Abstract
Uniquely for non-primate mammals, three classes of cone photoreceptors have been previously identified by microspectrophotometry in two marsupial species: the polyprotodont fat-tailed dunnart (Sminthopsis crassicaudata) and the diprotodont honey possum (Tarsipes rostratus). This report focuses on the genetic basis for these three pigments. Two cone pigments were amplified from retinal cDNA of both species and identified by phylogenetics as members of the short wavelength-sensitive 1 (SWS1) and long wavelength-sensitive (LWS) opsin classes. In vitro expression of the two sequences from the fat-tailed dunnart confirmed the peak absorbances at 363 nm in the UV for the SWS1 pigment and 533 nm for the LWS pigment. No additional expressed cone opsin sequences that could account for the middle wavelength cones could be amplified. However, amplification from the fat-tailed dunnart genomic DNA with RH1 (rod) opsin primer pairs identified two genes with identical coding regions but sequence differences in introns 2 and 3. Uniquely therefore for a mammal, the fat-tailed dunnart has two copies of an RH1 opsin gene. This raises the possibility that the middle wavelength cones express a rod rather than a cone pigment.
Keywords: visual pigments, trichromacy in marsupials, evolution of vision
1. Introduction
Visual pigments belong to the large family of G-protein-coupled receptors that share a common structure of seven α-helical transmembrane regions joined by cytoplasmic and luminal loops. They form a group of closely related proteins (opsins) that bind retinal, a derivative of vitamin A. Five classes of pigments are present in vertebrates, a rod class and four different cone classes distinguished on the basis of spectral sensitivity and amino acid sequence of their respective opsins: long wavelength-sensitive (LWS) with λmax 500–570 nm; middle wavelength-sensitive (MWS) with λmax 480–530 nm; and two short wavelength-sensitive classes, short wavelength-sensitive opsin 2 (SWS2) with λmax 400–470 nm and short wavelength-sensitive opsin 1 (SWS1) with λmax 355–445 nm. In monotreme and placental mammals, this complement is reduced to a rod and two cone classes: SWS2 and LWS in the former (Davies et al. 2007) and SWS1 and LWS in the latter (Yokoyama 2000). This loss of cone classes is believed to have resulted from a nocturnal lifestyle that mammals went through during their early evolution. As a result, most mammals are dichromats. In anthropoid or simian primates, however, trichromacy is generally present, and this is achieved by different genetic mechanisms in the two major primate lineages. In the Old World (catarrhine) primates from Africa and Asia, a duplication of the LWS gene is present with the two copies encoding a ‘red’ and ‘green’ variant with the λmax values at approximately 565 and 530 nm, respectively (Nathans et al. 1986; Dulai et al. 1999), whereas in most New World (platyrrhine) primates from Central and South America, the trichromacy is based on the polymorphism of the LWS gene, with different alleles encoding spectrally distinct red and green pigments (Mollon et al. 1984). In both cases, the major driving force behind the evolution of trichromacy, with its improved colour discrimination in the red/green region of the spectrum, is argued to be the detection and evaluation of ripe fruits (Mollon 1989; Osorio & Vorobyev 1996; Sumner & Mollon 2000; Regan et al. 2001) or young nutritious leaves (Dominy & Lucas 2001) against the green foliage of the rainforest.
A recent study of the spectral characteristics of photoreceptors in two marsupial species representative of the major marsupial taxonomic divisions, the arrhythmic and insectivorous polyprotodont fat-tailed dunnart (Sminthopsis crassicaudata) and the crepuscular and nectivorous diprotodont honey possum (Tarsipes rostratus), has identified in both species three classes of cone photoreceptors, with absorbance peaks as determined by microspectrophotometry (MSP) in the UV for the short wavelength-sensitive (SWS) class, at 509 and 505 nm, respectively, for the MWS class, and at 535 and 557 nm, respectively, for the LWS class (Arrese et al. 2002). The LWS and MWS visual pigments were found in both single and double cones, but not as LWS–MWS pairings in the latter.
Behavioural studies have indicated that trichromacy is present in these species (Arrese et al. 2006). The objective of the present study was to identify and characterize the opsin genes expressed in the different cone classes.
2. Material and methods
(a) Animals
Fat-tailed dunnarts (n=3) were obtained from a breeding colony established at the University of Adelaide (Animal Service Division), South Australia. Honey possums (n=3) were collected in the Mt Lesueur Nature Reserve, using pit traps, under licence from the Department of Conservation and Land Management. Animals were terminally anaesthetized with saffan (alphaxalone and alphadolone acetate, 0.1 ml/10 g body weight, i.p.). The study was approved by the Animal Ethics and Experimentation Committee of the University of Western Australia.
(b) Genomic DNA, retinal RNA and cDNA preparation
Genomic DNA was isolated from liver tissue using a standard phenol–chloroform method. Dissected retinae were collected in RNAlater (Ambion, Austin, Tex). Retinal mRNA was prepared from total RNA using either the EpiCentre MasterPure RNA Purification Kit followed by Qiagen Oligotex mRNA Purification Kit or the Quickprep Micro mRNA Purification Kit (Amersham Biosciences).
Single-stranded cDNA was synthesized using an oligo-d(T) anchor primer and Superscript III reverse transcriptase (Invitrogen) or transcriptor reverse transcriptase (Roche Diagnostics). 5′ and 3′ RACE was carried out using a 5′–3′ RACE Kit second generation (Roche Diagnostics), following the manufacturer's instructions.
(c) PCR amplification and cloning
Primers used in the amplification of opsin gene fragments are listed in tables 1 and 2. PCR products were visualized by agarose gel electrophoresis and cloned into pST-Blue-1 (Novagen) or pGEM-T easy cloning vector (Promega). After colony PCR screening, the inserts from positive colonies were sequenced using either vector- or opsin sequence-specific primers. Sequencing was carried out using a BigDye Terminator v. 3.1 Cycle Sequencing kit on either an ABI 3730 or ABI 3100 PRISM Genetic Analyser.
Table 1.
Oligonucleotide primers used for the amplification of SWS1 and LWS opsin sequences. (Numbers refer to position in coding sequence for gene-specific primers, + denotes a forward primer and − denotes a reverse primer.)
| primer | position | sequence (5′–3′) |
|---|---|---|
| oligo d(T) anchor | GACCACGCGTATCGATGTCGACTTTTTTTTTTTTTTTTV | |
| PCR anchor | GACCACGCGTATCGATGTCGAC | |
| opsin A+ | ACCACCCAGAAGGCAGAGAAG | |
| opsin B− | GACATAGATGATGGGGTTGTA | |
| SWS1 A+ | 158+ | GTGCTGGTGGCCACACTGC |
| SWS1 B+ | 637+ | CCTCATCTGCTTCTCCTAC |
| SWS1 E+ | 894+ | CCCATCATCTACTGCTTCATG |
| SWS1 F+ | 886+ | TCTACAAYCCCATCATCTACTG |
| SWS1 G− | 908− | GCAGTAGATGATGGGRTTGTAG |
| SWS1 H− | 993− | CTGAGAGCTGGTGGTTTCAG |
| SWS1 I− | 317− | CACAGACATGGCGGCCAAAG |
| SWS1 J+ | 56+ | TGGGATGGGCCTCAGTACCAC |
| LWS A+ | 938+ | CCACTATCTACAACCCCATC |
| LWS B− | 700− | TGATGATGCTGAGAGGAAGG |
| LWS C− | 646− | GAACGCCAGGGTCGGAGCTG |
| LWS D+ | 547+ | ACTGCACCACCCATCTTTGG |
| LWS E− | 737− | CGGATGGCCAGCCACACTTG |
| LWS F+ | 763+ | TCWGARTCYACCCAGAAGGC |
| LWS G+ | 776+ | AGAAGGCHGARAAGGAAGTG |
| LWS H− | 954− | GGGGTTGTAGATRGTGGCAC |
| LWS I− | 1028− | CCATCRTCMACCTTCTTCCC |
| LWS J− | 1071− | GACGGAAGAGACCTCTGTCC |
Table 2.
Oligonucleotide primers used for the amplification of RH1 exons and introns. (Numbers and +, − symbols are as given in the table legend 1.)
| primer | position | sequence (5′–3′) |
|---|---|---|
| rod A− | 157− | GGAAGCCCAGGACGATCAGC |
| rod A+ | 342+ | CTTCTTCGCCACCACAGGAG |
| rod B− | 394− | CCAAAACCACCAAGGCCCAG |
| rod B+ | 361+ | GGTGAAGTAGCCCTCTGGG |
| rod C− | 563− | CCACACGAACATTGCATTCC |
| rod C+ | 678+ | GGTCTTCACAGTCAAAGAG |
| rod D− | 714− | TTGCTGCTGGGCTGCGGC |
| rod E− | 958− | TGGTGATCATGCAGTTCCGG |
| rod F+ | 973+ | AAGAATCCATTGGGTGATGA |
| rod G+ | 939+ | CCGGAACTGCATGATCACCAC |
| rod H− | 69− | GGGCTCCGGACCACCCCAC |
| rod I− | 33− | GACGTAAAAGTTGGGTCCC |
| rod INT2 A+ | intron 2 | TGCCCCATCGCCAAAAGTTG |
| rod INT2 B+ | intron 2 | TGCCCCATCGCCAAAAGTGG |
| rod INT2 C+ | intron 2 | CCATCGCCAAAAGTTGAGAC |
| rod INT2 D+ | intron 2 | CCATCGCCAAAAGTGGAGGC |
| rod INT2 E+ | intron 2 | TTCCCTCCTATTTACCTCC |
| rod INT2 F+ | intron 2 | CCTTGGAAATCTATTTACCTCC |
(d) Phylogenetic analysis
Neighbour joining (Saitou & Nei 1987) was used to construct phylogenetic trees from opsin nucleotide sequences after alignment with ClustalW (Higgins et al. 1996). The degree of support for internal branching was assessed by bootstrapping with 1000 replicates using the MEGA2 computer package (Kumar et al. 2001).
(e) Expression of recombinant opsins
The entire coding sequences for the fat-tailed dunnart SWS1 and LWS opsins were amplified from retinal cDNA with Pfu DNA polymerase, using primer pairs SWS1F/SWS1R and LWSF/LWSR (table 3) containing EcoRI and SalI restriction sites. The resulting products were then cloned via these restriction sites into the expression vector pMT3 that contains the sequence for the 1D4 epitope from bovine rod opsin downstream of and in-frame with the SalI site (Franke et al. 1988). The opsin sequences were checked using vector-specific sequencing primers.
Table 3.
Oligonucleotide primers used for the production of expression constructs. (Coding sequences are shown in bold. Eco R1 and SalI restriction enzyme sites are italicized.)
| primer | sequence |
|---|---|
| SWS1F | 5′-GCGCGAATTCCACCATGTCAGGGGATGAGGAGTTC−3′ |
| SWS1R | 3′-CGGCGTCGACGCACTAGGCCCCACTTGGCTGGAG-5′ |
| LWSF | 5′-GCGCGAATTCCACCATGACACAGGCATGGGACCC−3′ |
| LWSR | 3′-CGGCGTCGACGCGGCAGGCGCCACAGAGGAGAC-5′ |
The pMT3 vector containing either the SWS1 or LWS coding sequences was transfected into HEK-293T cells with GeneJuice (Invitrogen) according to the manufacturer's instructions. Thirty 90 mm plates were used per transfection; the cells were harvested 48 hours post-transfection and washed with 1× PBS. The visual pigments were regenerated in 1× PBS with 40 μM 11-cis-retinal in the dark. Dodecyl maltoside (1% (w/v)) and PMSF (20 mg ml−1) were then added before passage over a CNBr-activated sepharose-binding column coupled to an anti-1D4 monoclonal antibody (Molday & MacKenzie 1983).
Absorption spectra were recorded in the dark using a dual path spectrophotometer (Spectronic Unicam, Cambridge, UK). The pigments were either bleached by exposure to light for 15 min (LWS) or acid denatured (SWS1) by the addition of 10.8 μl of 1 N HCl. The λmax value for each pigment was determined by subtracting the bleached or acid-denatured spectrum from the dark absorption spectrum to produce a difference spectrum. This was then fitted to a standard Govardovskii rhodopsin A1 template (Govardovskii et al. 2000) using an Excel spreadsheet to determine the λmax.
(f) In situ hybridization
Eye cups were fixed in 4% (w/v) paraformaldehyde in 1× PBS overnight, washed briefly with 1× PBS and cryoprotected by incubation overnight in 25% (w/v) sucrose. Ten micrometre thick sections were cut on a cryostat.
Riboprobes were generated from cloned coding sequences of RH1 and LWS opsin of the fat-tailed dunnart, RH2 opsin of the black bream (Acanthopagrus butcheri, a perciform fish from the family of Sparidae) and SWS2 of the platypus (Ornithorhynchus anatinus) by restriction enzyme digestion and ligation into pGEM-T easy. The probes were of 580, 568, 1100 and 1100 bp in length, respectively. Digoxigenin (DIG)-labelled antisense and sense RNA probes were then synthesized with SP6 and T7 primers using a DIG RNA labelling kit (Roche). Retinal sections were prepared for hybridization as follows: incubation in 1× PBS for 5 min at room temperature; 4% paraformaldehyde in 1× PBS for 15 min; 1× PBS for 5 min; 1× PBT (1×PBS+0.1% Tween 20)+proteinase K (5 μg ml−1) for 5 min; 1× PBT+glycine (2 mg ml−1) for 10 min; 1× PBT for 1 min; and hybridization solution (50% formamide, 5× SSC, 50 μg ml−1 tRNA, 1% SDS, 50 μg ml−1 heparin) for 15 min at 65°C. Denatured probe in hybridization solution was then added and the slides incubated overnight at 65°C. The slides were washed three times for 15 min at 65°C with wash solution I (50% formamide, 5× SSC, 1% SDS) followed by washing three times for 15 min with wash solution II (50% formamide, 2× SSC) and three times 10 min at room temperature in 1× TBST (140 mM NaCl, 2.7 mM KCl, 25 mM Tris–HCl (pH 7.5), 1% Tween 20). The sections were blocked with 1× TBST+10% sheep serum for 30 min at room temperature. Anti-DIG antibody solution (anti-DIG antibody 1 : 2000 in 1% sheep serum in 1× TBST) was added and the slides were incubated at room temperature for 2 hours followed by overnight incubation at 4°C. The slides were then washed four times at room temperature for 15 min with 1× TBST and three times for 10 min with NTMT (100 mM NaCl, 100 mM Tris–HCl (pH 9.5), 50 mM MgCl2, 1% Tween 20). The colour was developed by incubating the slides in 1× NTMT+NBT (4.5 μl ml−1) and BCIP (3.5 μl ml−1) for up to 7 days, replenishing the solution every 24 hours. The sections were fixed by washing the slides two times with 1× NTMT for 10 min, with PBT (pH 5.5) for 10 min, two times with 1× PBS for 10 min, with 4% paraformaldehyde in PBS for 30 min and with two times with 1× PBS for 10 min. The sections were finally mounted in 90% glycerol and sealed with a cover-slip for microscopy. The slides were viewed under a light microscope and images taken with a Nikon digital camera.
3. Results
The sequence of rod opsin from the fat-tailed dunnart has previously been reported (Hunt et al. 2003). Degenerate PCR primers (table 1) designed to the conserved regions of published sequences of opsin genes from a number of other vertebrate species were used to amplify the SWS1 and LWS coding sequences from retinal cDNA. These sequences have been deposited in GenBank with accession nos. AY442173, AY772472, AY772470 and EU232013.
(a) SWS cones
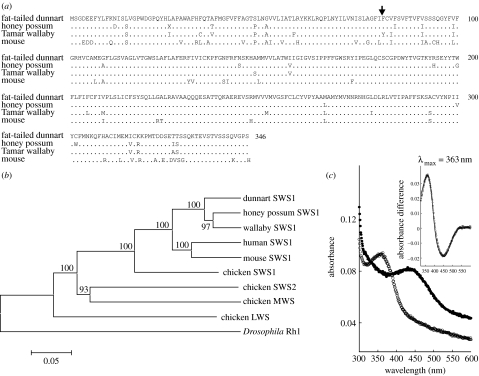
Previous MSP data (Arrese et al. 2002) have indicated the presence of UV-sensitive (UVS) receptors, although the low frequency of such cones and the limitations of the MSP instrumentation at such short wavelengths meant that the presence of the UVS class of visual pigment could not be fully confirmed. RT-PCR using the SWS1 primer pairs listed in table 1 generated fragments for both the fat-tailed dunnart and the honey possum, which were successfully extended by 5′ and 3′ RACE into full-length coding sequences. The deduced amino acid sequences are shown in figure 1a, aligned with Tamar wallaby violet-sensitive (VS) and mouse UVS sequences. Confirmation that the sequences belong to the SWS1 class was obtained from phylogenetic analysis where both sequences clade with other SWS1 sequences (figure 1b). To determine the λmax of the corresponding pigment, the sequence from the fat-tailed dunnart was subcloned into the pMT3 expression vector for in vitro expression and the resulting recombinant opsin was regenerated with 11-cis-retinal. As shown in figure 1c, the λmax of this pigment is in the UV at 363 nm.
Figure 1.
SWS1 opsin. (a) Deduced amino acid sequence for the fat-tailed dunnart and the honey possum aligned with sequences from the Tamar wallaby and mouse. (b) Opsin neighbour-joining tree of nucleotide sequences showing grouping of the fat-tailed dunnart and honey possum SWS1 sequences within the SWS1 clade. Accession nos. are as follows: AY772472 (honey possum SWS1); AY286017 (Tamar wallaby SWS1); AY442173 (fat-tailed dunnart SWS1); NM_007538 (mouse SWS1); NM_001708 (human SWS1); M92039 (chicken SWS1); M92037 (chicken SWS2); M92038 (chicken MWS); NM_205440 (chicken LWS); NM_008106 (mouse LWS); M62903 (chicken LWS); and X65877 (Drosophila Rh1). (c) Spectral analysis of in vitro expressed SWS1 sequence. Dark (open circles) and acid-denatured (filled circles) spectra, with difference spectrum and fitted Govardovskii template in the inset.
With the exception of avian UVS pigments, all vertebrate UVS pigments have Phe present at site 86 and site-directed mutagenesis has confirmed the central role played by this residue in conferring UV sensitivity (Cowing et al. 2002; Fasick et al. 2002). The SWS1 pigment of both marsupial species also has Phe86, consistent therefore with a λmax in the UV. By contrast, the Tamar wallaby has a VS pigment with Phe86 replaced by Tyr, as found in the bovine and porcine pigments (Cowing et al. 2002).
(b) LWS cones
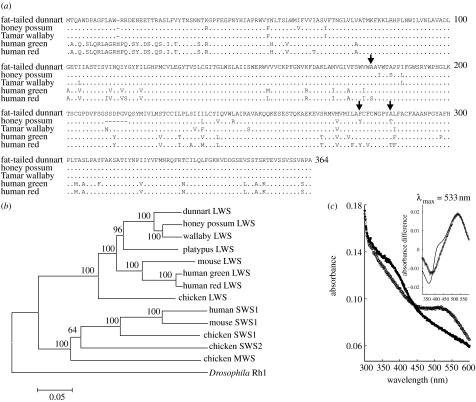
LWS opsin coding sequences were amplified for both species from retinal cDNA using the LWS primers listed in table 1. The deduced amino acid sequences are shown in figure 2a, aligned with the Tamar wallaby LWS sequences, and the human red and green LWS variants. The identity of these sequences as orthologues of the LWS sequences in other vertebrate species was obtained by phylogenetic analysis where both form a clade with other LWS sequences (figure 2b).
Figure 2.
LWS opsin. (a) Deduced amino acid sequence for the fat-tailed dunnart and honey possum aligned with LWS sequence from the Tamar wallaby and the L and M variants from human. (b) Neighbour-joining tree of nucleotide sequences showing grouping of the fat-tailed dunnart and the honey possum LWS sequences within the LWS clade. Accession nos. are as follows: EU232013 (fat-tailed dunnart LWS); AY286018 (Tamar wallaby LWS); AY772470 (honey possum LWS); NM_008106 (mouse LWS); M13300 (human red LWS); M13306 (human green LWS); EF050078 (platypus LWS); NM_205440 (chicken LWS); M92038 (chicken MWS); M92037 (chicken SWS2); M92039 (chicken SWS1); NM_007538 (mouse SWS1); NM_001708 (human SWS1); and X65877 (Drosophila Rh1). (c) Spectral analysis of in vitro expressed LWS sequence. Dark (open circles) and bleached (filled circles) spectra, with difference spectrum and fitted Govardovskii template in the inset.
The λmax values of LWS pigments across different mammalian species vary from 508 nm in the mouse, Mus musculus (Sun et al. 1997), to approximately 565 nm in primates (Bowmaker et al. 1991). Since the separate MWS and LWS classes of cone photoreceptors that have been identified by MSP in the retinae of both the honey possum and the fat-tailed dunnart show a similar range with the λmax values at 505 and 557 nm for the honey possum, and at 509 and 535 nm for the fat-tailed dunnart, in vitro expression of the fat-tailed dunnart sequence was used to determine the λmax of the encoded opsin. The difference spectrum for the corresponding pigment regenerated with 11-cis-retinal yielded a λmax at 533 nm (figure 2c), almost identical to the λmax of 535 nm determined by MSP for this species. This opsin gene encodes, therefore, the pigment present in the LWS cones.
A duplicated LWS opsin gene underlies the red and green pigments in Old World primates (Dulai et al. 1999) and the spectral shifts between these two pigments are largely attributable to substitutions at three sites, 180, 277 and 285, with polar residues Thr, Tyr and Thr, respectively, occupying these sites in the red pigment compared with non-polar residues Ala, Phe and Ala, respectively, in the green pigment (Neitz et al. 1991; Ibbotson et al. 1992). Significantly, the honey possum and the fat-tailed dunnart also differ at sites 277 and 285, with polar Tyr and Thr in the more LW-shifted honey possum pigment. Based on residue replacement experiments carried out by site-directed mutagenesis of the human pigments (Merbs & Nathans 1993; Asenjo et al. 1994), the residue differences at these sites are sufficient to account for the 22 nm shift between the LWS pigments in these two marsupial species.
(c) MWS cones
The above analysis demonstrates that SWS1 and LWS genes are responsible for the UVS and LWS pigments identified by MSP in the retinae of the honey possum and the fat-tailed dunnart. The pigment present in the MWS cones remains therefore to be identified.
The LWS pigment in the mouse, M. musculus, has a λmax at 508 nm, which is similar to the λmax values of the marsupial MWS pigments. This shortwave shift in the mouse pigment arises from a Ser rather than the more usual Ala residue at site 308 combined with the loss of chloride binding arising from a His197Tyr substitution in the chloride-binding site (Sun et al. 1997). A genomic fragment that encompasses intron 4 of 776 bp and the flanking regions of exons 3 and 4, the latter including the coding region for His197, was PCR amplified from the fat-tailed dunnart genomic DNA using the primer pair LWSD+ and LWSE− listed in table 1. The fragment was cloned into pGEM-T and 12 clones from two different PCRs were sequenced. In each case, the flanking coding sequences were identical to the sequence obtained from retinal cDNA and no variation was found in intron sequence. Therefore, there is no evidence for a second LWS gene with an altered chloride-binding site.
The possibility that a second LWS gene is present that encodes a MWS pigment with substitutions at the major tuning sites, 277, 285 and 308, was also examined by the PCR in the fat-tailed dunnart genomic DNA using gene-specific primers (table 1). As described above, the role of sites 277 and 285 in the tuning of primate LWS pigments have been well documented (Merbs & Nathans 1993; Asenjo et al. 1994), and Ser308 is responsible for a 30 nm shortwave shift in the LWS pigment in the dolphin (Fasick & Robinson 1998). A fragment was generated with primers LWSD+ and LWSJ−, which includes the coding regions for these key sites within exon 5. Six clones were sequenced but no variation in the coding or intronic regions was seen.
The RH2 class of opsins specifies pigments in non-mammalian species with the λmax values that vary from 480 to 550 nm. A large number of degenerate PCR primers designed to published RH2 sequences from a number of different species were therefore used with either dunnart retinal cDNA or genomic DNA as template, but all failed to amplify any RH2 gene fragments. Although less likely to specify a pigment with a λmax>500 nm, a similar screen for an SWS2 gene was undertaken using primers designed from the published sequences, but these also failed to generate any SWS2 opsin gene fragments.
(d) Rod opsin gene duplication
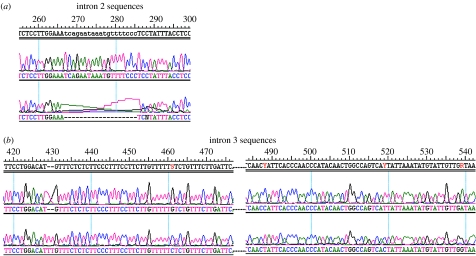
In the absence of either a duplicated and substituted LWS gene or the retention of an RH2 or SWS2 gene, the genome of the fat-tailed dunnart was screened for an RH1 rod opsin gene duplication. PCR amplifications using the primer pairs listed in table 2 generated fragments that included the 5′ and 3′ UTRs and introns 2, 3 and 4. In each case, the fragments were cloned and a number of clones from different PCRs were sequenced. No differences were found in either of the UTR regions, in intron 4, or in any of the sequenced regions of exons 2, 3 and 4. However, for both introns 2 and 3, the sequences fell into two classes, with an indel of 19 bp in intron 2 and four single bp substitutions and an indel of 2 bp in intron 3 (figure 3). Such major differences in intron sequence are generally associated with duplicated copies rather than alleles of a gene. However, to assess this further, intron 2 from a second unrelated animal was screened and the same two variants again recovered, suggesting that the variants identify different genes rather than alleles of the same gene. Uniquely therefore for a mammal, the fat-tailed dunnart has two copies of an RH1 rod opsin gene.
Figure 3.
Sequence variation obtained for the fat-tailed dunnart RH1 gene. Sequence electropherograms obtained from genomic DNA using RH1 opsin PCR primers showing (a) indel in intron 2 covering 19 bp and (b) 2 bp indel and four single nucleotide substitutions (shown in red) in intron 3.
(e) In situ hybridizations
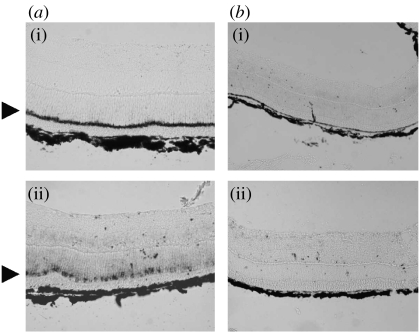
Homologous RH1 and LWS probes derived from the fat-tailed dunnart opsin sequences were used to assess the distribution of expression of these pigments in the fat-tailed dunnart retina. In all cases described below, the control sense probes failed to show any hybridization. The RH1 antisense probe showed a continuous band of hybridization to the outer nuclear layer (figure 4). This identifies the cell bodies of the rod photoreceptors and confirms the rod-dominated nature of the fat-tailed dunnart retina. The LWS antisense probe hybridized to substantially fewer cells in the outer nuclear layer than the RH1 probe. These cells that were present in patches around the retina are identified therefore as the LWS cone photoreceptors.
Figure 4.
In situ hybridization of opsin probes with fat-tailed dunnart retina: (a) antisense and (b) sense. (i) RH1 and (ii) LWS probes were obtained from the corresponding dunnart cDNA sequences. Positive staining is present only with the antisense probes where it is confined to the outer nuclear layer (marked by an arrowhead).
Hybridization with heterologous RH2 and SWS2 probes derived from the RH2 coding sequence of the black bream (Shand et al. in press) and the SWS2 coding region of the platypus (Davies et al. 2007) was also attempted. However, the antisense probes in both cases failed to show any evidence of hybridization (data not shown).
4. Discussion
The coding sequences for an SWS1 and an LWS cone opsin have been reported in a previous study of two species of the fat-tailed dunnart (Strachan et al. 2004). However, in this study, no data were presented that enabled these sequences to be correlated with the different cone classes identified by MSP (Arrese et al. 2002). It remained uncertain therefore whether the SWS1 sequence encoded a UVS pigment and the LWS gene encoded the MWS or LWS pigment. We have now shown by in vitro expression of the cloned SWS1 and LWS opsin sequences that they encode pigments that correlate precisely in λmax with the SWS and LWS clone classes as determined by MSP. The presence of a UVS SWS1 pigment is therefore confirmed; this pigment retains Phe86 as proposed for the ancestral vertebrate SWS1 pigment (Hunt et al. 2004, 2007) and contrasts with the VS SWS1 pigment of the Tamar wallaby (Deeb et al. 2003). This latter pigment has Tyr86 as found in the VS pigments of the cow, pig (Cowing et al. 2002) and squirrel (Carvalho et al. 2006), and represents therefore convergent evolution in the generation of violet sensitivity in metatherian and eutherian mammals.
The λmax values for the LWS cones of the fat-tailed dunnart and the honey possum differ by approximately 22 nm, and this difference can be adequately accounted for by substitution at sites 277 and 285 (Asenjo et al. 1994), with polar Tyr and Thr, respectively, present in the long-wavelength-shifted honey possum pigment. The same sites also account for the spectral shift between the red and green LWS pigments of primates (Neitz et al. 1991; Ibbotson et al. 1992; Williams et al. 1992) and are also substituted in the duplicated LWS genes of the blind cave fish, Astyanax (Yokoyama & Yokoyama 1990). These are all examples of convergent evolution in distantly related species and confirm the assertion (Hunt et al. 2001, 2004) that in many cases spectral tuning of a visual pigment can only be achieved by substitution at a limited number of sites that are able to interact with the chromophore to achieve the spectral shift and maintain a fully functional pigment.
The genetic basis for the MWS cone pigment originally identified by MSP (Arrese et al. 2002) remains uncertain. Despite an extensive series of PCR experiments with a large number of degenerate primer pairs designed to a highly conserved region of the RH2 and SWS2 coding sequences, none resulted in the amplification of either an SWS2 or RH2 opsin sequence. Strachan et al. (2004) similarly reported an inability to amplify any RH2 sequences. It is unlikely therefore that either has been retained in these marsupial species. Another possibility is that the MWS pigment arose from a duplicated LWS opsin gene that either lost its chloride-binding site or accumulated other substitutions that short wavelength shift the λmax to 505–509 nm. However, no evidence for a second LWS gene in the fat-tailed dunnart genome could be found, and despite many amplifications of LWS opsin sequences from retinal cDNA of both species, we have never found evidence for more than one expressed sequence. Finally, the possibility that the MWS pigment in these species is based on a cone opsin gene that has diverged so much that it is no longer possible to amplify it with degenerate primers is inherently unlikely, given the conserved nature of the amino acid sequences of opsins with very diverse spectral maxima across the vertebrate kingdom.
In the absence of a third cone opsin gene, we have to inevitably consider other alternatives, and one possibility that cannot be tested at present is that the MWS pigment is derived from the LWS pigment by post-translational modifications that shift the λmax to shorter wavelengths. It should be noted, however, that such a mechanism has yet to be described, so it remains unlikely. An alternative is that the MWS cones express a rod pigment and it may be significant that we have identified two RH1 rod opsin genes in the fat-tailed dunnart genome. These two genes encode identical pigments but differ in intron sequence. One copy is undoubtedly expressed in rod photoreceptors but the second copy may have acquired new regulatory sequences that target expression to a subset of cone photoreceptors. If so, the pigments in rods and MWS cones would be expected to have very similar λmax values, and this is exactly is what found with the values of 512 and 509 nm, respectively, in the fat-tailed dunnart and 502 and 505 nm, respectively, in the honey possum (Arrese et al. 2002). It would also account for the similar photochemical properties of the rods and MWS cones with a post-bleach build-up of photoproduct that absorbed below 430 nm (Arrese et al. 2002). Unfortunately, in the absence of any differences in the coding exons, it is not possible to confirm that both gene copies are expressed or to demonstrate that mRNA from one copy is present in cones and the other in rods. Such heterologous expression of visual pigments is not, however, unknown. The blue sensitive cones and green rods of the tiger salamander both express the same SWS2 cone pigment (Ma et al. 2001), thereby providing evidence that cone pigments can function with rod transducin, and the disparity in flash sensitivity could be attributed to a higher quantal photon catch by the larger rod outer segments. The converse situation proposed for the marsupial MWS cones expressing a rod pigment may be expected therefore to be fully functional and show a cone-like sensitivity.
Acknowledgments
The study was approved by the Animal Ethics and Experimentation Committee of the University of Western Australia.
This work was supported by a grant (03/100/256) from the Australian Research Council. We are grateful to Dr Rosalie Crouch for the generous gift of 11-cis-retinal, Dr Lynda Erskine for advice on in situ hybridization protocols, Prof. Glen Jeffery for advice and help with the histological analysis, and Ms Alison Oddy for technical assistance.
References
- Arrese C.A, Hart N.S, Thomas N, Beazley L.D, Shand J. Trichromacy in Australian marsupials. Curr. Biol. 2002;12:657–660. doi: 10.1016/s0960-9822(02)00772-8. doi:10.1016/S0960-9822(02)00772-8 [DOI] [PubMed] [Google Scholar]
- Arrese C.A, Beazley L.D, Neumeyer C. Behavioural evidence for marsupial trichromacy. Curr. Biol. 2006;16:R193–R194. doi: 10.1016/j.cub.2006.02.036. doi:10.1016/j.cub.2006.02.036 [DOI] [PubMed] [Google Scholar]
- Asenjo A.B, Rim J, Oprian D.D. Molecular determinants of human red/green color discrimination. Neuron. 1994;12:1131–1138. doi: 10.1016/0896-6273(94)90320-4. doi:10.1016/0896-6273(94)90320-4 [DOI] [PubMed] [Google Scholar]
- Bowmaker J.K, Astell S, Hunt D.M, Mollon J.D. Photosensitive and photostable pigments in the retinae of Old World monkeys. J. Exp. Biol. 1991;156:1–19. doi: 10.1242/jeb.156.1.1. [DOI] [PubMed] [Google Scholar]
- Carvalho L.d.S, Cowing J.A, Wilkie S.E, Bowmaker J.K, Hunt D.M. Shortwave visual sensitivity in tree and flying squirrels reflects changes in lifestyle. Curr. Biol. 2006;16:R81–R83. doi: 10.1016/j.cub.2006.01.045. doi:10.1016/j.cub.2006.01.045 [DOI] [PubMed] [Google Scholar]
- Cowing J.A, Poopalasundaram S, Wilkie S.E, Robinson P.R, Bowmaker J.K, Hunt D.M. The molecular mechanism for the spectral shifts between vertebrate ultraviolet- and violet-sensitive cone visual pigments. Biochem. J. 2002;367:129–135. doi: 10.1042/BJ20020483. doi:10.1042/BJ20020483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies W.L, Carvalho L.S, Cowing J.A, Beazley L.D, Hunt D.M, Arrese C.A. Visual pigments of the platypus: a novel route to mammalian colour vision. Curr. Biol. 2007;17:R161–R163. doi: 10.1016/j.cub.2007.01.037. doi:10.1016/j.cub.2007.01.037 [DOI] [PubMed] [Google Scholar]
- Deeb S.S, Wakefield M.J, Tada T, Marotte L, Yokoyama S, Marshall Graves J.A. The cone visual pigments of an Australian marsupial, the tammar wallaby (Macropus eugenii): sequence, spectral tuning, and evolution. Mol. Biol. Evol. 2003;20:1642–1649. doi: 10.1093/molbev/msg181. doi:10.1093/molbev/msg181 [DOI] [PubMed] [Google Scholar]
- Dominy N.J, Lucas P.W. Ecological importance of trichromatic vision to primates. Nature. 2001;410:363–366. doi: 10.1038/35066567. doi:10.1038/35066567 [DOI] [PubMed] [Google Scholar]
- Dulai K.S, von Dornum M, Mollon J.D, Hunt D.M. The evolution of trichromatic color vision by opsin gene duplication in New World and Old World primates. Genome Res. 1999;9:629–638. [PubMed] [Google Scholar]
- Fasick J.I, Robinson P.R. Mechanism of spectral tuning in the dolphin visual pigments. Biochemistry. 1998;37:433–438. doi: 10.1021/bi972500j. doi:10.1021/bi972500j [DOI] [PubMed] [Google Scholar]
- Fasick J.I, Applebury M.L, Oprian D.D. Spectral tuning in the mammalian short-wavelength sensitive cone pigments. Biochemistry. 2002;41:6860–6865. doi: 10.1021/bi0200413. doi:10.1021/bi0200413 [DOI] [PubMed] [Google Scholar]
- Franke R.R, Sakmar T.P, Oprian D.D, Khorana H.G. A single amino acid substitution in rhodopsin (lysine 248–leucine) prevents activation of transducin. J. Biol. Chem. 1988;263:2119–2122. [PubMed] [Google Scholar]
- Govardovskii V.I, Fyhrquist N, Reuter T, Kuzmin D.G, Donner K. In search of the visual pigment template. Vis. Neurosci. 2000;17:509–528. doi: 10.1017/s0952523800174036. doi:10.1017/S0952523800174036 [DOI] [PubMed] [Google Scholar]
- Higgins D.G, Thompson J.D, Gibson T.J. Using Clustal for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- Hunt D.M, Dulai K.S, Partridge J.C, Cottrill P, Bowmaker J.K. The molecular basis for spectral tuning of rod visual pigments in deep-sea fish. J. Exp. Biol. 2001;204:3333–3344. doi: 10.1242/jeb.204.19.3333. [DOI] [PubMed] [Google Scholar]
- Hunt D.M, et al. The rod opsin pigments from two marsupial species, the South American bare-tailed woolly opossum and the Australian fat-tailed dunnart. Gene. 2003;323:157–162. doi: 10.1016/j.gene.2003.09.016. doi:10.1016/j.gene.2003.09.016 [DOI] [PubMed] [Google Scholar]
- Hunt D.M, Cowing J.A, Wilkie S.E, Parry J, Poopalasundaram S, Bowmaker J.K. Divergent mechanisms for the tuning of shortwave sensitive visual pigments in vertebrates. Photochem. Photobiol. Sci. 2004;3:713–720. doi: 10.1039/b314693f. doi:10.1039/b314693f [DOI] [PubMed] [Google Scholar]
- Hunt D.M, Carvalho L.S, Cowing J.A, Parry J.W.L, Wilkie S.E, Davies W.L, Bowmaker J.K. Spectral tuning of shortwave-sensitive visual pigments in vertebrates. Photochem. Photobiol. 2007;83:303–310. doi: 10.1562/2006-06-27-IR-952. [DOI] [PubMed] [Google Scholar]
- Ibbotson R.E, Hunt D.M, Bowmaker J.K, Mollon J.D. Sequence divergence and copy number of the middle- and long-wave photopigment genes in Old World monkeys. Proc. R. Soc. B. 1992;247:145–154. doi: 10.1098/rspb.1992.0021. doi:10.1098/rspb.1992.0021 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen I.B, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. doi:10.1093/bioinformatics/17.12.1244 [DOI] [PubMed] [Google Scholar]
- Ma J.-X, et al. A visual pigment expressed in both rod and cone photoreceptors. Neuron. 2001;32:451–461. doi: 10.1016/s0896-6273(01)00482-2. doi:10.1016/S0896-6273(01)00482-2 [DOI] [PubMed] [Google Scholar]
- Merbs S.L, Nathans J. Role of hydroxyl-bearing amino acids in differentially tuning the absorption spectra of the human red and green cone pigments. Photochem. Photobiol. 1993;58:706–710. doi: 10.1111/j.1751-1097.1993.tb04956.x. doi:10.1111/j.1751-1097.1993.tb04956.x [DOI] [PubMed] [Google Scholar]
- Molday R.S, MacKenzie D. Monoclonal antibodies to rhodopsin: characterization, cross-reactivity, and application as structural probes. Biochemistry. 1983;22:653–660. doi: 10.1021/bi00272a020. doi:10.1021/bi00272a020 [DOI] [PubMed] [Google Scholar]
- Mollon J.D. “Tho' she kneel'd in that place where they grew.” The uses and origins of primate colour vision. J. Exp. Biol. 1989;146:21–38. doi: 10.1242/jeb.146.1.21. [DOI] [PubMed] [Google Scholar]
- Mollon J.D, Bowmaker J.K, Jacobs G.H. Variations of colour vision in a New World primate can be explained by polymorphism of retinal photopigments. Proc. R. Soc. B. 1984;222:373–399. doi: 10.1098/rspb.1984.0071. doi:10.1098/rspb.1984.0071 [DOI] [PubMed] [Google Scholar]
- Nathans J, Thomas D, Hogness D.S. Molecular genetics of human color vision: the genes encoding blue, green, and red pigments. Science. 1986;232:193–202. doi: 10.1126/science.2937147. doi:10.1126/science.2937147 [DOI] [PubMed] [Google Scholar]
- Neitz M, Neitz J, Jacobs G.H. Spectral tuning of pigments underlying red–green color vision. Science. 1991;252:971–974. doi: 10.1126/science.1903559. doi:10.1126/science.1903559 [DOI] [PubMed] [Google Scholar]
- Osorio D, Vorobyev M. Colour vision as an adaptation to frugivory in primates. Proc. R. Soc. B. 1996;263:593–599. doi: 10.1098/rspb.1996.0089. doi:10.1098/rspb.1996.0089 [DOI] [PubMed] [Google Scholar]
- Regan B.C, Julliot C, Simmen B, Vienot F, Charles-Dominique P, Mollon J.D. Fruits, foliage and the evolution of primate colour vision. Phil. Trans. R. Soc. B. 2001;356:229–283. doi: 10.1098/rstb.2000.0773. doi:10.1098/rstb.2000.0773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shand, J. et al In press. The influence of ontogeny and light environment on the expression of visual pigment opsins in the retina of the black bream, Acanthopagrus butcheri J. Exp. Biol [DOI] [PubMed]
- Strachan J, Chang L.Y, Wakefield M.J, Graves J.A, Deeb S.S. Cone visual pigments of the Australian marsupials, the stripe-faced and fat-tailed dunnarts: sequence and inferred spectral properties. Vis. Neurosci. 2004;21:223–229. doi: 10.1017/s0952523804213281. doi:10.1017/S0952523804213281 [DOI] [PubMed] [Google Scholar]
- Sumner P, Mollon J.D. Catarrhine photopigments are optimized for detecting targets against a foliage background. J. Exp. Biol. 2000;203(Pt 13):1963–1986. doi: 10.1242/jeb.203.13.1963. [DOI] [PubMed] [Google Scholar]
- Sun H, Macke J.P, Nathans J. Mechanisms of spectral tuning in the mouse green cone pigment. Proc. Natl Acad. Sci. USA. 1997;94:8860–8865. doi: 10.1073/pnas.94.16.8860. doi:10.1073/pnas.94.16.8860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A.J, Hunt D.M, Bowmaker J.K, Mollon J.D. The polymorphic photopigments of the marmoset: spectral tuning and genetic basis. EMBO J. 1992;11:2039–2045. doi: 10.1002/j.1460-2075.1992.tb05261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S. Molecular evolution of vertebrate visual pigments. Prog. Retin. Eye Res. 2000;19:385–419. doi: 10.1016/s1350-9462(00)00002-1. doi:10.1016/S1350-9462(00)00002-1 [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Yokoyama S. Convergent evolution of the red- and green-like visual pigment genes in fish, Astyanax fasciatus, and human. Proc. Natl Acad. Sci. USA. 1990;87:9315–9318. doi: 10.1073/pnas.87.23.9315. doi:10.1073/pnas.87.23.9315 [DOI] [PMC free article] [PubMed] [Google Scholar]