Abstract
We investigated sex-specific recombination rates in Hyla arborea, a species with nascent sex chromosomes and male heterogamety. Twenty microsatellites were clustered into six linkage groups, all showing suppressed or very low recombination in males. Seven markers were sex linked, none of them showing any sign of recombination in males (r=0.00 versus 0.43 on average in females). This opposes classical models of sex chromosome evolution, which envision an initially small differential segment that progressively expands as structural changes accumulate on the Y chromosome. For autosomes, maps were more than 14 times longer in females than in males, which seems the highest ratio documented so far in vertebrates. These results support the pleiotropic model of Haldane and Huxley, according to which recombination is reduced in the heterogametic sex by general modifiers that affect recombination on the whole genome.
Keywords: European tree frog, Haldane–Huxley rule, heterochiasmy, linkage map, nascent sex chromosome
1. Introduction
All processes affecting the evolutionary trajectories of populations may show striking differences among sexes. This holds true not only for the demographic processes resulting from the interactions between organisms and their environment (selection, dispersal and drift), but also for those that are considered part of the ‘genetic system’ (sensu Wright 1955), namely mutation and recombination (Hedrick 2007). Although the population-genetics consequences of such differences have been largely investigated, their proximal and ultimate causes are not yet fully understood (Hedrick 2007).
Regarding recombination, an important distinction is to be made between sex chromosomes and autosomes. In the first case, recombination is normally suppressed over part of the chromosome in the heterogametic sex, which allows preserving epistatic interactions with sex-antagonistic effects (Rice 1996). This non-recombining segment may be quite small in young sex chromosomes (e.g. less than 1% of the chromosome in the medaka fish Oryzias latipes, Kondo et al. 2004; 10% in Carica papaya, Liu et al. 2004), but encompass the whole chromosome (except for a tiny pseudo-autosomal region) in highly evolved ones such as those of birds or mammals. Scenarios for the evolution of sex chromosomes envision a progressive extension, and a concomitant differentiation, of this non-recombining region (e.g. Ohno 1967; Rice 1996; Charlesworth & Charlesworth 2000; Ayling & Griffin 2002; Fraser & Heitman 2005; Steinemann & Steinemann 2005).
Autosomes may also display striking sex differences in recombination. Two situations are to be distinguished here, referred to as achiasmy and heterochiasmy. The former refers to the complete suppression of recombination in one sex and has been described for a long time in Drosophila (in which males do not recombine; Morgan 1914) and in the silkworm Bombyx (in which females do not recombine; Tanaka 1914). These early observations suggested a correlation between achiasmy and heterogamety (flies are male heterogametic, while moths are female heterogametic), formalized into the so-called Haldane–Huxley rule. Haldane (1922) and Huxley (1928) further proposed a pleiotropic cause for this correlation, namely that genes causing achiasmy are selected because they prevent the recombination of sex chromosomes in the heterogametic sex. Since then, 29–34 independent origins of achiasmy have been documented (though none in vertebrates), with a clear pattern of correlation with heterogamety (Bell 1982; Burt et al. 1991), hence providing strong support for the Haldane–Huxley rule.
Heterochiasmy, by contrast, refers to a situation in which both sexes recombine, but at different rates, and is widespread throughout the living world. The ratios of female-to-male map length or chiasma numbers normally range 0.5–2 (Burt et al. 1991; Lenormand & Dutheil 2005; Lorch 2005). Higher values have been reported from a few fish species: 2.74 in the zebrafish Danio rerio (Singer et al. 2002), 3.18 in the catfish Ictalurus punctatus (Waldbieser et al. 2001), 3.25 in the rainbow trout Oncorhynchus mykiss (Sakamoto et al. 2000) and up to 8.26 in the Atlantic salmon Salmo salar (Moen et al. 2004). The lowest ratios are also found in fish (0.135 in the Japanese flounder Paralichthys olivaceus: Coimbra et al. 2003).
Also contrasting with achiasmy, the patterns of heterochiasmy do not seem to obey the Haldane–Huxley rule of a correlation with heterogamety. First, many exceptions occur in species with sex chromosomes. Though the ratio of female-to-male recombination rate often exceeds unity in mammals (as expected from the rule), ranging 1.05–1.65 (Archibald et al. 1995; Dietrich et al. 1996; Barendse et al. 1997; Kappes et al. 1997; Mellersh et al. 1997; Broman et al. 1998; Mikawa et al. 1999; Neff et al. 1999; Kong et al. 2002), exceptions occur among eutherians (e.g. 0.83 in sheep; Maddox et al. 2001), and the ratio is consistently below unity in marsupials, which are also male heterogametic (e.g. 0.5 in the short-tailed opossum Monodelphis domestica: Samollow et al. 2004). Ratios also often exceed unity in birds, which are female heterogametic (e.g. Stauss et al. 2003; Hansson et al. 2005). A second important problem is raised by significant differences in chiasma frequency between male and female meiosis in organisms with XO males (e.g. White 1976), no sex chromosomes (hermaphroditic animals or plants, e.g. Pastor & Callan 1952; Ved Brat 1966) or environmental sex determination (e.g. Isberg et al. 2006). Several alternative explanations to heterochiasmy have thus been proposed (see §4).
As for proximate causes, modifier genes with sex-specific expression are required to account for the sex differences in recombination on autosomes. Sex may affect not only the rate of recombination, but also the distribution of chiasmata, which are often localized closer to telomeres in males and to centromeres in females (e.g. Wallace et al. 1997; Broman et al. 1998; Sumida & Nishioka 2000; Kong et al. 2002; Singer et al. 2002; Hultén & Tease 2003). Selection experiments in Drosophila (e.g. Chinicci 1971; Charlesworth & Charlesworth 1985; Brooks & Marks 1986; Korol & Iliadi 1994; Korol et al. 1994; review in Otto & Barton 2001) have shown that modifiers of recombination can evolve rapidly, act in cis- or trans-configuration, have large or small effects and act either locally or genome wide (e.g. Brooks & Marks 1986; Brooks 1988).
In the case of sex chromosomes, by contrast, a central role is often assigned to chromosomal structural changes, such as inversion loops, duplications or accumulation of heterochromatin including repetitive sequences and retrotransposons (Ayling & Griffin 2002; Griffin et al. 2002; Peichel et al. 2004; Schartl 2004; Steinemann & Steinemann 2005). However, modifier genes might also be involved in the early stages of sex chromosome evolution. The 260 kb Y-specific region that has recently evolved in the medaka fish is already enriched in repetitive sequences (Nanda et al. 2002). Recombination is locally suppressed in genotypic (XY) males, but, remarkably, also in sex-reversed phenotypic (XX) males (Matsuda et al. 1998, 1999), hence implying the action of a local modifier with male-specific expression. The accumulation of repetitive sequences might thus be the consequence, rather than the cause, of recombination loss. Theoretical work by Nei (1967, 1968, 1969) indeed showed that a modifier located on any chromosome may quickly reduce recombination between genes situated on the Y chromosome (and involved in sex determination or differentiation), given a mechanism for sex-limited expression. Nei (1969) also suggested that selective pressure for reduced recombination on Y might actually implicate general modifiers controlling recombination over all chromosomes (in support of Haldane and Huxley's pleiotropic explanation for achiasmy).
The European tree frog (Hyla arborea) presents an interesting model in this context, owing to its sex chromosomes in statu nascendi (Berset-Brändli et al. 2006, 2007). Three sex-linked microsatellite markers showed the absence of recombination in males. Despite this lack of recombination, the Y chromosome displays neither morphological differentiation (Anderson 1991) nor evidence of genetic decay (the same primers amplify on both the X and Y chromosomes for all three markers), pointing to a very recent origin.
The first aim of this study was to investigate the patterns of recombination on additional sex-linked markers, in order to gain information on the evolution of sex chromosomes. If recombination is first suppressed in the immediate vicinity of the sex-determining genes, as assumed by classical models, then it should still occur on the major part of the sex chromosome (as it does in several incipient sex chromosomes; Naruse et al. 2000; Seefelder et al. 2000; Jamsari et al. 2004; Liu et al. 2004; Ma et al. 2004; Peichel et al. 2004). We thus expect a majority of markers on the sex chromosome to recombine somewhat in males, being located outside the differential segment.
The second aim was to extend the analysis to autosomal linkage groups, in order to document the patterns and intensity of heterochiasmy in this species, and thereby provide further material to investigate the several hypotheses regarding the evolution of heterochiasmy.
2. Material and methods
(a) Animals
The resource pedigree used in this study consisted of 478 individuals from 27 known family groups including the father, the mother and 2–22 (average 15.7) offspring per breeding pair. The adults were captured in amplexus from two ponds (Lavigny and Camp Romain) close to Lausanne University (Western Switzerland) during the 2006 breeding season and brought back to the laboratory. After egg laying, individuals were sampled for buccal cells (two sterile buccal swabs per individual; Pidancier et al. 2003; Broquet et al. 2007) and then released at the place of capture. Tadpoles were reared in the laboratory until the tail had grown enough for the tip of the veil (ca 2 mm2) to be sampled without damage. Animals were kept a few more days in the laboratory until the velum regenerated and then they were released at the same place their parents had been caught. Tail veils and buccal swabs were stored dry at −80°C before analysis.
DNA was extracted using a QIAgen DNeasy Tissue Kit (QIAgen), following the manufacturer's protocol, with a few additional steps for the buccal swabs: samples were incubated overnight at 56°C in proteinase K, and after incubation, a QIA Shredder was used according to the manufacturer's conditions. DNA was eluted in a 200 μl volume (QIAgen Buffer AE) and stored at −18°C.
(b) Microsatellite markers
Analyzes were based on 26 specific markers, among which 7 (Ha 1-60, Ha 5-201, Ha 5-22, Ha 1-67, Ha 1-20, Ha 1-25 and Ha 1-103) were developed by Arens et al. (2000), 11 (Ha A-11, Ha A-119, Ha A-127, Ha A-130, Ha B-5R3, Ha B-12, Ha D-3R3, Ha D-115, Ha E-2, Ha H-107 and Ha H-108) by Berset-Brändli et al. (in press) and 8 developed anew for the present study (table 1). Genetic libraries were built by Genetic Identification Services (GIS, http://www.genetic-id-services.com; Chatsworth, CA, USA) using a pooled sample containing 100 μg of genomic DNA extracted from 10 tadpoles collected in Western Switzerland, and enriched for CA, GA, CAG, AAC, AAT, TAGA, CATC and TACA following the method of Jones et al. (2002). Primers were designed using DesignerPCR v. 1.03 (Research Genetics, Inc.).
Table 1.
Characterization of eight polymorphic microsatellite loci for the European tree frog H. arborea. (Reported are locus name; repeat motif, sequences for forward (F) and reverse (R) primers; optimal annealing temperature, T (°C); allele size range in bases pairs; number of alleles (Na) excluding null alleles; observed (Ho) and expected (He) heterozygosity calculated on adults; and GenBank accession number.)
| locus | motif | primers | T (°C) | allele sizes | Na | Ho | He | accession no. |
|---|---|---|---|---|---|---|---|---|
| Ha A-103 | (GT)17 | F: HEX-GCCTAGAAATGTGCAGTGATC | 60 | 235 | 1 | na | na | EU525921 |
| R-CAATTCACACCCAAATCAGAT | ||||||||
| Ha A-110 | (CA)15 | F: NED-AAGGGTTAAATCACCTATCC | 56 | 183–189 | 3 | 0.39 | 0.47 | EU525922 |
| R-ACGCAAAAAACATCTGTG | ||||||||
| Ha A-136 | (CA)15 | F: HEX-CCACTGTAAGTAAAATGTGTGC | 56 | 155–165 | 5 | 0.36 | 0.53 | EU525923 |
| R-TAAAATCCACCAAGAAACCTAC | ||||||||
| Ha A-139 | (GT)21 | F: HEX-GTTTCCAGATAGCGAAAACTG | 58 | 272–280 | 4 | 0.21 | 0.30 | EU525924 |
| R-CACTGCTCCCAGTATCAGAA | ||||||||
| Ha D-104 | (TAGA)7 | F: FAM-GCTGGCTGACTTATTCTTTG | 58 | 256–268 | 4 | 0.65 | 0.70 | EU525925 |
| R-TCTTCTCTCCACGGTCTTC | ||||||||
| Ha D-106 | (TAGA)9 | F: FAM-CACCATAGCTGTATAGCTCTCC | 60 | 231–247 | 5 | 0.56 | 0.74 | EU525926 |
| R-CAAAGATTAAGGCTGTGTTCA | ||||||||
| Ha D-110 | (TAGA)42 | F: HEX-AACTGCATGTTCATGTTTCAC | 60 | 334–360 | 6 | 0.24 | 0.47 | EU525927 |
| R-CCTGACTTCTTAAATGTGCTTT | ||||||||
| Ha H-116 | (TACA)17 | F: NED-AATGGGGGTGAGTAAGGGTTA | 58 | 223–235 | 4 | 0.17 | 0.16 | EU525928 |
| R-CAGGTCCTGACACTGTGACAC |
(c) Microsatellite amplification
The microsatellites developed in this study and those from Berset-Brändli et al. (in press) were amplified in 20 μl reaction volumes each containing 0.2 mM dNTP, 0.5 μM of each primer, 1× QIAgen PCR Buffer (with MgCl2 15 mM), 0.2 mM MgCl2 (0.5 mM MgCl2 for Ha A-139 and Ha H-116, no MgCl2 for Ha H-108, Ha E-2, Ha D-115, Ha A-136, Ha A-110, Ha D-104, Ha D-106 and Ha D-110), 1U QIAgen Taq and 1 or 2 μl of extracted DNA depending on amplified loci. The PCR programmes were performed on GeneAmp PCR Systems 2700 and 9700 (Perkin Elmer, Norwalk, CT), according to the following thermal profiles: initial denaturation at 94°C for 5 min, followed by 40 cycles at 94°C for 45 s (45 cycles for Ha A-139 and Ha H-116), annealing at optimal primer temperature (table 1) for 45 s, elongation at 72°C for 1 min and a final elongation step at 72°C for 5 min. For the microsatellites developed by Arens et al. (2000), each 25 μl amplification volume contained, depending on the amplified loci, between 1.5 and 3.0 μl extraction product, 0.25 mM dNTP, 0.3 μM of each primer, 2× QIAgen PCR Buffer (with MgCl2 15 mM), 0.25 MgCl2 for Wha 5-201 and Wha 1-60 and 0.5 mM for Wha 1-103, and 0.625 U QIAgen Taq (0.75 U for Wha 1-67 and Wha 5-201). PCR was performed on GeneAmp PCR Systems 2700 and 9700 (Perkin Elmer), according to the following thermal profiles: initial denaturation at 94°C for 5 min, followed by 45 cycles at 94°C for 45 s, annealing at optimal primer temperature (between 55 and 61°C) for 45 s, elongation at 72°C for 90 s, and a final elongation step at 72°C for 5 min. The templates were run on an ABI Prism 3100 (Applied Biosystems) automated DNA sequencer. Alleles were scored with GeneMapper v. 3.7 (Applied Biosystems).
(d) Genetic analyses
Analyses of genetic diversity were carried out with Fstat v. 2.9.3.2 (Goudet 2001). Null alleles were easily identified from our pedigrees and assigned identification numbers for linkage analyses. These were performed using Crimap v. 5.0 (Green et al. 1990). Pairwise linkage analysis identified linkages between any two loci, assuming both equal and unequal recombination rates in the two sexes. Pairwise linkage was considered significant if the logarithm of odds (LOD) score was greater than 3. The ALL option was used to select, for linkage groups with three or more linked loci, the order with the highest log likelihood. This likelihood was further compared with that of alternative orders by pairwise flipping of neighbouring loci (FLIPS option), while the BUILD option (multipoint analysis) was used to estimate the recombination rates and Kosambi mapping distances for the sex-specific maps. Linkage maps were drawn using MapChart v. 2.1 (Voorrips 2002). The significance of the sex average versus sex-specific linkage groups was tested following Ott (1991, p. 196). The correlation between male and female map segment lengths was calculated in R (R development Core Team 2006).
3. Results
Repeat motives, primer sequences, annealing temperatures, allele numbers (excluding null alleles), heterozygosities and accession numbers are provided in table 1 for the eight microsatellites developed anew for the present study.
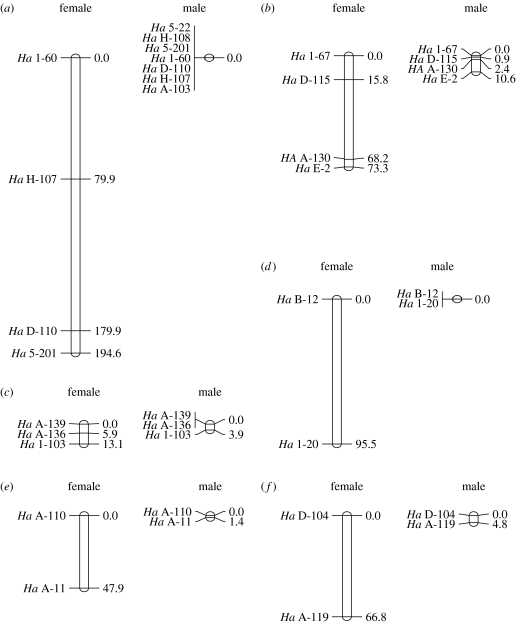
Among the 26 microsatellite markers used, 20 turned out to belong to six linkage groups. The first group (LG1) was sex linked and comprised seven markers, including the three ones (Ha 5-22, Ha 1-60 and Ha 5-201) already identified in Berset-Brändli et al. (2007), two markers (Ha H-107 and Ha H-108) from the set developed by Berset-Brändli et al. (in press) and two from the new set (Ha D-110 and Ha A-103). Ha D-110 had a null allele fixed on Y, so that all males displayed one allele only, inherited from their mother. Conversely, Ha A-103 had a null allele fixed on X, so that it did not amplify in females, and males displayed one allele only, inherited from their father. Overall, allele frequencies differed markedly between the X and the Y chromosomes (table 2), suggesting the absence of exchange. Pairwise recombination rates in this group (table 3a) were indeed always zero in males, and varied from 0.14 to 0.5 in females (average r=0.43) for the four markers showing polymorphism on the X. Accordingly, map length was 0.0 cM in males and greater than 194.6 cM in females (figure 1a).
Table 2.
Allele frequencies on X and Y chromosomes for the seven sex-linked markers used in this study.
| locus | allele | X | Y | locus | allele | X | Y | locus | allele | X | Y |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ha 1-60 | 116 | 0.15 | 0 | Ha D-110 | null | 0 | 1 | Ha 5-201 | 230 | 0.05 | 0 |
| 120 | 0.31 | 0 | 334 | 0.07 | 0 | 233 | 0.02 | 1 | |||
| 122 | 0.40 | 0 | 338 | 0.70 | 0 | 236 | 0.22 | 0 | |||
| 124 | 0.10 | 0 | 342 | 0.15 | 0 | 242 | 0.70 | 0 | |||
| 126 | 0.05 | 0 | 345 | 0.01 | 0 | Ha H-107 | 267 | 0.69 | 0.96 | ||
| 130 | 0 | 0.15 | 353 | 0.05 | 0 | 269 | 0.31 | 0.04 | |||
| 137 | 0 | 0.48 | 360 | 0.01 | 0 | Ha A-103 | null | 1 | 0 | ||
| 139 | 0 | 0.33 | Ha H-108 | 250 | 1 | 0 | 253 | 0 | 1 | ||
| 141 | 0 | 0.04 | 252 | 0 | 1 | Ha 5-22 | 230 | 1 | 0 | ||
| 236 | 0 | 1 |
Table 3.
The six linkage groups identified from the 20 H. arborea microsatellites. (The recombination fractions (r) are presented separately for males (above diagonal) and females (below diagonal). Female recombination rate could not be calculated (na) for comparisons involving sex-linked markers fixed on the X chromosome (Ha A-103, Ha 5-22 and Ha H-108).)
| (a) LG1 | A-103 | 5-22 | H-108 | 1-60 | H-107 | D-110 | 5-201 |
|---|---|---|---|---|---|---|---|
| A-103 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 5-22 | na | 0 | 0 | 0 | 0 | 0 | |
| H-108 | na | na | 0 | 0 | 0 | 0 | |
| 1-60 | na | na | na | 0 | 0 | 0 | |
| H-107 | na | na | na | 0.47 | 0 | 0 | |
| D-110 | na | na | na | 0.50 | 0.45 | 0 | |
| 5-201 | na | na | na | 0.50 | 0.50 | 0.14 |
| (b) LG2 | 1-67 | D-115 | A-130 | E-2 | |||
|---|---|---|---|---|---|---|---|
| 1-67 | 0.01 | 0.01 | 0.14 | ||||
| D-115 | 0.15 | 0.02 | 0.09 | ||||
| A-130 | 0.41 | 0.38 | 0.08 | ||||
| E-2 | 0.31 | 0.21 | 0.05 | ||||
| (c) LG3 | A-139 | A-136 | 1-103 | |||||
|---|---|---|---|---|---|---|---|---|
| A-139 | 0 | 0.03 | ||||||
| A-136 | 0.08 | 0.03 | ||||||
| 1-103 | 0.05 | 0.08 | ||||||
| (d) LG4 | 1-20 | B-12 | |||||
|---|---|---|---|---|---|---|---|
| 1-20 | 0.00 | ||||||
| H B-12 | 0.49 | ||||||
| (e) LG5 | A-11 | A-110 | |||||
|---|---|---|---|---|---|---|---|
| A11 | 0.01 | ||||||
| A-110 | 0.37 | ||||||
| (f) LG6 | A-119 | D-104 | |||||
|---|---|---|---|---|---|---|---|
| A119 | 0.05 | ||||||
| D-104 | 0.44 | ||||||
Figure 1.
Female- and male-specific linkage-group maps for H. arborea: (a) LG1, (b) LG2, (c) LG3, (d) LG4, (e) LG5 and (f) LG6. Provided is the order with the highest log likelihood (Crimap v. 5.0, option ALL). Locus positions are indicated by their distance in centimorgans from the ‘top’ of the linkage group.
The five other groups were not sex linked and comprised four (LG2), three (LG3) and two (LG4 to LG6) markers (table 3b–f). Finally, six loci were not linked to any others (Ha D-106, Ha H-116, Ha A-127, Ha 1-25, Ha B-5 and Ha D-3). Null alleles were present at low frequencies (3–7%) at loci Ha 1-67 (LG 2), Ha A-136 and Ha A-139 (LG3), Ha A-119 (LG5) as well as Ha D-106 and Ha D-3 (both unlinked). Pairwise recombination rates within linkage groups ranged 0.00–0.14 in males and 0.05–0.49 in females. Average rates per linkage group were always lower in males (0.00–0.06) than in females (0.07–0.49). The grand mean over groups was 0.028 in males versus 0.324 in females, providing a ratio of autosomal recombination rate of 11.6. Accordingly, map lengths were always much shorter in males (range 0.0–10.6 cM) than in females (range 13.1–95.5 cM; figure 1b–f).
For all the six linkage groups, sex-specific models of recombination fitted the data significantly better than a model assuming equal recombination in males and females (p<0.0001 in all the cases; Ott 1991). The loci order that was assigned the highest log likelihood by the ALL option (figure 1) was confirmed by the FLIPS option, but alternative orders had very similar likelihoods when pairs of neighbouring loci were very close (LG1 to 3). Summing up over the six linkage groups, the total map length was 24 times larger in females (greater than 491.2 cM) than in males (20.7 cM). Over the five autosomal linkage groups (LG2 to LG6), map length was 14.33 times larger in females (296.6 cM) than in males (20.7 cM), which appears to be the highest ratio reported up to now, notwithstanding achiasmate species. The correlation between male and female map segment lengths over the 11 pairwise neighbouring loci available for both sexes was slightly negative, but not significantly so (r2=0.14, p=0.25).
4. Discussion
Twenty of the 26 markers investigated here clustered into six linkage groups, possibly corresponding to six chromosomes out of the 12 characterizing H. arborea (Anderson 1991). The six remaining markers displayed no significant linkage disequilibrium and might have been distributed among the six remaining autosomes. However, we obviously cannot exclude that several linkage groups map to the same chromosome.
All seven sex-linked markers were non-recombining in males. Thus, the Y-specific region is unlikely to be restricted to a small segment around the sex-determining gene, and more probably encompasses the largest part of the sex chromosome. This result points to a scenario that differs markedly from classical models, which envision an initially small non-recombining segment that progressively expands along the Y chromosome, driven by structural changes. Our findings are actually more in line with Nei's (1969) assumption of general modifiers with sex-specific expression, which may repress recombination at once over whole chromosomes (or even the whole genome).
A second surprising result was the extreme heterochiasmy. Recombination was strongly female biased, and very low in males (or even suppressed for LG4) on all five presumably autosomal linkage groups. The ratio of female-to-male map length (14.33) seems to be the highest value documented so far in vertebrates. The direction of the bias is in line with the Haldane–Huxley rule, since male is the heterogametic sex. As already mentioned, however, this rule suffers many exceptions. A comparative approach by Burt et al. (1991) of 54 species of plants and animals failed to show any significant effect of gamety on heterochiasmy. Lenormand & Dutheil (2005) confirmed these negative results with an updated set and also showed heterochiasmy to be a fast-evolving trait, so that phylogenetic inertia is unlikely to play a significant role in the patterns observed. Alternative interpretations have therefore been proposed, stressing a role for mechanistic effects (Berstein et al. 1988; Plomion & Omalley 1996), sexual selection (Trivers 1988), imprinting or haploid selection (Lenormand 2003) and neutral drift (Burt et al. 1991).
The mechanistic hypothesis assigns a role to sex differences in physiological or molecular processes, such as higher metabolic rate or oxidative stress in females, requiring higher rate of recombinational repair. This hypothesis certainly cannot be generalized, e.g. to hermaphroditic species in which male and female meioses occur simultaneously. In our specific case, breeding also imposes a tremendous physiological stress on males, whose metabolism exceeds its basal rate by 40 times during calling activity (Grafe & Thein 2001).
Trivers (1988) argued that, since recombination dissipates the epistatic component of fitness traits (which can be high; Merilä & Sheldon 1999), it should be more costly to the sex under stronger selection. Owing to their lek-breeding habits (Friedl & Klump 2005), male European tree frogs are certainly under strong sexual selection. However, Burt et al. (1991) rejected Trivers' hypothesis based on their comparative review of animal and plant data. Moreover, Lenormand (2003) showed that heterochiasmy cannot result from selection on diploids, unless some very specific mechanisms are invoked, namely different cis–trans effects on epistasis or sex-of-origin effects (i.e. imprinting).
Differently imprinted regions in the human genome seem to show sex differences in recombination rate (Lercher & Hurst 2003), but differences are slight, and gene imprinting remains rare and limited in vertebrates (ca 0.1% of genes in mammals; Burns et al. 2001). Genomic-wide imprinting has been suggested in the hybridogenetic Rana esculenta (Tunner 2000), but evidence is controversial at best (Vorburger 2001).
Lenormand (2003) also showed a potential role for haploid selection (more precisely, sex differences in the epistatic component of haploid fitness), supported by empirical evidence from plants (in which heterochiasmy seems to correlate with the potential for haploid selection), but not from animals (Lenormand & Dutheil 2005). The haploid phase is indeed much reduced in animals compared with that in plants (Christians et al. 1999; Xu et al. 1999), but also shows sex differences. Female meiosis is achieved at fertilization, leaving no scope for haploid gene expression. By contrast, some genes are expressed in spermatids, involved, for example, in meiotic drive or sperm competition (Joseph & Kirkpatrick 2004). Such genes might contribute to the observed bias towards lower male recombination in animals (Joseph & Kirkpatrick 2004), assuming that they lie on the same chromosome and interact epistatically. There is no evidence, however, for a particularly developed haploid phase in H. arborea, or for a particularly intense sperm competition (Friedl & Klump 2005).
Burt et al. (1991) proposed a role for neutral drift because no general explanation emerged from their comparative analysis. More specifically, selection would still determine the specific average rate of recombination, but sex differences would remain more or less neutral. Supporting this idea, the X chromosome seemed to recombine more than autosomes in female mice (Jagiello & Fang 1987), possibly compensating for the absence of recombination in males (but see Shifman et al. 2006). In line with this interpretation, the average recombination rate is particularly high in female H. arborea (r=0.35), and especially so (r=0.46) for the two linkage groups, LG1 and LG4, which did not recombine in males (for comparison, average values were 0.101 in females and 0.052 in males of Acrocephalus arundinaceus, a species considered to display substantial heterochiasmy; Hansson et al. 2005). However, the correlation between male and female map lengths, though negative, was not significant, providing no statistical support for a within-genome compensation mechanism.
There is clearly no single general interpretation to heterochiasmy, and the factors involved are probably species specific. As for H. arborea, the association of extreme heterochiasmy with nascent sex chromosomes seems to support a role for general modifiers in the initial steps of the building of sex chromosomes, in line with the pleiotropic model of Haldane (1922), Huxley (1928) and Nei (1969). This might account for the two striking patterns documented here, namely (i) suppression of male recombination on all seven sex-linked markers investigated, and (ii) strongly depressed male recombination over the whole genome. Up to now, strong heterochiasmy has been mostly documented in fish, a group also known for generalized absence of heteromorphic sex chromosomes and high lability of sex determination mechanisms (e.g. Devlin & Nagahama 2002; Volff et al. 2007).
According to a possible scenario, the recent apparition of a master sex-determining gene on Hyla's proto-Y chromosome could have induced selective pressures on general modifiers, resulting in the general regression or loss of recombination in the heterogametic sex (and possibly a concomitant increase in the homogametic sex as compensation). An alternative scenario would be that non-recombination could have in fact preceded the origin of a new master sex determination gene. Heterogamety was simply more likely to appear in the sex that initially had no recombination, benefiting from epistatic interactions right from the beginning. One way to decide among these alternatives would be to identify and investigate in Hyla phylogeny the closest sister species still possessing the ancestral sex determination mechanism. A much lower heterochiasmy is expected in such species if the evolution of general modifiers follows (rather than precedes) the change in sex determination mechanisms. This comparison might also allow testing whether heterochiasmy in H. arborea results only from a decreased recombination in males, or also from a concomitant increase in females, as predicted by the compensation theory.
Acknowledgments
C. Aguilar, G. Emaresi, J. Guélat, E. Rattey and J. Yearsley provided much help for the field work, and C. Berney, N. Duvoisin and N. DiMarco for the laboratory work. The presentation of our results benefited from discussions with J. Goudet, L. J. Lawson Handley, L. Keller, G. Kerth, M. Stöck and J. Yearsley. B. Charlesworth, T. Lenormand and P. D. Lorch kindly answered our e-mail questions regarding the evolutionary models of heterochiasmy. This study was supported by Swiss National Fund for Scientific Research (grant 3100A0-108100 to NP).
References
- Anderson K. Chromosome evolution in Holarctic Hyla treefrogs. In: Green D.M, Sessions S.K, editors. Amphibian cytogenetics and evolution. Academic Press; San Diego, CA: 1991. pp. 299–312. [Google Scholar]
- Archibald A.L, et al. The pigmap consortium linkage map of the pig (Sus scrofa) Mamm. Genome. 1995;6:157–175. doi: 10.1007/BF00293008. doi:10.1007/BF00293008 [DOI] [PubMed] [Google Scholar]
- Arens P, Van't Westende W, Butger R, Smulders M.J.M, Vosman B. Microsatellite markers for the European tree frog Hyla arborea. Mol. Ecol. 2000;9:1919–1952. doi: 10.1046/j.1365-294x.2000.01095-15.x. doi:10.1046/j.1365-294x.2000.01095-15.x [DOI] [PubMed] [Google Scholar]
- Ayling L.J, Griffin D.K. The evolution of sex chromosomes. Cytogenet. Genome Res. 2002;99:125–140. doi: 10.1159/000071584. doi:10.1159/000071584 [DOI] [PubMed] [Google Scholar]
- Barendse W, et al. A medium-density genetic linkage map of the bovine genome. Mamm. Genome. 1997;8:21–28. doi: 10.1007/s003359900340. doi:10.1007/s003359900340 [DOI] [PubMed] [Google Scholar]
- Bell G. The evolution and genetics of sexuality. Croom Helm; London, UK; Canberra, Australia: 1982. The masterpiece of nature. [Google Scholar]
- Berset-Brändli L, Jaquiéry J, Dubey S, Perrin N. A sex-specific marker reveals male heterogamety in European tree frogs. Mol. Biol. Evol. 2006;23:1104–1106. doi: 10.1093/molbev/msk011. doi:10.1093/molbev/msk011 [DOI] [PubMed] [Google Scholar]
- Berset-Brändli L, Jaquiéry J, Perrin N. Recombination is suppressed and variability reduced in a nascent Y chromosome. J. Evol. Biol. 2007;20:1182–1188. doi: 10.1111/j.1420-9101.2006.01278.x. doi:10.1111/j.1420-9101.2006.01278.x [DOI] [PubMed] [Google Scholar]
- Berset-Brändli, L., Jaquiéry, J., Broquet, T. & Perrin, N. In press. Isolation and characterization of microsatellite loci for the European tree frog (Hyla arborea). Mol. Ecol. Resources [DOI] [PubMed]
- Berstein H, Hopf F, Michod R. Is meiotic recombination an adaptation for repairing DNA, producing genetic variation, or both? In: Michod R, Levin B, editors. The evolution of sex. Sinauer; Sunderland, MA: 1988. pp. 139–160. [Google Scholar]
- Broman K.W, Murray J.C, Sheffield V.C, White R.L, Weber J.L. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am. J. Hum. Genet. 1998;63:861–869. doi: 10.1086/302011. doi:10.1086/302011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks L.D. The evolution of recombination rates. In: Michod R.E, Levin B.R, editors. The evolution of sex: an examination of current ideas. Sinauer; Sunderland, MA: 1988. pp. 87–105. [Google Scholar]
- Brooks L.D, Marks R.W. The organization of genetic variation for recombination in Drosophila melanogaster. Genetics. 1986;114:525–547. doi: 10.1093/genetics/114.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broquet T, Berset-Brändli L, Emaresi G, Fumagalli L. Buccal swabs allow efficient and reliable microsatellite genotyping in amphibians. Conserv. Genet. 2007;8:509–511. doi:10.1007/s10592-006-9180-3 [Google Scholar]
- Burns J.L, Jackson D.A, Hassan A.B. A view through the clouds of imprinting. FASEB J. 2001;15:1694–1703. doi: 10.1096/fj.010085rev. doi:10.1096/fj.010085rev [DOI] [PubMed] [Google Scholar]
- Burt A, Graham B, Harvey P.H. Sex differences in recombination. J. Evol. Biol. 1991;4:259–277. doi:10.1046/j.1420-9101.1991.4020259.x [Google Scholar]
- Charlesworth B, Charlesworth D. Genetic variation in recombination in Drosophila. I. Response to selection and preliminary genetic analysis. Heredity. 1985;54:71–83. doi:10.1038/hdy.1985.10 [Google Scholar]
- Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Phil. Trans. R. Soc. B. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. doi:10.1098/rstb.2000.0717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinicci J.P. Modification of recombination frequency in Drosophila. I. Selection for increased and decreased recombination. Genetics. 1971;69:71–83. doi: 10.1093/genetics/69.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians E, Boiani M, Garagna S, Dessy C, Redi C.A, Renard J.P, Zuccotti M. Gene expression and chromatin organization during mouse oocyte growth. Dev. Biol. 1999;207:76–85. doi: 10.1006/dbio.1998.9157. doi:10.1006/dbio.1998.9157 [DOI] [PubMed] [Google Scholar]
- Coimbra M.R.M, Kobayashi K, Koretsugu S, Hasegawa O, Ohara E, Ozaki A. A genetic linkage map of the Japanese flounder, Paralichtys olivaceus. Aquaculture. 2003;220:203–218. doi:10.1016/S0044-8486(02)00353-8 [Google Scholar]
- Devlin R.H, Nagahama Y. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture. 2002;208:191–364. doi:10.1016/S0044-8486(02)00057-1 [Google Scholar]
- Dietrich W.F, et al. A comprehensive genetic map of the mouse genome. Nature. 1996;380:149–152. doi: 10.1038/380149a0. doi:10.1038/380149a0 [DOI] [PubMed] [Google Scholar]
- Fraser J.A, Heitman J. Chromosomal sex-determining regions in animals, plants and fungi. Curr. Opin. Genet. Dev. 2005;15:645–651. doi: 10.1016/j.gde.2005.09.002. doi:10.1016/j.gde.2005.09.002 [DOI] [PubMed] [Google Scholar]
- Friedl T.W.P, Klump G.M. Sexual selection in the lek-breeding European treefrog: body size, chorus attendance, random mating and good genes. Anim. Behav. 2005;70:1141–1154. doi:10.1016/j.anbehav.2005.01.017 [Google Scholar]
- Goudet, J. 2001 Fstat 2.9.3, a program to estimate and test gene diversities and fixation indices. See http://www2.unil.ch/popgen/softwares/fstat.htm
- Grafe T.U, Thein J. Energetics of calling and metabolic substrate use during prolonged exercise in the European treefrog Hyla arborea. J. Comp. Physiol. B: Biochem. Syst. Environ. Physiol. 2001;171:69–76. doi: 10.1007/s003600000151. doi:10.1007/s003600000151 [DOI] [PubMed] [Google Scholar]
- Green, P., Falls, K. & Crooks, S. 1990 Documentation for Crimap, version 2.4. St Louis, MO: Washington University School of Medicine.
- Griffin D.K, Harvey S.A, Campos-Ramos R, Ayling L.J, Bromage N.R, Masabanda J.S, Penman D.J. Early origins of the X and Y chromosomes: lessons from tilapia. Cytogenet. Genome Res. 2002;99:157–163. doi: 10.1159/000071588. doi:10.1159/000071588 [DOI] [PubMed] [Google Scholar]
- Haldane J.B.S. Sex ratio and unisexual sterility in hybrid animals. J. Genet. 1922;12:101–109. [Google Scholar]
- Hansson B, Akesson M, Slate J, Pemberton J.M. Linkage mapping reveals sex-dimorphic map distances in a passerine bird. Proc. R. Soc. B. 2005;272:2289–2298. doi: 10.1098/rspb.2005.3228. doi:10.1098/rspb.2005.3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick P.W. Sex: differences in mutation, recombination, selection, gene flow, and genetic drift. Evolution. 2007;61:2750–2771. doi: 10.1111/j.1558-5646.2007.00250.x. doi:10.1111/j.1558-5646.2007.00250.x [DOI] [PubMed] [Google Scholar]
- Hultén, M. & Tease, C. 2003 Genetic mapping: comparison of direct and indirect approaches. In Nature encyclopedia of the human genome, vol. 2 (ed. D. N. Cooper), pp. 876–881. London, UK: Nature Publishing Group.
- Huxley J.S. Sexual differences of linkage in Gammarus chevreuxi. J. Genet. 1928;20:145–156. [Google Scholar]
- Isberg S.R, Johnston S.M, Chen Y, Moran C. First evidence of higher female recombination in a species with temperature-dependent sex determination: the saltwater crocodile. J. Hered. 2006;97:599–602. doi: 10.1093/jhered/esl035. doi:10.1093/jhered/esl035 [DOI] [PubMed] [Google Scholar]
- Jagiello G, Fang J.S. Observations on chiasmata in mouse diplotene oocytes and spermatocytes. Cytologia. 1987;52:283–292. [Google Scholar]
- Jamsari A, Nitz I, Reamon-Buttner S.M, Jung C. BAC-derived diagnostic markers for sex-determination in asparagus. Theor. Appl. Genet. 2004;108:1140–1146. doi: 10.1007/s00122-003-1529-0. doi:10.1007/s00122-003-1529-0 [DOI] [PubMed] [Google Scholar]
- Jones K.C, Levine K.F, Banks J.D. Characterization of 11 polymorphic tetranucleotide microsatellites for forensic applications in California elk (Cervus elaphus canadensis) Mol. Ecol. Notes. 2002;2:425–427. doi:10.1046/j.1471-8286.2002.00264.x [Google Scholar]
- Joseph S.B, Kirkpatrick M. Haploid selection in animals. Trends Ecol. Evol. 2004;19:592–597. doi:10.1016/j.tree.2004.08.004 [Google Scholar]
- Kappes S.M, Keele J.W, Stone R.T, McGraw R.A, Sonstegard T.S, Smith T.P.L, Lopez-Corrales N.L, Beattie C.W. A second-generation linkage map of the bovine genome. Genome Res. 1997;7:235–249. doi: 10.1101/gr.7.3.235. doi:10.1101/gr.7.3.235 [DOI] [PubMed] [Google Scholar]
- Kondo M, Nanda I, Hornung U, Schmid M, Schartl M. Evolutionary origin of the medaka Y chromosome. Curr. Biol. 2004;14:1664–1669. doi: 10.1016/j.cub.2004.09.026. doi:10.1016/j.cub.2004.09.026 [DOI] [PubMed] [Google Scholar]
- Kong A, et al. A high-resolution recombination map of the human genome. Nat. Genet. 2002;31:241–247. doi: 10.1038/ng917. doi:10.1038/ng917 [DOI] [PubMed] [Google Scholar]
- Korol A.B, Iliadi K.G. Recombination increase resulting from directional selection for geotaxis in Drosophila. Heredity. 1994;72:64–68. doi: 10.1038/hdy.1994.7. doi:10.1038/hdy.1994.7 [DOI] [PubMed] [Google Scholar]
- Korol A.B, Preygel I.A, Preygel S.I. Chapman and Hall; London, UK: 1994. Recombination variability and evolution. [Google Scholar]
- Lenormand T. The evolution of sex dimorphism in recombination. Genetics. 2003;163:811–822. doi: 10.1093/genetics/163.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand T, Dutheil J. Recombination difference between sexes: a role for haploid selection. PLoS Biol. 2005;3:396–403. doi: 10.1371/journal.pbio.0030063. doi:10.1371/journal.pbio.0030063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher M.J, Hurst L.D. Imprinted chromosomal regions of the human genome have unusually high recombination rates. Genetics. 2003;165:1629–1632. doi: 10.1093/genetics/165.3.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, et al. A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature. 2004;427:348–352. doi: 10.1038/nature02228. doi:10.1038/nature02228 [DOI] [PubMed] [Google Scholar]
- Lorch P.D. Sex differences in recombination and mapping adaptations. Genetica. 2005;123:39–47. doi: 10.1007/s10709-003-2706-4. doi:10.1007/s10709-003-2706-4 [DOI] [PubMed] [Google Scholar]
- Ma H, Moore P.H, Liu Z, Kim M.S, Yu Q, Fitch M.M, Sekioka T, Paterson A.H, Ming R. High-density linkage mapping revealed suppression of recombination at the sex-determining locus in papaya. Genetics. 2004;166:419–436. doi: 10.1534/genetics.166.1.419. doi:10.1534/genetics.166.1.419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox J.F, et al. An enhanced linkage map of the sheep genome comprising more than 1000 loci. Genome Res. 2001;11:1275–1289. doi: 10.1101/gr.135001. doi:10.1101/gr.GR-1350R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, Matsuda C, Hamaguchi S, Sakaizumi M. Identification of the sex chromosomes of the medaka, Oryzias latipes, by fluorescence in situ hybridization. Cytogenet. Cell Genet. 1998;82:257–262. doi: 10.1159/000015113. doi:10.1159/000015113 [DOI] [PubMed] [Google Scholar]
- Matsuda M, Sotoyama S, Hamaguchi S, Sakaizumi M. Male-specific restriction of recombination frequency in the sex chromosomes of the medaka, Oryzias latipes. Genet. Res. 1999;73:225–231. doi:10.1017/S0016672399003754 [Google Scholar]
- Mellersh C.S, et al. A linkage map of the canine genome. Genomics. 1997;46:326–336. doi: 10.1006/geno.1997.5098. doi:10.1006/geno.1997.5098 [DOI] [PubMed] [Google Scholar]
- Merilä J, Sheldon B.C. Genetic architecture of fitness and non-fitness traits—empirical patterns and development of ideas. Heredity. 1999;83:103–109. doi: 10.1046/j.1365-2540.1999.00585.x. doi:10.1038/sj.hdy.6885850 [DOI] [PubMed] [Google Scholar]
- Mikawa S, et al. A linkage map of 234 DNA markers in an intercross of Gottingen miniature and Meishan pigs. Anim. Genet. 1999;30:407–417. doi: 10.1046/j.1365-2052.1999.00493.x. doi:10.1046/j.1365-2052.1999.00493.x [DOI] [PubMed] [Google Scholar]
- Moen T, Hoyheim B, Munck H, Gomez-Raya L. A linkage map of Atlantic salmon (Salmo salar) reveals an uncommonly large difference in recombination rate between the sexes. Anim. Genet. 2004;35:81–92. doi: 10.1111/j.1365-2052.2004.01097.x. doi:10.1111/j.1365-2052.2004.01097.x [DOI] [PubMed] [Google Scholar]
- Morgan T.H. No crossing over the male Drosophila of genes in the second and third pair of chromosomes. Biol. Bull. 1914;26:195–204. doi:10.2307/1536193 [Google Scholar]
- Nanda I, et al. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc. Natl Acad. Sci. USA. 2002;99:11 778–11 783. doi: 10.1073/pnas.182314699. doi:10.1073/pnas.182314699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruse K, Fukamachi F, Mitani H, Kondo M, Matsuoka T. A detailed linkage map of medaka, Oryzias latipes: comparative genomics and genome evolution. Genetics. 2000;154:1773–1784. doi: 10.1093/genetics/154.4.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff M.W, Broman K.W, Mellersh C.S, Ray K, Acland G.M, Aguirre G.D, Ziegle J.S, Ostrander E.A, Rine J. A second-generation genetic linkage map of the domestic dog, Canis familiaris. Genetics. 1999;151:803–820. doi: 10.1093/genetics/151.2.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Modification of linkage intensity by natural selection. Genetics. 1967;57:625–641. doi: 10.1093/genetics/57.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Evolutionary change of linkage intensity. Nature. 1968;218:1160–1161. doi: 10.1038/2181160a0. doi:10.1038/2181160a0 [DOI] [PubMed] [Google Scholar]
- Nei M. Linkage modification and sex difference in recombination. Genetics. 1969;63:681–699. doi: 10.1093/genetics/63.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Springer; New York, NY: 1967. Sex chromosomes and sex-linked genes. [Google Scholar]
- Ott J. John Hopkins University Press; Baltimore, MD: 1991. Analysis of human genetic linkage. [Google Scholar]
- Otto S.P, Barton N.H. Selection for recombination in small populations. Evolution. 2001;55:1921–1931. doi: 10.1111/j.0014-3820.2001.tb01310.x. [DOI] [PubMed] [Google Scholar]
- Pastor J.B, Callan H.G. Chiasma formation in spermatocytes and oocytes of the turbellarian Dendroceolum lacteum. J. Genet. 1952;50:449–454. [Google Scholar]
- Peichel C.L, et al. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosomes. Curr. Biol. 2004;14:1416–1424. doi: 10.1016/j.cub.2004.08.030. doi:10.1016/j.cub.2004.08.030 [DOI] [PubMed] [Google Scholar]
- Pidancier N, Miquel C, Miaud C. Buccal swabs as a non-destructive tissue sampling method for DNA analysis in amphibians. Herpetol. J. 2003;13:175–178. [Google Scholar]
- Plomion C, Omalley D.M. Recombination rate differences for pollen parents and seed parents in Pinus pinaster. Heredity. 1996;77:341–350. doi:10.1038/hdy.1996.152 [Google Scholar]
- R Development Core Team 2006 R: a language and environment for statistical computing, reference index, version 2.2.1. Vienna, Austria: R Foundation for Statistical Computing.
- Rice W.R. Evolution of the Y sex chromosome in animals. Bioscience. 1996;46:331–343. doi:10.2307/1312947 [Google Scholar]
- Sakamoto T, et al. A microsatellite linkage map of rainbow trout (Oncorhynchus mykiss) characterized by large sex-specific differences in recombination rates. Genetics. 2000;155:1331–1345. doi: 10.1093/genetics/155.3.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samollow P.B, Kammerer C.M, Mahaney S.M, Schneider J.L, Westenberger S.J, VandeBerg J.L, Robinson E.S. First-generation linkage map of the gray, short-tailed opossum, Monodelphis domestica, reveals genome-wide reduction in female recombination rates. Genetics. 2004;166:307–329. doi: 10.1534/genetics.166.1.307. doi:10.1534/genetics.166.1.307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schartl M. Sex chromosome evolution in non-mammalian vertebrates. Curr. Opin. Genet. Dev. 2004;14:634–641. doi: 10.1016/j.gde.2004.09.005. doi:10.1016/j.gde.2004.09.005 [DOI] [PubMed] [Google Scholar]
- Seefelder S, Ehrmaier G, Schweizer G, Seigner E. Male and female genetic linkage map of hops, Humulus lupulus. Plant Breed. 2000;119:249–255. doi:10.1046/j.1439-0523.2000.00469.x [Google Scholar]
- Shifman S, Bell J.T, Copley R.R, Taylor M.S, Williams R.W, Mott R, Flint J. A high-resolution single nucleotide polymorphism genetic map of the mouse genome. PLoS Biol. 2006;4:2227–2237. doi: 10.1371/journal.pbio.0040395. doi:10.1371/journal.pbio.0040395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer A, Perlman H, Yan Y.L, Walker C, Corley-Smith G, Brandhorst B, Postlethwait J. Sex-specific recombination rates in zebrafish (Danio rerio) Genetics. 2002;160:649–657. doi: 10.1093/genetics/160.2.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauss M, Tomiuk J, Segelbacher G, Driesel S, Fietz J, Bachmann L, Kompf J. Sex-specific recombination rates in Parus major and P. caeruleus, an exception to Huxley's rule. Hereditas. 2003;139:199–205. doi: 10.1111/j.1601-5223.2003.01698.x. doi:10.1111/j.1601-5223.2003.01698.x [DOI] [PubMed] [Google Scholar]
- Steinemann S, Steinemann M. Y chromosomes: born to be destroyed. Bioessays. 2005;27:1076–1083. doi: 10.1002/bies.20288. doi:10.1002/bies.20288 [DOI] [PubMed] [Google Scholar]
- Sumida M, Nishioka M. Genetic linkage groups in the Japanese brown frog (Rana japonica) J. Hered. 2000;91:1–7. doi: 10.1093/jhered/91.1.1. doi:10.1093/jhered/91.1.1 [DOI] [PubMed] [Google Scholar]
- Tanaka Y. A study of Mendelian factors in the silkworm, Bombyx mori. Mol. Gen. Genet. 1914;12:161. [Google Scholar]
- Trivers R. Sex differences in rates of recombination and sexual selection. In: Michod R, Levin B, editors. The evolution of sex. Sinauer; Sunderland, MA: 1988. pp. 270–286. [Google Scholar]
- Tunner H.G. Evidence for genomic imprinting in unisexual triploid hybrid frogs. Amphib. Reptil. 2000;21:135–141. doi:10.1163/156853800507327 [Google Scholar]
- Ved Brat S. Genetic systems in Allium. 2. Sex differences in meiosis. Chromosomes Today. 1966;1:31–40. [Google Scholar]
- Volff J.N, Nanda I, Schmid M, Schartl M. Governing sex determination in fish: regulatory putches and ephemeral dictators. Sex. Dev. 2007;1:85–99. doi: 10.1159/000100030. doi:10.1159/000100030 [DOI] [PubMed] [Google Scholar]
- Voorrips R.E. MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. doi:10.1093/jhered/93.1.77 [DOI] [PubMed] [Google Scholar]
- Vorburger C. Genomic imprinting or mutation and interclonal selection in triploid hybrid frogs? A comment on Tunner. Amphib. Reptil. 2001;22:263–265. doi:10.1163/15685380152030472 [Google Scholar]
- Waldbieser G.C, Bosworth B.G, Nonneman D.J, Wolters W.R. A microsatellite-based genetic linkage map for channel catfish, Ictalurus punctatus. Genetics. 2001;158:727–734. doi: 10.1093/genetics/158.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace H, Wallace B.M, Badawy G.M. Lampbrush chromosomes and chiasmata of sex-reversed crested newts. Chromosoma. 1997;106:526–533. doi: 10.1007/s004120050275. doi:10.1007/s004120050275 [DOI] [PubMed] [Google Scholar]
- White, M. J. D. 1976 Blattodea, Mantodea, Isoptera, Grylloblattodea, Phasmatodea, Dermaptera and Embioptera. Animal cytogenetics Insecta, vol. 3, pp. 1–75. Berlin, Germany: Gebrüder Borntraeger.
- Wright S. Classification of the factors of evolution. Cold Spring Harbor Symp. Quant. Biol. 1955;20:16–24. doi: 10.1101/sqb.1955.020.01.004. [DOI] [PubMed] [Google Scholar]
- Xu H.L, Swoboda I, Bhalla P.L, Singh M.B. Male gametic cell-specific gene expression in flowering plants. Proc. Natl Acad. Sci. USA. 1999;96:2554–2558. doi: 10.1073/pnas.96.5.2554. doi:10.1073/pnas.96.5.2554 [DOI] [PMC free article] [PubMed] [Google Scholar]