Abstract
Knowledge of the evolutionary history of plants that are ecologically dominant in modern ecosystems is critical to understanding the historical development of those ecosystems. Metrosideros is a plant genus found in many ecological and altitudinal zones throughout the Pacific. In the Hawaiian Islands, Metrosideros polymorpha is an ecologically dominant species and is also highly polymorphic in both growth form and ecology. Using 10 non-coding chloroplast regions, we investigated haplotype diversity in the five currently recognized Hawaiian Metrosideros species and an established out-group, Metrosideros collina, from French Polynesia. Multiple haplotype groups were found, but these did not match morphological delimitations. Alternative morphologies sharing the same haplotype, as well as similar morphologies occurring within several distinct island clades, could be the result of developmental plasticity, parallel evolution or chloroplast capture. The geographical structure of the data is consistent with a pattern of age progressive island colonizations and suggests de novo intra-island diversification. If single colonization events resulted in a similar array of morphologies on each island, this would represent parallel radiations within a single, highly polymorphic species. However, we were unable to resolve whether the pattern is instead explained by ancient introgression and incomplete lineage sorting resulting in repeated chloroplast capture. Using several calibration methods, we estimate the colonization of the Hawaiian Islands to be potentially as old as 3.9 (−6.3) Myr with an ancestral position for Kaua'i in the colonization and evolution of Metrosideros in the Hawaiian Islands. This would represent a more ancient arrival of Metrosideros to this region than previous studies have suggested.
Keywords: chloroplast, Hawaiian Islands, Metrosideros, parallel evolution, plasticity, polymorphism
1. Introduction
Hawaiian Islands are renowned for their many endemic lineages and adaptive radiations of plants and animals. The islands themselves are formed of a series of volcanoes that decrease in age from northwest to southeast. Molecular phylogenies of some of the more spectacular adaptive radiations have revealed the history of diversification of these lineages in the archipelago, often with progressive colonization of new islands as they emerged from the sea (Wagner & Funk 1995; e.g. in the amakihi complex of Hawaiian honeycreepers (Fleischer et al. 1998), the silversword complex (Baldwin 1997), and the plant genus Schiedea (Wagner et al. 2005)). While adaptive radiations have attracted researchers' attention, until now no definitive phylogeographical studies have been done on ecologically dominant plant species, or species groups that are widespread throughout native habitats in the islands but have not diversified to produce large complexes of endemic species. Knowledge of whether these species are recent dispersers or are ancient lineages in the archipelago, or whether there have been single or multiple colonization events (e.g. Howarth et al. 2003; Harbaugh & Baldwin 2007), is crucial to understanding the evolutionary history of the Hawaiian terrestrial ecosystem.
In particular, we often observe specialized interactions and apparent co-diversification of animals with the plants they depend on. A classic example is Metrosideros (Myrtaceae), a geographically widespread genus of plants that, in the Hawaiian Islands, is a major component of many plant communities, from tall forest to bog. Many of the archipelago's endemic nectarivorous birds rely heavily on Metrosideros as a nectar source, including the now-extinct Hawaiian honeyeaters (Aves: Passeriformes incertae sedis) and members of the adaptive radiation of Hawaiian honeycreepers (Aves: Drepanidini). Among insects, several endemic groups have members that are highly specialized on Metrosideros (e.g. cerambycid beetles, bud and leaf miners, leafhoppers, planthoppers and psyllids; Gruner 2004a,b), and in at least one case the insects appear to have undergone diversification on this host plant over several million years. The arrival of Metrosideros in the archipelago and its dispersal to new islands may have been a stimulus to evolutionary diversification in those lineages, in which case evolutionary events in the bird and insect lineages will correlate with the dispersal history of Metrosideros. Alternatively, there may have been independent evolution of plant and animal lineages, with the development of specialized interactions occurring only post diversification (e.g. Percy et al. 2004). Unravelling these histories would enhance understanding of the processes that underlie adaptive radiation and the assembly of highly endemic biological communities on islands.
Hawaiian Metrosideros (commonly called ohi'a in the islands) is of further interest to systematists and developmental biologists owing to its extreme phenotypic variability; species limits for Hawaiian Metrosideros have not been confidently resolved, and some taxonomies recognize only one highly polymorphic species in the archipelago, while a more recent treatment recognizes five (Dawson & Stemmermann 1990). One of the five species is particularly notable for its ecological dominance, i.e. Metrosideros polymorpha (ohi'a lehua). This taxon is the dominant woody species in many different ecological zones (e.g. as a tall emergent forest tree, a dwarf shrub in bogs, and the primary woody pioneer of new lava flows) and occurs over a wide altitudinal range from sea level to tree line at 2500 m (Mueller-Dombois 1987; Mueller-Dombois 1992; Cordell et al. 2000). The patterns of morphological diversity in M. polymorpha are complex, both within and among the Hawaiian Islands (e.g. leaf shape, pubescence and growth habit; Rock 1917; Dawson & Stemmermann 1990). Over several decades, different methods have been applied to the analysis of this complexity, including morphometric, ecological and physiological approaches (Corn 1979; Stemmermann 1983; Stemmermann & Ihsle 1993; Cordell et al. 1998; Cordell et al. 2000; Melcher et al. 2000), as well as allozyme and RAPD marker analyses (Aradhya et al. 1991, 1993; James et al. 2004). The results in all cases suggest a mixture of genetically controlled and plastic traits. However, there remains considerable dispute about the role of plasticity versus genetic determination in the phenotypic traits that are expressed.
Elsewhere in the Pacific, the genus is widely distributed on islands from New Caledonia and New Zealand in the west, to the French Polynesian archipelagos and the Hawaiian Islands in the central and eastern Pacific basin. The family Myrtaceae is well represented in continental regions adjacent to the Pacific, and the family's earliest diversification was in the Australian dry-fruited clades (e.g. Melaleuca and Callistemon; Sytsma et al. 2004). The genus Metrosideros (also dry fruited) has centres of diversity in tropical New Caledonia and temperate New Zealand with variable growth forms from montane shrubs to forest trees. A previous molecular study (Wright et al. 2001) used nuclear ITS and ETS regions to assess phylogenetic and biogeographical patterns in Pacific Metrosideros. Although this study was able to determine some clear patterns, in particular a close association between Hawaiian, Marquesan and New Zealand taxa, there was insufficient variation to distinguish the five Hawaiian species or to clearly differentiate Marquesan and Hawaiian taxa. Wright et al. estimated the time of colonization of the Hawaiian Islands by Metrosideros at less than 1 Myr ago, a relatively recent age in relation to the more than five million year history of the main Hawaiian Islands.
To better resolve the biogeographical history and systematics of Hawaiian Metrosideros, we increased the potential analytical power by sequencing 10 highly variable non-coding chloroplast regions. We also greatly increased the extent of sampling (both within taxa and within island populations) across all of the main Hawaiian Islands, and included samples from several other Polynesian island groups as out-group taxa. As well as being the first Hawaiian Metrosideros study to generate a large molecular dataset, ours is the first molecular study to sample extensively from more than one island, which has enabled us to better estimate the age of colonization of the Hawaiian Islands by Metrosideros, and to compare observed differences in haplotype and phenotypic diversity among islands with the ages of the islands (Fleischer et al. 1998).
2. Material and methods
(a) Taxonomic sampling and selection of chloroplast regions
The genus Metrosideros consists of two subgenera (namely, subg. Metrosideros and subg. Mearnsia) with the centre of diversity for both in the western Pacific (Dawson 1976). The central and eastern Pacific species are classified in the subg. Metrosideros that includes all the taxa studied here. The Hawaiian taxa have variously been treated as members of a single widespread species, Metrosideros collina, with a range from the south Pacific to the Hawaiian Islands and French Polynesia (Rock 1917), a single endemic Hawaiian species, M. polymorpha (Skottsberg 1936) or more recently as a complex of five endemic Hawaiian species, with M. polymorpha subdivided into eight varieties and Metrosideros waialeale into two varieties (Dawson & Stemmermann 1990).
We sampled 97 individuals from five Hawaiian Islands: Kaua'i (17 individuals); O'ahu (30 individuals); Moloka'i (13 individuals); Maui (12 individuals); and Hawai'i (25 individuals; see electronic supplementary material, appendix A). Multiple individuals were sampled from the five Hawaiian species as follows: Metrosideros macropus (3); M. polymorpha (80); Metrosideros rugosa (6); Metrosideros tremuloides (4); and M. waialealae (4). This sampling includes seven of the eight varieties of M. polymorpha and the two varieties of M. waialealae. Our aim was to cover the phenotypic range within islands and particularly at locations where multiple phenotypes occur sympatrically. We also covered a broad range of ecological sites, sampling at different elevations from 275 to 2060 m within the Hawaiian Islands, and from 150 to 600 m in the Austral, Society and Marquesas Islands. On the Hawaiian island of Maui, a roughly equal number of specimens came from eastern and western parts of the island, ranging in elevation from 875 to 2060 m. Twelve morphological characters, including those frequently used for taxonomic identification and classification, were scored for 75 Hawaiian and Marquesan individuals (see electronic supplementary material, appendix B).
We sampled 10 individuals representing out-group taxa as follows: M. collina from three French Polynesian archipelagos (Austral, Society and Marquesan islands); Metrosideros excelsa from New Zealand; and Metrosideros nervulosa from Lord Howe Island. These were selected for their close relationship to Hawaiian taxa based on the study of Wright et al. (2001). Sequences from GenBank of Eucalyptus globulus (Myrtaceae) were used as a further out-group for rooting the trees. Collection details and GenBank numbers are given in electronic supplementary material, appendices A, B and C.
We sequenced 10 of the non-coding chloroplast (cpDNA) regions identified by Shaw et al. (2005) as having high numbers of parsimony informative characters useful for resolving relationships at low taxonomic levels, as follows: the rpS16 intron; the trnS–trnG spacer; the trnG intron; the three spacers in the trnC–ycf6–psbM–trnD region; the three spacers in the trnD–trnE–trnY–trnT region; and the rpL16 intron (see electronic supplementary material, appendix C). Primer combinations from Shaw et al. (2005) are given in electronic supplementary material, appendix C.
(b) DNA Extraction, amplification and sequencing
DNA was extracted from silica-dried leaf material using a QIAGEN DNeasy Plant Mini Kit. The PCR conditions were: 5 min at 80°C, 33 cycles of 55 s at 95°C, 1 min at 52°C, 3 min at 72°C, with an extension of 5 min at 72°C. In a few cases, amplification was improved by using the region specific PCR conditions in Shaw et al. (2005). Amplified fragments were sequenced in both directions on an ABI 3100 automated sequencer. The sequences were edited manually using Sequencher v. 3.1.1 and Se-Al 2.0a11 (Rambaut 1996). Sequence fragments for all cpDNA regions were highly homologous and easily aligned by eye.
(c) Phylogenetic and comparative analyses
We performed several phylogenetic analyses such as maximum parsimony (MP), maximum likelihood (ML) and Bayesian analysis (BA). The MP search was performed using PAUP* v. 4.0b10 (Swofford 2002), the ML with GARLI 0.941 (Zwickl 2006), and the BA with MrBayes v. 3.1.2 (Huelsenbeck & Ronquist 2001). For the MP search, we employed a parsimony ratchet approach (Nixon 1999); 10 batches of parsimony ratchet with 200 interactions each were run in PAUP* using PAUPRat 1 (Sikes & Lewis 2001). The ratchet approach is a more efficient tree search strategy for large datasets (Sikes & Lewis 2001). For a comparison with the ML analysis using GARLI, we also ran a likelihood ratchet analysis using PAUPRat with a modified input file following Vos (2003). The BA searches were run with four chains for two million generations, saving every 100 generations. After excluding 25% of the trees as burn-in, the majority-rule consensus tree was used to determine posterior probability support values. Appropriate models for the ML and BA analyses were determined with Modeltest v. 3.7 (Posada & Crandall 1998). Support values, such as bootstrap, jackknife and posterior probability, were generated in PAUP* (1000 replicates) and MrBayes. Non-parametric bootstrap analysis in a likelihood framework was performed with GARLI (100 replicates). To test for significant deviations from a molecular clock hypothesis, we used the Χ2 likelihood ratio test with and without the molecular clock enforced. We also tested for local rate variation across lineages using r8s 1.71 (Sanderson 2002).
For the comparison of genetic, geographical and morphological distances, we obtained for each individual genetic distance (uncorrected p value) from PAUP*; geographical distance determined by converting decimal degree units to km using the R-Package v. 3.02 (Casgrain & Legendre 2001) GeoDistances application, and morphological distance estimated using the R-Package SIMIL function to create a similarity matrix from a matrix of 12 binary and quantitative characters (see electronic supplementary material, appendix B) implementing the Gower (symmetrical) coefficient (option S15). The similarity matrix was then converted to a distance matrix using the CONVERT function. The 12 morphological characters were also plotted using a principal coordinates (PCO) analysis in order to establish that these 12 characters recovered recognized phenotypic groups.
We performed Mantel tests (999 iterations) with PopTools v. 2.6.9 (Hood 2005) to assess the strength of correlation between genetic, geographical and morphometric distances, for the whole dataset, and regression analysis for each island independently to see if the pattern on each island reflected that of other islands and the data as a whole.
(d) Population genetic analyses
The program DnaSP v. 4.10.7 (Rozas et al. 2003) was used to calculate genetic diversity measures for each island. Because some islands comprised complex population structures, we took ancestral polymorphism into account by estimating net divergence: the pairwise divergence between populations minus the average divergence within each population (Nei & Li 1979). This allows estimation of divergence between populations that have not yet achieved reciprocal monophyly. In order to capture the deeper levels of divergence, the Nei & Li method was used with ML distances using parameters estimated from the data using PAUP*. For this dataset, branch length estimates using ML and ML distances were virtually identical. The Nei & Li net distances were imported into PAUP* as a user-defined distance matrix to infer minimum-evolution population relationships with the estimated branch lengths.
(e) Rate calibration along proposed dispersal route
We used the ages of volcanic islands to date nodes of the phylogeny within the Hawaiian Islands, and along the most likely dispersal route from New Zealand (Clague 1996; Craig et al. 2001; Price & Clague 2002; Bonneville et al. 2006). The ages of islands along the most likely dispersal route allow an independent test of a calibration based only on ages within the Hawaiian Islands. Our calibrations were calculated by two different approaches: phylogenetic rate smoothing (Sanderson 2002) that relaxes the assumption of the molecular clock and the Nei & Li distance method (Nei & Li 1979). We ran the rate smoothing program r8s v. 1.71 (Sanderson 2002) using two phylogenies estimated by ML as described previously. The first included all individuals sequenced in the study and the second included one representative from each population in the phylogeny. The optimal rate smoothing parameter for each phylogeny was chosen by a cross-validation procedure implemented in r8s using penalized likelihood with the TN (truncated Newton method) search algorithm (Sanderson 2002).
3. Results
(a) Phylogenetic and comparative analyses
The dataset is 7892 bp in length and includes 213 variable characters, of which 134 are phylogenetically informative, and 49 code for the indels and repeats (see electronic supplementary material, appendix C). PsbM–trnD is the only region that provides no phylogenetically informative characters within the Hawaiian Islands (see electronic supplementary material, appendix C). This region also has the largest number of indels (25% of the variation), including a 156 bp indel that distinguishes E. globulus (Australia), M. excelsa (New Zealand) and M. nervulosa (Lord Howe Island) from the eastern Pacific Metrosideros taxa (Hawai'i and French Polynesia).
The level of homoplasy in our data is low. The consistency indices (CI) indicate that indels are highly conserved (CI: 1.0), and repeat characters are more homoplasious (CI: 0.68) than the single nucleotide polymorphism characters (CI: 0.9). The MP tree that we compared with other analyses is the majority rule consensus of 1667 shortest trees obtained from 10 batches of parsimony ratchet (tree length: 273; CI: 0.857; RI: 0.928). The two ML analyses, using GARLI and the likelihood ratchet with PAUPRat, generated identical trees (log-likelihood: −12604.36), are shown in figure 1. The model selected with Modeltest based on both Akaike information criterion and Bayesian information criterion was F81 (Felsenstein 1981). This model was selected for each of the chloroplast regions independently and for the combined chloroplast dataset, but the model for the combined data also included a proportion of invariable sites and a gamma-shaped distribution of rates across sites (F81+I+G).
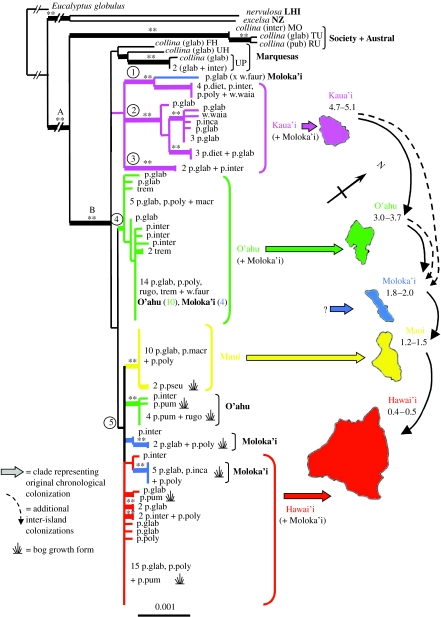
Figure 1.
Maximum-likelihood tree showing the five main haplotype groups (circled numbers). Interrupted branches at the base of the tree are not to scale. Thick branches indicate ML bootstrap greater than 50; asterisk indicates greater than 75 and double asterisk indicates greater than 85 in at least one of ML or MP analyses (table 1). Hawaiian and French Polynesian taxa fall into two main phenotypic groups: glabrous and pubescent, which can be differentiated using a multivariate analysis of the 12 morphological characters (see electronic supplementary material, appendix B). Taxa shown here can be partitioned as follows: glabrous group—M. macropus (macr); M. polymorpha var. glaberrima (p.glab); M. polymorpha var. macrophylla (p.macr); M. tremuloides (trem); M. waialealae var. fauriei (w.faur); M. waialealae var. waialealae (w.waia); pubescent group—M. polymorpha var. dieteri (p.diet); M. polymorpha var. incana (p.inca); M. polymorpha var. polymorpha (p.poly); M. polymorpha var. pseudorugosa (p.pseu); M. polymorpha var. pumilla (p.pum); M. rugosa (rugo); p.inter designates a phenotype intermediate between glabrous and pubescent. Where a haplotype is shared by multiple individuals the number of individuals is given followed by the taxa/phenotypes; bog growth form is also indicated. LHI, Lord Howe Island and NZ, New Zealand. Society, Austral and Marquesas Islands are abbreviated as follows: MO, Moorea; TU, Tubuai; RU, Rurutu; FH, Fatu Hiva; UH, Ua Huka; UP, Ua Pou. Hawaiian Islands are shown with ages of emergence.
The MP tree topology was highly consistent with the ML tree (shown in figure 1). The MP analysis differed only in the absence of monophyly of the Marquesan samples. Support values for each analysis are shown in figure 1 and table 1.
Table 1.
Support values for the four main haplotype groups (H) that include multiple haplotypes, monophyly of the island of Kaua'i (i.e. H1, H2 and H3, figure 1), monophyly of the Hawaiian archipelago, and monophyly of a Hawaiian–Marquesan lineage of Metrosideros. (Presence of a clade but with less than 50% support is indicated by yes/−. The MP support values are determined from a jackknife analysis, the BA values are posterior probabilities, and the ML values are from a non-parametric bootstrap analysis.)
| dataset | analysis | H1 | H2 | H4 | H5 | monophyletic Kauai | monophyletic Hawaiian archipelago | monophyletic Hawaiian–Marquesan |
|---|---|---|---|---|---|---|---|---|
| all data, all taxa | MP | 63 | 89 | yes/− | yes/− | yes/− | — | 90 |
| BA | 1.00 | 1.00 | 1.00 | 0.97 | 0.95 | — | 1.00 | |
| ML | 59 | 97 | 56 | yes/− | yes/− | — | 65 | |
| without gap coding | MP | 88 | 75 | 54 | — | — | — | 100 |
| BA | 1.00 | 1.00 | 1.00 | 0.63 | 0.66 | — | 1.00 | |
| ML | 89 | 90 | 55 | — | — | — | yes/− | |
| without E. globulus M. excelsa M. nervulosa | MP | 56 | 86 | yes/− | yes/− | yes/− | yes/− | 100 |
| BA | 1.00 | 1.00 | 0.97 | 0.92 | 0.99 | 0.68 | 1.00 | |
| ML | 57 | 93 | 53 | — | — | yes/− | 100 |
The Χ2 likelihood ratio test with and without the molecular clock enforced indicated that a molecular clock could not be rejected (p=0.99), both with and without E. globulus. A test of local rate variation with r8s found a non-significant slower local clock (p=0.081) within the Marquesas–Hawaiian lineage versus the Austral–Society lineage.
A Mantel test of the molecular and geographical distances indicated a highly significant positive correlation (p=<0.001), and, by contrast, a significant negative correlation between molecular and morphological datasets (p=0.024). The multivariate PCO analysis of the 12 morphological characters clearly distinguished the split between two primary morphotypic groups, glabrous and pubescent, but within those groups taxa were not clearly differentiated.
(b) Geographical structure and haplotype diversity
We found 45 haplotypes, with 35 occurring in the Hawaiian Islands. In only one case is a haplotype found on more than one island (the most common haplotype on O'ahu is also found on Moloka'i; figure 1).
Correlations found in the data as a whole are also apparent within the Hawaiian Islands and to some extent on individual islands. There is a significant positive correlation between molecular and geographical distances for all islands combined (p=<0.001), and the same positive association is found within each island, though it is non-significant for O'ahu, Maui and Hawai'i. A negative association between molecular and morphological distance is found for all islands combined and within each island, but these relationships are non-significant except within Kaua'i and Moloka'i (p=<0.05). On each island there is a mixture of unique and shared haplotypes, with the most common haplotype being shared between 4 (Kaua'i) and 15 (Hawai'i) individuals. In all cases, these common haplotypes are shared by individuals with phenotypes that range across the morphological and taxonomic spectra, representing both glabrous and pubescent morphological groups (figure 1).
Measures of both nucleotide (Pi) and haplotype diversity (Hd) per island, regardless of population structure, are the greatest on Kaua'i, the oldest of the major islands, and decrease on progressively younger islands (see electronic supplementary material, appendix D; correlation of Pi and island age: within the Hawaiian Islands, p=0.02; including Marquesas, p=0.004). A significant correlation is not found between Hd and island age (p=0.072) owing to the complex population structure found on some islands. Moloka'i has a higher than expected haplotype diversity (Hd=0.78) for its age (1.8–2 Myr) due to the occurrence of multiple haplotypes that may have derived from both older and younger islands. By contrast, Maui, with a monophyletic, nearly homogeneous haplotype group, has a lower than expected haplotype diversity (Hd=0.3) for its age (1.2–1.5 Myr).
We identified five main haplotype groups, although groups 4 and 5 are only well supported in a BA (table 1). Three of the five main haplotype groups are found on the island of Kaua'i. Two of these are unique to Kaua'i, while a third (group 1 in figure 1) also includes a single individual from Moloka'i. Of the 13 haplotypes found on the island of O'ahu, 10 are in group 4 (figure 1). This group is exclusive to O'ahu, with the exception of the most common O'ahu haplotype that is shared by four individuals from Moloka'i. Three additional haplotypes found on O'ahu, representing a clade of six individuals from the Ka'ala bog in the Wai'anae Mountains (approx. 1220 m elevation), are found in group 5, which mainly contains individuals from Maui and Hawai'i. Two small clades of taxa which are exclusively found in or near bogs on Moloka'i (approx. 1250 m elevation) are also found in group 5. The two bogs on Moloka'i are less than 1 km apart and in both cases a single haplotype found within each bog is shared by both pubescent and glabrous phenotypes. Owing to a basal polytomy in haplotype group 5, the origin of these small bog-associated clades remains ambiguous.
Two alternative biogeographical scenarios for haplotype group 5 are a recent sequential colonization (O'ahu, Moloka'i, Maui and Hawai'i) or back colonizations from Hawai'i to older islands.
All individuals from the youngest islands, Maui and Hawai'i, are in the poorly supported haplotype group 5. However, the Hawai'i haplotypes may still represent a monophyletic island lineage, as seen for the Maui haplotypes. Only Moloka'i and O'ahu show a clear evidence of being colonized multiple times. Moloka'i may be unusual in the possible repeated colonizations of this island (as many as three to four colonizations) from two to three different source islands (Kaua'i, O'ahu and possibly Hawai'i). It is also the only island for which evidence of an original haplotype stock consistent with a chronological colonization of the islands is less clear (but see population analyses below). Although they may be absent from Moloka'i, a well-defined ‘original’ or ‘indigenous’ haplotype group, which the other islands seem to possess, a sequential colonization of Moloka'i from O'ahu is not ruled out because the poor resolution in haplotype group 5 precludes a definitive interpretation of the direction of colonizations in this group.
The observed haplotype diversity on Maui is low, with only two haplotypes found on the island: one that is nearly ubiquitous on both the eastern and western parts of the island and another in the bog growth forms of M. polymorpha var. pseudorugosa on West Maui.
(c) Population genetic analyses among the Hawaiian Islands
Given the sometimes complex pattern of distribution, populations were defined as follows: Kaua'i haplotype groups 2 and 3 were treated as distinct populations since they are found in the north and south of the island, respectively. Group 1 is geographically mixed from the north and south of Kaua'i, and also includes a single individual from Moloka'i; it was therefore excluded from the Nei & Li divergence estimates below (including it as a separate population does not alter results). On O'ahu, all individuals from haplotype group 4 were considered a single population, as were the small monophyletic clade of O'ahu individuals in group 5. All Maui individuals were considered a single population as they compose a monophyletic lineage. Moloka'i has the most complex history, being colonized by other haplotype groups: 1 (from Kaua'i), 4 (from O'ahu) and possibly 5. The Moloka'i individuals in group 5 fall into two small clades, but when these were considered as separate populations, the population tree showed them clustering together, so these clades were treated as a single population. Figure 2 illustrates how the distances between these populations correlate with island age.
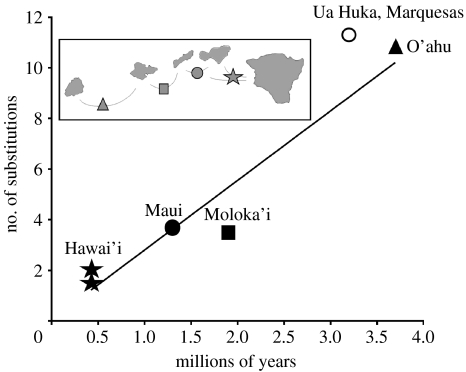
Figure 2.
Regression forced through zero for comparisons among populations on the Hawaiian Islands. The points are as follows: emergence of O'ahu 3.7 Myr ago plotted against the average distance between two Kaua'i populations and the populations on O'ahu and the younger islands; emergence of Moloka'i 1.9 Myr ago plotted against the average distance between the main O'ahu population (haplotype group 4) and populations on Moloka'i and the younger islands; emergence of Maui 1.3 Myr ago plotted against the divergence between the main Moloka'i population (in haplotype group 5) and the Maui population; emergence of Hawai'i 0.43 Myr ago plotted against the divergence between Hawai'i and the main Moloka'i and Maui populations, respectively. The slope is 2.7485 and the R2 is 0.92. The white circle is an estimate within the Marquesas Islands, based on the emergence of Ua Huka 3.23 Myr ago. Inset illustrates the node position in a sequential colonization scenario.
(d) Rate calibration within the Hawaiian Islands and along the most likely dispersal route
The ages of emergence of the Hawaiian Islands are significantly correlated (p=0.01) with the Nei & Li net divergences estimated between islands (figure 2).
The greater sampling and phylogenetic resolution in our chloroplast phylogeny is consistent with a dispersal route proposed by Wright et al. (2001) from the western Pacific (e.g. New Zealand) to the eastern Pacific (e.g. French Polynesia and Hawaiian Islands). Our data also confirm the close relationship between Metrosideros on the Marquesas and Hawaiian Islands, and identify Metrosideros on the Austral/Society Islands as the probable sister group to the Marquesas/Hawaiian clade. Central Pacific Metrosideros (e.g. from Fiji and Samoa) were shown to be distinct and an unlikely origin of eastern Pacific Metrosideros (Wright et al. 2001). The Austral Islands (maximum age of approx. 58 Myr) are a probable first stepping stone, being geographically located between New Zealand and the much younger Marquesas Islands (maximum age of approx. 6 Myr; Craig et al. 2001; Bonneville et al. 2006). The Austral–Cook geological region has a complex history of volcanism and seamount formation over a wide temporal range (Bonneville et al. 2006). Three periods of hot spot activity have been established for this region dating from 58 Myr ago to the present. Calibrating the ancestral node that corresponds to the deep split in the chloroplast phylogeny between Metrosideros in New Zealand, and the Polynesian clade that includes populations from the Austral/Society Islands (node A, figure 1), with the maximum age of the most recent of these hot spots (0–20 Myr) produces colonization dates of the Marquesas and Hawaiian Islands consistent with the ages of these islands. However, given the existence of even older islands in the Austral region, we also shifted the calibration of node A back to 48 Myr ago. The estimated ages of other nodes obtained from this analysis would require the colonization of now submerged islands in the Hawaiian chain by Metrosideros more than 13 Myr ago, as well as inferring an extremely slow rate of sequence evolution in the Metrosideros lineage.
With node A calibrated to the maximum age of the most recent of the Austral hot spots (approx. 19 Myr), the emergence of the Marquesas corresponds to the split between the Austral/Society clade and the Marquesas/Hawai'i clade (node B, figure 1). The phylogenetic rate smoothing approach gave similar results when applied to the phylogeny (i) with all sequences included, (ii) with one sequence from each population, and (iii) the tree based on Nei/Li distances between populations. When node A was fixed at 19 Myr ago, the predicted ages of node B were consistent with emergence of the Marquesas Islands, with point estimates of 4.9–6.1 Myr ago (95% CIs from 3.3 to 8.3 Myr ago). The colonization of the island of Kaua'i in the Hawaiian Islands was estimated to range between 2.4 and 3.9 Myr ago (95% CIs from 1.4 to 6.3 Myr ago; figure 2, table 2). When the calibration point is shifted to within the Hawaiian Islands, at the node representing the split between Kaua'i and O'ahu lineages (3.7 Myr ago), the ancestral node A dates to 21.9–22.9 Myr ago; colonization of the Marquesas to 6.2–6.5 Myr ago; O'ahu–Moloka'i to 2.1–2.2 Myr ago; and Moloka'i–Maui to 1.3–1.4 Myr ago. These similar age estimates obtained whether the calibration point is within or outside the Hawaiian Islands reflects the internal consistency of the data and lack of significant rate heterogeneity.
Table 2.
Estimated ages using the r8s program with 95% CIs. (The TN search algorithm was used under penalized likelihood with a smoothing parameter of 2.0 for all sequences and 1.0 for the other analyses.)
| method for inferring phylogeny branch lengths | all sequences ML | one sequence per population ML | Nei/Li distances based on ML distances |
|---|---|---|---|
| colonization of Austral Islands | 19 Myr ago (date fixed) | 19 Myr ago (date fixed) | 19 Myr ago (date fixed) |
| colonization of Marquesas Islands | 6.1 (4.6–8.2) | 4.9 (3.3–7.3) | 5.6 (3.7–8.3) |
| colonization of Hawaiian Islands | 3.9 (2.6–6.3) | 2.4 (1.4–4.7) | 2.6 (1.4–5.3) |
A number of assumptions are associated with a molecular calibration of island lineages (detailed in Fleischer et al. 1998). In particular, there is an assumption that islands are populated at the time or shortly after they become subaerial. In addition, maximum ages are necessarily used for calibration points because minimum ages are not available when island chains are still in the process of being formed. The use of maximum ages in our calibrations provides estimates of a maximally oldest lineage given the assumption above, but does not preclude the existence of younger lineages with faster rates of sequence evolution, resulting in colonization of all islands some period after their formation. However, under this scenario of a young/recent lineage, a pattern of progressive island colonization that is correlated with island age, as we see in the Metrosideros data, is not expected because all emerged islands would be available for colonization from the start. An independent estimate of the rate of sequence evolution for Metrosideros would help assess the plausibility of a younger Hawaiian Metrosideros lineage versus, as we are proposing, an early island colonizer that played an important role in shaping the ecology of the islands soon after their emergence.
(e) Taxonomic implications
Our analysis confirms that the Marquesan members of M. collina are closer to the Hawaiian taxa than to other M. collina from the Society and Austral Islands (Wright et al. 2001). The Hawaiian taxa were at one time included within M. collina as endemic Hawaiian subspecies (Rock 1917). However, it would be more appropriate to place the Hawaiian plus Marquesan taxa in a species separate from M. collina (i.e. M. polymorpha). The chloroplast data do not support the recognition of five distinct species in the Hawaiian Islands.
4. Discussion
(a) Ancient arrival and progressive island colonization
Wright et al. (2001) proposed that a recent Pleistocene dispersal out of New Zealand accounted for a Hawaiian/Marquesan colonization, dating an arrival of Metrosideros in the Hawaiian Islands to less than 1 Myr ago. The chloroplast data suggest an older presence of Metrosideros in the Hawaiian Islands, which we now estimate to be as old as 3.9 (−6.3 with 95% confidence) Myr. Supporting a more ancient history of Metrosideros in Hawai'i are the correlations of molecular distances with island age, the stepping-stone pattern of colonization from older to younger islands, and, in particular, the higher haplotype diversity on the oldest emergent island of Kaua'i compared with the younger islands. An accelerated rate of substitution could explain the higher haplotype diversity on Kaua'i, but this is unlikely given that a molecular clock cannot be rejected and the best-fit maximum-likelihood models indicate a lack of rate heterogeneity in the chloroplast data. Thus, Kaua'i may have relatively high haplotype diversity because it was colonized comparatively early with a temporal gap before the remainder of the Hawaiian Islands were colonized.
Our analysis of molecular rates is consistent with colonization of the Marquesas soon after these islands emerged (approx. 6 Myr ago), followed by dispersal from the Marquesas to the Hawaiian Islands shortly thereafter. However, some ambiguity remains because the molecular data provide insufficient resolution to clearly distinguish a monophyletic Hawaiian group from the closest sister group in the Marquesas or to establish that Kaua'i was colonized from the Marquesas and not vice versa. In addition, shifting the calibration age of node A to some of the oldest Austral Island ages (Bonneville et al. 2006) would require colonization of now submerged islands in the Hawaiian chain by Metrosideros more than 13 Myr ago. More plausibly, the location of the Marquesas between the Hawaiian Islands and the probable source populations for the Hawaiian–Marquesan clade in central and southern French Polynesia makes a northward direction of colonization probable.
The presence of Metrosideros throughout the Pacific region has long been considered the result of highly effective dispersal via small wind dispersed seeds that can be transported by air currents over long distances and endure periods of immersion in salt water (Corn 1972; Carlquist 1974; Wright et al. 2000, 2001). Such efficient dispersal should make the repeated colonization of islands more likely. However, our data show a number of monophyletic island lineages, which does not support a pattern of numerous repeated colonizations. Although there is evidence that at least one Hawaiian island (Moloka'i) was colonized more than once, the general pattern of island-based clades is consistent with single island colonization events followed by in situ diversification. This geographical structure together with the correlation between substitution rate and island age suggests that there has been successive, chronological colonization within the Hawaiian Islands followed by limited additional inter-island colonizations (e.g. the possible recolonization of bog habitats).
(b) Surprising results from central islands and bogs
Within the Hawaiian Islands, there are two anomalies to a proposed pattern of sequential colonization and monophyletic island lineages. The first is the multiple origins of haplotypes on Moloka'i. There is a higher than expected extra-island (i.e. polyphyletic) colonization rate for Moloka'i, which could be explained by this island's central position in the Hawaiian island chain, although this fails to explain the distinct contrast with Maui that is also centrally positioned and is the closest neighbouring island to Moloka'i. Maui has a monophyletic and almost invariant haplotype profile. Given that the two islands have a Pleistocene history of land connections (Price 2004; Price & Elliott-Fisk 2004), it is surprising that we found less haplotype diversity on Maui and that this island does not appear to be the source of any of the haplotypes found on Moloka'i. More extensive sampling would be required to test whether this anomaly really indicates a lack of shared haplotypes, or haplotype groups, which is counter to expectations given the lengthy historical geographical linkage between these two islands.
The other anomaly is the origins of bog growth forms. The bog growth forms on O'ahu and Moloka'i appear to have extra-island origins, while those of Hawai'i and Maui appear to have intra-island origins. In addition, all bog haplotypes (those from Hawai'i, Maui, Moloka'i and O'ahu) have a sister relationship with a haplotype from a non-bog growth form. This pattern suggests that (i) there are multiple origins for the bog growth form and (ii) bogs may be more prone than other habitats to repeated colonization by Metrosideros. Both the scenarios may be related to the limited patchy distribution of Hawaiian bogs and their brief geological lifespan (Mueller-Dombois 2006). Studies of pollen and radiocarbon in bog sediments reveal that modern Hawaiian bogs date primarily to the Holocene, and that the relative abundance of Metrosideros within and surrounding the bogs shows peaks and valleys that may correlate with wetter and drier climate phases during the latest Pleistocene through Holocene (Selling 1948; Burney et al. 1995; Hotchkiss & Juvik 1999; Burney 2002). Most bogs apparently did not exist at their current locations at the Last Glacial Maximum, but other bogs may have existed at sites where environmental conditions were suitable at the time.
Newly formed bog habitats could have provided opportunities for multiple shifts to the bog habitat in Metrosideros. In a genetic study of E. globulus, this usually tall forest tree has recurrently given rise to a dwarf ecotype found parapatrically on coastal cliffs (Foster et al. 2007). However, if Metrosideros bog growth forms are repeatedly derived from non-bog forms, an indigenous origin of bog plants from surrounding areas on the same island would seem more likely. But our data suggest that bogs may equally likely be colonized from other islands. This raises the question: if Metrosideros propagules are as likely to have origins from other islands as from within an island, what mechanism(s) is responsible for maintaining the integrity of the island lineages that we see in the data? The frequently observed pioneer habit of Metrosideros (such as the colonization of new lava flows; Drake 1992) suggests that habitats other than bogs would also be available for extra-island colonization, but we find no evidence that haplotypes representing other pioneer growth forms are derived from off-island. It is therefore possible that where Metrosideros is already established, in situ competition effectively limits the establishment of propagules from other islands.
(c) Developmental plasticity versus genetic determination
The reconciling of phenotypic and genotypic patterns requires more data to support either of two primary hypotheses: parallel evolution and chloroplast capture. As our data are from a maternally inherited organelle genome (the chloroplast), an argument can be made that the lack of correlation between morphotypes and genotypes is due to hybridization and chloroplast capture (resulting in reticulate inheritance of the chloroplast genome). Several studies have found evidence for extensive chloroplast capture (Steane et al. 1998; Palme et al. 2004; Bänfer et al. 2006; Jakob & Blattner 2006) that in some cases explains the apparent non-monophyly of morphotypes. A study of New Zealand Metrosideros found that chloroplast capture within climatic refugia could entirely explain apparent non-monophyly of those species (Gardner et al. 2004). Under this scenario, evidence for multiple colonizations of each of the Hawaiian Islands (perhaps representing different morphotypes) would be masked by the spread of one or a few dominant chloroplast haplotypes within each island. However, since we have a complex pattern of multiple matching haplotypes and unique haplotypes representing the same morphotype on each island, a hypothesis of recent post-colonization chloroplast capture is insufficient to explain the total diversity of haplotypes. The evocation of earlier, ancient hybridization events followed by introgression and incomplete lineage sorting, such as that proposed for Eucalyptus (McKinnon et al. 2004) would also be required.
In summary, the lack of a positive correlation between chloroplast and morphological divergence may have several explanations: (i) hybridization between morphotypes may be confounding the phylogenetic signal due to chloroplast capture within islands, (ii) ancient introgression and incomplete lineage sorting may be sufficient to confound the signal reflecting the history of the morphological phenotype, but not to mask the geographical signature of sequential colonization, (iii) morphological divergence in Metrosideros, if genetically based, may be controlled by a few nuclear genes and the chloroplast data may have insufficient variation to reflect this, (iv) similar Metrosideros morphologies may have evolved independently, and repeatedly, on different islands, and (v) morphological variation in Metrosideros may not be genetically determined but developmentally plastic.
In at least one study of a morphological cline between two Eucalyptus species, hybridization was rejected in favour of a single evolutionary lineage that had undergone directional selection and diversification (Holman et al. 2003). If hybridization is not the dominant process in our system, the observed data could be explained by either lack of genetic basis for the diversity of phenotypes (e.g. environmental or developmental plasticity), or rapid and frequent parallel evolution of different phenotypes via a fast evolving and highly mutable nuclear gene region. Under a scenario of environmental or developmental plasticity, the frequent occurrence of multiple alternative morphotypes growing side by side within the same environments (such as in bogs, forests and lava fields) is puzzling, and would require an explanatory mechanism. West-Eberhard (2005) has argued that islands, and island radiations, are particularly fertile environments for the developmental expression of alternative phenotypes. Under this scenario, lineages with a developmentally and adaptively versatile ancestral phenotype may give rise to novel phenotypes by developmental recombination rather than genetic mutation (West-Eberhard 2003, 2005).
One mechanism that can result in phenotypic variation, even in the absence of genetic polymorphisms, is the effect of frequency-dependent selection. Studies have shown that disruptive selection, when extreme phenotypes have a fitness advantage, can increase phenotypic variance without affecting genetic variance (Rueffler et al. 2006). Studies of arthropod communities on Metrosideros in the Hawaiian Islands have shown that foliar structure and pubescence are critical in determining arthropod communities (Gruner et al. 2005), and several herbivorous groups preferentially feed on glabrous morphotypes (Alyokhin et al. 2004; Gruner et al. 2005). Selection to avoid herbivory could therefore have favoured repeated shifts to a pubescent morphotype. If shifts between morphotypes occur locally, this could also explain the negative correlation that is found between genotype and phenotype. An ancestral glabrous morphotype in the Hawaiian Islands is supported by character-state mapping and Bayesian ancestral character state reconstruction, but an ancestral colonizer that was heterozygous for phenotype is also possible, as both glabrous and pubescent forms occur in the Marquesas and other Polynesian islands.
Under the scenario of genetic control of morphotypes, the relevant loci could well be in a small number of nuclear genes. Thus far, the published nuclear allozyme and RAPD data for Hawaiian Metrosideros have shown that genetic diversity is not predictive of morphological diversity within populations (Aradhya et al. 1991; James et al. 2004). It therefore seems likely that if the presence of alternative phenotypes (e.g. those in sympatry and those sharing the same chloroplast haplotype) is genetically determined, comparative data from the putative nuclear genes controlling the phenotypic diversity are required to assess the extent of historical consistency in the chloroplast data.
(d) Conclusion
The chloroplast data present a distinct geographical pattern that supports a hypothesis of sequential colonization of progressively younger islands. Metrosideros therefore appears to have diversified in the Hawaiian Islands following the geological succession of islands and shows a signature pattern consistent with the rule of progression found in a number of classic Hawaiian radiations (Funk & Wagner 1995; Roderick & Gillespie 1998; Hormiga et al. 2003; Nepokroeff et al. 2003). We found that Metrosideros may have a more ancient history in the Hawaiian Islands than previous studies suggested, with important implications for the development and evolution of the Hawaiian biota, particularly of those endemic organisms that may have co-evolved with Metrosideros. As with other island systems where diversification and radiation processes have taken place, there remain many challenges to elucidating the temporal and spatial patterns. Additional data in future studies which add increased phylogenetic resolution and greater Pacific-wide sampling should help resolve more precisely the colonization history in this group.
Acknowledgments
We thank suppliers of leaf material or DNAs for this study: Cliff Morden (University of Hawai'i); Margaret Wright (University of Colorado); David Lorence, Ken Wood, and Steve Perlman (National Tropical Botanical Garden, NTBG); Jon Price (Smithsonian Institution, SI); and Peter Wilson (Royal Botanic Garden Sydney). D.M.P. was supported by a Smithsonian Institution Postdoctoral Fellowship. Laboratory work was funded by the Genetics programme, SI. Field collections and the contribution by W.L.W. were supported by the McBryde Chair programme, NTBG. We are grateful to Sara Hotchkiss, Queutin Cronk and two anonymous referees for comments.
Supplementary Material
Collection information or GenBank accession number. Collection numbers beginning HI are supplied by the Hawaii DNA Library (Morden et al., 1996)
Voucher and phenotype data for the Metrosideros specimens, including the matrix of 12 morphological characters. The location of vouchers is given by the following codes: BISH, Bishop Museum; COLO, University of Colorado; HAW, University of Hawaii; NSW, National Herbarium of New South Wales; PTBG, National Tropical Botanical Garden, USA; US, Smithsonian Institution; † indicates no voucher available; where the phenotype represented is clearly distinct and geographically restricted (i.e. for M. macropus, M. rugosa and M. tremuloides) we used other specimens from the same localities for morphological characters, and the voucher details are given for these specimens. All other taxa lacking vouchers were excluded from the morphological analysis. Morphological characters are as follows: (i) bud bracts: 0=small, 1=medium, 2=large; (ii) upper leaf pubescence: 0=glabrous, 1=slight pubescence, 2=pubescent; (iii) lower leaf pubescence: 0=glabrous, 1=slight pubescence, 2=pubescent; (iv) young leaf pubescence: 0=glabrous, 1=slight pubescence, 2=pubescent, 3=densely pubescent; (v) inflorescence pubescence: 0=glabrous, 1=slight pubescence, 2=pubescent, 3=densely pubescent; (vi) leaf length: quantitative; (vii) leaf width: quantitative; (viii) petiole length: quantitative; (ix) internode length: quantitative; (x) leaf margin revolute: 0=absent, 1=slight, 2=present, 3=pronounced; (xi) leaf margin: 0=flat, 1=slight crinkle, 2=pronounced crinkle; (xii) leaf apex: 0=blunt, 1=bluntly-acute, 2=blunt and acute, 3=acute; ? indicates missing data. M. excelsa and M. nervulosa are not included in the morphological analysis
For each region the number of variable and phylogenetically informative characters (PICs) are given, as well as the type of variable character–whether single nucleotide polymorphisms (SNPs), simple sequence repeats (SSRs) [number of bases per repeat], mononucleotide repeats [the base nucleotide that is repeated], and indels; *indicates that one 24 bp difference in the outgroup taxa only is treated here as a single character. The short spacer region trnY-trnE is within a 131 bp region amplified with both trnD-trnE and trnY-trnT primers and contains one variable SNP character.GenBank numbers are given for all sequences generated in this study.
Summary of diversity measures for the Marquesas (three islands) and each of the Hawaiian islands; n=number of individuals sampled, h=number of haplotypes, Hd=haplotype diversity, Pi=nucleotide diversity, ‘p’=uncorrected genetic distance
References
- Alyokhin, A. V., Yang, P. & Messing, R. H. 2004 Oviposition of the invasive two-spotted leafhopper on an endemic tree: effects of an alien weed, foliar pubescence, and habitat humidity. J. Insect Sci 4:13, 7. (insectscience.org/4.13) [DOI] [PMC free article] [PubMed]
- Aradhya K.M, Mueller-Dombois D, Ranker T.A. Genetic evidence for recent and incipient speciation in the evolution of Hawaiian Metrosideros (Myrtaceae) Heredity. 1991;67:129–138. doi:10.1038/hdy.1991.72 [Google Scholar]
- Aradhya K.M, Mueller-Dombois D, Ranker T.A. Genetic structure and differentiation in Metrosideros polymorpha (Myrtaceae) along altitudinal gradients in Maui, Hawaii. Genet. Res. 1993;61:159–170. [Google Scholar]
- Baldwin B.G. Adaptive radiation of the Hawaiian Silversword alliance: congruence and conflict of phylogenetic evidence from molecular and non-molecular investigations. In: Givnish T.J, Sytsma K.J, editors. Molecular evolution and adaptive radiation. Cambridge University Press; New York, NY: 1997. pp. 104–128. [Google Scholar]
- Bänfer G, Moog U, Fiala B, Mohamed M, Weising K, Blattner F.R. A chloroplast genealogy of myrmecophytic Macaranga species (Euphorbiaceae) in Southeast Asia reveals hybridization, vicariance and long-distance dispersals. Mol. Ecol. 2006;15:4409–4424. doi: 10.1111/j.1365-294X.2006.03064.x. doi:10.1111/j.1365-294X.2006.03064.x [DOI] [PubMed] [Google Scholar]
- Bonneville A, Dosso L, Hildenbrand A. Temporal evolution and geochemical variability of the South Pacific superplume activity. Earth Planet. Sci. Lett. 2006;244:251–269. doi:10.1016/j.epsl.2005.12.037 [Google Scholar]
- Burney D.A. Late Quaternary chronology and stratigraphy of twelve sites on Kauai. Radiocarbon. 2002;44:13–44. [Google Scholar]
- Burney D.A, DeCandido R.V, Burney L.P, Kostel-Hughes F.N, Stafford T.W, Jr, James H.F. A Holocene record of climate change, fire ecology and human activity from montane Flat Top Bog, Maui. J. Paleolimnol. 1995;13:209–217. doi:10.1007/BF00682765 [Google Scholar]
- Carlquist S. Columbia University Press; New York, NY: 1974. Island biology. [Google Scholar]
- Casgrain, P. & Legendre, P. 2001 The R-package for multivariate and spatial analysis, version 4.0 d5. Montreal, Canada: University of Montreal. See http://www.bio.umontreal.ca/casgrain/en/labo/R/
- Clague D.A. The growth and subsidence of the Hawaiian-Emperor volcanic chain. In: Keast A, Miller S.E, editors. The origin and evolution of Pacific Island Biotas, New Guinea to eastern Polynesia: patterns and processes. SPB Academic Publishing; Amsterdam, The Netherlands: 1996. pp. 35–50. [Google Scholar]
- Cordell S, Goldstein G, Mueller-Dombois D, Webb D, Vitousek P.M. Physiological and morphological variation in Metrosideros polymorpha, a dominant Hawaiian tree species, along an altitudinal gradient: the role of phenotypic plasticity. Oecologia. 1998;113:188–196. doi: 10.1007/s004420050367. doi:10.1007/s004420050367 [DOI] [PubMed] [Google Scholar]
- Cordell S, Goldstein G, Melcher P.J, Meinzer F.C. Photosynthesis and freezing avoidance in ohia (Metrosideros polymorpha) at treeline in Hawaii. Arct. Antarct. Alp. Res. 2000;32:381–387. doi:10.2307/1552386 [Google Scholar]
- Corn C.A. Seed dispersal methods in Hawaiian Metrosideros. In: Behnke J.A, editor. Challenging biological problems: directions toward their solution. Oxford University Press; New York, NY: 1972. pp. 422–435. [Google Scholar]
- Corn, C. A. 1979 Variation in Hawaiian Metrosideros PhD dissertation, University of Hawaii, Honolulu.
- Craig D.A, Currie D.C, Joy D.A. Geographical history of the central–western Pacific black fly subgenus Inseliellum (Diptera: Simuliidae: Simulium) based on a reconstructed phylogeny of the species, hot-spot archipelagoes and hydrological considerations. J. Biogeogr. 2001;28:1101–1127. doi:10.1046/j.1365-2699.2001.00619.x [Google Scholar]
- Dawson J.W. Pacific capsular Myrtaceae XI: redefinition of Metrosideros Banks ex Gaertn. and definition of infrageneric categories. Blumea. 1976;23:7–11. [Google Scholar]
- Dawson, J. W. & Stemmermann, L. 1990 Metrosideros Banks ex Gaertn. In Manual of the flowering plants of Hawaii, vol. 1 (eds W. L. Wagner, D. R. Herbst & S. H. Sohmer), pp. 964–970. Honolulu, HI: University of Hawaii Press.
- Drake D.R. Seed dispersal of Metrosideros polymorpha (Myrtaceae): a pioneer tree of Hawaiian lava flows. Am. J. Bot. 1992;79:1224–1228. doi:10.2307/2445048 [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 1981;17:368–376. doi: 10.1007/BF01734359. doi:10.1007/BF01734359 [DOI] [PubMed] [Google Scholar]
- Fleischer R.C, McIntosh C.E, Tarr C.L. Evolution on a volcanic conveyor belt: using phylogeographic reconstructions and K–Ar-based ages of the Hawaiian Islands to estimate molecular evolutionary rates. Mol. Ecol. 1998;7:533–545. doi: 10.1046/j.1365-294x.1998.00364.x. doi:10.1046/j.1365-294x.1998.00364.x [DOI] [PubMed] [Google Scholar]
- Foster S.A, McKinnon G.E, Steane D.A, Potts B.M, Vaillancourt R.E. Parallel evolution of dwarf ecotypes in the forest tree Eucalyptus globulus. New Phytol. 2007;175:370–380. doi: 10.1111/j.1469-8137.2007.02077.x. doi:10.1111/j.1469-8137.2007.02077.x [DOI] [PubMed] [Google Scholar]
- Funk V.A, Wagner W.L. Biogeographic patterns in the Hawaiian Islands. In: Wagner W.L, Funk V.A, editors. Hawaiian biogeography: evolution on a hot-spot archipelago. Smithsonian Institution Press; Washington, DC: 1995. pp. 379–419. [Google Scholar]
- Gardner R.C, de Lange P.J, Keeling D.J, Bowala T, Brown H.A, Wright S.D. A Late Quaternary phylogeography for New Zealand inferred from chloroplast DNA haplotypes in Metrosideros (Myrtaceae) Biol. J. Linn. Soc. 2004;83:399–412. doi:10.1111/j.1095-8312.2004.00398.x [Google Scholar]
- Gruner D.S. Arthropods from ohi'a lehua (Myrtaceae: Metrosideros polymorpha), with new records for the Hawaiian Islands. Bish. Mus. Occas. Pap. 2004a;78:33–52. [Google Scholar]
- Gruner D.S. Attenuation of top-down and bottom-up forces in a complex terrestrial community. Ecology. 2004b;85:3010–3022. doi:10.1890/04-0020 [Google Scholar]
- Gruner D.S, Taylor A.D, Forkner R.E. The effects of foliar pubescence and nutrient enrichment on arthropod communities of Metrosideros polymorpha (Myrtaceae) Ecol. Entomol. 2005;30:428–443. doi:10.1111/j.0307-6946.2005.00710.x [Google Scholar]
- Harbaugh D.T, Baldwin B.G. Phylogeny and biogeography of the sandalwoods (Santalum, Santalaceace): repeated dispersals throughout the Pacific. Am. J. Bot. 2007;94:1028–1040. doi: 10.3732/ajb.94.6.1028. doi:10.3732/ajb.94.6.1028 [DOI] [PubMed] [Google Scholar]
- Holman J.E, Hughes J.M, Fensham R.J. A morphological cline in Eucalyptus: a genetic perspective. Mol. Ecol. 2003;12:3013–3025. doi: 10.1046/j.1365-294x.2003.01970.x. doi:10.1046/j.1365-294X.2003.01970.x [DOI] [PubMed] [Google Scholar]
- Hood, G. M. 2005 PopTools, Version 2.6.9. Canberra, Australia: CSIRO. See http://www.cse.csiro.au/poptools/
- Hormiga G, Arnedo M, Gillespie R.G. Speciation on a conveyor belt: sequential colonization of the Hawaiian islands by Orsonwelles spiders (Araneae, Linyphiidae) Syst. Biol. 2003;52:70–88. doi: 10.1080/10635150390132786. doi:10.1080/10635150390132786 [DOI] [PubMed] [Google Scholar]
- Hotchkiss S, Juvik J.O. A Late-Quaternary pollen record from Ka'au Crater, O'ahu, Hawai'i. Quat. Res. 1999;52:115–128. doi:10.1006/qres.1999.2052 [Google Scholar]
- Howarth D.G, Gustafsson M.H.G, Baum D.A, Motley T.J. Phylogenetics of the genus Scaevola (Goodeniaceae): implication for dispersal patterns across the Pacific Basin and colonization of the Hawaiian Islands. Am. J. Bot. 2003;90:915–923. doi: 10.3732/ajb.90.6.915. doi:10.3732/ajb.90.6.915 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. doi:10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Jakob S.S, Blattner F.R. A chloroplast genealogy of Hordeum (Poaceae): long-term persisting haplotypes, incomplete lineage sorting, regional extinction, and the consequences for phylogenetic inference. Mol. Biol. Evol. 2006;23:1602–1612. doi: 10.1093/molbev/msl018. doi:10.1093/molbev/msl018 [DOI] [PubMed] [Google Scholar]
- James S.A, Puttock C.F, Cordell S, Adams R.P. Morphological and genetic variation within Metrosideros polymorpha (Myrtaceae) on Hawai'i. N. Zeal. J. Bot. 2004;42:263–270. [Google Scholar]
- McKinnon G.E, Vaillancourt R.E, Steane D.A, Potts B.M. The rare silver gum, Eucalyptus cordata, is leaving its trace in the organellar gene pool of Eucalyptus globulus. Mol. Ecol. 2004;13:3751–3762. doi: 10.1111/j.1365-294X.2004.02364.x. doi:10.1111/j.1365-294X.2004.02364.x [DOI] [PubMed] [Google Scholar]
- Melcher P.J, Cordell S, Jones T.J, Scowcroft P.G, Niemczura W, Giambelluca T.W, Goldstein G. Supercooling capacity increases from sea level to tree line in the Hawaiian tree species Metrosideros polymorpha. Int. J. Plant Sci. 2000;161:369–379. doi: 10.1086/314271. doi:10.1086/314271 [DOI] [PubMed] [Google Scholar]
- Mueller-Dombois D. Forest dynamics in Hawaii. Trends Ecol. Evol. 1987;2:216–220. doi: 10.1016/0169-5347(87)90024-3. doi:10.1016/0169-5347(87)90024-3 [DOI] [PubMed] [Google Scholar]
- Mueller-Dombois D. Distributional dynamics in the Hawaiian vegetation. Pac. Sci. 1992;46:221–231. [Google Scholar]
- Mueller-Dombois D. Long-term rain forest succession and landscape change in Hawai'i: the ‘Maui Forest Trouble’ revisited. J. Veg. Sci. 2006;17:685–692. doi:10.1658/1100-9233(2006)17[685:LRFSAL]2.0.CO;2 [Google Scholar]
- Nei M, Li W.H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl Acad. Sci. USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. doi:10.1073/pnas.76.10.5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepokroeff M, Sytsma K.J, Wagner W.L, Zimmer E.A. Reconstructing ancestral patterns of colonization and dispersal in the Hawaiian understory tree genus Psychotria (Rubiaceae): a comparison of parsimony and likelihood approaches. Syst. Biol. 2003;52:820–838. doi:10.1080/10635150390251072 [PubMed] [Google Scholar]
- Nixon K.C. The Parsimony Ratchet, a new method for rapid parsimony analysis. Cladistics. 1999;15:407–414. doi: 10.1111/j.1096-0031.1999.tb00277.x. doi:10.1111/j.1096-0031.1999.tb00277.x [DOI] [PubMed] [Google Scholar]
- Palme A.E, Su Q, Palsson S, Lascoux M. Extensive sharing of chloroplast haplotypes among European birches indicates hybridization among Betula pendula, B. pubescens and B. nana. Mol. Ecol. 2004;13:167–178. doi: 10.1046/j.1365-294x.2003.02034.x. doi:10.1046/j.1365-294X.2003.02034.x [DOI] [PubMed] [Google Scholar]
- Percy D.M, Page R.D.M, Cronk Q.C.B. Plant–insect interactions: double-dating associated insect and plant lineages reveals asynchronous radiations. Syst. Biol. 2004;53:120–127. doi: 10.1080/10635150490264996. doi:10.1080/10635150490264996 [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall K.A. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. doi:10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Price J.P. Floristic biogeography of the Hawaiian Islands: influences of area, environment and paleogeography. J. Biogeogr. 2004;31:487–500. doi:10.1046/j.0305-0270.2003.00990.x [Google Scholar]
- Price J.P, Clague D.A. How old is the Hawaiian biota? Geology and phylogeny suggest recent divergence. Proc. R. Soc. B. 2002;269:2429–2435. doi: 10.1098/rspb.2002.2175. doi:10.1098/rspb.2002.2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J.P, Elliott-Fisk D. Topographic history of the Maui Nui complex, Hawai'i and its implications for biogeography. Pac. Sci. 2004;58:27–45. doi:10.1353/psc.2004.0008 [Google Scholar]
- Rambaut, A. 1996 Se-Al: Sequence Alignment Editor, version 2. Oxford, UK: Oxford University. See http://evolve.zoo.ox.ac.uk/
- Rock, J. F. 1917 The ohia lehua trees of Hawaii. A revision of the Hawaiian species of the genus Metrosideros Banks, with special reference to the varieties and forms of Metrosideros collina (Forster) A. Gray subspecies polymorpha (Gaud.) Rock. Hawaii Board Agric. Forest. Bot. Bull [Botanical Bulletin. Territory of Hawaii, Board of Agriculture, Division of Forestry] 4, 1–76.
- Roderick G.K, Gillespie R.G. Speciation and phylogeography of Hawaiian terrestrial arthropods. Mol. Ecol. 1998;7:519–531. doi: 10.1046/j.1365-294x.1998.00309.x. doi:10.1046/j.1365-294x.1998.00309.x [DOI] [PubMed] [Google Scholar]
- Rozas J, Sánchez-DelBarrio J.C, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. doi:10.1093/bioinformatics/btg359 [DOI] [PubMed] [Google Scholar]
- Rueffler C, Van Dooren T.J.M, Leimar O, Abrams P.A. Disruptive selection and then what? Trends Ecol. Evol. 2006;21:238–245. doi: 10.1016/j.tree.2006.03.003. doi:10.1016/j.tree.2006.03.003 [DOI] [PubMed] [Google Scholar]
- Sanderson M.J. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol. Biol. Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- Selling, O. H. 1948 Studies in Hawaiian pollen statistics, Part III: on the Late Quaternary history of the Hawaiian vegetation. Bishop Mus. Special Pub. 39.
- Shaw J, Lickey E.B, Beck J.T, Farmer S.B, Liu W.S, Miller J, Siripun K.C, Winder C.T, Schilling E.E, Small R.L. The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 2005;92:142–166. doi: 10.3732/ajb.92.1.142. doi:10.3732/ajb.92.1.142 [DOI] [PubMed] [Google Scholar]
- Sikes, D. S. & Lewis, P. O. 2001 PAUPRat: PAUP* implementation of the parsimony ratchet, version 1. Distributed by the authors. Storrs, CT: University of Connecticut.
- Skottsberg C. Vascular plants from the Hawaiian Islands. II. Acta. Horti. Gothob. 1936;10:97–193. [Google Scholar]
- Steane D.A, Byrne M, Vaillancourt R.E, Potts B.M. Chloroplast DNA polymorphism signals complex interspecific interactions in Eucalyptus (Myrtaceae) Aust. Syst. Bot. 1998;11:25–40. doi:10.1071/SB96028 [Google Scholar]
- Stemmermann L. Ecological studies of Hawaiian Metrosideros in a successional context. Pac. Sci. 1983;37:361–373. [Google Scholar]
- Stemmermann L, Ihsle T. Replacement of Metrosideros polymorpha, Ohia, in Hawaiian dry forest succession. Biotropica. 1993;25:36–45. doi:10.2307/2388977 [Google Scholar]
- Swofford, D. L. 2002 PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4.0. Sunderland, MA: Sinauer Associates. See http://paup.csit.fsu.edu/
- Sytsma K.J, Litt A, Zjhra M.L, Pires J.C, Nepokroeff M, Conti E, Walker J, Wilson P.G. Clades, clocks, and continents: Historical and biogeographical analysis of Myrtaceae, Vochysiaceae, and relatives in the Southern Hemisphere. Int. J. Plant Sci. 2004;165:S85–S105. doi:10.1086/421066 [Google Scholar]
- Vos R.A. Accelerated likelihood surface exploration: the likelihood ratchet. Syst. Biol. 2003;52:368–373. doi: 10.1080/10635150390196993. doi:10.1080/10635150309330 [DOI] [PubMed] [Google Scholar]
- Wagner W.L, Funk V.A, editors. Hawaiian biogeography: evolution on a hot spot archipelago. Smithsonian Institution Press; Washington, DC: 1995. [Google Scholar]
- Wagner W.L, Weller S.G, Sakai A.K. Monograph of Schiedea (Caryophyllaceae subfam. Alsinoideae) Syst. Bot. Monogr. 2005;72:1–169. [Google Scholar]
- West-Eberhard M.J. Oxford University Press; New York, NY: 2003. Developmental plasticity and evolution. [Google Scholar]
- West-Eberhard M.J. Developmental plasticity and the origin of species differences. Proc. Natl Acad. Sci. USA. 2005;102:6543–6549. doi: 10.1073/pnas.0501844102. doi:10.1073/pnas.0501844102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S.D, Yong C.G, Dawson J.W, Whittaker D.J, Gardner R.C. Riding the ice age El Niño? Pacific biogeography and evolution of Metrosideros subg. Metrosideros (Myrtaceae) inferred from nuclear ribosomal DNA. Proc. Natl Acad. Sci. USA. 2000;97:4118–4123. doi: 10.1073/pnas.050351197. doi:10.1073/pnas.050351197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S.D, Yong C.G, Wichman S.R, Dawson J.W, Gardner R.C. Stepping stones to Hawaii: a trans-equatorial dispersal pathway for Metrosideros (Myrtaceae) inferred from nrDNA (ITS+ETS) J. Biogeogr. 2001;28:769–774. doi:10.1046/j.1365-2699.2001.00605.x [Google Scholar]
- Zwickl, D. J. 2006 GARLI: genetic algorithm for rapid likelihood inference, version 0.9. Austin, TX: University of Texas. See http://www.bio.utexas.edu/grad/zwickl/web/garli.html
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Collection information or GenBank accession number. Collection numbers beginning HI are supplied by the Hawaii DNA Library (Morden et al., 1996)
Voucher and phenotype data for the Metrosideros specimens, including the matrix of 12 morphological characters. The location of vouchers is given by the following codes: BISH, Bishop Museum; COLO, University of Colorado; HAW, University of Hawaii; NSW, National Herbarium of New South Wales; PTBG, National Tropical Botanical Garden, USA; US, Smithsonian Institution; † indicates no voucher available; where the phenotype represented is clearly distinct and geographically restricted (i.e. for M. macropus, M. rugosa and M. tremuloides) we used other specimens from the same localities for morphological characters, and the voucher details are given for these specimens. All other taxa lacking vouchers were excluded from the morphological analysis. Morphological characters are as follows: (i) bud bracts: 0=small, 1=medium, 2=large; (ii) upper leaf pubescence: 0=glabrous, 1=slight pubescence, 2=pubescent; (iii) lower leaf pubescence: 0=glabrous, 1=slight pubescence, 2=pubescent; (iv) young leaf pubescence: 0=glabrous, 1=slight pubescence, 2=pubescent, 3=densely pubescent; (v) inflorescence pubescence: 0=glabrous, 1=slight pubescence, 2=pubescent, 3=densely pubescent; (vi) leaf length: quantitative; (vii) leaf width: quantitative; (viii) petiole length: quantitative; (ix) internode length: quantitative; (x) leaf margin revolute: 0=absent, 1=slight, 2=present, 3=pronounced; (xi) leaf margin: 0=flat, 1=slight crinkle, 2=pronounced crinkle; (xii) leaf apex: 0=blunt, 1=bluntly-acute, 2=blunt and acute, 3=acute; ? indicates missing data. M. excelsa and M. nervulosa are not included in the morphological analysis
For each region the number of variable and phylogenetically informative characters (PICs) are given, as well as the type of variable character–whether single nucleotide polymorphisms (SNPs), simple sequence repeats (SSRs) [number of bases per repeat], mononucleotide repeats [the base nucleotide that is repeated], and indels; *indicates that one 24 bp difference in the outgroup taxa only is treated here as a single character. The short spacer region trnY-trnE is within a 131 bp region amplified with both trnD-trnE and trnY-trnT primers and contains one variable SNP character.GenBank numbers are given for all sequences generated in this study.
Summary of diversity measures for the Marquesas (three islands) and each of the Hawaiian islands; n=number of individuals sampled, h=number of haplotypes, Hd=haplotype diversity, Pi=nucleotide diversity, ‘p’=uncorrected genetic distance