Abstract
Batesian mimics gain protection from predation through the evolution of physical similarities to a model species that possesses anti-predator defences. This protection should not be effective in the absence of the model since the predator does not identify the mimic as potentially dangerous and both the model and the mimic are highly conspicuous. Thus, Batesian mimics should probably encounter strong predation pressure outside the geographical range of the model species. There are several documented examples of Batesian mimics occurring in locations without their models, but the evolutionary responses remain largely unidentified. A mimetic species has four alternative evolutionary responses to the loss of model presence. If predation is weak, it could maintain its mimetic signal. If predation is intense, it is widely presumed the mimic will go extinct. However, the mimic could also evolve a new colour pattern to mimic another model species or it could revert back to its ancestral, less conspicuous phenotype. We used molecular phylogenetic approaches to reconstruct and test the evolution of mimicry in the North American admiral butterflies (Limenitis: Nymphalidae). We confirmed that the more cryptic white-banded form is the ancestral phenotype of North American admiral butterflies. However, one species, Limenitis arthemis, evolved the black pipevine swallowtail mimetic form but later reverted to the white-banded more cryptic ancestral form. This character reversion is strongly correlated with the geographical absence of the model species and its host plant, but not the host plant distribution of L. arthemis. Our results support the prediction that a Batesian mimic does not persist in locations without its model, but it does not go extinct either. The mimic can revert back to its ancestral, less conspicuous form and persist.
Keywords: character evolution, Lepidoptera, parametric bootstrap, wing pattern evolution
1. Introduction
Many organisms have evolved defensive colour patterns to mitigate the persistent risk of predation. Aposematic, or warning, coloration is one such defensive strategy used by noxious organisms to visually communicate their toxicity or distastefulness to potential predators (Cott 1940; Guilford 1990). An aposematic pattern confers survival benefits to the prey because it is both easier for the predator to learn and less likely to be forgotten (Gittleman & Harvey 1980; Roper & Redstone 1987; Roper 1990; Alatalo & Mappes 1996; Prudic et al. 2007). These benefits are believed to have facilitated the evolution of aposematic coloration from ancestrally cryptic patterns (Fisher 1930; Sherratt & Beatty 2003). Closely related to aposematic coloration is visual defensive mimicry that is often a deceptive signal, defined as a close physical resemblance among unrelated species (Bates 1862). Batesian mimicry is a specialized signal with complex interactions between the model, the mimic and the predators (Ruxton et al. 2004 and references therein). In this mimicry system, an unprotected species (mimic) evolves a physical resemblance to an aposematic species (model), and the mimic gains protection from predation by deceiving the predators. This relationship is often considered parasitic because the unprotected species undermines the effectiveness of the aposematic signal.
Much theoretical and empirical research has focused on the conditions necessary for Batesian mimicry to arise and how it functions once it exists, but much less is known about how these relationships change through time, especially when the model is no longer present (Ruxton et al. 2004; Mappes et al. 2005). It is presumed that the protection afforded to the mimic should break down in locations where the model is absent because the predators do not recognize either the model or mimic as unprofitable prey and the mimic has a highly conspicuous phenotype (Pfennig et al. 2001, 2007; Ruxton et al. 2004). The geographical range of the mimic is predicted to be limited by the geographical range of the model even though suitable habitat for the mimic may exist outside that range. Yet, there are examples where a mimic has been documented occurring in locations without its model (Ritland 1995; Pfennig et al. 2001; Prudic et al. 2002). The mimetic lineage has several potential outcomes in response to this change in model presence. This species could remain a Batesian mimic in locations without its model, especially when predation is weak (Pfennig et al. 2007). A mimic could also go extinct in these locations due to intense predation (Pfennig et al. 2001). Less explored are the expectations that the mimic could evolve a new colour pattern to mimic another model species or revert back to its ancestral, non-mimetic phenotype (Ruxton et al. 2004). The latter two scenarios are traditionally considered unlikely and have the least empirical support because it is assumed that considerable genetic change is responsible for the specialized phenotype of a Batesian mimic (Fisher 1930; Ruxton et al. 2004; Pfennig et al. 2007).
The North American admiral butterflies (Nymphalidae: Limenitis) provide a unique opportunity to investigate the evolution and maintenance of Batesian mimicry relationships. The ancestral wing pattern of a black ground colour and dorsal–lateral white band is commonly found in Eurasian congeners (Platt 1983; Mullen 2006). This wing pattern is regarded as an example of disruptive coloration because the butterfly is difficult to detect and attack especially when it is flying through its native riparian habitat (Platt & Brower 1968; Platt 1983). This type of cryptic pattern reduces prey detectability by visually inhibiting the predator from seeing the prey's actual body outline (Cuthill et al. 2005; Stevens & Cuthill 2006). However, in North America, there has been a divergence away from this cryptic defensive strategy (Platt 1983; Mullen 2006). Three of the four Limenitis species are conspicuously coloured mimics, each of a different model species, and all with very specialized wing pattern morphologies (Prudic et al. 2002 and references therein). Unlike other well-studied butterfly mimicry systems, such as Heliconius, these three mimetic species also have different larval host plant requirements than their respective models (Opler et al. 2004). Since the geographical distribution of the host species plays a large role in determining the geographical distribution of a phytophagous insect species (Bernays & Chapman 1994; Loxdale & Lushai 1999), disjunctions in the geographical distributions of model and mimic phytophagous insects may arise due to differences in larval host plant requirements. Mimetic herbivorous insects may exist in locations without their respective model, if hosts are available for the mimetic but not the model species. Of particular interest are the different phenotypes possessed by admiral subspecies of Limenitis arthemis. The red-spotted purple (Limenitis arthemis astyanax and Limenitis arthemis arizonensis) is a conspicuously black and purple-coloured Batesian mimic of the pipevine swallowtail (Papilionidae: Battus philenor; Platt et al. 1971). The white admiral (Limenitis arthemis arthemis and Limenitis arthemis rubrofasciata) exhibits the ancestral white-banded cryptic form of the Eurasian congeners and is a close non-mimetic relative of the red-spotted purple. Based on wing colour information, it has been hypothesized that the white admiral or its nearest relative spread east out of Eurasia across Beringia and south to its current range. Subsequently, the red-spotted purple arose in sympatry with the model, the pipevine swallowtail (Remington 1958; Platt & Brower 1968; Platt 1983). However, recent phylogenetic evidence indicates that this assumption of evolutionary history may not be the case. The white admiral may have arisen from the red-spotted purple lineage, but this hypothesis warrants further investigation (Mullen 2006).
Here we reconstructed the evolutionary history of North American Limenitis using molecular phylogenetic methods. We documented the direction of the evolutionary transitions from cryptic to mimetic forms and vice versa. Using parametric bootstrapping, we also evaluated the likelihood that a mimetic form changed back to the ancestral cryptic form. We then investigated if Batesian mimicry transitions from mimetic to cryptic were correlated with the absence of the model species, based on the geographical distribution of the model species.
2. Material and methods
(a) Taxon sampling
We sampled all the North American members of the genus Limenitis and three species of Palaearctic Limenitis (electronic supplementary material, table). We sampled all subspecies of L. arthemis and most subspecies of Limenitis archippus and Limenitis weidemeyerii (Hodges et al. 1983). Putative hybrids of all North American Limenitis have been observed in the field, but the extent to which they are viable is currently unknown (Platt 1983). We did not include any specimens from overlap or hybrid zones, as classification of hybrid individuals is difficult and may hinder phylogenetic inference (Grant & Grant 1998). Additionally, we included samples from two closely related genera, Neptis and Adelpha, as out-groups (Willmott 2003).
(b) DNA extraction and sequencing
We inferred the evolutionary relationships among North American Limenitis using one mitochondrial gene, cytochrome oxidase subunit II (COII), and two nuclear genes, elongation factor 1 alpha (EF1a) and wingless. We extracted total genomic DNA from leg or thoracic tissue using DNeasy tissue extraction kit (Qiagen, Inc., CA, USA) according to the manufacturer's protocol. PCRs (50 μl) were performed on a Mastercycler (Eppendorf, NY, USA), in 1X Eppendorf Hotmaster Taq Buffer containing 0.2 μM of each primer, 0.1 mM dNTPs, 2.5 mM MgCl2 and 1 unit of Eppendorf Hotmaster Taq DNA polymerase. We amplified COII using the amplification primers Pierre and Eva (Caterino & Sperling 1999). The thermal cycle profile was an initial 1.5 min denaturation at 94°C; 32 cycles of 40 s at 94°C, 40 s at 45°C and 45 s at 72°C; a 7 min final extension at 72°C. To amplify EF1a, we used the amplification primers ef44f and efrcM4r (Monteiro & Pierce 2001) with the profile: initial 2 min denaturation at 94°C; 35 cycles of 20 s at 94°C, 15 s at 50°C and 60 s at 65°C; a 5 min final extension at 65°C. We amplified wingless using the amplification primers LepWG1 and LepWG2 (Brower & DeSalle 1998) with the profile: initial 2 min denaturation at 94°C; 15 cycles of 30 s at 94°C, 30 s at 65°C −1°C per cycle, 1 min at 65°C; 25 cycles of 30 s at 94°C, 30 s at 50°C, 1 min at 65°C; and a 10 min final extension at 65°C. PCR products were purified with a Millipore size-exclusion filtration (Millipore, MA, USA) prior to direct sequencing. Both strands were sequenced using an Applied Biosystems BigDye Terminator v. 3.1 cycle sequencing kit (Applied Biosystems, CA, USA) and run on an Applied Biosystems 3730XL DNA Analyzer (Applied Biosystems). We generated consensus sequences from the two strands and aligned these sequences by eye with the aid of the program BioEdit (Hall 1999). Representative sequences for all alleles of each locus have been deposited in GenBank (accession numbers: DQ117851–DQ117897, EF643261–EF643361 and EU433934–EU433946).
(c) Phylogenetic analyses
We performed maximum parsimony and Bayesian MCMC on each gene separately. For each gene, we began by removing redundant sequences from the alignment. Maximum parsimony analyses were performed in PAUP* v. 4.0b10 (Swofford 2001). We initially performed a heuristic tree search with 1000 random-addition replicates, saving all the most parsimonious trees per replicate. To assess node support, we performed 1000 non-parametric bootstrap pseudoreplicates, each with 10 stepwise addition, nearest-neighbour interchange heuristic search replicates, saving the first 1000 most parsimonious trees per heuristic search replicate.
We used a hierarchical likelihood ratio test to estimate the model of evolution for the Bayesian analyses (Huelsenbeck & Rannala 1997). COII, EF1a and wingless all fit a GTR+Γ model, with empirical base frequencies. Using this model in MrBayes v. 3.01 (Huelsenbeck & Ronquist 2001), we analysed each gene separately; in each analysis, we ran four chains (three hot and one cold) for 2 000 000 generations, sampled trees every 100 generations, and discarded the trees sampled in the first 1 000 000 generations (burnin). To estimate the posterior probability of clades, we used the post-burnin sampled trees to generate a 50% majority rule consensus tree using the ‘sumt’ command in MrBayes.
We used the three gene trees to generate a species tree in Mesquite (Maddison & Maddison 2006) to infer the relationships among North American Limenitis. Briefly, because the three genes are evolving independently, and may not always accurately reflect the history of the species' relationships, we used the gene trees to search for the ‘best-fit’ species tree that minimizes the number of deep coalescences of the gene trees (Maddison 1997). Gene trees of recently diverged taxa may not be monophyletic, due to incomplete lineage sorting (Hudson & Coyne 2002; Funk & Omland 2003), although a significant phylogenetic signal may still be present (Maddison & Knowles 2006). By accounting for the possibility of incomplete lineage sorting, this approach infers of species' relationships without requiring the gene trees to be monophyletic. We used the heuristic search function in Mesquite to find the optimal species tree that minimizes the number of deep coalescences of the gene trees, using a nearest-neighbour interchange branch swapping algorithm, treating gene trees as unrooted, and allowing polytomies to be automatically resolved before the coalescence cost is assessed (Maddison & Maddison 2006). This approach has proved useful for inferring true species relationships when gene trees do not recover the monophyly of species (Maddison & Knowles 2006).
(d) Character evolution
We coded wing morphology into three states: 0=white-banded (L. a. arthemis, L. a. rubrofasciata, L. weidemeyerii, Limenitis lorquini, Limenitis camilla, Limenitis populi, Limenitis reducta); 1=black, Battus mimic (L. a. astyanax, L. a. arizonensis); and 2=Danaus mimic (L. archippus). Ancestral character states were reconstructed on the species tree in Mesquite (Maddison & Maddison 2006), using a parsimony model of unordered states.
(e) Parametric bootstrapping
Phylogenetic reconstruction of COII demonstrated strong support against a clade of the black mimetic phenotype (see §3). To determine whether our reconstructions were significantly better than those reflecting monophyly of the mimetic phenotype, we performed phylogenetic parametric bootstrapping (Huelsenbeck et al. 1996) on the COII gene tree. Briefly, we compared parsimony tree lengths of an unconstrained search to tree lengths of a search constrained to retain only those trees in which the black mimetic taxa formed an exclusive clade. This difference in tree lengths represented our observed test statistic (δobs). From the most parsimonious constrained trees, we used the tree with the highest likelihood to estimate the model of evolution using hierarchical likelihood ratio tests (Huelsenbeck & Rannala 1997). This model (GTR+Γ+I) was then implemented in Mesquite v. 1.12 (Maddison & Maddison 2006) to simulate 1000 character matrices on the most parsimonious reconstruction with an exclusive black mimetic clade. For each matrix, we performed two parsimony tree searches, one unconstrained and one constrained as above, and the difference in tree lengths were used to generate a distribution of the test statistic, δ. We determined the significance of our results by comparing δobs with the simulated distribution of δ. A significant value of δobs leads to the rejection of the hypothesis that the group tested is monophyletic.
(f) Distribution and geographical range of L. arthemis forms, the model and the host plants
We used published geographical information of North American butterfly and host plant county records to determine whether the transition from mimetic to cryptic L. arthemis form was correlated with the absence of its Batesian model, B. philenor, or some other ecological factor such as host plant availability. We evaluated this question at two spatial scales: at the level of our taxonomic sampling used to reconstruct the gene trees and at the level of the entire geographical range of L. arthemis. For the butterfly information, we used two sources of county records (Layberry et al. 1998; Opler et al. 2004) and for the plant information, we used one source of county records (USDA 2007). By combining information from these databases, we were able to document the presence or absence of (i) L. arthemis' larval host plant, (ii) the model B. philenor, and (iii) B. philenor's larval host plant (Aristolochia spp.) for two sets of locations: (a) the exact locations where we sampled our specimens used to reconstruct the phylogeny and (b) across the entire range of L. arthemis in North America. For the smaller geographical analysis, 17 different counties were examined, six with the white-banded form and 11 with the black mimetic form. For the larger geographical analysis, 1509 out of 3135 counties in North America (excluding Hawaii) had records of L. arthemis: 144 counties with the white-banded form only; 1166 counties with the black mimetic form only; and 199 counties with both forms together. We used linear regression in JMP-In (JMP 2002) to evaluate factors predicting the geographical distributions of the mimetic and cryptic forms of L. arthemis.
3. Results
(a) Phylogenetics and character evolution
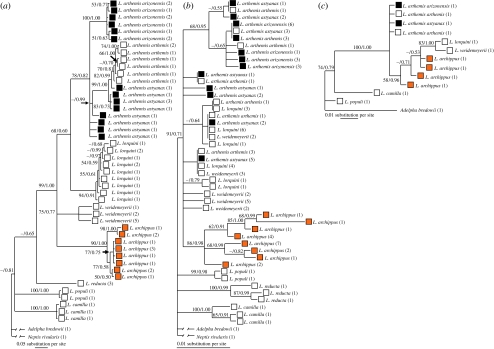
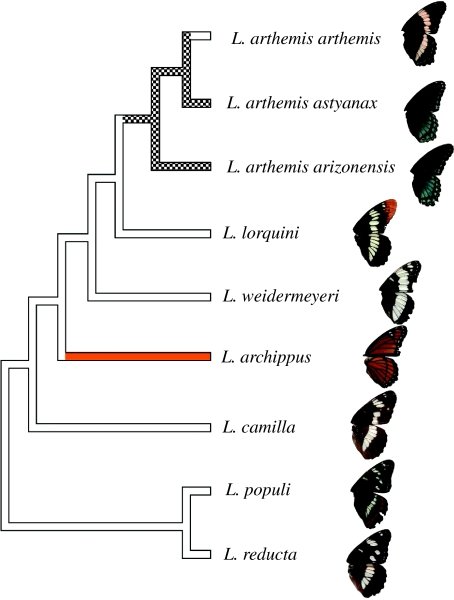
We reconstructed the gene trees for all North American Limenitis (figure 1). Parsimony and Bayesian reconstructions for each gene were topologically congruent, although Bayesian reconstructions showed support for some clades that were not supported in the parsimony analyses. For COII, there were no shared haplotypes among the species of Limenitis. Within L. arthemis, the white-banded cryptic form and the black mimetic form also shared no haplotypes. There were no shared haplotypes between the two mimetic subspecies (L. a. astyanax and L. a. arizonensis). In contrast, many EF1a haplotypes were found in more than one species, and only L. archippus was recovered as monophyletic. The wingless data showed the least phylogenetic resolution, although there was significant support for the monophyly of North American Limenitis. The four specimens of L. arthemis (two L. a. arthemis, one L. a. arizonensis and one L. a. astyanax) all possessed identical wingless haplotypes. In the species tree search, minimizing the number of deep coalescences of the COII, EF1a and wingless Bayesian gene trees, we recovered two best-fit trees, which differed only in the arrangement of out-group taxa (Adelpha bredowii and Neptis rivularis). The inferred relationships among North American Limenitis species are shown in figure 2, along with ancestral reconstructions of wing morphology. Our results confirm that the white-banded cryptic form is the ancestral phenotype of North American Limenitis (Mullen 2006). The two equally parsimonious ancestral reconstructions of the wing pattern in L. arthemis on this species tree are: (i) the black mimetic form (L. a. astyanax and L. a. arizonensis) evolved once from a white-banded cryptic ancestor, and reverted back to the white-banded cryptic form in the lineage leading to L. a. arthemis or (ii) the black mimetic form evolved twice from a white-banded cryptic ancestor (figure 2).
Figure 1.
Consensus trees from Bayesian inference of (a) COII, (b) EF1a and (c) wingless. Branch labels show maximum parsimony bootstrap support/Bayesian posterior probability. Wing morphology character states for terminal taxa are shown. White, white-banded; black, Battus mimic; orange, Danaus mimic.
Figure 2.
Species tree of North American Limenitis inferred from minimizing deep coalescences of gene trees. Branch colours show maximum parsimony character reconstructions of ancestral states (white, non-mimetic disruptive coloration; black, Battus mimic; orange, Danaus mimic; checkered, equivocal between white and black).
The most parsimonious trees in unconstrained searches of the COII data were 276 steps and the most parsimonious trees in constrained searches were 284 steps (δobs=284–276=8). This falls well outside of our simulated distribution of δ (mean δ=0.242, 95% CI=0–3), so we reject an exclusive black mimetic clade at p<0.001.
(b) Distribution and geographical range of L. arthemis forms, the model and the host plants
At the smaller geographical scale using only the locations of the specimens used for phylogenetic analyses, the black mimetic form was positively correlated with the presence of the model (N=11, r2=0.75, p<0.001), whereas the presence of the white-banded cryptic form of L. arthemis was negatively correlated with the presence of the model, B. philenor (N=6, r2=0.45, p=0.004; table 1a). This was also explained by the distribution of the model's host plant. Aristolochia spp. were positively correlated with the presence of the black mimetic form of L. arthemis (N=11, r2=0.33, p=0.024), but were negatively correlated with the presence of the white-banded cryptic form (N=6, r2=0.31, p=0.025; table 1a). However, the willow, the major L. arthemis larval host plant, was present in every county in which L. arthemis was found. The presence of the host plant did not explain the different geographical distributions of the two L. arthemis forms (N=17).
Table 1.
Presence and absence county records of the model B. philenor and its larval host plant, Aristolochia spp., in relation to county records of the mimetic and non-mimetic forms of L. arthemis. (a) The data from the specimens used in the molecular phylogeny; (b) the data across the entire geographical range of L. arthemis.
| county presence of L. a. astyanax and L. a. arizonensis (mimetic black form) | county presence of L. a. arthemis and L. a. rubrofasciata (non-mimetic white-banded form) | ||
|---|---|---|---|
| (a) | |||
| Battus philenor Batesian model | present | 10 | 0 |
| absent | 1 | 6 | |
| Aristolochia spp. (Battus host) | present | 8 | 0 |
| absent | 3 | 6 | |
| (b) | |||
| Battus philenor Batesian model | present | 897 | 90 |
| absent | 465 | 256 | |
| Aristolochia spp. (Battus host) | present | 788 | 35 |
| absent | 574 | 311 |
Focusing on the entire geographical range of L. arthemis, the presence of the white-banded cryptic form was again negatively correlated with the presence of the model, B. philenor (N=346, r2=0.15, p<0.001), and the black mimetic form was positively correlated with the model (N=1362, r2=0.75, p<0.001; table 1b). These patterns were also explained by the distribution of the model's larval host plant (white-banded form: N=346, r2=0.22, p<0.001, slope negative; black mimetic form: N=1362, r2=0.27, p<0.001, slope positive; table 1b). However, willow always occurred in the same county as any L. arthemis and did not explain the different geographical distributions of the L. arthemis forms (N=1509), and thus there are suitable host plants for L. arthemis in locations both with and without the Batesian model, B. philenor.
4. Discussion
Batesian mimicry is a highly specialized interaction between the model, the mimic and the predator. The mimic gains protection from predation by resembling a dangerous or unpalatable species. Since it is dependent on the honest signal of the model, the Batesian mimic should not occur in locations without its model. The predators will not recognize the mimic as potentially dangerous in areas without the model and will predate on the mimics more intensely because this prey is highly conspicuous and palatable (Ruxton et al. 2004; Pfennig et al. 2007). Yet, there are documented examples of a Batesian mimic occurring in locations without its model in ecological time (Ritland 1995; Pfennig et al. 2001; Prudic et al. 2002). Here we examined whether a Batesian mimic could persist outside the range of its model from a phylogenetic perspective considering the four potential evolutionary outcomes outlined in the literature. A Batesian mimetic species could go extinct if predation was intense or it could persist as a Batesian mimic if predation was weak (Pfennig et al. 2007). Although thought to be more unlikely owing to genetic constraints, a mimic could also evolve a new colour pattern to mimic another model species; or it could revert back to its ancestral non-mimetic phenotype (Fisher 1930; Ruxton et al. 2004). The data presented here support the hypothesis that a Batesian mimic does not persist in locations without its model, but it does not go extinct either (figure 1; table 1). Instead, in locations where the model does not occur, the mimic may have reverted back to its ancestral form that is less conspicuous to predators.
Previous studies have hypothesized that the black mimetic form of L. arthemis evolved from the white-banded cryptic form of L. arthemis (Platt & Brower 1968; Platt 1983). However, our results from the phylogenetic analyses are consistent with the hypothesis that the white-banded cryptic form evolved from a black mimetic form. The inferred species tree and parametric bootstrapping results demonstrate that cryptic L. a. arthemis did not give rise to the mimetic subspecies L. a. astyanax and L. a. arizonensis; rather, the lineages leading to the two mimetic subspecies diverged before the evolution of L. a. arthemis (figure 2). Although the nuclear gene trees revealed little resolution among the North American species of Limenitis, there was no uncertainty about these relationships in the species tree inference, demonstrating the phylogenetic signal present in gene trees (Maddison & Knowles 2006). Parsimony character reconstructions of wing phenotypes are equivocal, but it is unlikely that the black mimetic phenotype evolved twice given current distributions of North American Limenitis species. When the model, B. philenor, co-occurs with Limenitis species, those Limenitis species are always mimics, of B. philenor or other distasteful model species over a broad geographical scale (Platt & Brower 1968). Thus, the hypothesis of two independent origins of the black mimetic phenotype would have required long-term coexistence of B. philenor and a non-mimetic ancestor of L. a. arthemis and L. a. astyanax. This pattern of coexistence of B. philenor and white-banded cryptic Limenitis is not observed across the contemporary permanent range of B. philenor. Given the inferred history of North American Limenitis (Mullen 2006, this study), the lack of any cryptic Limenitis co-occurring with B. philenor, and the rejection of a black mimetic clade by parametric bootstrapping, we propose that a black mimetic lineage gave rise to the white-banded cryptic lineage observed in L. arthemis.
Based on the adaptive function for this evolutionary pattern, we predicted that the ecological data of both model and mimic would also link strongly with our phylogenetic results. The reversion to a white-banded cryptic form is consistent with the geographical distributions of the model (B. philenor) whose distribution is limited by its larval host plant (Aristolochia spp.). The geographical distribution of the white-banded cryptic L. arthemis is negatively correlated and the black mimetic form is positively correlated with the geographical range of the model (table 1 and electronic supplementary material, figure). A similar geographical pattern is found in another Batesian mimic of B. philenor, the eastern tiger swallowtail (Papilionidae: Papilio glaucus), which is a female-limited Batesian mimic of B. philenor (Brower 1958). Both mimetic and non-mimetic female forms of P. glaucus are present in locations where B. philenor occurs. However, in the same geographical region where L. arthemis changes from the black mimetic form to the white-banded cryptic form and B. philenor becomes increasingly rarer, P. glaucus females also shift from their mimetic to non-mimetic form (Brower & Brower 1962).
Reversion to a cryptic wing phenotype in L. arthemis is not unexpected given our knowledge of the genetic architecture underlying wing pattern variation. Even though butterfly wing patterns are a complex trait, the genetic mechanisms may be easy to change (Nijhout 1991). In butterfly Batesian mimicry, the wing pattern of the mimic is thought to be produced by a single gene of major effect with a suite of modifier genes that refine the wing pattern (Turner 1977). Although this mechanism has not been demonstrated definitively in Limenitis (but see Platt 1983), it has been shown in other Batesian mimics such as Papilio dardanus (Nijhout 2003). Also, in Heliconius butterflies, a homologous gene or complex of genes regulates wing pattern diversity in three different species with different phenotypes (Joron et al. 2006). Rather than a constraining role, this locus seems to provide wing phenotype flexibility across species and environments. It presumably functions by responding to a wide range of selection pressures, including predation, to produce radically divergent, locally adapted wing patterns from a very similar genetic mechanism. The genes regulating butterfly wing patterns do not appear to become constrained when these lineages become either Batesian or Müllerian mimics.
Our results also relate to a previously unreported phenomenon regarding butterfly distribution and range. Butterfly species, like many phytophagous insects, are limited by the distribution of their larval host plants (Bernays & Chapman 1994; Loxdale & Lushai 1999). Host acceptance and selection are determined by a wide variety of ecological and physiological factors (e.g. Futuyma & Moreno 1988; Bernays 1989; Thompson 2005). However, all of these factors relate to the interaction between insect species and host plant species. Our results suggest that the distribution of the black mimetic form of L. arthemis is determined not by the distribution of its own larval host plant (Salix spp.), but by the distribution of its model's larval host plant (Aristolochia spp.; table 1). Thus, the broad geographical distributions of phytophagous insect species and subspecies also depend on other indirect interactions in their communities, in addition to the relationship with their own larval host plant(s). This distributional pattern has been shown many times at a smaller community scale (e.g. Bernays 1989), but not across the larger geographical range of a species or subspecies.
To conclude, our results demonstrate that a Batesian mimic does not persist in locations without its model. The distribution of the mimic is correlated with the presence of the model and, in this system, the model's host plants. Batesian mimicry is a specialized signal with complex interactions between the model, the mimic and the predators across a broad geographical range. Thus, the distribution of the mimic may be limited by the distribution of the model species in many cases. However, our findings also demonstrate that a mimic does not necessarily go extinct without its model. When the model is absent, the mimic may re-evolve an ancestral defensive strategy and become less conspicuous to predators thereby persisting in locations without its Batesian model.
Acknowledgments
This study was supported by an NSF DDIG grant and a University of Arizona BIO5 fellowship to K.L.P. Many thanks to C. Nice, S. Nyland, B. Platt, A. Porter, D. Ritland and F.A.H. Sperling for specimens; R. Guralnick and D. Maddison for facilities; and K. Lotts for access to the North American butterfly county records.
Supplementary Material
Specimen information for molecular phylogeny
Approximate geographic ranges of white-banded L. arthemis phenotypes (hatched), mimetic L. arthemis phenotypes (purple) and B. philenor (grey). Ranges after Platt & Brower (1968) and Opler et al. (2004)
References
- Alatalo R.V, Mappes J. Tracking the evolution of warning signals. Nature. 1996;382:485–503. doi:10.1038/382708a0 [Google Scholar]
- Bates H.W. Contributions to an insect fauna of the Amazon valley. Lepidoptera: Heliconidae. Trans. Linn. Soc. Lond. 1862;23:495–566. [Google Scholar]
- Bernays E.A. Host range in phytophagous insects: the potential role of generalist predators. Evol. Ecol. 1989;3:299–311. doi:10.1007/BF02285261 [Google Scholar]
- Bernays E.A, Chapman R.F. Chapman and Hall; New York, NY: 1994. Host–plant selection by phytophagous insects. [Google Scholar]
- Brower J.V.Z. Experimental studies of mimicry in some North American butterflies. Part II. Battus philenor and Papilio troilus, P. polyxenes, and P. glaucus. Evolution. 1958;12:123–136. doi:10.2307/2406023 [Google Scholar]
- Brower L.P, Brower J.V.Z. The relative abundance of model and mimic butterflies in natural populations of the Battus philenor mimicry complex. Ecology. 1962;43:154–158. [Google Scholar]
- Brower A.V.Z, DeSalle R. Patterns of mitochondrial versus nuclear DNA sequence divergence among nymphalid butterflies: the utility of wingless as a source of characters for phylogenetic inference. Insect Mol. Biol. 1998;7:73–82. doi: 10.1046/j.1365-2583.1998.71052.x. doi:10.1046/j.1365-2583.1998.71052.x [DOI] [PubMed] [Google Scholar]
- Caterino M.S, Sperling F.A.H. Papilio phylogeny based on mitochondrial cytochrome oxidase I and II genes. Mol. Phylogenet. Evol. 1999;11:122–137. doi: 10.1006/mpev.1998.0549. doi:10.1006/mpev.1998.0549 [DOI] [PubMed] [Google Scholar]
- Cott H.B. Methuen & Co. Ltd; London, UK: 1940. Adaptive coloration in animals. [Google Scholar]
- Cuthill I.C, Stevens M, Sheppard J, Maddocks T, Párraga C.A, Troscianko T.S. Disruptive coloration and background pattern matching. Nature. 2005;434:72–74. doi: 10.1038/nature03312. doi:10.1038/nature03312 [DOI] [PubMed] [Google Scholar]
- Fisher R.A. 1st edn. Dover Publications; New York, NY: 1930. The genetical theory of natural selection. [Google Scholar]
- Funk D.J, Omland K.E. Species-level paraphyly and polyphyly: frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 2003;34:397–423. doi:10.1146/annurev.ecolsys.34.011802.132421 [Google Scholar]
- Futuyma D.J, Moreno G. The evolution of ecological specialization. Annu. Rev. Ecol. Syst. 1988;19:207–234. doi:10.1146/annurev.es.19.110188.001231 [Google Scholar]
- Gittleman J, Harvey P. Why are distasteful prey not cryptic? Nature. 1980;286:149–150. doi:10.1038/286149a0 [Google Scholar]
- Grant P.R, Grant B.R. Speciation and hybridization of birds on islands. In: Grant P.R, editor. Evolution on islands. Oxford University Press; Oxford, UK: 1998. pp. 142–162. [Google Scholar]
- Guilford T. The evolution of aposematism. In: Evans D.L, Schmidt J.O, editors. Insect defenses: adaptive mechanism and strategies of prey and predators. State University of New York Press; Albany, NY: 1990. pp. 23–64. [Google Scholar]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Hodges R.W, Dominic T, Davis D.R, Ferguson D.C, Franclemont J.G, Monroe E.G, Powell J.A. E. W. Classey Ltd; London, UK: 1983. Check list of the Lepidoptera of America North of Mexico. [Google Scholar]
- Hudson R.R, Coyne J.A. Mathematical consequences of the genealogical species concept. Evolution. 2002;56:1557–1565. doi: 10.1111/j.0014-3820.2002.tb01467.x. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P, Rannala B. Phylogenetic methods come of age: testing hypotheses in an evolutionary context. Science. 1997;276:227–232. doi: 10.1126/science.276.5310.227. doi:10.1126/science.276.5310.227 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P, Ronquist F. MrBayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. doi:10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P, Hillis D.M, Nielsen R. A likelihood-ratio test of monophyly. Syst. Biol. 1996;45:546–558. doi:10.2307/2413530 [Google Scholar]
- JMP 2002 JMP-In statistical exploration software, version 5. Cary, NC: SAS Institute, Inc.
- Joron M, et al. A conserved supergene locus controls colour pattern diversity in Heliconius butterflies. PLoS Biol. 2006;4:e303. doi: 10.1371/journal.pbio.0040303. doi:10.1371/journal.pbio.0040303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layberry R.A, Hall P.W, Lafontaine D. University of Toronto Press; Toronto, Canada: 1998. The butterflies of Canada. [Google Scholar]
- Loxdale H.D, Lushai G. Slaves of the environment: the movement of insects in relation to their ecology and genotype. Phil. Trans. R. Soc. B. 1999;354:1479–1495. doi:10.1098/rstb.1999.0492 [Google Scholar]
- Maddison W.P. Gene trees in species trees. Syst. Biol. 1997;46:523–536. doi:10.2307/2413694 [Google Scholar]
- Maddison W.P, Knowles L.L. Inferring phylogeny despite incomplete lineage sorting. Syst. Biol. 2006;55:21–30. doi: 10.1080/10635150500354928. doi:10.1080/10635150500354928 [DOI] [PubMed] [Google Scholar]
- Maddison, W. P. & Maddison, D. R. 2006 Mesquite: a modular system for evolutionary analysis, version 1.12. See http://mesquiteproject.org
- Mappes J, Marples N, Endler J.A. The complex business of survival by aposematism. Trends Ecol. Evol. 2005;20:593–603. doi: 10.1016/j.tree.2005.07.011. doi:10.1016/j.tree.2005.07.011 [DOI] [PubMed] [Google Scholar]
- Monteiro A, Pierce N.E. Phylogeny of Bicyclus (Lepidoptera: Nymphalidae) inferred from COI, COII and EF-1α gene sequences. Mol. Phylogenet. Evol. 2001;18:264–281. doi: 10.1006/mpev.2000.0872. doi:10.1006/mpev.2000.0872 [DOI] [PubMed] [Google Scholar]
- Mullen S.P. Wing pattern evolution and the origins of mimicry among North American admiral butterflies (Nymphalidae: Limenitis) Mol. Phylogenet. Evol. 2006;39:747–758. doi: 10.1016/j.ympev.2006.01.021. doi:10.1016/j.ympev.2006.01.021 [DOI] [PubMed] [Google Scholar]
- Nijhout H.F. Smithsonian Institution Press; Washington, DC: 1991. The development and evolution of butterfly wing patterns. [Google Scholar]
- Nijhout H.F. Polymorphic mimicry in Papilio dardanus: mosaic dominance, big effects, and origins. Evol. Dev. 2003;5:579–592. doi: 10.1046/j.1525-142x.2003.03063.x. doi:10.1046/j.1525-142X.2003.03063.x [DOI] [PubMed] [Google Scholar]
- Opler, P. A., Pavulaan, H. & Stanford, R. E. 2004 Butterflies of North America Jamestown, ND: Northern Prairie Wildlife Research Center. See http://www.npwrc.usgs.gov/resource/distr/lepid/bflyusa/bflyusa.htm
- Pfennig D.W, Harcombe W.R, Pfennig K.S. Frequency dependant Batesian mimicry. Nature. 2001;410:323. doi: 10.1038/35066628. doi:10.1038/35066628 [DOI] [PubMed] [Google Scholar]
- Pfennig D.W, Harper G.R, Jr, Brumo A.F, Harcombe W.R, Pfenning K.S. Population differences in predation on Batesian mimics in allopatry with their model: selection against mimics is strongest when they are common. Behav. Ecol. Sociobiol. 2007;61:505–511. doi:10.1007/s00265-006-0278-x [Google Scholar]
- Platt A.P. Evolution of North American admiral butterflies. Bull. Entomol. Soc. Am. 1983;29:10–22. [Google Scholar]
- Platt A.P, Brower L.P. Mimetic versus disruptive coloration in intergrading populations of Limenitis arthemis and astyanax butterflies. Evolution. 1968;22:699–718. doi: 10.1111/j.1558-5646.1968.tb03471.x. doi:10.2307/2406897 [DOI] [PubMed] [Google Scholar]
- Platt A.P, Coppinger R.P, Brower L.P. Demonstration of the selective advantage of mimetic Limenitis butterflies presented to caged avian predators. Evolution. 1971;25:692–701. doi: 10.1111/j.1558-5646.1971.tb01927.x. doi:10.2307/2406950 [DOI] [PubMed] [Google Scholar]
- Prudic K.L, Shapiro A.M, Clayton N.S. Evaluating a putative mimetic relationship between two butterflies, Adelpha bredowii and Limenitis lorquini (Lepidoptera: Nymphalidae) Ecol. Entomol. 2002;27:68–75. doi:10.1046/j.0307-6946.2001.00384.x [Google Scholar]
- Prudic K.L, Skemp A.K, Papaj D.R. Aposematic coloration, luminance contrast, and the benefits of conspicuousness. Behav. Ecol. 2007;18:41–46. doi:10.1093/beheco/arl046 [Google Scholar]
- Remington C.L. Genetics of populations of Lepidoptera. Proc. Tenth Int. Cong. Entomol. 1958;2:787–805. [Google Scholar]
- Ritland D.B. Comparative unpalatability of mimetic viceroy butterflies (Limenitis archippus) from four southeastern US populations. Oecologia. 1995;103:327–336. doi: 10.1007/BF00328621. doi:10.1007/BF00328621 [DOI] [PubMed] [Google Scholar]
- Roper T.J. Responses of domestic chicks to artificially colored insect prey: effects of previous experience and background color. Anim. Behav. 1990;39:466–473. doi:10.1016/S0003-3472(05)80410-5 [Google Scholar]
- Roper T.J, Redstone S. Conspicuousness of distasteful prey affects the strength and durability of one-trial avoidance learning. Anim. Behav. 1987;35:739–747. doi:10.1016/S0003-3472(87)80110-0 [Google Scholar]
- Ruxton G.D, Sherratt T.N, Speed M.P. Oxford University Press; Oxford, UK: 2004. Avoiding attack: the evolutionary ecology of crypsis, warning signals and mimicry. [Google Scholar]
- Sherratt T.N, Beatty C.D. The evolution of warning signals as reliable indicators of prey defense. Am. Nat. 2003;162:377–389. doi: 10.1086/378047. doi:10.1086/378047 [DOI] [PubMed] [Google Scholar]
- Stevens M, Cuthill I.C. Disruptive coloration, crypsis and edge detection in early visual processing. Proc. R. Soc. B. 2006;273:2141–2147. doi: 10.1098/rspb.2006.3556. doi:10.1098/rspb.2006.3556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D. L. 2001 PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sunderland, MA: Sinauer Associates.
- Thompson J.N. University of Chicago Press; Chicago, IL: 2005. The geographic mosaic of coevolution. [Google Scholar]
- Turner J.R.G. Butterfly mimicry: the genetical evolution of an adaptation. Evol. Biol. 1977;10:163–206. [Google Scholar]
- USDA 2007 The plants database. Baton Rouge, LA: National Plant Data Center. See http://plants.usda.gov
- Willmott K.R. Cladistic analysis of the neotropical butterfly genus Adelpha (Lepidoptera: Nymphalidae), with comments on the subtribal classification of Limenitidini. Syst. Entomol. 2003;28:279–322. doi:10.1046/j.1365-3113.2003.00209.x [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Specimen information for molecular phylogeny
Approximate geographic ranges of white-banded L. arthemis phenotypes (hatched), mimetic L. arthemis phenotypes (purple) and B. philenor (grey). Ranges after Platt & Brower (1968) and Opler et al. (2004)