion bodies are removed by autophagy, it is not known whether a similar mechanism is responsible for how other substrates, including soluble proteins and organelles, are selectively targeted by the autophagic pathway. Here, we systematically investigate the role of ubiquitin modifications in autophagic targeting of diverse types of substrates, including long-lived cytoplasmic proteins and membrane-bound organelles. Our findings define a general mechanism for substrate-specific autophagy in mammalian cells in which mono-ubiquitination is itself sufficient to signal autophagy through a pathway involving p62 binding.
Results
To test whether ubiquitin can serve as a general signal to specifically target substrates for autophagy, we attached it to monomeric RFP, a long-lived, cytosolic protein (i.e., 4.6-day half-life) (15). The fusion construct, called UB-RFP, was generated by linking ubiquitin with a G76V mutation (glycine at position 76 replaced with valine) in its C terminus to the N terminus of RFP through a peptide bond. This modified (G76V) version of ubiquitin was used because it prevents UB-RFP from being deubiquitinated by cellular factors without affecting UB-RFP susceptibility for degradation by proteasomes (16–18).
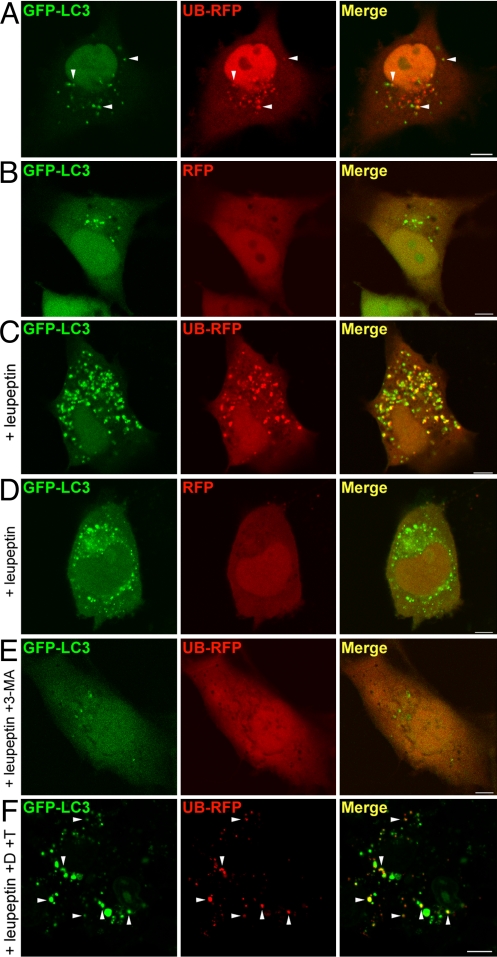
Confocal microscopic imaging in COS-7 cells transiently expressing UB-RFP and the autophagic marker GFP-LC3 (consisting of the green fluorescent protein fused to the N terminus of LC3) revealed UB-RFP colocalized with GFP-LC3 in small punctate structures scattered throughout the cytoplasm (arrowheads, Fig. 1A) in addition to diffusely distributing in the cytosol and nucleus. By contrast, in cells coexpressing RFP and GFP-LC3, all of the RFP was diffusely distributed throughout the cytosol and nucleus with no significant colocalization with GFP-LC3-containing punctate structures (Fig. 1B). The number and size of structures containing UB-RFP and/or GFP-LC3 increased under conditions in which degradation of autophagic substrates within lysosomes via lysosomal enzymes was inhibited by leupeptin (19) (Fig. 1C), in contrast to that in RFP and GFP-LC3 coexpressing cells treated with leupeptin, wherein RFP was absent from the GFP-LC3-containing structures (Fig. 1D). Finally, no structures containing both UB-RFP and/or GFP-LC3 were formed in leupeptin-treated cells that were incubated with the autophagy inhibitor 3-MA, which blocks the PI3K activity necessary for autophagosome formation (20) (Fig. 1E) indicating that the structures containing UB-RFP and GFP-LC3 were autophagosomes.
Fig. 1.
Ubiquitin targets cytosolic RFP to autophagosomes for degradation. (A and B) COS-7 cells transiently cotransfected with GFP-LC3 and UB-RFP (A), or GFP-LC3 and RFP (B). Cells were imaged 24 h after transfection. Arrowheads in (A) indicate obvious examples of colocalized GFP-LC3 and UB-RFP. (C and D) COS-7 cells transiently cotransfected with either GFP-LC3 and UB-RFP (C) or GFP-LC3 and RFP (D), and treated with 0.25 mM leupeptin for 20 h before imaging. (E) COS-7 cells transiently coexpressing GFP-LC3 and UB-RFP, and treated with 0.25 mM leupeptin and 10 mM 3-MA for 20 h before imaging. (F) Fluorescence Protease Protection assay of COS-7 cells coexpressing GFP-LC3 and UB-RFP. 24 h after cotransfection and leupeptin treatment, cells were washed and then treated with 0.6% [vol/vol] digitonin (D) for 10 min and than with 0.005% [wt/vol] trypsin (T), followed by imaging. Arrowheads indicate obvious examples of colocalized GFP-LC3 and UB-RFP. (Scale bars, 10 μm.)
When the plasma membrane of cells expressing UB-RFP-expressing cells was selectively permeabilized with digitonin, and then trypsin was applied to digest all fluorescent protein moieties facing the cytosol (21), all of the cytoplasmic and nuclear pools of UB-RFP were degraded, whereas the signal attributable to UB-RFP associated with autophagosomes (marked with coexpressed GFP-LC3) remained undiminished (Fig. 1F). This indicated that UB-RFP was specifically sequestered by autophagosomes rather than merely associated with autophagosomal membranes.
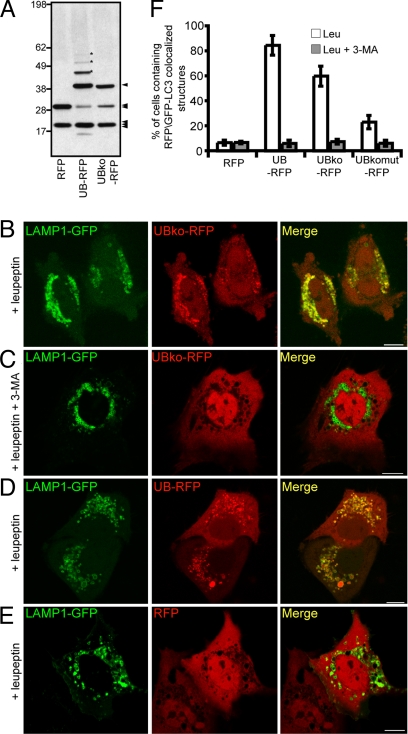
To test what type of ubiquitination (i.e., mono- versus polyubiquitination) was relevant for targeting UB-RFP for autophagy, we used a modified version of UB-RFP, termed UBko-RFP, where all seven lysine residues within UB were mutated to arginine so that the fusion protein could not be polyubiquitinated (22). Protein immunoblot analysis (with anti-RFP antibodies) of COS-7 cells expressing RFP, UB-RFP, or UBko-RFP revealed that UBko-RFP did not undergo polyubiquitination (Fig. 2A). Furthermore, UBko-RFP was not readily degraded by proteasomes, as expected for a protein that is not polyubiquitinated [supporting information (SI) Fig. S1]. When UBko-RFP was expressed in leupeptin-treated cells, significant colocalization was observed with the coexpressed lysosomal marker, LAMP1-GFP (consisting of the lysosomal associated membrane protein 1 fused to the N terminus of the GFP) (23) (Fig. 2B). No significant lysosomal accumulation of UBko-RFP occurred in cells treated with leupeptin and the autophagy inhibitor 3-MA (Fig. 2C). UB-RFP (which can be either mono- or polyubiquitinated) showed a similar colocalization with LAMP1-GFP in cells treated only with leupeptin (Fig. 2D), whereas RFP alone (i.e., with no ubiquitin modification) did not colocalize with the lysosomal marker protein in leupeptin-treated cells (Fig. 2E). These data are quantified in Fig. 2F. Mono-ubiquitination is thus minimally sufficient to target RFP to the autophagic pathway.
Fig. 2.
Monomeric ubiquitin is sufficient to target RFP for degradation by autophagy. (A) Immunoblot of cell lysate from COS-7 cells expressing RFP, UB-RFP, or UBko-RFP. 25 μg of total protein was resolved by SDS/PAGE and immunoblot using rabbit anti-RFP antibodies and donkey anti-rabbit IgG conjugated to horseradish peroxidase. Asterisks indicate the position of three higher molecular weight species of polyubiquitinated UBko-RFP. The single arrowhead indicates the position of mono-ubiquitinated RFP, whereas the double and triple arrowheads indicate the position of RFP alone and a cross reacting band, respectively. Molecular masses (in kDa) are indicated on the left side of the blot. (B–E) COS-7 cells cotransfected with LAMP1-GFP and either UB-RFP (B and C), UB-RFP (D), or RFP (E). Leupeptin (0.25 mM) was added 20 h before imaging. Cells shown in (C) were treated also with 10 mM 3-MA. (Scale bars, 10 μm.) (F) Quantification of the percentage of cells with five or more punctate RFP signals that colocalized with GFP-LC3 in cells coexpressing GFP-LC3 and various RFP constructs as indicated. Cells were also incubated with either leupeptin alone (white bar), or with leupeptin and 3-MA (dark gray bars). Shown are the averages ± standard deviations from three independent experiments with each experiment including at least 50 cells scored.
In other ubiquitin-targeting systems, a hydrophobic patch domain in the ubiquitin polypeptide, including a leucine at position 8, isoleucine-44, and valine-70, is essential for its interactions with ubiquitin-binding proteins (24). To determine whether this patch is also required for the autophagic targeting of ubiquitinated substrates, we mutated to alanine each of the three hydrophobic residues within the patch domain of UBko-RFP yielding UBkomut-RFP and then looked at the extent of its autophagic targeting compared with UB-RFP and UBkoRFP. As shown in Fig. 2F, expression of UBkomut-RFP resulted in significantly fewer cells having punctate structures containing the RFP protein and GFP-LC3 (P < 0.01). Hence, the hydrophobic patch on ubiquitin appears to be necessary for ubiquitinated RFP substrates to be targeted for autophagy.
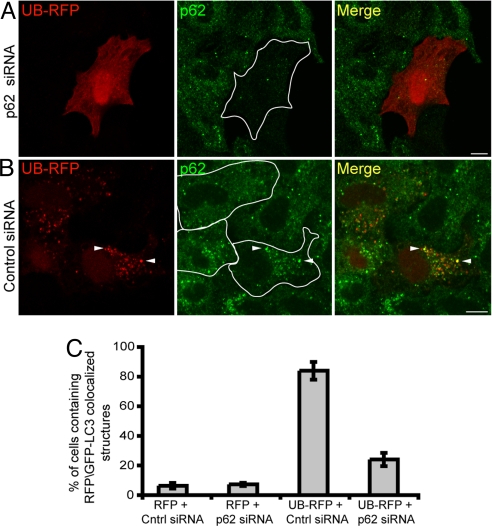
The signaling adaptor scaffold protein known as p62 is one type of ubiquitin-binding protein that binds to the hydrophobic patch on ubiquitin (25). To test whether p62 is involved in targeting UB-RFP to autophagosomes, we depleted p62 from HeLa cells by using small inhibitory RNA (siRNA) (Fig. S2) and then examined the intracellular distribution of UB-RFP. No punctate autophagic structures containing UB-RFP were observed in most p62 siRNA-depleted cells (Fig. 3 A and C) compared with control siRNA-treated cells (Fig. 3 B and C), suggesting that p62 is required for UB-RFP to become sequestered within autophagosomes. Furthermore, because the distribution of p62 in the control siRNA-treated cells (arrowheads, Fig. 3B) or non-siRNA-treated cells (Fig. S3) overlapped significantly with the punctate structures containing UB-RFP, the data further suggested that UB-RFP targets to autophagosomes in a complex with p62.
Fig. 3.
p62 is required for sequestering UB-RFP into punctate structures. (A and B) HeLa cells transfected with either siRNA pools directed against p62 (A) or control siRNA (B). Twenty-four hours later, cells were transfected again with the respective siRNA and also with a plasmid encoding UB-RFP. Four hours after the second transfection, leupeptin (0.25 mM) was added to the cells, and 20 h after that, cells were fixed and stained with anti-p62 and Alexa 543-goat anti-rabbit antibodies. White lines in micrographs illustrating endogenous p62 immunostaining frame the outline of cells coexpressing UB-RFP. Arrowheads in (B) indicate obvious examples of colocalized UB-RFP and endogenous p62. (Scale bars, 10 μm.) (C) Quantification of the percentage of cells with 5 or more punctate structures containing either RFP or UB-RFP and GFP-LC3 in cells transfected with also control siRNA or p62 siRNA and treated with leupeptin as in (A and B). Shown are the averages ± standard deviations from three independent experiments with each experiment including at least 50 cells scored.
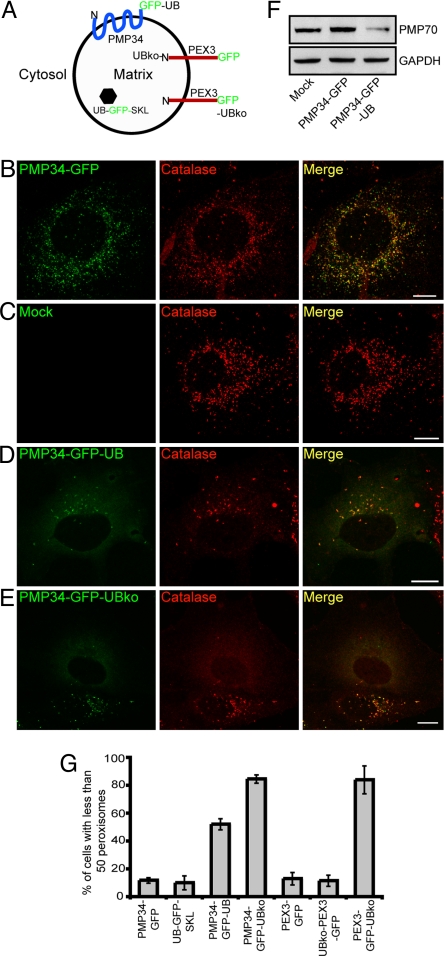
We investigated next whether attaching ubiquitin to a larger structure in the cytoplasm, such as a membrane-bound organelle, targeted it for autophagy. As a test case, we chose the peroxisome, a normally long-lived organelle with hundreds of copies per cell that can be readily quantified via light microscopy (26, 27). We attached GFP or GFP-UB (containing the G76V mutation to prevent further conjugation of ubiquitin moieties) to the cytoplasmically exposed C terminus tail of the human 34-kDa, multispanning (Ncytosol-Ccytosol) peroxisomal integral membrane protein (PMP34) (28, 29). The predicted orientation of PMP34-GFP-UB is shown in Fig. 4A. When expressed individually in COS-7 cells the fluorescent pattern attributable to PMP34-GFP overlapped with that attributable to immunostained endogenous peroxisomal matrix catalase (Fig. 4B) and there was no obvious change in the overall number of peroxisomes in these cells relative to mock-transfected cells (Fig. 4C), indicating the GFP tag did not affect either PMP34's ability to target to peroxisomes, nor the steady-state number of peroxisomes within PMP34-GFP-transformed cells. In PMP34-GFP-UB-expressing cells, however, the total number of catalase-containing structures was dramatically decreased (Fig. 4D). Similar results were observed with a mono-ubiquitinated form of PMP34-GFP-UB, called PMP34-GFP-UBko, in which the lysines in ubiquitin were mutated to arginines (Fig. 4E). This loss of peroxisomes in cells expressing PMP34-GFP-UB was further verified by quantifying the amount of an endogenous PMP (PMP70) (30) using protein immunoblot analysis (Fig. 4F) and by quantifying the number of peroxisomes within PMP34-GFP-UB- and PMP34-GFP-UBko-expressing cells (Fig. 4G).
Fig. 4.
Ubiquitination of a PMP results in a decrease in the number of peroxisomes within a cell. (A) Schematic illustration of the predicted topological orientation of PMP34-GFP-UBko, UB-GFP-SKL, UBko-PEX3-GFP, and PEX3-GFP-UBko. (B–E) COS-7 cells transiently expressing either PMP34-GFP (A), empty vector (mock) (B), PMP34-GFP-UB (C), or PMP34-GFP-UBko (D) were fixed and stained with anti-catalase and Alexa 543-goat anti-rabbit antibodies 48 h after transfection. Note that two PMP34-GFP-UBko-transformed cells can be seen in (D), both of which display a reduced number of peroxisomes relative to cells transformed with the empty vector (B) or PMP34-GFP (A). (F) Immunoblot of cell lysate from COS-7 cells expressing PMP34-GFP, PMP34-GFP-UB or mock treated. Cells were lysed, and 25 μg of total protein was subjected to SDS/PAGE and then immunoblotted with antibodies against PMP70 or the cytosolic protein glyceraldehydes phosphate dehydrogenase (GAPDH) serving as a protein loading control. (G) Percentage of total number of transfected cells with less than 50 peroxisomes per cell expressing various proteins as indicated 48 h after transfection. Shown are the averages ± standard deviations from three independent samples with each experiment including at least 100 cells scored.
To determine whether the ubiquitin moiety on peroxisomes needs to face the cytosol to facilitate a reduction in the number of peroxisomes within a cell, we tested the effect of expressing UB-GFP-SKL, which targets ubiquitin to the matrix of peroxisomes via a C-terminal-appended -SKL sequence, also known as a type 1 matrix peroxisomal targeting signal (31) (Fig. 4A). No significant loss of peroxisomes was observed in cells transfected with UB-GFP-SKL (Fig. 4G; P > 0.8). We also attached monomerized ubiquitin (i.e., UBko) to the matrix-facing, N-terminal of PEX3, a (Nmatrix-Ccytosol) peroxisomal integral PMP (32) (Fig. 4A). Both PEX3-GFP and UBko-PEX3-GFP targeted efficiently to peroxisomes in transiently transformed COS-7 cells and there was also no obvious reduction in the number of peroxisomes in either set of cells (Fig. 4G and Fig. S4 A and B). By contrast, expression of a PEX3 construct with ubiquitin attached to its cytosol-facing C terminus (PEX3-GFP-UBko) led to significant loss of peroxisomes within cells (Fig. 4G and Fig. S4C; P < 0.01). Hence, for ubiquitin-mediated degradation of peroxisomes to occur, the ubiquitin modification must be on the cytosolic face of the peroxisomal boundary membrane.
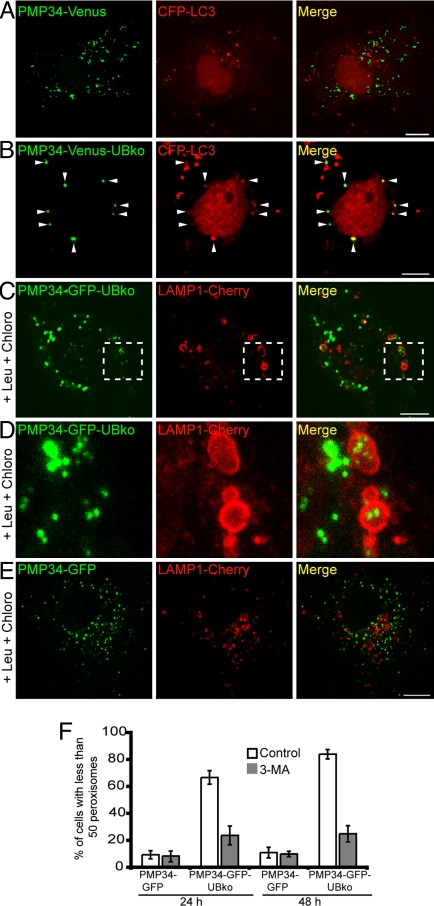
The degradation of peroxisomes via the autophagic pathway is referred to as pexophagy (7, 33, 34). To determine whether the loss of peroxisomes observed in the above experiments with PMP34-GFP-UB or PMP34-GFP-UBko occurred by pexophagy, we examined whether ubiquitinated peroxisomes became sequestered in autophagosomes. Toward that end, cells were cotransfected with a peroxisome marker (i.e., PMP34 fused to Venus and the UBko moiety, PMP34-Venus-UBko, or PMP34-Venus alone) and with the autophagosomal marker protein, CFP-LC3. We found that none of the PMP34-Venus-labeled structures were colocalized with the autophagosome marker protein, CFP-LC3, two after days of transfection (Fig. 5A), whereas virtually all of the PMP34-Venus-UBko-labeled structures colocalized with CFP-LC3 (arrowheads, Fig. 5B). Hence, peroxisomes modified with ubiquitin show an increased association with autophagosomes.
Fig. 5.
The autophagic pathway mediates a decrease in ubiquitinated peroxisomes. (A and B) COS-7 cells coexpressing CFP-LC3 and either PMP34-Venus (A) or PMP34-Venus-UBko (B). Arrowheads in (B) indicate obvious examples of colocalized PMP34-Venus-UBko and CFP-LC3. (C–E) COS-7 cells coexpressing LAMP1-Cherry and either PMP34-GFP-UBko (C and D) or PMP34-GFP (E). Cells were also treated with leupeptin (0.25 mM) and chloroquine (0.1 mM) 8 h after transfection. The higher magnified images shown in (D) are a maximum intensity projection of a z series of the magnified area of the single slice image of the cell outlined in (C). (F) Percentage of the total number of transfected cells with less than 50 peroxisomes per cell expressing either PMP34-GFP or PMP34-GFP-UB, and with or without 3-MA, after 24-h or 48-h post-transfection. Shown are averages ± standard deviation from three independent samples with each experiment including at least 100 cells scored. (Scale bars, 10 μm.)
We determined next whether the autophagosomes containing ubiquitin-modified peroxisomes fused with lysosomes. Specifically, we coexpressed PMP34-GFP-UBko or PMP34-GFP with LAMP1 fused to the modified RFP Cherry (35) (LAMP1-Cherry), and then treated the cells with leupeptin and chloroquine (Fig. 5 C–E). Chloroquine was used along with leupeptin as this lysosomotropic amine raises the pH of acidic lysosomes, thereby preventing the quenching of the pH-sensitive GFP (36). In these cells, a significant fraction of PMP34-GFP-UBko-containing peroxisomes associated with LAMP1-Cherry-containing lysosomes (Fig. 5C). Magnification of a portion of the PMP34-GFP-UBko transfected cell revealed that some of the peroxisomes actually resided inside the lysosomes (Fig. 5D), consistent with these peroxisomes having been delivered to lysosomes by an autophagosome-lysosome fusion event. This targeting to lysosomes was determined also to be specific for PMP34-GFP-UBko, because no similar colocalizations were observed for PMP34-GFP and LAMP1-Cherry (Fig. 5E).
To further verify that ubiquitinated peroxisomes were lost by autophagy, we examined the effect of inhibiting autophagy by 3-MA treatment on cells expressing PMP34-GFP-UBko. The loss of peroxisomes in PMP34-GFP-UBko expressing cells was sensitive to 3-MA (Fig. 5F). Hence, the ubiquitinated peroxisomes are degraded within cells by a pathway that depends on Class III PI3 Kinase activation of autophagy.
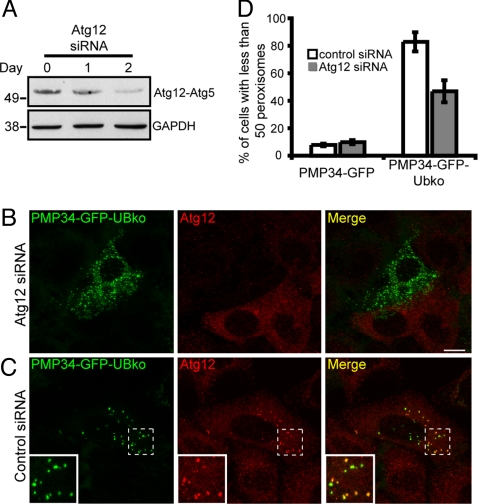
Autophagy is also inhibited by knocking down the Atg12 expression by siRNA treatment, as Atg12 conjugates with Atg5 to form an essential component of autophagy machinery required for the elongation of the nascent autophagosomal membranes (37). To test a role of Atg12 in the peroxisome loss phenotype induced by PMP34-GFP-UBko expression, we depleted Atg12 by siRNA in HeLa Cells (Fig. 6A). The peroxisome loss phenotype was much less severe in these cells (Fig. 6 B and D) compared with PMP34-GFP-UBko cells treated with control siRNA (Fig. 6 C and D). This result, together with the fact that Atg12 colocalized with PMP34-GFP-UBko containing peroxisomes in control siRNA-treated cells (where there is peroxisome loss due to PMP34-GFP-UBko expression) (see inset in Fig. 6C), suggested that the peroxisome decrease occurred by autophagy.
Fig. 6.
Silencing of Atg12 expression prevents ubiquitin-mediated peroxisome degradation. (A) Immunoblot of cell lysate from HeLa cells before Atg12 siRNA treatment (day 0), or 1 and 2 days after Atg12 siRNA treatment as indicated. Cells were lysed and 25 μg of total protein was subjected to SDS/PAGE and then immunoblotted with antibodies against either Atg12 or GAPDH serving as a loading control. Depletion in Atg12 expression was indicated by the decrease in the Atg12-Atg5 protein conjugate. (B–C) HeLa cells were transfected with either siRNA pool directed against Atg12 (B), or a nontargeting control siRNA (C). Twenty-four hours after the initial transfection, cell were transfected again with the appropriate siRNA along with a plasmids encoding PMP34-GFP-UBko. Forty-eight hours after the second transfection cells were fixed and stained with anti-Atg12 and Alexa 543-goat anti-rabbit antibodies. (D) Percentage of the total number of cells with less than 50 peroxisomes per cell expressing either PMP34-GFP or PMP34-GFP-UBko and treated with either control siRNA or Atg12 siRNA. Shown are the averages ± standard deviations from three independent samples with each experiment including at least 100 cells scored. (Scale bars, 10 μm.)
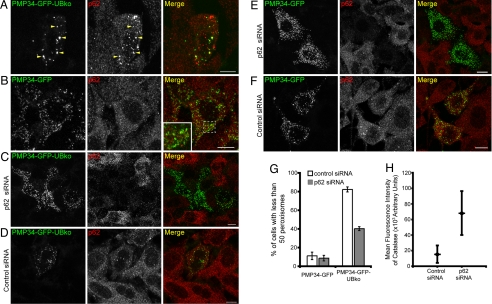
To determine whether p62 is involved in ubiquitin-mediated autophagy of peroxisomes, HeLa cells expressing PMP34-GFP-UBko were immunolabeled with endogenous p62. A significant portion of p62 colocalized with ubiquitinated PMP34-GFP-UBko-labeled peroxisomes (arrowheads, Fig. 7A). In cells expressing PMP34-GFP, by contrast, p62 did not colocalize with peroxisomes (Fig. 7B). Hence, p62 is specifically recruited to ubiquitinated peroxisomes. Upon p62 depletion by siRNA, the peroxisome loss phenotype observed in PMP34-GFP-UBko cells was alleviated (Fig. 7 C, D, and G). This suggested that delivery of ubiquitinated peroxisomes to autophagosomes requires p62, similar to ubiquitinated aggregates (13, 14) and ubiquitinated soluble proteins shown in Fig. 3. Interestingly, a careful examination of p62-depleted cells expressing PMP34-GFP (Fig. 7E) suggested they had more peroxisomes compared with cells not depleted of p62 (Fig. 7F). This was confirmed by quantifying the levels of immunodetected endogenous catalase: A significant increase in fluorescence intensity (P < 0.01) attributable to immunostained catalase was found in p62-depleted cells compared with that in cells treated with nontargeting (control) siRNA (Fig. 7H). Peroxisome levels within cells, therefore, appear to be regulated through an autophagic pathway involving p62.
Fig. 7.
p62 is required for the degradation of peroxisomes. (A and B) HeLa cells transiently expressing either PMP34-GFP-UBko (A) or PMP34-GFP (B) were fixed and stained with anti-p62 and Alexa 543-goat anti-rabbit antibodies 48 h after transfection. Arrowheads in (A) indicate obvious examples of colocalized PMP34-GFP-UBko and endogenous p62. The area of the cell outlined in the merge image in (B) is shown at higher magnification in the inset; note the lack of any obvious colocalizations of expressed PMP34-GFP and endogenous p62 in this cell. (C–F) HeLa cells transfected with either control siRNA or a siRNA pool for p62 (p62 siRNA) as indicated. Twenty-four hours after the initial transfection, cells were retransfected with siRNA and also with a plasmids encoding either PMP34-GFP-UBko (C and D) or PMP34-GFP (E and F). Cells were fixed and stained as above either 48 h after first transfection treatment. All images shown are maximum projections of z-series. (G) Percentage of the total number of transfected cells with less than 50 peroxisomes expressing either PMP34-GFP or PMP34-GFP-UBko and treated with either control siRNA or p62 siRNA at 72-h post-transfection. Shown are the averages ± standard deviations from three independent samples with each experiment including at least 75 cells scored. (H) The mean fluorescence intensity of the immunolabeled endogenous catalase from at least 25 cells treated with either control siRNA or a siRNA pool for p62. Also shown are the standard deviations of the mean values (P < 0.01). (Scale bars, 10 μm.)
Discussion
Here, we report a general mechanism for substrate-specific autophagy that involves ubiquitin and the ubiquitin-binding protein, p62. We showed that when long-lived cytoplasmic substrates (including RFP and peroxisomes) were linked to ubiquitin, they became selectively sequestered and degraded by the autophagic pathway. That is, these ubiquitinated substrates were first sequestered into Atg12- and LC3-positive structures that were sensitive to 3-MA and were subsequently degraded by lysosomal hydrolytic enzymes upon autophagosome-lysosome fusion. We showed also that mono-ubiquitination was sufficient to target these substrates to autophagosomes, and that this process depended on the ubiquitin-binding protein, p62. Finally, we presented evidence that the normal turnover of peroxisomes requires p62, which suggests that the normal turnover of this organelle may involve ubiquitination of one of its peroxisomal membrane proteins.
Until now, p62 has been studied primarily in the context of protein aggregates and inclusion bodies, where it plays an architectural role in the formation of these structures. Specifically, p62 was shown to bind ubiquitinated proteins within aggregates, to mediate polymerization of these aggregates through its ability to self-associate into a scaffold, and to cause autophagic vesicles to envelope the aggregates through its affinity for LC3, ultimately leading to the formation of a lysosomal-destined inclusion body (38). Given our results of p62 involvement in autophagic degradation of UB-RFP and ubiquitin-modified peroxisomes, the possibility emerges that p62 has a similar architectural role in the degradation of these substrates. For instance, we showed that UB-RFP became concentrated in autophagic bodies at much higher levels than that found in the cytoplasm, suggestive of an aggregation event during autophagic targeting. UB-RFP was also colocalized with p62 within these autophagosomes, consistent with p62 having a dual role in concentrating UB-RFP and recruiting LC3-positive membranes. Conversely, no autophagic sequestration of UB-RFP occurred in p62-depleted cells, indicating that p62 was essential for this process. Hence, p62's role may be both for cross-linking (scaffolding) UB-RFP into aggregates and for attracting autophagic vesicles.
Our results suggest p62 could serve a similar set of functions in the autophagic targeting of ubiquitin-modified peroxisomes. Once ubiquitinated membrane proteins located on the cytosolic surface of the peroxisome reach high enough levels, the association of these proteins with p62 could recruit LC3-containing membranes to cause autophagic engulfment of the organelle. Furthermore, our finding that depletion of p62 causes an increase in peroxisome number in cells suggests that peroxisomes are normally degraded in a p62-dependent matter.
Ubiquitinated forms of RFP that can be polyubiquitinated were more efficiently targeted for autophagy compared with forms that were only mono-ubiquitinated, whereas the reverse was true for peroxisomes. A possible explanation for this relates to possible differences in the attraction of nascent autophagosomes to small proteins versus the exposed surfaces of large organelles, and to the competitive interplay between autophagy and the proteasome for ubiquitinated substrates. Because p62 has been shown to bind single ubiquitin units (39), polyubiquitinated RFP should bind more p62 molecules than mono-ubiquitinated RFP and, thus, should more readily aggregate and attract LC3-containing membranes onto the protein aggregate's surface, or perhaps simply target individual proteins to nascent autophagosomes more readily. In the case of peroxisomes, however, ubiquitinated proteins on the cytosolic surface of the organelle are already stabilized by being associated with the organelle's boundary membrane. Thus, aggregation of polyubiquitinated species by p62 molecules might be less important for attracting autophagic vesicles to peroxisomes. Furthermore, because a portion of the polyubiquitinated, nascent PMP pool could be destroyed by the proteasome before being inserted posttranslationally into peroxisomal membranes, more monoubiquitinated PMPs would associate with peroxisomal membranes than polyubiquitinated PMPs at identical expression levels. Consequently, peroxisomes containing monoubiquitinated PMPs should be more efficiently targeted for autophagy compared with peroxisomes containing their polyubiquitinated counterparts.
Which endogenous PMP(s) is/are ubiquitinated to activate selective pexophagy? Although the answer to this question remains to be addressed experimentally, possible candidates for pexophagy-mediated peroxisome ubiquitination are PEX4 (a ubiquitin conjugation enzyme) (40) and PEX5 (the soluble receptor responsible for recognizing, targeting and subsequently importing the majority of nascent peroxisomal matrix-destined proteins) (5). In the process of translocating a PTS1-containing protein into the peroxisomal matrix, PEX5 is inserted into the organelle's membrane. After release of its cargo, the receptor is then ubiquitinated by PEX4 and retro-translocated out of the bilayer by the action of an AAA-ATPase complex consisting of PEX1, PEX6, and PEX26 (41). Given this recycling pathway, either ubiquitin-bound PEX4 or ubiquitinated PEX5 could accumulate on peroxisomes under conditions that either down-regulate components of the PMP insertion machinery (i.e., decreased PEX3 expression) and/or inhibit one or more components comprising the above-mentioned AAA-ATPase complex (e.g., PEX1, PEX6, and/or PEX26). Interestingly, pexophagy in the yeast Hansenula polymorpha is preceded by a decrease in the expression of Pex3p (a homologue of mammalian PEX3) (42). Likewise, the absence of Pex14p, which docks Pex5p (the homologes of PEX14 and PEX5, respectively) onto the peroxisomal membrane (43) prevented pexophagy in H. polymorpha (44), possibly because of the lack of Pex5p on the peroxisomal membrane. It is conceivable, therefore, that either or both PEX4 and PEX5 are the principal endogenous peroxisomal proteins that are ubiquitinated to serve as a signal for the degradation of the peroxisome in mammalian cells. However, it is not known whether an ubiquitin/p62 dependent pathway is involved in selective pexophagy in yeast (45).
That ubiquitin modification and p62 binding cause diverse substrates, including both small soluble proteins and large membrane-bound organelles, to be specifically targeted to autophagosomes in mammalian cells has important implications for understanding how these cells regulate protein and organelle turnover. In particular, conditions within cells that modulate levels of ubiquitinated substrates and p62 expression may have key regulatory effects on protein and organelle turnover. Consistent with this possibility, the endoplasmic reticulum (ER) in yeast is well known to undergo dramatic changes in size and undergoes autophagy in response to ER stress-induced conditions during which ubiquitinated ER proteins accumulate (46). Likewise, sperm cell mitochondria are known to be ubiquitinated before being degraded in fertilized eggs (47). Finally, as demonstrated in this study, depletion of p62 leads to an increase in the number of peroxisomes within cells, implying that the molecular machinery regulating their normal turnover includes p62.
In summary, our findings suggest a general mechanism for substrate-specific autophagy involving ubiquitin modification and p62 binding that can regulate the turnover of small molecules, as well as entire organelles, within cells. Future work in this area needs to address how ubiquitin modification on these and other cellular substrates is regulated, what proteins undergo ubiquitination, and how p62 ultimately distinguishes these various ubiquitinated substrates.
Methods
Cell Lines and Reagents.
COS-7 and HeLa cells were obtained from American Type Culture Collection (ATCC). Rabbit anti-catalase (Calbiochem), mouse anti-p62 (BD Biosciences), rabbit anti-Atg12 (Cell Signaling), rabbit anti-GAPDH (Abcam), rabbit anti-PMP70 (Zymed) and Alexa Fluor 546 goat anti-rabbit and anti-mouse (Invitrogen) antibodies were used according to manufacturers' recommendations. Rabbit anti-RFP antibodies were kindly provided by Dr. Ramanujan Hegde [National Institute of Child Health and Human Develoment/National Institutes of Health, Bethesda, MD]. Leupeptin, chloroquine, and 3-MA were purchased from Sigma–Aldrich Ltd.
Plasmid Construction.
Detail of the plasmids used and their construction are described in SI Methods.
Cell Cultures, DNA Transfections, and siRNA.
COS-7 and HeLa cells were cultured in DMEM (Biosource International) in 10% (wt/vol) FBS (FBS) at 5% CO2 in a 37°C incubator. For imaging, cells were plated on 4-chambered Lab-Tek glass coverslips (Nalge Nunc Int.). Transfections of plasmids into COS-7 and HeLa cells were performed by using FuGENE 6 (Roche Molecular Biochemicals) according to the manufacturer's instructions.
The p62 siRNA and control siRNA (purchased from Santa Cruz Biotechnology) were transfected twice in a 24-h interval into HeLa cells by using Santa Cruz Biotechnology siRNA Transfection Reagent according to the manufacturer's instructions. Plasmids were cotransfected with siRNA during the second transfection. For Atg12 silencing experiments, the same protocol was repeated with Atg12 siRNA ON-TARGETplus SMART-pool (purchased from Dharmacon).
For catalase quantification, any potential variations in the number of peroxisomes during the cell cycle in HeLa cells was abrogated by synchronizing them using the double thymidine-block protocol (48). Specifically, cells were plated to 50% confluency in DMEM, 10% FBS containing 2 mM thymidine for 16 h. After, the cells were washed and replated in fresh media without thymidine for 12 h and cells were then treated again with 2 mM thymidine for an additional 16 h. Finally, the cells were washed to remove thymidine and then transfected with either p62 siRNA or control siRNA as mentioned above. To identify transfected cells, p62 and control siRNA were also (co)-transfected with pECFP-N1 (Clontech). Ten hours after the last transfection, the cells were then treated again with 2 mM thymidine for 16 h before being, fixed, and immunostained for endogenous p62 and catalase.
For immunofluorescence microscopy, a standard immunofluorescence staining protocol was used (49). For live-cell imaging experiments, cells were imaged in phenol-free Leibovitz's L-15 medium (Gibco). Cells were maintained at 37°C by using either a customized temperature-regulated air blower or an enclosed heated stage.
Microscopy and Image Analysis.
All fluorescence images were acquired with a Carl Zeiss LSM 510 laser-scanning confocal microscope and either a 63 × 1.4 NA Plan-Apochromat oil objective or 40 × 1.3 NA Plan-Neofluar oil objective ( Carl Zeiss MicroImaging, Inc.). All images are maximum intensity projection seven optical slices in z-dimension unless otherwise stated in the legend. Each optical slice is 1-μm thick with 0.5-μm overlap between the splices. All images were adjusted for brightness and contrast and compiled into figures by using Photoshop CS (Adobe).
ImageJ software (National Institutes of Health) was used to calculate the number of peroxisomes and to determine the total fluorescence intensity attributable to endogenous catalase within whole cells. Briefly, to calculate the total number of peroxisomes, images were converted into threshold images that were then analyzed with the ImageJ “analyze particle” macro (26). To calculate total fluorescence density from the endogenous catalase within a cell, z-sections of the whole cell was complied into a single image by summing all of the images in the z plane. To normalize the signal, the background of each summed image was then subjected to threshold and the total fluorescence intensity of the fluorescent signal attributable to immunostained endogenous catalase was quantified by summing the fluorescent signal above the background set by the threshold.
Immunoblotting.
Cell lysate for immunoblotting were prepared by lysing the cells with 1% (wt/vol) sodium dodecylsulphate (SDS)/0.1 M Tris·HCl (pH 8.5) buffer and quickly heating it to 100°C to denature all proteins. After shearing the nucleic acids by repeated vortexing the total protein concentration of the lysate sample was determined by using standard BCA protein assay kit (Thermo Scientific). Each lysate were analyzed by standard SDS/PAGE followed by transfer to a nitrocellulose membrane. Membranes were probed with the appropriate primary and secondary antibodies as indicated in the legends. Immunoreactive bands were then visualized by using an enhanced chemiluminescent detection system (Thermo Scientific). A duplicate immunoblot was probed for endogenous GAPDH serving as a protein loading control.
Supplementary Material
Acknowledgments.
We thank Dr. Satinder Gidda (University of Guelph) for assistance with the construction of PEX3-GFP-UBko and UBko-RFP. This work was supported by grants from the National Institutes of Health (to J.L.-S.) and the Natural Sciences and Engineering Research Council of Canada (to R.T.M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810611105/DCSupplemental.
References
- 1.Mizushima N. Autophagy: Process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 2.Liang XH, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 3.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 4.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Platta HW, Erdmann R. Peroxisomal dynamics. Trends Cell Biol. 2007;17:474–484. doi: 10.1016/j.tcb.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakai Y, Oku M, van der Klei IJ, Kiel JA. Pexophagy: Autophagic degradation of peroxisomes. Biochim Biophys Acta. 2006;1763:1767–1775. doi: 10.1016/j.bbamcr.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 8.Mijaljica D, Prescott M, Klionsky DJ, Devenish RJ. Autophagy and vacuole homeostasis: A case for self-degradation? Autophagy. 2007;3:417–421. doi: 10.4161/auto.4441. [DOI] [PubMed] [Google Scholar]
- 9.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 10.Deretic V. Autophagy as an immune defense mechanism. Curr Opin Immunol. 2006;18:375–382. doi: 10.1016/j.coi.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 11.Schmid D, Dengjel J, Schoor O, Stevanovic S, Munz C. Autophagy in innate and adaptive immunity against intracellular pathogens. J Mol Med. 2006;84:194–202. doi: 10.1007/s00109-005-0014-4. [DOI] [PubMed] [Google Scholar]
- 12.Kaniuk NA, Kiraly M, Bates H, Vranic M, Volchuk A, Brumell JH. Ubiquitinated-protein aggregates form in pancreatic beta-cells during diabetes-induced oxidative stress and are regulated by autophagy. Diabetes. 2007;56:930–939. doi: 10.2337/db06-1160. [DOI] [PubMed] [Google Scholar]
- 13.Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 14.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 15.Verkhusha VV, et al. High stability of Discosoma DsRed as compared to Aequorea EGFP. Biochemistry. 2003;42:7879–7884. doi: 10.1021/bi034555t. [DOI] [PubMed] [Google Scholar]
- 16.Butt TR, Khan MI, Marsh J, Ecker DJ, Crooke ST. Ubiquitin-metallothionein fusion protein expression in yeast. A genetic approach for analysis of ubiquitin functions. J Biol Chem. 1988;263:16364–16371. [PubMed] [Google Scholar]
- 17.Johnson ES, Bartel B, Seufert W, Varshavsky A. Ubiquitin as a degradation signal. EMBO J. 1992;11:497–505. doi: 10.1002/j.1460-2075.1992.tb05080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat Biotechnol. 2000;18:538–543. doi: 10.1038/75406. [DOI] [PubMed] [Google Scholar]
- 19.Kominami E, Hashida S, Khairallah EA, Katunuma N. Sequestration of cytoplasmic enzymes in an autophagic vacuole-lysosomal system induced by injection of leupeptin. J Biol Chem. 1983;258:6093–6100. [PubMed] [Google Scholar]
- 20.Seglen PO, Gordon PB. 3-Methyladenine: Specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci USA. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lorenz H, Hailey DW, Wunder C, Lippincott-Schwartz J. The fluorescence protease protection (FPP) assay to determine protein localization and membrane topology. Nat Protoc. 2006;1:276–279. doi: 10.1038/nprot.2006.42. [DOI] [PubMed] [Google Scholar]
- 22.Johnson ES, Ma PC, Ota IM, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- 23.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 24.Fisher RD, et al. Structure and ubiquitin binding of the ubiquitin-interacting motif. J Biol Chem. 2003;278:28976–28984. doi: 10.1074/jbc.M302596200. [DOI] [PubMed] [Google Scholar]
- 25.Seibenhener ML, Geetha T, Wooten MW. Sequestosome 1/p62–more than just a scaffold. FEBS Lett. 2007;581:175–179. doi: 10.1016/j.febslet.2006.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim PK, Mullen RT, Schumann U, Lippincott-Schwartz J. The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J Cell Biol. 2006;173:521–532. doi: 10.1083/jcb.200601036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schrader M, Fahimi HD. Growth and division of peroxisomes. Int Rev Cytol. 2006;255:237–290. doi: 10.1016/S0074-7696(06)55005-3. [DOI] [PubMed] [Google Scholar]
- 28.Honsho M, Fujiki Y. Topogenesis of peroxisomal membrane protein requires a short, positively charged intervening-loop sequence and flanking hydrophobic segments. study using human membrane protein PMP34. J Biol Chem. 2001;276:9375–9382. doi: 10.1074/jbc.M003304200. [DOI] [PubMed] [Google Scholar]
- 29.Jones JM, Morrell JC, Gould SJ. Multiple distinct targeting signals in integral peroxisomal membrane proteins. J Cell Biol. 2001;153:1141–1150. doi: 10.1083/jcb.153.6.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imanaka T, Shiina Y, Takano T, Hashimoto T, Osumi T. Insertion of the 70-kDa peroxisomal membrane protein into peroxisomal membranes in vivo and in vitro. J Biol Chem. 1996;271:3706–3713. doi: 10.1074/jbc.271.7.3706. [DOI] [PubMed] [Google Scholar]
- 31.Gould SJ, Keller GA, Hosken N, Wilkinson J, Subramani S. A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol. 1989;108:1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soukupova M, Sprenger C, Gorgas K, Kunau WH, Dodt G. Identification and characterization of the human peroxin PEX3 Eur. J Cell Biol. 1999;78:357–374. doi: 10.1016/S0171-9335(99)80078-8. [DOI] [PubMed] [Google Scholar]
- 33.Kim J, Klionsky DJ. Autophagy, cytoplasm-to-vacuole targeting pathway, and pexophagy in yeast and mammalian cells. Annu Rev Biochem. 2000;69:303–342. doi: 10.1146/annurev.biochem.69.1.303. [DOI] [PubMed] [Google Scholar]
- 34.Yan M, Rayapuram N, Subramani S. The control of peroxisome number and size during division and proliferation. Curr Opin Cell Biol. 2005;17:376–383. doi: 10.1016/j.ceb.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 35.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 36.Wu Y, Termine DJ, Swulius MT, Moremen KW, Sifers RN. Human endoplasmic reticulum mannosidase I is subject to regulated proteolysis. J Biol Chem. 2007;282:4841–4849. doi: 10.1074/jbc.M607156200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizushima N, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bjorkoy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paine MG, Babu JR, Seibenhener ML, Wooten MW. Evidence for p62 aggregate formation: Role in cell survival. FEBS Lett. 2005;579:5029–5034. doi: 10.1016/j.febslet.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 40.van der Klei IJ, Hilbrands RE, Kiel JA, Rasmussen SW, Cregg JM, Veenhuis M. The ubiquitin-conjugating enzyme Pex4p of Hansenula polymorpha is required for efficient functioning of the PTS1 import machinery. EMBO J. 1998;17:3608–3618. doi: 10.1093/emboj/17.13.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Platta HW, Debelyy MO, El Magraoui F, Erdmann R. The AAA peroxins Pex1p and Pex6p function as dislocases for the ubiquitinated peroxisomal import receptor Pex5p. Biochem Soc Trans. 2008;36:99–104. doi: 10.1042/BST0360099. [DOI] [PubMed] [Google Scholar]
- 42.Bellu AR, Salomons FA, Kiel JA, Veenhuis M, Van Der Klei IJ. Removal of Pex3p is an important initial stage in selective peroxisome degradation in Hansenula polymorpha. J Biol Chem. 2002;277:42875–42880. doi: 10.1074/jbc.M205437200. [DOI] [PubMed] [Google Scholar]
- 43.Komori M, et al. The Hansenula polymorpha PEX14 gene encodes a novel peroxisomal membrane protein essential for peroxisome biogenesis. EMBO J. 1997;16:44–53. doi: 10.1093/emboj/16.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bellu AR, Komori M, van der Klei IJ, Kiel JA, Veenhuis M. Peroxisome biogenesis and selective degradation converge at Pex14p. J Biol Chem. 2001;276:44570–44574. doi: 10.1074/jbc.M107599200. [DOI] [PubMed] [Google Scholar]
- 45.Farre JC, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell. 2008;14:365–376. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yorimitsu T, Nair U, Yang Z, Klionsky DJ. Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006;281:30299–30304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sutovsky P, et al. Ubiquitin tag for sperm mitochondria. Nature. 1999;402:371–372. doi: 10.1038/46466. [DOI] [PubMed] [Google Scholar]
- 48.Harper JV. Synchronization of cell populations in G1/S and G2/M phases of the cell cycle. Methods Mol Biol. 2005;296:157–166. doi: 10.1385/1-59259-857-9:157. [DOI] [PubMed] [Google Scholar]
- 49.Donaldson J. In: Current Protocols in Immunology. Coligan J, editor. New York: John Wiley and Sons, Inc.; 2003. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.