Abstract
Self/non-self recognition mechanisms underlie the development, immunology and social behaviour of virtually all living organisms, from bacteria to humans. Indeed, recognition processes lie at the core of how social cooperation evolved. Much evidence suggests that the major histocompatibility complex (MHC) both facilitates nepotistic interactions and promotes inbreeding avoidance. Social discrimination based on MHC differences has been demonstrated in many vertebrates but whether the labels used in discrimination are directly associated with the MHC, rather than with other genes with which it covaries, has remained problematic. Furthermore, effects of familiarity on natural preferences have not been controlled in most previous studies. Here we show that African clawed frog (Xenopus laevis) tadpoles discriminate among familiar full siblings based on MHC haplotype differences. Subjects (N=261) from four parental crosses preferred siblings with which they shared MHC haplotypes to those with no MHC haplotypes in common. Using only full siblings in experimental tests, we controlled for genetic variation elsewhere in the genome that might influence schooling preferences. As test subjects were equally familiar with stimulus groups, we conclude that tadpole discrimination involves a self-referent genetic recognition mechanism whereby individuals compare their own MHC type with those of conspecifics.
Keywords: altruism, immunology, kin recognition, major histocompatibility complex, phenotype matching, recognition alleles
1. Introduction
Major histocompatibility complex (MHC) genes facilitate not only adaptive immune function but also the social behaviour of vertebrates (reviewed in Piertney & Oliver 2006). Social discrimination based on MHC differences may be favoured by kin selection (Manning et al. 1992; Yamazaki et al. 2000) and fitness benefits that accrue to parents that outbreed, including enhanced immunocompetence of MHC-variable offspring (Landry et al. 2001; Piertney & Oliver 2006). MHC loci exhibit extraordinary polymorphism, so labels that they encode should uniquely identify individuals and serve as markers to map their genetic relationships (Thomas 1975). However, whether discrimination is elicited by the MHC, rather than by other genes with which it normally covaries, has been difficult to ascertain.
Frogs were among the first vertebrates shown to demonstrate kin-recognition abilities, most extensively in the context of larval schooling (Waldman & Adler 1979; Waldman 2005) and also in mate choice (Waldman et al. 1992; Waldman & Tocher 1998). Tadpoles can recognize their siblings and paternal half-siblings even without previous experience of them (Blaustein & O'Hara 1981; Waldman 1981; Cornell et al. 1989), which suggests the involvement of genetic labels. Yet, tadpoles' social preferences can be influenced by their social experience (Waldman 1981, 2005) especially during early embryonic development (Hepper & Waldman 1992). Genetic recognition systems, such as those mediated by the MHC or major urinary proteins (Hurst et al. 2005), are known to incorporate influences of individuals' previous social environment (Yamazaki et al. 1988; Penn & Potts 1998a), so these results are not contradictory.
MHC loci play a key role in the immune system by producing molecular markers that regulate cellular self/non-self recognition (Salter-Cid et al. 1998; Gantress et al. 2003). They also uniquely determine individuals' odour profiles that facilitate self/non-self social recognition (Manning et al. 1992; Penn & Potts 1998b; Yamazaki et al. 2000; Penn 2002; Rajakaruna et al. 2006) and are discernible even by heterospecifics (Gilbert et al. 1986). Therefore, MHC molecules should be able to serve as effective kin-recognition labels. Indeed, Atlantic salmon, sand lizards, savannah sparrows, mice and humans all show evidence of MHC-disassortative mating preferences (Piertney & Oliver 2006). Furthermore, MHC differences correlate with nepotistic female choice of communal nesting partners (Manning et al. 1992) and parent–progeny recognition (Yamazaki et al. 2000) in mice, as well as schooling preferences of salmonids (Olse´n et al. 1998, 2002; Rajakaruna et al. 2006).
While intriguing, studies to date have failed to resolve the role of the MHC because either MHC type has been confounded by genome-wide similarities (Manning et al. 1992; Penn & Potts 1998a) or genetic differences have been limited to the MHC (Yamazaki et al. 2000). Here we disentangle MHC effects on tadpoles' association preferences from those attributable to genetic background. Using robust sample sizes, we studied the ability of subjects to discriminate between groups of their siblings based solely on whether they shared with them particular MHC alleles even though, as expected among outbred siblings, they otherwise had half their genes in common.
Owing to its primordial MHC organization, Xenopus laevis differs from all other vertebrates examined to date, as its four MHC loci (one MHC class I locus and three MHC class II loci; Liu et al. 2002) are in complete linkage disequilibrium (Nonaka et al. 1997). Although X. laevis are tetraploid, duplicated MHC genes have become silenced to a diploid number (Flajnik et al. 1999b). The high level of sequence polymorphism observed at the X. laevis MHC class I locus, which exceeds that typical for vertebrates (Bos & Waldman 2006), may compensate for the limiting of MHC gene numbers. Consequently, X. laevis serves as a model organism for examining MHC type discrimination.
We typed tadpoles based on polymorphisms in the MHC class I peptide-binding region (PBR) of four defined haplotypes (Flajnik et al. 1999a; Liu et al. 2002). Owing to the linkage between the MHC loci, we thus were able to test whether they discriminated among siblings based on genetic similarity in the entire MHC region. As we examined only responses of full siblings to one another, any behavioural discrimination must be attributable solely to MHC haplotype differences.
2. Material and methods
(a) Subjects
We used X. laevis that we bred in our colony from stock with known sequences for MHC class I and class II alleles. The haplotypes were defined as f, g, j and r (GenBank: class Ia accession numbers AF185579, AF185580, AF185582 and AF185586; class II accession numbers AF454374–AF454382). The laboratory frog strains originated from the Basel Institute for Immunology and had been bred for several generations in our laboratory. Periodically, stock were outcrossed and phenotypic variation was markedly apparent within and among sibships.
We crossed pairs of MHC-heterozygous frogs that shared haplotypes (i.e. rj×rj, rg×rg, fg×fg and fr×fr). The progeny comprised mixtures of homozygotes and heterozygotes (e.g. the rj×rj cross produced rr, rj and jj progeny). We reared tadpoles with their siblings in groups of 200 within 40 l tanks for two to three weeks. We fed all tadpoles by maintaining a suspension of finely ground nettle. We determined the MHC haplotypes of all stimulus and subject tadpoles by the polymerase chain reaction (PCR) from tail tip tissue before we tested them. After we clipped their tails, we isolated tadpoles in 1 l polypropylene cups for one to four weeks, during which time tadpoles' tails fully regenerated, and we then tested them. At the time of testing, tadpoles had not yet begun developing hind limbs (stage 54; Nieuwkoop & Faber 1956). Only MHC-homozygous tadpoles were used as subjects and stimulus animals in this study.
(b) Sequence-specific primer PCR MHC genotyping
We extracted genomic DNA from tail tips using PrepMan Ultra sample preparation reagent (Applied Biosystems, Foster City, CA, USA). We MHC-typed tadpoles by touchdown PCR using sequence-specific primers that anneal to polymorphic sequences within the MHC class I-α1 domain (encoding the PBR) for each of the four haplotypes (f, g, j and r). In each reaction, we included primers that amplify DNA from a conserved region of the MHC (class I-α3 domain) to control for any failed PCRs that otherwise would be falsely scored as negative (Krausa et al. 1993). We designed primers using Primer3 (Rozen & Skaletsky 2000) from the known X. laevis sequences (Flajnik et al. 1999a; see table 1).
Table 1.
Primer details.
| haplotype | locus | primer direction | primer sequence | amplicon length (bp) | primer concentrations (pmol) | |
|---|---|---|---|---|---|---|
| haplotype specific | control | |||||
| f | class l-α1 | sense | GTCTCAGATCGAGCCTTTGG | 106 | 16.5 | 3.5 |
| antisense | TTGCAGGTTCATCTCTACCAGT | 16.5 | ||||
| g | class l-α1 | sense | GTCTCAGATCGAGCCTTTGG | 178 | 12.5 | 1 |
| antisense | GCTCTGATCCCTTGGCAAT | 20 | ||||
| j | class l-α1 | sense | GTCTCAGATCGAACCTTTGG | 178 | 15 | 0.8 |
| antisense | CCTCTTCTCCTTTCGCTTT | 30 | ||||
| r | class l-α1 | sense | AGATAGAGCATTTGGGCTGC | 134 | 21.2 | 2.5 |
| antisense | ATTCAGGTCCTGCTTTGTCC | 21.2 | ||||
| control | class l-α3 | sense | TCACCCTCATGTAAGAATTTCAGA | 236 | n.a. | n.a. |
| antisense | GCTCCACATGACAGGCATAA | |||||
Sequences were amplified on 96-well PCR plates (Axygen Scientific, PCR-96-C) in 12.5 μl PCRs, each containing 50 ng of template DNA, PCR buffer (63.6 mM KCl, 127.2 mM Tris–HCl (pH 8.3), 1.9 mM MgCl2), 180 μM dNTP (100 mM, Eppendorf) and 0.2 units Taq polymerase (Roche Diagnostics). Primer concentrations varied depending on the haplotype being assessed (table 1).
The conditions for touchdown PCR in a thermocycler (Eppendorf Mastercycler Gradient; Eppendorf, Hamburg, Germany) are as follows: denaturation for 90 s at 94°C, followed by five cycles of denaturation for 30 s at 94°C; annealing for 45 s at 70°C and primer extension for 30 s at 72°C, followed by 20 cycles of denaturation for 30 s at 94°C; annealing for 50 s at 65°C and primer extension for 45 s at 72°C, followed by five cycles of denaturation for 30 s at 94°C; annealing for 1 min at 56°C and primer extension for 2 min at 72°C.
We electrophoresed PCR products next to known positives and negatives for 40 min at 70 V in horizontal 2% agarose gels. Gels were visualized by ethidium bromide fluorescence.
(c) Association preference tests
We simultaneously exposed subjects to two stimulus groups of 10 of their siblings on either side of a testing apparatus, separated by mesh net enclosures. Subjects shared MHC haplotypes with one of the stimulus groups but not with the other. We measured time spent by subjects associating with each of the groups.
Tests were conducted in polypropylene tanks (210×140×45 mm), with removable grey PVC-coated fibreglass (0.028 cm diameter) mesh (7.1×5.5 threads cm−1) nets (43×140×45 mm) at each end, filled with 1.2 l of filtered deep aquifer water at 21°C. We placed 10 size- and stage-matched stimulus tadpoles into each of the mesh nets. A line drawn along the centre of each tank was used to demarcate the two halves of the test arena (124×140×45 mm). Lighting was diffused, achieved by reflecting two 100 W incandescent lamps off the ceiling of the test room.
We introduced test subjects by perforated spoon (to limit water transfer) into the centre of the apparatus. We allowed tadpoles to acclimate for 5 min and then tested them for 40 min. We tested each subject twice, reversing the stimulus groups to eliminate any side bias. Consequently, each tadpole was tested for a total of 80 min. Tadpole association tests were recorded with a CCTV camera (Panasonic WV-BP330/G) using a variable focal lens (Panasonic WV-LZF61/2) positioned 1 m above the testing apparatus and a time-lapse (1/5 speed) VHS recorder (Panasonic AG-TL350). The movements of subject tadpoles were tracked from videotape using EthoVision v. 3.0 (Noldus Information Technology, Wageningen, The Netherlands) and time spent on either side of the centre line was computed for each subject.
Sample sizes varied between genotypes within families depending on the availability of genotyped progeny of appropriate developmental stage (table 2). All subjects were simultaneously presented with MHC-identical siblings and those with which they shared no MHC haplotypes.
Table 2.
Individual preferences by sibship.
| sibship | genotype | N | no. of individuals spending more time near | p | |
|---|---|---|---|---|---|
| MHC-identical siblings | MHC-different siblings | ||||
| rj×rj | rr | 19 | 13 | 6 | 0.073 |
| jj | 18 | 11 | 7 | ||
| rg×rg | rr | 41 | 27 | 14 | 0.005 |
| gg | 31 | 21 | 10 | ||
| fg×fg | ff | 36 | 23 | 13 | 0.031 |
| gg | 41 | 25 | 16 | ||
| fr×fr | ff | 40 | 24 | 16 | 0.015 |
| rr | 35 | 24 | 11 | ||
Within each family, we compared the time spent by subjects of one of the MHC-homozygous genotypes associating with MHC-identical siblings to that spent by subjects of the other MHC-homozygous genotype associating with MHC-different siblings. As time associating with these stimulus groups sum to unity, this permits comparison of time spent oriented to each group while preserving statistical independence. After testing the distribution of time that subjects spent associating with MHC-identical siblings for normality using the Kolmogorov–Smirnov test, we evaluated these differences by two-sample t-tests.
We tested the overall effect of MHC similarity on tadpole association preferences by hierarchically nested analysis of variance. To distinguish between association preferences of the genotypes within families, we compared alternate subjects of each genotype for the time spent associating with MHC-identical siblings and that spent associating with MHC-different siblings. The effects of MHC similarity, family nested within MHC similarity, and genotype nested within family and MHC similarity were included as factors in the analysis. We compared the number of subjects that spent more time on the side of the tank near the MHC-identical stimulus groups to the number of subjects that spent more time near the MHC-dissimilar stimulus groups using the binomial distribution. All statistical inferences were based on two-tailed distributions. Analyses were conducted with Statistica v. 7.1 (Statsoft, Tulsa, OK, USA).
3. Results
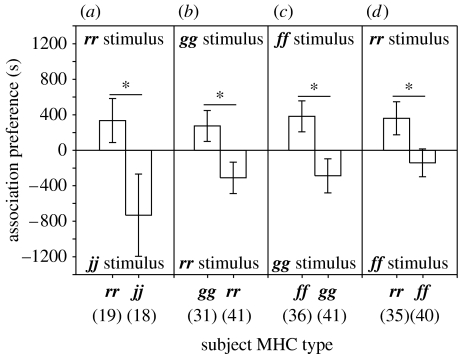
Subjects discriminated among siblings based on their MHC haplotypes. They spent more time with siblings with which they shared MHC haplotypes than with those with which they shared no MHC haplotypes (figure 1). These preferences were consistent among families (rj×rj: t35=2.06, p=0.047; rg×rg: t70=2.31, p=0.024; fg×fg: t75=2.56, p=0.012; and fr×fr: t73=2.08, p=0.041), so we pooled the results. Overall, the effect of MHC similarity on schooling preference was highly significant (F1,245=22.41, p<0.001), whereas variation in MHC-assortative preferences among families (F6,245=0.83, p=0.54) and between MHC types within families (F8,245=1.13, p=0.34) was not significant. Thus, tadpoles' association with siblings bearing their own MHC haplotypes was not confounded by any preference for particular haplotypes that might be more generally ‘attractive’ than others. Across all the families, most subjects preferred MHC-identical siblings to MHC-different siblings (table 2).
Figure 1.
Association preferences (time spent near MHC-identical stimulus group minus time spent near MHC-dissimilar stimulus group) of subjects from four families: (a) rj×rj progeny, (b) rg×rg progeny, (c) fg×fg progeny, (d) fr×fr progeny. Means±s.e.m. are shown and sample sizes are indicated in brackets. *p<0.05 (two tailed).
4. Discussion
Tadpoles' association preferences correlated with, and appear to have been determined by, loci within shared MHC haplotypes. In previous studies, MHC-correlated behaviours corresponded to overall genetic similarity (Manning et al. 1992; Penn & Potts 1998a) or, conversely, owing to limited genetic diversity among subjects, phenotypic differences were restricted to those determined by the MHC (Yamazaki et al. 1988, 2000). As we tested only groups of full siblings in this study, we controlled for overall genetic similarity among subjects and stimulus animals while maintaining genetic diversity, as expected among siblings, elsewhere in the genome. Hence, our results demonstrate that X. laevis tadpoles can recognize cues associated with the genotype of the specific MHC class I-α1 domain (encoding the PBR) or other closely linked loci.
Our results further suggest that tadpoles discriminated MHC similarity by self-referencing. Subjects had interacted freely with their siblings bearing every combination of MHC haplotypes prior to being tested. Previous studies suggest that response biases in anuran larvae are based on templates that incorporate aspects of the early embryonic environment (Waldman 1988, 2005). Tadpoles reared in social isolation from eggs can discriminate between siblings and non-siblings (Blaustein & O'Hara 1981; Waldman 1981, 2005). Furthermore, tadpoles imprint on odorants that are present in their embryonic environment and subsequently orient towards these odours (Hepper & Waldman 1992). In the current study, however, early social interactions could not have contributed to the formation of recognition templates used for discriminating among disparate MHC types as subjects shared their embryonic and early social environments with siblings bearing all the haplotypes. The ability of X. laevis tadpoles to discriminate among siblings based on MHC haplotype is based either on learning one's own MHC type or an inherent ability to recognize MHC similarity rather than on sharing a common embryonic environment.
The immune system of Xenopus tadpoles appears to function without MHC class I molecules expressed on cell surfaces (Salter-Cid et al. 1998; Flajnik et al. 1999a). This suggests that loci in linkage disequilibrium with the MHC class I, such as the MHC class II that is expressed on cell surfaces in tadpoles, contribute to the cues involved in directing the observed association preferences. However, MHC class I mRNA transcripts are expressed in the lungs, gills and intestine of tadpoles (Salter-Cid et al. 1998). Despite the limited expression of the MHC class Ia in tadpoles, its expression in organs with epithelial surfaces in contact with the environment may be sufficient for the production of MHC-determined odours.
Both MHC class I (in terrestrial vertebrates) and class II loci (in fishes) have been shown to influence behavioural discrimination (Piertney & Oliver 2006). MHC class I molecules bind peptides generated in the cytosol, whether endogenously or virally derived (Bernatchez & Landry 2003). By contrast, MHC class II molecules are mainly responsible for the immune presentation of extracellular pathogens (Bernatchez & Landry 2003). Class II molecules are more likely to influence the microbial flora of their vertebrate hosts, which may contribute to individual odour profiles (Schellinck et al. 1995; Lanyon et al. 2007). Nonetheless, both class I (Leinders-Zufall et al. 2004) and class II (Milinski et al. 2005) peptide ligands have been found to influence chemosensory signals. The relative roles of these functionally distinct loci on MHC type discrimination are not understood. Because the expression of the MHC class I changes ontogenetically (Salter-Cid et al. 1998), X. laevis may prove to be a model organism for elucidating the mechanism by which MHC type discrimination is achieved—whether through the release of MHC class I molecules from epithelial tissues in contact with the environment, or of volatile aromatics or peptide products associated with either MHC class I or class II expression (Penn 2002).
Our experiment allowed us to identify behavioural preferences attributable solely to the MHC within families that shared a normal complement of half of their non-MHC genes. These results suggest that the MHC is also used in social discrimination between relatives and non-relatives, and among non-relatives, but further tests are needed to validate these conclusions.
The MHC discrimination demonstrated here is likely to be kin-selected rather than an incidental consequence of MHC expression. Unlike MHC-biased mating preferences, which may confer direct fitness benefits on offspring by increasing their immunocompetence (Landry et al. 2001; Piertney & Oliver 2006), MHC-assortative schooling is more likely to decrease tadpoles' direct fitness as MHC-similar individuals share disease susceptibilities (Gantress et al. 2003; Barribeau 2007). While tadpoles might be less likely to become infected by their siblings (Lewis 1998), any new pathogen to which they have no specific immunity will probably spread throughout the group and cause more mortality than would be the case if tadpoles were schooling with non-siblings bearing a diversity of MHC alleles. The inclusive fitness benefits associated with kin discrimination should therefore outweigh the decreased direct fitness consequences of MHC-assortative schooling.
This study provides robust evidence of social discrimination based on MHC similarity without the possibility of confounding environmental and genetic factors. Furthermore, because tadpole association preferences correlate with MHC similarity, even when subjects are equally familiar with all sibling MHC types, our results provide evidence for self-referent matching of MHC-determined phenotypes. Tadpoles use highly polymorphic matching loci to socially discriminate among conspecifics, which should permit the effective discrimination of kin by genetic similarity detection (Grafen 1990).
Acknowledgments
All protocols involving animals were approved by the University of Canterbury Animal Ethics Committee.
We thank Andrew Bagshaw, Seth Barribeau, Louis Du Pasquier, Martin Flajnik, Neil Gemmell, Marie Hale, Tia Neha and two anonymous referees for their comments on the manuscript; Louis Du Pasquier and Martin Flajnik for supplying us with the X. laevis frog lines used; and Kelly Lock and Sandra Negro for their help in processing tissue samples for genotyping tadpoles. This work was supported by the Marsden Fund (Royal Society of New Zealand) and Noldus Information Technology. B.W. was supported by a University of Canterbury travelling fellowship while preparing the manuscript.
References
- Barribeau, S. M. 2007 The environmental, social, and genetic factors predisposing Xenopus laevis tadpoles to infection. PhD thesis, University of Canterbury.
- Bernatchez L, Landry C. MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J. Evol. Biol. 2003;16:363–377. doi: 10.1046/j.1420-9101.2003.00531.x. doi:10.1046/j.1420-9101.2003.00531.x [DOI] [PubMed] [Google Scholar]
- Blaustein A.R, O'Hara R.K. Genetic control for sibling recognition? Nature. 1981;290:246–248. doi: 10.1038/290246a0. doi:10.1038/290246a0 [DOI] [PubMed] [Google Scholar]
- Bos D.H, Waldman B. Polymorphism, natural selection, and structural modeling of class Ia MHC in the African clawed frog (Xenopus laevis) Immunogenetics. 2006;58:433–442. doi: 10.1007/s00251-006-0114-5. doi:10.1007/s00251-006-0114-5 [DOI] [PubMed] [Google Scholar]
- Cornell T.J, Berven K.A, Gamboa G.J. Kin recognition by tadpoles and froglets of the wood frog Rana sylvatica. Oecologia. 1989;78:312–316. doi: 10.1007/BF00379103. doi:10.1007/BF00379103 [DOI] [PubMed] [Google Scholar]
- Flajnik M.F, Ohta Y, Greenberg A.S, Salter-Cid L, Carrizosa A, Du Pasquier L, Kasahara M. Two ancient allelic lineages at the single classical class I locus in the Xenopus MHC. J. Immunol. 1999a;163:3826–3833. [PubMed] [Google Scholar]
- Flajnik M.F, Ohta Y, Namikawa-Yamada C, Nonaka M. Insight into the primordial MHC from studies in ectothermic vertebrates. Immunol. Rev. 1999b;167:59–67. doi: 10.1111/j.1600-065x.1999.tb01382.x. doi:10.1111/j.1600-065X.1999.tb01382.x [DOI] [PubMed] [Google Scholar]
- Gantress J, Maniero G.D, Cohen N, Robert J. Development and characterization of a model system to study amphibian immune responses to iridoviruses. Virology. 2003;311:254–262. doi: 10.1016/s0042-6822(03)00151-x. doi:10.1016/S0042-6822(03)00151-X [DOI] [PubMed] [Google Scholar]
- Gilbert A.N, Yamazaki K, Beauchamp G.K, Thomas L. Olfactory discrimination of mouse strains (Mus musculus) and major histocompatibility types by humans (Homo sapiens) J. Comp. Psychol. 1986;100:262–265. doi:10.1037/0735-7036.100.3.262 [PubMed] [Google Scholar]
- Grafen A. Do animals really recognize kin? Anim. Behav. 1990;39:42–54. doi:10.1016/S0003-3472(05)80724-9 [Google Scholar]
- Hepper P.G, Waldman B. Embryonic olfactory learning in frogs. Q. J. Exp. Psychol. B. 1992;44:179–197. doi: 10.1080/02724999208250611. [DOI] [PubMed] [Google Scholar]
- Hurst J.L, Thom M.D, Nevison C.M, Humphries R.E, Beynon R.J. MHC odours are not required or sufficient for recognition of individual scent owners. Proc. R. Soc. B. 2005;272:715–724. doi: 10.1098/rspb.2004.3004. doi:10.1098/rspb.2004.3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krausa P, Moses J, Bodmer W, Bodmer J, Browning M. HLA-A locus alleles identified by sequence specific PCR. Lancet. 1993;341:121–122. doi: 10.1016/0140-6736(93)92605-s. doi:10.1016/0140-6736(93)92605-S [DOI] [PubMed] [Google Scholar]
- Landry C, Garant D, Duchesne P, Bernatchez L. ‘Good genes as heterozygosity’: the major histocompatibility complex and mate choice in Atlantic salmon (Salmo salar) Proc. R. Soc. B. 2001;268:1279–1285. doi: 10.1098/rspb.2001.1659. doi:10.1098/rspb.2001.1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanyon C.V, Rushton S.P, O'Donnell A.G, Goodfellow M, Ward A.C, Petrie M, Jensen S.P, Gosling L.M, Penn D.J. Murine scent mark microbial communities are genetically determined. Microbiol. Ecol. 2007;59:576–583. doi: 10.1111/j.1574-6941.2006.00252.x. doi:10.1111/j.1574-6941.2006.00252.x [DOI] [PubMed] [Google Scholar]
- Leinders-Zufall T, et al. MHC class I peptides as chemosensory signals in the vomeronasal organ. Science. 2004;306:1033–1037. doi: 10.1126/science.1102818. doi:10.1126/science.1102818 [DOI] [PubMed] [Google Scholar]
- Lewis K. Pathogen resistance as the origin of kin altruism. J. Theor. Biol. 1998;193:359–363. doi: 10.1006/jtbi.1998.0725. doi:10.1006/jtbi.1998.0725 [DOI] [PubMed] [Google Scholar]
- Liu Y, Kasahara M, Rumfelt L.L, Flajnik M.F. Xenopus class II A genes: studies of genetics, polymorphism, and expression. Dev. Comp. Immunol. 2002;26:735–750. doi: 10.1016/s0145-305x(02)00034-4. doi:10.1016/S0145-305X(02)00034-4 [DOI] [PubMed] [Google Scholar]
- Manning C.J, Wakeland E.K, Potts W.K. Communal nesting patterns in mice implicate MHC genes in kin recognition. Nature. 1992;360:581–583. doi: 10.1038/360581a0. doi:10.1038/360581a0 [DOI] [PubMed] [Google Scholar]
- Milinski M, Griffiths S, Wegner K.M, Reusch T.B.H, Haas-Assenbaum A, Boehm T. Mate choice decisions of stickleback females predictably modified by MHC peptide ligands. Proc. Natl Acad. Sci. USA. 2005;102:4414–4418. doi: 10.1073/pnas.0408264102. doi:10.1073/pnas.0408264102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop P.D, Faber J. North-Holland; Amsterdam, The Netherlands: 1956. Normal table of Xenopus laevis (Daudin) [Google Scholar]
- Nonaka M, Namikawa C, Kato Y, Sasaki M, Salter-Cid L, Flajnik M.F. Major histocompatibility complex gene mapping in the amphibian Xenopus implies a primordial organization. Proc. Natl Acad. Sci. USA. 1997;94:5789–5791. doi: 10.1073/pnas.94.11.5789. doi:10.1073/pnas.94.11.5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olse´n K.H, Grahn M, Lohm J, Langefors A. MHC and kin discrimination in juvenile Arctic char, Salvelinus alpinus (L.) Anim. Behav. 1998;56:319–327. doi: 10.1006/anbe.1998.0837. doi:10.1006/anbe.1998.0837 [DOI] [PubMed] [Google Scholar]
- Olse´n K.H, Grahn M, Lohm J. Influence of MHC on sibling discrimination in Arctic char, Salvelinus alpinus (L.) J. Chem. Ecol. 2002;28:783–795. doi: 10.1023/a:1015240810676. doi:10.1023/A:1015240810676 [DOI] [PubMed] [Google Scholar]
- Penn D.J. The scent of genetic compatibility: sexual selection and the major histocompatibility complex. Ethology. 2002;108:1–21. doi:10.1046/j.1439-0310.2002.00768.x [Google Scholar]
- Penn D, Potts W. MHC-disassortative mating preferences reversed by cross-fostering. Proc. R. Soc. B. 1998a;265:1299–1306. doi: 10.1098/rspb.1998.0433. doi:10.1098/rspb.1998.0433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn D, Potts W.K. Untrained mice discriminate MHC-determined odors. Physiol. Behav. 1998b;64:235–243. doi: 10.1016/s0031-9384(98)00052-3. doi:10.1016/S0031-9384(98)00052-3 [DOI] [PubMed] [Google Scholar]
- Piertney S.B, Oliver M.K. The evolutionary ecology of the major histocompatibility complex. Heredity. 2006;96:7–21. doi: 10.1038/sj.hdy.6800724. doi:10.1038/sj.hdy.6800724 [DOI] [PubMed] [Google Scholar]
- Rajakaruna R.S, Brown J.A, Kaukinen K.H, Miller K.M. Major histocompatibility complex and kin discrimination in Atlantic salmon and brook trout. Mol. Ecol. 2006;15:4569–4575. doi: 10.1111/j.1365-294X.2006.03113.x. doi:10.1111/j.1365-294X.2006.03113.x [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics methods and protocols: methods in molecular biology. Humana Press; Totowa, NJ: 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Salter-Cid L, Nonaka M, Flajnik M.F. Expression of MHC class Ia and class Ib during ontogeny: high expression in epithelia and coregulation of class Ia and lmp7 genes. J. Immunol. 1998;160:2853–2861. [PubMed] [Google Scholar]
- Schellinck H.M, Rooney E, Brown R.E. Odors of individuality of germfree mice are not discriminated by rats in a habituation–dishabituation procedure. Physiol. Behav. 1995;57:1005–1008. doi: 10.1016/0031-9384(94)00353-7. doi:10.1016/0031-9384(94)00353-7 [DOI] [PubMed] [Google Scholar]
- Thomas L. Symbiosis as an immunologic problem: the immune system and infectious diseases. In: Neter E, Milgrom F, editors. Fourth international congress of immunology. S. Karger; Basel, Germany: 1975. pp. 2–11. [Google Scholar]
- Waldman B. Sibling recognition in toad tadpoles: the role of experience. Z. Tierpsychol. 1981;56:341–358. [Google Scholar]
- Waldman B. The ecology of kin recognition. Annu. Rev. Ecol. Syst. 1988;19:543–571. doi:10.1146/annurev.es.19.110188.002551 [Google Scholar]
- Waldman B. Kin recognition in amphibians. In: Hepper P.G, editor. Kin recognition. Cambridge University Press; Cambridge, UK: 2005. pp. 162–219. [Google Scholar]
- Waldman B, Adler K. Toad tadpoles associate preferentially with siblings. Nature. 1979;282:611–613. doi:10.1038/282611a0 [Google Scholar]
- Waldman B, Tocher M. Behavioral ecology, genetic diversity, and declining amphibian populations. In: Caro T, editor. Behavioral ecology and conservation biology. Oxford University Press; New York, NY: 1998. pp. 394–443. [Google Scholar]
- Waldman B, Rice J.E, Honeycutt R.L. Kin recognition and incest avoidance in toads. Am. Zool. 1992;32:18–30. doi:10.1093/icb/32.1.18 [Google Scholar]
- Yamazaki K, Beauchamp G.K, Kupniewski D, Bard J, Thomas L, Boyse E.A. Familial imprinting determines H-2 selective mating preferences. Science. 1988;240:1331–1332. doi: 10.1126/science.3375818. doi:10.1126/science.3375818 [DOI] [PubMed] [Google Scholar]
- Yamazaki K, Beauchamp G.K, Curran M, Bard J, Boyse E.A. Parent–progeny recognition as a function of MHC odortype identity. Proc. Natl Acad. Sci. USA. 2000;97:10 500–10 502. doi: 10.1073/pnas.180320997. doi:10.1073/pnas.180320997 [DOI] [PMC free article] [PubMed] [Google Scholar]