Abstract
Assemblage-level phylogenies carry the signature of ecological and evolutionary processes, which may provide useful information on modes of assemblage formation. We present a global-scale analysis of the emergent phylogenetic properties of mammal assemblages on islands, in which we compared the structure of 595 island assemblages with null models constructed under four alternative definitions of regional source pools. Although most assemblages had a structure indistinguishable from random samples, for some mammal taxa, up to 40% of island assemblages were phylogenetically overdispersed. This suggests that in at least some cases, the processes that shape island faunas are not independent of phylogeny. Furthermore, measures of phylogenetic structure were associated in some cases with island geographical features (size, maximum elevation and habitat diversity). Our results suggest that part of the signal of assemblage formation processes is detectable in the phylogenies of contemporary island mammal faunas, though much is obscured by the complexity of these processes.
Keywords: assembly rules, community ecology, competitive exclusion, null models, phylogenetic overdispersion, phylogenetic clustering
1. Introduction
The prediction that the structure of species assemblages should be influenced by species relatedness has been made since the earliest days of ecology. Because closely related species are often ecologically similar, they are expected to compete more strongly for limited resources than more distant relatives, and may therefore be less likely to coexist (Darwin 1859; Elton 1946). This idea was first examined quantitatively through analyses of species–genus ratios (Elton 1946; Grant 1966; Simberloff 1970), and, more recently, phylogenies have been used to test the association between relatedness, ecological similarity and coexistence (Brooks & McLennan 1993; Losos 1996; Losos et al. 2003; Silvertown 2004; Silvertown et al. 2006). A recent extension of this idea has been the analysis of emergent properties of assemblage-level phylogenies, which are predicted to carry the signature of assembly processes. Different assembly processes predict phylogenies that are clustered (species are more closely related than expected by chance), overdispersed (species are less closely related than expected) or randomly dispersed, within the source-pool phylogeny (Haydon et al. 1993; Webb 2000; Webb et al. 2002).
So far, the analysis of phylogenetic emergent properties has been applied mostly at the scale of local communities, where both phylogenetically clustered and overdispersed communities have been found (Webb 2000; Anderson et al. 2004; Cavender-Bares et al. 2004, 2006; Kembel & Hubbell 2006; Slingsby & Verboom 2006; Swenson et al. 2006; Helmus et al. 2007). At local scales, the main predictions are that (i) habitat filtering (where close relatives are brought together by shared habitat preferences) should give rise to phylogenetically clustered assemblages or (ii) competitive exclusion among close relatives should give rise to phylogenetically overdispersed assemblages (Haydon et al. 1993; Webb 2000; Webb et al. 2002). Less is known about phylogenetic structuring of assemblages delimited at larger spatial scales, although there is evidence that scale does influence the patterns observed (Cavender-Bares et al. 2006; Swenson et al. 2006). At larger scales, assemblage structure may be shaped not only by local-scale ecological processes, but also by broader biogeographic and evolutionary mechanisms such as in situ speciation or character displacement (Brown et al. 2000). To what extent are such processes detectable from the phylogenies of large assemblages?
Island systems have long been considered natural testing grounds for investigating the structure and dynamics of species assemblages (Grant 1966; MacArthur & Wilson 1967; Lack 1969; Simberloff 1970; Diamond 1975; Losos 1995), and, in this paper, we present a global analysis of patterns in the phylogenetic structure of island mammal assemblages. We ask two basic questions. First, do island assemblages show non-random phylogenetic structure when compared with null models constructed from regional source pools? Hypotheses for mechanisms that might produce non-random structure on islands include the following.
Competition. There is evidence that, even at large geographical scales, mammal assemblages may be shaped by interspecific competition (Letcher et al. 1994; Brown et al. 2000; Davies et al. 2007). Furthermore, if extinction rates are higher in insular assemblages, species sorting by competitive exclusion may be more pronounced. Hence, if competition is more common among close relatives, this mechanism predicts that island assemblages should often be phylogenetically overdispersed within the regional source pool.
Colonization and endemic radiation. Assemblages on oceanic islands (those never attached to continental land masses) arise largely by colonization and endemic radiation. If successful colonization is influenced by phylogenetically conserved biological traits that affect dispersal ability and suitability to island environments (Ebenhard 1991), both of these mechanisms predict assemblages on oceanic islands that are more phylogenetically clustered than expected from the regional source pool.
Selective extinction. On land-bridge islands (those separated from continental land masses after the last glacial period), assemblages may be shaped largely by extinctions, as isolated faunas ‘relax’ to lower equilibrium species-richness levels (Harcourt & Schwartz 2001). The probability of extinction appears to be related to phylogenetically conserved life-history or ecological traits (Brown & Lomolino 1998; Purvis et al. 2005), so this mechanism predicts an increase in phylogenetic clustering on islands as close relatives sharing detrimental traits become extinct.
Our second question is whether the phylogenetic structure of island mammal assemblages is influenced by the geographical features of islands. Again, there are several alternative hypotheses, which include the following.
Endemic speciation. In situ speciation may be promoted by increased island size (Losos & Schluter 2000) or by greater topographic or habitat diversity (Rosenzweig 1995). Under this mechanism, more phylogenetically clustered assemblages are more likely to be found on larger, more topographically diverse or more habitat-diverse islands.
Coexistence of competitors. Increased island size or greater topographic diversity may also facilitate stable coexistence of close relatives (Grant 1966); hence, the prediction is that phylogenetically overdispersed assemblages are more likely to be found on smaller or flatter islands.
2. Material and methods
(a) Datasets
Islands were defined as all discrete land masses that feature in the vector basemap of ArcGIS v. 9 (ESRI 2002), excluding the six continents and Greenland. For each island, we measured the following features: (i) area, (ii) latitude (degrees from the equator of the island's geographical centroid), (iii) maximum elevation, from the ETOPO5 gridded elevation dataset (NOAA 1988), (iv) number of habitat types, from the global land cover classification (Hansen et al. 1998), and (v) whether ‘land-bridge’ or ‘oceanic’, based on its separation from a continental land mass by below 120 m or above 120 m water depth, respectively (Heaney 1986). To avoid measurement distortions associated with different map projections, island areas were calculated from a Behrmann equal-area projection of the ArcGIS basemap and latitudinal centroids were calculated from an unprojected map with a geographical coordinate system.
Analyses were performed separately for each of six broad taxonomic guilds of mammals (table 1): terrestrial Carnivora; primates; ungulates (Artiodactyla + Perissodactyla, without Cetacea); Rodentia; microbats (Microchiroptera); and megabats (Megachiroptera); other mammal taxa were distributed too narrowly or recorded from too few islands for powerful tests. Although these taxa do not necessarily correspond to natural ecological guilds (Simberloff & Dayan 1991), it was necessary to choose groups of broad ecological equivalence that were large enough for robust global-scale analyses. A species list was obtained for each island by overlaying polygon geographical range maps (Sechrest 2003; Grenyer et al. 2006). The recorded presence of a species on islands in this database was usually based on records from field surveys (Sechrest 2003), so we felt that species lists obtained in this way give a reasonable indication of the sets of species maintaining breeding populations on each island. In total, there were 595 islands in our dataset with at least two species from any taxon; a list of islands and their geographical features is given in the electronic supplementary material (table S1). The global mammal phylogeny was a dated composite tree of 4510 species constructed by combining previously published and new supertrees built by matrix representation with parsimony (Bininda-Emonds et al. 2007). Because many of the node ages in this phylogeny were affected by a software bug, we use a version with corrected node ages.
Table 1.
Summary of data for the six mammal taxa included in this study.
| taxon | species in the database | land-bridge islands with more than one species | oceanic islands with more than one species | mean species per island |
|---|---|---|---|---|
| carnivores | 238 | 206 | 19 | 7.2 |
| ungulates | 238 | 105 | 13 | 3.5 |
| primates | 233 | 62 | 11 | 4.6 |
| rodents | 2015 | 164 | 47 | 7.7 |
| megabats | 166 | 38 | 130 | 5.5 |
| microbats | 759 | 149 | 176 | 13.8 |
(b) Measuring phylogenetic assemblage structure
We used two measures of phylogenetic assemblage structure, the net relatedness index (NRI) and nearest taxon index (NTI) of Webb et al. (2002), which measure the degree of phylogenetic clustering or dispersion relative to a specified source-pool phylogeny. NRI is calculated as
where is the mean phylogenetic distance (measured as branch lengths) between all pairs of species, and and s(Xn) are, respectively, the mean and standard deviation of all pairwise distances for multiple random draws of n species from the source pool. An increase in the NRI value indicates increasing phylogenetic clustering (or decreasing overall relatedness) of a set of species relative to the source pool. For the calculation of NTI, X is replaced by Y, the distance from each species to its nearest relative. NRI therefore reflects patterns of dispersion throughout the phylogeny, while NTI reflects patterns near the tips.
The designation of source pools of potential island inhabitants is one of the most contentious issues in the study of island ecology. Most null-model-based studies of island assemblage formation have considered species inhabiting other islands in the same archipelago, and coastal areas of adjacent continental land masses, as part of the source pool for a given island (e.g. Diamond 1975; Grant & Abbott 1980), but there is little to indicate what the correct size or shape of the ‘catchment area’ should be. We used a definition of source-pool catchment areas based on the general assumption that the potential for a given species to inhabit an island is determined primarily by (i) the distance separating the island from land masses on which the species occurs and (ii) the species' proximity to the coastlines of adjacent land masses. Source pools were thus defined as the set of species occurring within a buffer of specified size drawn around the coast of each island and within 50 km of any coastline. To test the sensitivity of results to the size of the catchment area, analyses were repeated using alternative source pools based on buffer sizes of 500, 1000 and 2000 km. A potential problem in defining the pool for a given island is that assemblages on adjacent islands may be shaped by processes similar to those on the island of interest; if these islands are included as part of the pool, this could make it more difficult to detect non-random structure (Colwell & Winkler 1984). We therefore included an additional set of source pools in our analyses, using a 1000 km buffer and including only species occurring on land masses larger than 50 000 km2. Calculation of source pools and other GIS operations were performed using ArcGIS v. 9.
For each mammal taxon, emergent phylogenetic structure was calculated for the phylogeny specified by the set of species found on each island. NRI and NTI were measured for every island with at least two species, using 1000 sets of species drawn randomly from the island's source pool. We then attached significance values to the phylogenetic structure measures for each island using randomization tests, by sampling the appropriate number of species 1000 times from the island's source pool, and comparing the observed values of NRI and NTI with the distributions of random values. These procedures were performed using functions written by M.C. in R (R Development Core Team 2007).
(c) Associations between phylogenetic assemblage structure and island features
To test associations between phylogenetic assemblage structure and geographical features of islands, we fitted generalized least-squares (GLS) models, with NRI and NTI as response variables and island species richness, area, maximum elevation and habitat diversity as predictors. All predictor variables were log-transformed. To account for spatial autocorrelation in the species composition of islands, we included in the models a spherical spatial correlation structure based on the latitude and longitude values of each island's geographical centroid. The distance over which spatial correlation was measured was estimated by the inspection of variograms of GLS model residuals. We ran the models separately for land-bridge and oceanic islands for each mammal taxon and each source-pool definition. For each model, we simplified the list of predictors to a minimum adequate model by backwards deletion from a full model (Crawley 2002). At each step of the model-fitting procedure, we examined diagnostic plots of fitted values against standardized residuals, and, if necessary, retested models after omitting any extreme outliers on these plots. We checked for collinearity among variables by calculating variance inflation factors (VIFs). Although area, elevation and habitat diversity were often positively correlated, all VIF values were less than 3.01, lower than is normally considered likely to bias regression coefficients. The spatial GLS models were run using the ‘gls’ function in the R library nlme (Pinheiro et al. 2005).
(d) Testing for methodological biases
Two potential sources of bias in our methods are as follows. First, phylogenies of island assemblages may tend to be less well known, and hence more poorly resolved, than those of mainland source pools, which could bias results towards overdispersion on islands (because phylogenetic distances between species are overestimated in a poorly resolved phylogeny). Second, NRI and NTI could be intrinsically biased towards detecting overdispersion, particularly when there are few species on an island and in its source pool (Simberloff 1970). To test for the first potential bias, we used Fisher's exact probability tests to determine whether the likelihood of significant clustering or overdispersion was contingent on whether island phylogenies were more or less well resolved than the phylogenies of their corresponding source pools. Resolution of phylogenies was measured as the number of nodes/(number of tips−1). This test was performed separately for each taxon.
To test for the second potential bias, we carried out the following simulation. We first randomly generated 1000 ten-species ‘source-pool’ phylogenies under a Yule (pure-birth) process. From each of these, we randomly selected a four-species ‘island assemblage’. We calculated NRI for each random island assemblage and tested its significance by resampling another 1000 four-species assemblages from the same source pool. We thus obtained a distribution of 1000 p-values, each representing a test of Ho (random phylogenetic structure) for a randomly selected assemblage. The proportion of these p(NRI) values that rejected Ho at α=0.05 in either tail of the distribution (0.025≥p≥0.975) gave a measure of the type I error rate of NRI and an indication of bias in the direction of clustering or overdispersion. We did not test statistical power (type II error) for NRI because we lack an explicit model to generate expected levels of clustering or overdispersion.
3. Results
(a) Prevalence of islands with significant phylogenetic clustering and overdispersion
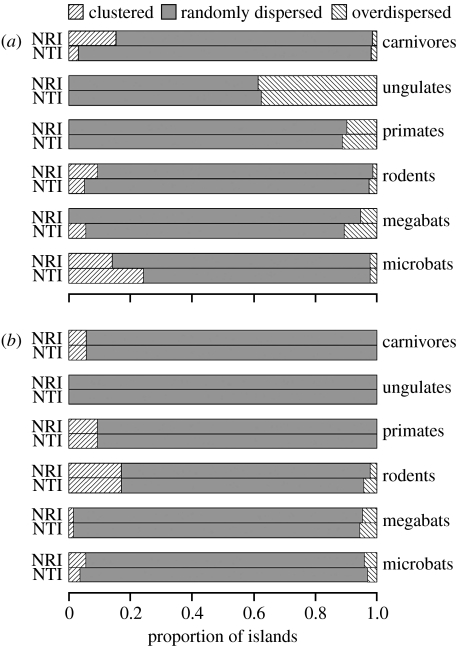
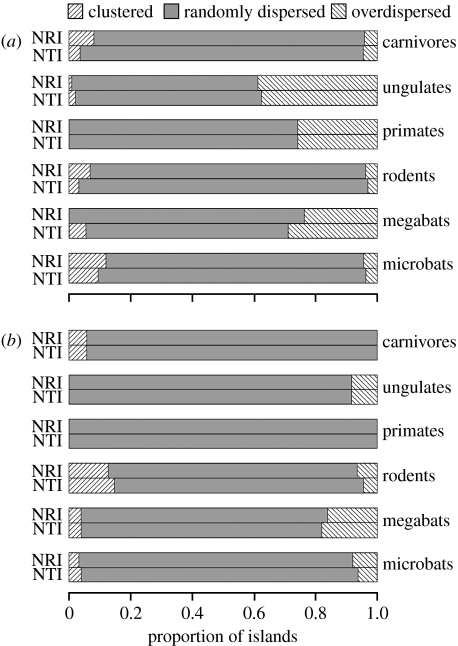
The effect of varying the size of source pools was relatively minor, so here we present only a comparison of results for 1000 km source pools with and without small islands included in the source-pool definition. Full results for all four source-pool types are provided in the electronic supplementary material.
Overall, the phylogenetic structure of assemblages on the majority of islands was not significantly different from sets of species drawn randomly from the surrounding source pools (figures 1 and 2; figures S2–S5 in the electronic supplementary material). Using 1000 km source pools, only ungulates had a substantial proportion of non-random assemblages (approx. 40% of the land-bridge islands were significantly overdispersed; figure 1a). However, when the source-pool definition included only species occurring on land masses larger than 50 000 km2, significant overdispersion became more common, with a relatively high proportion of ungulate, primate and megabat assemblages on land-bridge islands being overdispersed (figure 2a). On the other hand, in carnivores, rodents and microbats, clustering was more common than overdispersion.
Figure 1.
Proportion of islands on which the phylogenetic structure of mammal assemblages showed significant clustering, random dispersion and significant overdispersion, using 1000 km source pools. (a) Land-bridge islands and (b) oceanic islands.
Figure 2.
Proportion of islands on which the phylogenetic structure of mammal assemblages showed significant clustering, random dispersion and significant overdispersion, using 1000 km source pools, excluding islands smaller than 50 000 km2. (a) Land-bridge islands and (b) oceanic islands.
(b) Associations between phylogenetic assemblage structure and island geography
Although island mammal assemblages with non-random phylogenetic structure were in the minority, several island geographical features were nevertheless associated with phylogenetic structure (tables 2 and 3; table S2 in the electronic supplementary material). For land-bridge islands (tables 2a and 3a), the models reveal patterns of increasing phylogenetic dispersion (decreasing NRI and NTI values) on islands with increasing maximum elevation (carnivores, megabats and microbats), islands with decreasing area (megabats) and islands with decreasing habitat diversity (ungulates). On oceanic islands (tables 2b and 3b), only the associations with maximum elevation in carnivores and habitat diversity in ungulates remain significant. Island species richness appears in nearly all of the final models. On land-bridge islands, the pattern is for increasing species richness to be associated with increasing phylogenetic dispersion; on oceanic islands, increasing richness is usually associated with decreasing dispersion.
Table 2.
Associations between island geographical features and the phylogenetic structure (NRI and NTI) of island mammal assemblages. (Coefficients for best-fitting spatial GLS models using 1000 km source pools; see text for details of model-fitting procedures. (a) Land-bridge islands and (b) oceanic islands. *p≤0.001; **p≤0.01; ***p≤0.05; #p≤0.06.)
| group | carnivores | primates | ungulates | rodents | megabats | microbats | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| metric | NRI | NTI | NRI | NTI | NRI | NTI | NRI | NTI | NRI | NTI | NRI | NTI |
| (a) | ||||||||||||
| d.f. | 191 | 191 | 60 | 60 | 98 | 162 | 34 | 35 | 147 | 143 | ||
| intercept | 1.98* | 1.891* | 0.265 | 0.718 | −0.461 | 0.78*** | −4.046*** | −8.839* | 1.377* | 1.189* | ||
| species richness | −0.439* | −0.618* | −0.573*** | −0.937* | −0.21# | −0.618** | −0.322** | −0.222*** | ||||
| area | 0.253** | 0.453* | ||||||||||
| elevation | −0.098* | −0.077* | −0.183** | −0.297** | −0.101* | |||||||
| habitat diversity | 0.3*** | |||||||||||
| (b) | ||||||||||||
| d.f. | 15 | 15 | 9 | 9 | 10 | 10 | 45 | 45 | 127 | 127 | 169 | |
| intercept | 0.63 | 0.799 | −2.316*** | −2.522 | −1.839 | −2.303 | 0.096 | −0.339 | 0.818 | 0.59*** | 0.464# | |
| species richness | 1.558* | 1.431** | 2.207* | 1.397* | 1.225* | 1.986* | 0.74** | 0.722** | −0.501* | −0.385** | −0.177# | |
| area | ||||||||||||
| elevation | −0.224** | −0.27** | ||||||||||
| habitat diversity | ||||||||||||
Table 3.
Associations between island geographical features and the phylogenetic structure (NRI and NTI) of island mammal assemblages. (Coefficients for best-fitting spatial GLS models using 1000 km source pools with islands smaller than 50 000 km2 excluded; see text for details of model-fitting procedures. (a) Land-bridge islands and (b) oceanic islands. *p≤0.001; **p≤0.01; ***p≤0.05; #p≤0.06.)
| group | carnivores | primates | ungulates | rodents | megabats | microbats | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| metric | NRI | NTI | NRI | NTI | NRI | NTI | NRI | NTI | NRI | NTI | NRI | NTI |
| (a) | ||||||||||||
| d.f. | 191 | 191 | 60 | 60 | 98 | 162 | 162 | 35 | 35 | 140 | 136 | |
| intercept | 1.025 | 1.768* | 0.623 | 0.494 | −0.592 | 1.333* | 1.326* | −3.955# | −8.091* | 1.279** | 0.629* | |
| species richness | −0.302*** | −0.479* | −0.507*** | −0.867* | −0.443* | −0.483* | ||||||
| area | 0.194# | 0.403* | ||||||||||
| elevation | −0.046# | −0.058*** | −0.14# | −0.262** | −0.09** | |||||||
| habitat diversity | 0.296*** | −0.625*** | ||||||||||
| (b) | ||||||||||||
| d.f. | 15 | 15 | 9 | 9 | 10 | 10 | ||||||
| intercept | 0.211 | 0.83 | 0.666 | −1.685 | −1.108*** | −2.459 | ||||||
| species richness | 2.188* | 1.421** | −0.402# | 0.613# | 2.348* | |||||||
| area | ||||||||||||
| elevation | −0.24** | −0.261*** | ||||||||||
| habitat diversity | 0.438# | |||||||||||
(c) Tests for methodological bias
Phylogenies of island assemblages did not tend to be less well resolved than those of their source pools. On the contrary, for every taxon, more than half of the island phylogenies were better resolved than their source-pool phylogenies. Furthermore, only in the carnivores did the likelihood of island phylogenies being overdispersed depend significantly on the island phylogeny being less well resolved than the source-pool phylogeny (table 4). In the other taxa, there was no significant association between phylogenetic resolution and non-randomness of phylogenetic assemblage structure.
Table 4.
Results of Fisher's exact probability tests for associations between significance of clustering or dispersion of island assemblages and resolution of island phylogenies relative to source-pool phylogenies. (The p-values are shown for the phylogenetic structure measured with NRI and NTI.)
| mammal group | two-tailed p (NRI) | two-tailed p (NTI) |
|---|---|---|
| carnivores | 0.004 | 0.004 |
| primates | 0.39 | 0.39 |
| ungulates | 1 | 1 |
| rodents | 0.062 | 0.325 |
| microbats | 0.587 | 0.785 |
| megabats | 1 | 1 |
The simulated distributions of p(NRI) values, based on four-species assemblages selected at random from 10-species source pools, showed acceptable type I error rates and no evidence of bias towards clustering or overdispersion. Approximately equal proportions of simulated assemblages rejected Ho at α=0.05 in the direction of clustering (0.019) and overdispersion (0.024). Although we did not test type II error, the non-zero proportions of p(NRI) values do at least indicate that the method has some power to detect non-randomness in a small assemblage.
4. Discussion
Our study represents the first global analysis of the phylogenetic emergent properties of island assemblages. The rationale for our approach is similar to that for studies of species–genus ratios (Elton 1946; Grant 1966; Simberloff 1970): the notion that relatedness among species may give rise to non-random assemblage structure, thereby offering clues to the processes by which assemblages are formed. The phylogenetic approach can be considered a more sophisticated development of species–genus ratio analyses, because phylogenetic branch lengths represent evolutionary relatedness more precisely, and often more accurately, than the sometimes arbitrary groupings of taxonomy. The emergent property approach also has motivations similar to analyses that have examined patterns of island species coexistence and ecological similarity in a phylogenetic context (e.g. Losos 1995; Losos et al. 2003). However, whereas such analyses have usually sought to identify particular ecological traits that play a key role in governing species coexistence, the emergent property approach effectively assumes that phylogeny encapsulates the many dimensions of a species' ecological niche, and is thus a reasonable proxy for the overall ecological similarity of species (Wiens 2004; Wiens & Graham 2005).
In island mammal assemblages, emergent phylogenetic structure is usually indistinguishable from that of regional source pools. This could suggest that the processes of assemblage formation in island mammals are largely independent of phylogeny (e.g. phylogenetically random colonization and extinction, and no interspecific competition). Alternatively, the phylogenetic signal of such processes may have become obscured or weakened. For example, despite general evidence in favour of phylogenetic conservatism in ecological traits (Wiens 2004; Wiens & Graham 2005), it could be the case that divergence in key ecological traits between sympatric close relatives is common. A classic example of this process comes from Caribbean islands, where communities of Anolis lizards show little phylogenetic structure but strong ecological dispersion, suggesting that ecological differentiation following island colonization has allowed close relatives to coexist (Losos et al. 2003). Similar evidence for mammals comes from a recent study of large-scale distribution patterns in carnivores: closely related species that overlap more in geographical distributions also differ more in carnassial tooth length (Davies et al. 2007). This suggests that competition for food resources can lead to ecological divergence among close relatives, allowing them to coexist at large spatial scales. Although Davies et al. (2007) did include a small sample of island carnivores in their analysis, it remains unclear whether a similar mechanism would apply in island mammals. On the one hand, there is some evidence that rates of morphological evolution are accelerated in island mammals (Millien 2006), so it is plausible that ecological divergence may have occurred in the relatively short time periods that many islands have been isolated from continental land masses. On the other hand, the small size of many islands means that extinction rates are elevated, possibly speeding the process of competitive exclusion and the consequent structuring of island assemblages by species sorting. Tests for the influence of ecological lability on phylogenetic assemblage structure would require a large amount of island-specific data on key ecological traits of mammal populations. Unfortunately, sufficient data of this kind are unavailable at present and an investigation of this issue must be left for future studies.
Despite the predominance of randomly structured assemblages, in some taxa, a relatively high proportion of assemblages on land-bridge islands were significantly overdispersed. Furthermore, measures of phylogenetic dispersion (NRI and NTI) were associated in some cases with island elevation, area and habitat diversity. This suggests that there is at least some predictable regularity in island phylogenetic structure, and that, in many cases, processes of island assemblage formation are not independent of phylogeny. One factor complicating the interpretation of these results is that different processes could produce the same pattern. For example, the negative associations between NRI/NTI and elevation could be produced by a tendency for phylogenetically clustered assemblages to be found on flat islands, or for phylogenetically overdispersed assemblages to be found on mountainous islands (or both). In some cases, these two alternatives can be distinguished by the inspection of bivariate scatterplots, although in other cases the trends in the data are not very clear.
Interpreting the results must also be done with reference to the different processes by which land-bridge and oceanic islands are formed, and by which their mammal faunas are likely to have been assembled. On land-bridge islands, we expect many assemblages to represent ‘relaxation’ faunas: relict sets of species isolated by rising sea level at the end of the Pleistocene, and subsequently whittled down by selective or random extinction. In this kind of assemblage, strong competitive pressure between close relatives could accelerate extinctions (thus increasing phylogenetic dispersion) on islands with fewer opportunities for competitors to coexist by spatial or ecological segregation—islands that are flat, small or have low habitat diversity. The patterns for ungulates and megabats may be consistent with this interpretation: in ungulates, phylogenetically overdispersed assemblages are more common on islands with few habitat types, while overdispersed megabat assemblages are more common on the smallest islands. On the other hand, negative associations between NRI/NTI and elevation in carnivores and bats seem to run counter to this explanation. Inspection of scatterplots suggests that these latter patterns are produced mostly by a tendency for increased phylogenetic clustering on the flattest islands; a possible explanation is that close relatives are usually restricted to similar elevational zones.
Assemblages on oceanic islands are likely to be shaped predominantly by colonization and endemic speciation, although extinction may also play an important role. If colonization success is phylogenetically selective, reflecting conserved biological traits (such as wing length or swimming ability) that influence dispersal ability, we would expect to see increased phylogenetic clustering on oceanic islands, irrespective of geography. The data are only partly consistent with this scenario: oceanic islands have a higher proportion of significantly clustered assemblages among the primates and rodents, but not in the other taxa (figure 1). The different slopes of the associations between island species richness and NRI/NTI on land-bridge and oceanic islands may also indicate different assembly processes. In the carnivores, primates and ungulates, the positive slopes on oceanic islands may simply be an artefact of low species richness, but, in rodents (for which species-richness levels are higher), it may indicate a greater contribution of endemic speciation to assemblage structure on oceanic islands. A similar pattern does not occur for the bats: the signal of endemic speciation may be obscured by their greater powers of dispersal.
A final point to consider is that patterns of mammal assemblage structure on some islands may have become obscured in the recent past by human-induced extinctions and introductions. In regions such as the Mediterranean, a long history of human impact has left a strong mark on present-day island mammal assemblages, to the extent that these may represent highly modified faunas that show little influence of natural ecological processes (Blondel & Vigne 1993). Furthermore, in many parts of the world, human impact has been most severe in coastal regions, corresponding to the areas of the source pools we have used in our analyses. In such cases, detailed reconstructions of prehistoric island and coastal mammal faunas may help reveal any distorting effects that humans have had on phylogenetic assemblage structure.
Acknowledgments
We thank Jonathan Davies for his helpful comments on the manuscript. Funding for this work was provided by a NERC research fellowship (NE/C517992/1) to M.C.
Supplementary Material
Phylogenetic structure results for all source pools
List of islands in the database with their geographic features
GLS results for all source pools
References
- Anderson T.M, Lachance M.A, Starmer W.T. The relationship of phylogeny to community structure: the cactus yeast community. Am. Nat. 2004;164:709–721. doi: 10.1086/425372. doi:10.1086/425372 [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds O.R.P, et al. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. doi:10.1038/nature05634 [DOI] [PubMed] [Google Scholar]
- Blondel J, Vigne J.-D. Space, time and man as determinants of diversity of birds and mammals in the Mediterranean region. In: Ricklefs R.E, Schluter D, editors. Species diversity in ecological communities. University of Chicago Press; Chicago, IL: 1993. pp. 135–146. [Google Scholar]
- Brooks D.R, McLennan D.A. Historical ecology: examining phylogenetic components of community evolution. In: Ricklefs R.E, Schluter D, editors. Species diversity in ecological communities. University of Chicago Press; Chicago, IL: 1993. pp. 267–280. [Google Scholar]
- Brown J.H, Lomolino M.V. Sinauer Associates; Sunderland, MA: 1998. Biogeography. [Google Scholar]
- Brown J.H, Fox B.J, Kelt D.A. Assembly rules: desert rodent communities are structured at scales from local to continental. Am. Nat. 2000;156:314–321. doi: 10.1086/303385. doi:10.1086/303385 [DOI] [PubMed] [Google Scholar]
- Cavender-Bares J, Ackerly D.D, Baum D.A, Bazzaz F.A. Phylogenetic overdispersion in Floridian oak communities. Am. Nat. 2004;163:823–843. doi: 10.1086/386375. doi:10.1086/386375 [DOI] [PubMed] [Google Scholar]
- Cavender-Bares J, Keen A, Miles B. Phylogenetic structure of floridian plant communities depends on taxonomic and spatial scale. Ecology. 2006;87:S109–S122. doi: 10.1890/0012-9658(2006)87[109:psofpc]2.0.co;2. doi:10.1890/0012-9658(2006)87[109:PSOFPC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Colwell R.K, Winkler D.W. A null model for null models in biogeography. In: Strong D.R, Simberloff D, Abele L.G, Thistle A.B, editors. Ecological communities: conceptual issues and the evidence. Princeton University Press; Princeton, NJ: 1984. pp. 344–359. [Google Scholar]
- Crawley M.J. Wiley; Chichester, UK: 2002. Statistical computing: an introduction to data analysis using S-plus. [Google Scholar]
- Darwin C. John Murray; London, UK: 1859. On the origin of species. [Google Scholar]
- Davies T.J, Meiri S, Barraclough T.G, Gittleman J.L. Species co-existence and character divergence across carnivores. Ecol. Lett. 2007;10:146–152. doi: 10.1111/j.1461-0248.2006.01005.x. doi:10.1111/j.1461-0248.2006.01005.x [DOI] [PubMed] [Google Scholar]
- Diamond J.M. Assembly of species communities. In: Cody M.L, Diamond J.M, editors. Ecology and evolution of communities. Belknap Press; Cambridge, MA: 1975. pp. 342–444. [Google Scholar]
- Ebenhard T. Colonization in metapopulations—a review of theory and observations. Biol. J. Linn. Soc. 1991;42:105–121. doi:10.1111/j.1095-8312.1991.tb00554.x [Google Scholar]
- Elton C.S. Competition and the structure of animal communities. J. Anim. Ecol. 1946;15:54–68. doi:10.2307/1625 [Google Scholar]
- ESRI 2002 ArcMap version 9. San Diego, CA: ESRI.
- Grant P.R. Ecological compatibility of bird species on islands. Am. Nat. 1966;100:451–462. doi:10.1086/282438 [Google Scholar]
- Grant P.R, Abbott I. Interspecific competition, island biogeography, and null hypotheses. Evolution. 1980;34:332–341. doi: 10.1111/j.1558-5646.1980.tb04822.x. doi:10.2307/2407397 [DOI] [PubMed] [Google Scholar]
- Grenyer R, et al. Global distribution and conservation of rare and threatened vertebrates. Nature. 2006;444:93–96. doi: 10.1038/nature05237. doi:10.1038/nature05237 [DOI] [PubMed] [Google Scholar]
- Hansen M., Defries R. S., Townshend J. R. G. & Sohlberg R. 1998 UMD global land cover classification, 1 km. See http://glcf.umiacs.umd.edu/data/landcover/
- Harcourt A.H, Schwartz M.W. Primate evolution: a biology of holocene extinction and survival on the Southeast Asian Sunda shelf islands. Am. J. Phys. Anthropol. 2001;114:4–17. doi: 10.1002/1096-8644(200101)114:1<4::AID-AJPA1001>3.0.CO;2-6. doi:10.1002/1096-8644(200101)114:1<4::AID-AJPA1001>3.0.CO;2-6 [DOI] [PubMed] [Google Scholar]
- Haydon D, Radtkey R.R, Pianka E.R. Experimental biogeography: interactions between stochastic, historical, and ecological processes in a model archipelago. In: Ricklefs R.E, Schluter D, editors. Species diversity in ecological communities. University of Chicago Press; Chicago, IL: 1993. pp. 267–280. [Google Scholar]
- Heaney L.R. Biogeography of mammals in SE Asia: estimates of rates of colonization, extinction and speciation. Biol. J. Linn. Soc. 1986;28:127–165. doi:10.1111/j.1095-8312.1986.tb01752.x [Google Scholar]
- Helmus M.R, Bland T.J, Williams C.K, Ives A.R. Phylogenetic measures of biodiversity. Am. Nat. 2007;169:E68–E83. doi: 10.1086/511334. doi:10.1086/511334 [DOI] [PubMed] [Google Scholar]
- Kembel S.W, Hubbell S.P. The phylogenetic structure of a neotropical forest tree community. Ecology. 2006;87:S86–S99. doi: 10.1890/0012-9658(2006)87[86:tpsoan]2.0.co;2. doi:10.1890/0012-9658(2006)87[86:TPSOAN]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Lack D. The number of bird species on islands. Bird Study. 1969;16:193–209. [Google Scholar]
- Letcher A.J, Purvis A, Nee S, Harvey P.H. Patterns of overlap in the geographic ranges of Palearctic and British Mammals. J. Anim. Ecol. 1994;63:871–879. doi:10.2307/5264 [Google Scholar]
- Losos J.B. Community evolution in greater antillean Anolis lizards—phylogenetic patterns and experimental tests. Phil. Trans. R. Soc. B. 1995;349:69–75. doi:10.1098/rstb.1995.0092 [Google Scholar]
- Losos J.B. Phylogenetic perspectives on community ecology. Ecology. 1996;77:1344–1354. doi:10.2307/2265532 [Google Scholar]
- Losos J.B, Schluter D. Analysis of an evolutionary species-area relationship. Nature. 2000;408:847–850. doi: 10.1038/35048558. doi:10.1038/35048558 [DOI] [PubMed] [Google Scholar]
- Losos J.B, Leal M, Glor R.E, de Queiroz K, Hertz P.E, Schettino L.R, Lara A.C, Jackman T.R, Larson A. Niche lability in the evolution of a Caribbean lizard community. Nature. 2003;424:542–545. doi: 10.1038/nature01814. doi:10.1038/nature01814 [DOI] [PubMed] [Google Scholar]
- MacArthur R.H, Wilson E.O. Princeton University Press; Princeton, NJ: 1967. The theory of island biogeography. [Google Scholar]
- Millien V. Morphological evolution is accelerated among island mammals. PloS Biol. 2006;4:1863–1868. doi: 10.1371/journal.pbio.0040321. doi:10.1371/journal.pbio.0040321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NOAA N.G.D.C. 1988 Data Announcement 88-MGG-02, Digital relief of the surface of the Earth. See http://www.ngdc.noaa.gov/mgg/global/etopo5.HTML
- Pinheiro, J., Bates, D., DebRoy, S. & Sarkar, D. 2005 nlme: linear and nonlinear mixed effects models. R package version, pp. 31–66. Vienna, Austria: R Development Core Team.
- Purvis A, Cardillo M, Grenyer R, Collen B. Correlates of extinction risk: phylogeny, biology, threat and scale. In: Purvis A, Gittleman J.L, Brooks T, editors. Phylogeny and conservation. Cambridge University Press; Cambridge, UK: 2005. pp. 295–316. [Google Scholar]
- R Development Core Team. R Development Core Team; Vienna, Austria: 2007. R: a language and environment for statistical computing. [Google Scholar]
- Rosenzweig M.L. Cambridge University Press; Cambridge, UK: 1995. Species diversity in space and time. [Google Scholar]
- Sechrest, W. 2003 Global diversity, endemism and conservation of mammals, PhD dissertation. University of Virginia, Charlottesville, Virginia.
- Silvertown J. The ghost of competition past in the phylogeny of island endemic plants. J. Ecol. 2004;92:168–173. doi:10.1111/j.1365-2745.2004.00853.x [Google Scholar]
- Silvertown J, McConway K, Gowing D, Dodd M, Fay M.F, Joseph J.A, Dolphin K. Absence of phylogenetic signal in the niche structure of meadow plant communities. Proc. R. Soc. B. 2006;273:39–44. doi: 10.1098/rspb.2005.3288. doi:10.1098/rspb.2005.3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simberloff D.S. Taxonomic diversity of island biotas. Evolution. 1970;24:23–47. doi: 10.1111/j.1558-5646.1970.tb01738.x. doi:10.2307/2406712 [DOI] [PubMed] [Google Scholar]
- Simberloff D, Dayan T. The guild concept and the structure of ecological communities. Annu. Rev. Ecol. Syst. 1991;22:115–143. doi:10.1146/annurev.es.22.110191.000555 [Google Scholar]
- Slingsby J.A, Verboom G.A. Phylogenetic relatedness limits co-occurrence at fine spatial scales: evidence from the schoenoid sedges (Cyperaceae: Schoeneae) of the Cape Floristic region, South Africa. Am. Nat. 2006;168:14–27. doi: 10.1086/505158. doi:10.1086/505158 [DOI] [PubMed] [Google Scholar]
- Swenson N.G, Enquist B.J, Pither J, Thompson J, Zimmerman J.K. The problem and promise of scale dependency in community phylogenetics. Ecology. 2006;87:2418–2424. doi: 10.1890/0012-9658(2006)87[2418:tpapos]2.0.co;2. doi:10.1890/0012-9658(2006)87[2418:TPAPOS]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Webb C.O. Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. Am. Nat. 2000;156:145–155. doi: 10.1086/303378. doi:10.1086/303378 [DOI] [PubMed] [Google Scholar]
- Webb C.O, Ackerly D.D, McPeek M.A, Donoghue M.J. Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 2002;33:475–505. doi:10.1146/annurev.ecolsys.33.010802.150448 [Google Scholar]
- Wiens J.J. Speciation and ecology revisited: phylogenetic niche conservatism and the origin of species. Evolution. 2004;58:193–197. doi: 10.1111/j.0014-3820.2004.tb01586.x. [DOI] [PubMed] [Google Scholar]
- Wiens J.J, Graham C.H. Niche conservatism: integrating evolution, ecology, and conservation biology. Annu. Rev. Ecol. Evol. Syst. 2005;36:519–539. doi:10.1146/annurev.ecolsys.36.102803.095431 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic structure results for all source pools
List of islands in the database with their geographic features
GLS results for all source pools