Abstract
Extinction risk varies among species, and comparative analyses can help clarify the causes of this variation. Here we present a phylogenetic comparative analysis of species-level extinction risk across nearly the whole of the class Mammalia. Our aims were to examine systematically the degree to which general predictors of extinction risk can be identified, and to investigate the relative importance of different types of predictors (life history, ecological, human impact and environmental) in determining extinction risk. A single global model explained 27.3% of variation in mammal extinction risk, but explanatory power was lower for region-specific models (median R2=0.248) and usually higher for taxon-specific models (median R2=0.383). Geographical range size, human population density and latitude were the most consistently significant predictors of extinction risk, but otherwise there was little evidence for general, prescriptive indicators of high extinction risk across mammals. Our results therefore support the view that comparative models of relatively narrow taxonomic scope are likely to be the most precise.
Keywords: Red List, phylogenetically independent contrasts, supertree
1. Introduction
Why are some species at greater risk of extinction than others? Identifying the underlying causes of high extinction risk is an important step in understanding the processes contributing to current species declines, and predicting the probable future declines in the face of escalating human pressure on natural habitats. Although the set of circumstances contributing to extinction risk is unique for each species (and often for populations), comparative studies have begun to reveal general patterns and correlates of extinction risk within large groups of species. In general, the distribution of extinction risk among species is phylogenetically non-random, with some taxonomic groups more likely to contain threatened species than others (McKinney 1997; Purvis et al. 2000b). This implies that biological differences among taxa are at least partly responsible for the differences in extinction risk. Indeed, ecological and life-history traits are often associated strongly with extinction risk in comparative analyses (Fisher & Owens 2004; Reynolds et al. 2005).
However, comparative studies have rarely been able to explain the greater part of variation in extinction risk among species; models typically have R2 values less than 0.5, leaving over half of the variation unaccounted for. Furthermore, comparative models for different taxa often give idiosyncratic, and sometimes conflicting, results (Purvis et al. 2000b; Fisher & Owens 2004). For these reasons, it has been suggested that the most powerful and informative comparative models of extinction risk will be those of narrow scope, restricted to single regions and relatively small taxonomic groups (Fisher & Owens 2004). As with all models, however, there is an obvious trade-off between predictive power and generality. Tightly focused extinction risk models may be the most powerful, but the results may not be applicable beyond the particular region or taxon in question. Furthermore, comparative studies of narrow focus often necessarily deal with small datasets, limiting the scope for testing complex hypotheses without the risk of overparametrizing models. An alternative approach is to broaden the scope of the models by analysing global datasets for large taxa, while explicitly testing and accounting for heterogeneity among phylogenetic and geographical subsets. In this way, the generality of the results may be maximized while permitting the possibility that the correlates of extinction risk vary among regions and taxa. Indeed, such heterogeneity is itself of interest, both for understanding extinction processes and for mitigating human impacts. By applying a common methodology to a global dataset, this approach also reduces the problems involved with synthesizing results of disparate studies, which is often confounded by the differences in analytical methods and the types of response and predictor variables selected for study (Fisher & Owens 2004; Purvis et al. 2005).
In this paper, we apply such a global approach to analysing correlates of species-level extinction risk in non-marine mammals. Part of the reason that predictive power of comparative studies has been limited may be that with few exceptions (e.g. Fisher et al. 2003; Cardillo et al. 2004, 2005), previous studies have focused on intrinsic (i.e. ecological and life history) traits as extinction risk predictors. Intrinsic predictors of extinction risk only tell part of the story: they represent the degree to which different species are able to withstand external, usually anthropogenic, threatening processes. A species' risk of extinction must be influenced not only by its biology but also by the severity of the impacts to which it is exposed, and by the interaction between external and intrinsic factors (Reynolds 2003; Purvis et al. 2005; Price & Gittleman 2007). Hence, the threatening processes themselves must also be part of the extinction risk equation. We therefore include in our study both intrinsic and external factors, and their interactions. Our list of putative predictors includes intrinsic traits shown in previous studies to be associated with extinction risk in mammals, including body size, home range size, trophic level, population density, geographical range size and life-history indicators of maximum rate of population increase, such as gestation length, interbirth interval and litter size (e.g. Purvis et al. 2000a; Johnson et al. 2002; Fisher et al. 2003; Jones et al. 2003; Isaac & Cowlishaw 2004; O'Grady et al. 2004; Cardillo et al. 2005, 2006; Price & Gittleman 2007). For example, large body size has been linked to elevated extinction risk because larger mammal species are more likely targets for hunting, but also because larger species usually have lower reproductive rates and slower population growth (Cardillo et al. 2005). We also include external factors that might mediate species' responses to human impact. Mammal population sizes may be related to temperature or rainfall, and in the regions of low productivity or resource availability, populations may be highly variable, leaving them more vulnerable to extinction (Owen-Smith 1990; Owen-Smith et al. 2005). We examine the degree to which the sets of extinction risk predictors are consistent across the mammals, both phylogenetically and geographically. Our primary aim is to examine the degree to which future mammal declines and extinctions are predictable, by providing the most thorough analysis to date of the correlates of mammal extinction risk.
2. Material and methods
(a) Databases
Our measure of extinction risk was derived from the IUCN Red List categories (IUCN 2004), converted to an ordinal index from 0 to 5, following several previous studies (Purvis et al. 2000a; Jones et al. 2003; Cardillo et al. 2004). We excluded from all our analyses (including the calculation of external threat index (ETI); see below) those threatened species not listed under criterion A of the Red List (a measurable recent decline in distribution or population size), to avoid any circularity inherent in predicting extinction risk from geographical range size or population density. Our extinction risk index therefore corresponds to a coarse measure of the rate of recent and ongoing decline in population size or the extent of geographical distribution, but one based on objective, quantitative criteria (Rodrigues et al. 2006).
We compiled information on four kinds of variables as putative predictors of extinction risk: life history; ecological; human impact; and environmental (definitions and descriptions of the variables included are provided in the electronic supplementary material). Biological data came from the ‘PanTheria’ database, a compilation of data on ecological and life-history traits for up to 4030 mammal species, from over 3300 published literature sources. Before use in the analyses, data values were put through an error checking procedure to identify and remove extreme outliers that were likely to have represented data entry errors. Multiple data values for a single trait for each species were then summarized to a single measure of central tendency. Full details of these procedures and the construction of the PanTheria database, and the database itself, will be provided in a forthcoming publication. For phylogenetic comparative analyses, we used a dated, composite supertree phylogeny of 4510 mammal species (the ‘best-dates’ tree of Bininda-Emonds et al. 2007).
Species geographical range maps (Sechrest 2003; Grenyer et al. 2006) were used to calculate range sizes and generate derived variables summarizing human impact and environmental conditions within each species' range. We used three indirect measures of human impact as follows: (i) mean human population density (HPD) within the geographical range of each species, from the 1995 Gridded Population of the World database (CIESIN 2000), (ii) the 5th percentile of the distribution of HPD values within a species' range, which represents the amount of ‘people-free space’ available to each species, e.g. a value of 50 means that HPD is below 50 people km−2 in only 5% of a species' range and (iii) the ETI (Cardillo et al. 2005). ETI is calculated for a given species i as
where j is a species that shares part of its geographical range with species i; rj is the extinction risk index value of species j; and w is the size of the geographical range shared by species i and j. ETI is intended to summarize the set of threatening processes that may affect mammals generally but are not necessarily captured by measuring HPD. For each species' geographical range, we also calculated the absolute latitude of the geographical centroid, the median values of actual evapotranspiration, mean annual temperature and precipitation, and whether or not the species is island endemic (restricted to non-continental landmasses). Climate data were from the GRID databases of the United Nations Environment Program (UNEP 2003). Geographical data were processed in ArcGIS (ESRI 2002), and the calculations were corrected for latitudinal distortions in the area of grid cells.
(b) Statistical models
To avoid statistical problems arising from the phylogenetic signal in extinction risk and many of the putative predictors (Fisher & Owens 2004; Purvis et al. 2005), all analyses were done using phylogenetically independent contrasts. Contrasts were calculated after transforming phylogenetic branch lengths by raising them to a power (κ), with the value of κ optimized for each variable to minimize the correlation between absolute scaled contrasts and their standard deviations (Garland et al. 1992). Soft polytomies were resolved arbitrarily into a series of bifurcations separated by zero-length branches, and contrasts computed at each resulting bifurcation; these contrasts were then given reduced weight to ensure a single degree of freedom for each polytomy. Different arbitrary resolutions give exactly the same result, and the downweighting ensures that the tests will be statistically conservative (Purvis & Garland 1993).
All models were run using linear regression forced through the origin (Garland et al. 1992). We began by finding a global minimum adequate model (MAM) from the full set of predictor variables, for all mammal species in the dataset. Because there were a large number of missing values in the dataset, varying the set of predictors in the model usually changed the number of species represented, making it difficult to guarantee finding the best-fitting model. To search for the best model as effectively as possible, we followed the heuristic procedures described by Purvis et al. (2000a) and Cardillo et al. (2004). At each step in fitting the model, we tested model robustness by identifying and removing any strongly influential contrasts, defined as values with studentized residuals of three or more (Jones & Purvis 1997).
Having thus identified a set of predictors that independently contribute to mammal extinction risk, we then tested whether the slopes of these predictors varied among mammal taxa and geographical regions. We divided the mammals into the following 10 groups (hereafter ‘taxa’): Carnivora; Primates; Rodentia; marsupials; ungulates (Artiodactyla+Perissodactyla); Lagomorpha; Afrotheria (Proboscidea+Hyracoidea+Macroscelidea+Tubulidentata+Afrosoricida); Chiroptera; and ‘minor clades’ (Xenarthra+Scandentia+Eulipotyphla+Monotremata). The minor clades were pooled for analysis because each had too few data values for reliable model fitting; all the other groups form monophyletic clades in the mammal supertree, although the ungulates exclude Cetaceans from the Cetartiodactyla. We divided the world into the following geographical regions based on the continental subdivisions in the ArcGIS basemap: Africa; Australasia (Australia+New Zealand+New Guinea and the surrounding islands); Eurasia; North America; and South America. We then fitted two extra models: one that included the terms in the MAM together with the terms describing the interactions between taxon and each main effect, and another with interactions between geographical region and each main effect. In the latter model, we included only those mammal species endemic to each region to ensure that we were using exclusive, non-overlapping sets of species. This avoided the need to either assign wide-ranging species to one region or another, or to include them multiple times in the analysis. In the models with interactions, we set the value of κ to zero to ensure equal branch lengths before calculating phylogenetically independent contrasts; this was necessary because the optimization procedure renders slope estimates non-comparable (Garland et al. 1992). Finally, we identified the sets of independent predictors for each mammal taxon and each region separately, by finding a MAM for each, using the procedures outlined above. We did not fit a separate model for Afrotheria, because only two species were listed as threatened under criterion A in this clade, giving very few informative contrasts. Calculating independent contrasts, optimizing κ and fitting regression models were done with a set of functions written by A.P. in R (R Development Core Team 2007).
3. Results
(a) Global models of extinction risk
In presenting and discussing our results, we use adjusted R2 values as a guide to the explanatory power of the various models. We emphasize, however, that R2 values for regressions forced through the origin may be biased (Eisenhauer 2003) and should be interpreted cautiously. A global MAM based on the full mammal dataset explained 27.3% of the variation in extinction risk. This identified five variables as significant independent predictors of extinction risk (table 1): weaning age (positive effect); population density (negative); home range size (positive); geographical range size (negative); and 5th percentile of HPD (positive). Significant quadratic terms for population density and HPD were included in the model, and HPD was involved in two significant interaction terms, one with geographical range size and one with population density.
Table 1.
Global minimum adequate models (MAMs) of mammal extinction risk, using phylogenetically independent contrasts. (Model with no taxon or region interaction terms: , d.f.=302. This model includes 372 species; there are fewer degrees of freedom than this because multispecies polytomies contribute a single degree of freedom each to the model. Model including terms from the MAM plus a taxon–HPD interaction: , d.f.=277. Note that this model excludes nine between-taxon contrasts that are included in the MAM. Model including terms from the MAM plus region–population density and region–geographical range size interactions: , d.f.=206. Degrees of freedom are lower for this model because it excludes species found across multiple regions. #p≤0.05; *p≤0.1; **p≤0.01; ***p≤0.001; HPD=5th percentile value of human population density within a species' distribution.)
| no taxon or region interactions | taxon interaction | region interaction | |||
|---|---|---|---|---|---|
| predictor | t value | F value | partial R2 | F value | partial R2 |
| weaning age | 2.26# | 6.87** | 0.018 | 20.46*** | 0.068 |
| home range size | 4.02*** | 1.2 | 0.003 | 4.63# | 0.015 |
| population density | −1.83* | 0.39 | 0.001 | 0.06 | 0.0002 |
| population density2 | 2.17# | 10.08** | 0.024 | 7.33** | 0.024 |
| geographical range size | −3.23** | 63.2*** | 0.151 | 39.57*** | 0.131 |
| HPD | 4.35*** | 1.5 | 0.004 | 0.02 | 0.0001 |
| HPD2 | −3.9*** | 1.7 | 0.004 | 2.1 | 0.007 |
| geographical range size–HPD | −3.39*** | 15.4*** | 0.04 | 0.99 | 0.003 |
| population density–HPD | −2.9** | 0.59 | 0.001 | 0.47 | 0.002 |
| taxon–HPD | 5.47*** | 0.068 | |||
| region–population density | 2.63# | 0.035 | |||
| region–geographical range size | 2.55# | 0.034 | |||
(b) Variation in extinction risk models among taxa
To test for variation among taxa in the slopes of these five predictors, we added to the MAM terms describing the interactions between taxon and each main effect. Only the interaction with HPD was significant (table 1). Including this interaction term added a small amount of predictive power to the MAM (adjusted R2=0.282), although the sets of data points used in the two models were non-identical, because nine between-taxon contrasts were omitted from the model with the interaction. Nevertheless, when these nine contrasts were removed from the MAM, the model with a taxon–HPD interaction term was still preferred over a model without this interaction (AIC=63.74 and 86.76, respectively).
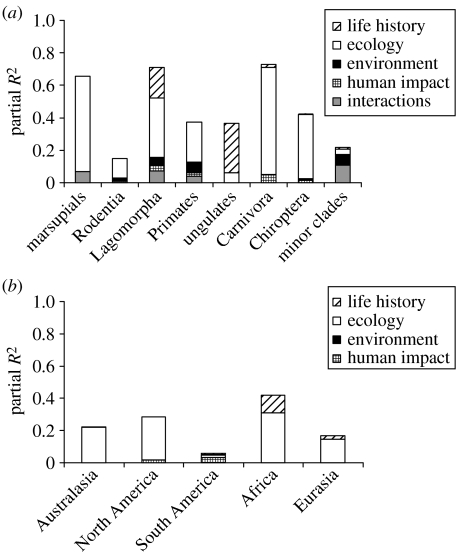
To examine further the variation among taxa, we found a separate MAM for each (table 2). Subdividing the mammals in this way produced models with substantially higher explanatory power, in most cases, than the global MAM: with the exception of the rodents (adjusted R2=0.156) and minor clades (adjusted R2=0.195), R2 values ranged from 0.33 for ungulates to 0.7 for Carnivora. However, there was little qualitative consistency between the models. Geographical range size was the most common predictor, appearing in every model except that for ungulates. In the representation of other predictors, however, the models differed considerably. There were also substantial differences among taxa in the proportion of variation in extinction risk explained by the different types of predictors (figure 1a): for example, life-history variables accounted for none of the variation in risk for rodents, but a large proportion of the variation in risk for ungulates and minor clades.
Table 2.
Minimum adequate models of extinction risk for mammal taxa. (Values shown are t values. HPD, human population density; AET, mean annual actual evapotranspiration. #p≤0.05; *p≤0.1; **p≤0.01; ***p≤0.001.)
| Carnivora | ungulates | Chiroptera | marsupials | Primates | Rodentia | Lagomorpha | minor clades | |
|---|---|---|---|---|---|---|---|---|
| R2 | 0.7 | 0.326 | 0.419 | 0.649 | 0.346 | 0.156 | 0.678 | 0.186 |
| species in model | 62 | 60 | 761 | −191 | 181 | 1644 | 51 | 208 |
| degrees of freedom | 49 | 46 | 368 | 145 | 129 | 590 | 44 | 99 |
| life history | ||||||||
| adult mass | −1.87* | −4.13*** | 0.74 | 3.15** | ||||
| adult mass2 | 2.19# | |||||||
| weaning age | 3.47** | |||||||
| litter size | 4.55*** | |||||||
| litters per year | −3.04** | |||||||
| gestation length | 3.21*** | |||||||
| ecology | ||||||||
| population density | −3.28** | −2.13# | ||||||
| geographical range size | −4.59*** | 0.55 | −15.57*** | −5.67*** | −7.25*** | 0.97 | 1.38 | |
| geographical range size2 | 2.96*** | −2.86** | ||||||
| environmental | ||||||||
| precipitation | 3.05** | 2.47* | ||||||
| latitude | 2.49# | −2.19# | 3.82*** | |||||
| AET | −2.93** | |||||||
| human impact | ||||||||
| HPD 5th percentile | −1.52 | −1.53 | 2.52# | |||||
| HPD 5th percentile2 | 2.75** | −2.1# | ||||||
| HPD mean | 2.99** | 3.75*** | ||||||
| interactions | ||||||||
| adult mass–latitude | 2.86** | |||||||
| geographical range size–adult mass | 5.39*** | −2.74** | ||||||
| geographical range size–HPD mean | −3.28** | |||||||
Figure 1.
Partitioning of variance in extinction risk among four different types of predictors. (a) Taxon-specific models and (b) region-specific models.
(c) Variation in extinction risk models among regions
We next examined geographical variation in extinction risk models. In a model that included interactions between predictors and region, the interactions between region and population density and those between region and geographical range size were significant (table 1). This model had slightly lower predictive power than the MAM (R2=0.262), although again, the set of contrasts used was not identical to the MAM, as species occurring across multiple regions were excluded. When fitted to identical sets of contrasts, however, the model with region–population density and region–geographical range size interactions was preferred to the model without these interactions (AIC=249.91 and 258.6, respectively). Separate MAMs for the different geographical regions had lower predictive power, in general, than those for the separate taxa: R2 values ranged from 0.114 for South America to 0.406 for Africa (table 3). Although geographical range size appeared in every region-specific model, there was, again, considerable variation between the models for different regions, both in the predictors represented and in the partitioning of variance among the different predictor types (figure 1b).
Table 3.
Minimum adequate models of mammal extinction risk for geographical regions. (Values shown are t values. HPD, human population density; ETI, external threat index. #p≤0.05; *p≤0.1; **p≤0.01; ***p≤0.001.)
| Australasia | North America | South America | Africa | Eurasia | |
|---|---|---|---|---|---|
| R2 | 0.248 | 0.277 | 0.114 | 0.406 | 0.162 |
| species in model | 262 | 451 | 607 | 220 | 517 |
| degrees of freedom | 162 | 284 | 266 | 160 | 321 |
| life history | |||||
| adult mass | −2.6# | 3.84*** | 2.79** | 2.87** | |
| weaning age | 2.99** | ||||
| ecology | |||||
| geographical range size | −5.84*** | −10.24*** | 1.69* | −5.21*** | −7.52*** |
| geographical range size2 | 4.09*** | ||||
| environmental | |||||
| island status | |||||
| human impact | |||||
| HPD 5th percentile | −0.81 | ||||
| HPD 5th percentile2 | 2.3# | ||||
| HPD rate of increase | |||||
| ETI | 1.57 | ||||
| ETI2 | −1.96# | ||||
| interactions | |||||
| geographical range size–adult mass | 2.55# | −3.73*** | |||
4. Discussion
Our comparative analyses show that the chance of a mammal species surviving far into the future will depend on multiple, interacting aspects of its biology, geography and the external threats to which it is exposed. If we think of global mammal diversity as passing through a filter induced by human activity (Balmford 1996), these results offer an insight into the nature of this filter. Thus, among mammals generally the species most likely to go extinct will be those that wean at a late age, require large home ranges, live at low population densities, have narrow geographical distributions and share the great majority of their distributions with large human populations. Furthermore, there appear to be synergistic effects that indicate a ‘double jeopardy’ for species with small distributions or low population densities that also inhabit regions densely populated by people. For example, the blackbuck Antilope cervicapra (currently listed as Near Threatened) has a value for HPD 5th percentile of 80.25 people km−2 (in the top 5% of values among all species), but has itself a mean population density of only 4.31 individuals km−2 (in the bottom 5% of values among species). The Javan gibbon Hylobates moloch (currently listed as Critically Endangered) is in a similar position, sharing its distribution with 645 people km−2 but having itself a population density of only 6.18 individuals km−2. Species such as these will have little access to people-free refuge areas where they can maintain viable populations large enough for long-term survival.
Although it explains 27% of variation in extinction risk among species, the global model masks a great deal of heterogeneity among mammal taxonomic groups and geographical regions, both in the sets of predictors included and the overall explanatory power of the final models. A noteworthy exception is the near-ubiquitous negative association between extinction risk and geographical range size. This association has often been found in previous studies of extinction risk in separate mammal taxa (Purvis et al. 2000a; Johnson et al. 2002; Jones et al. 2003; Cardillo et al. 2004; Price & Gittleman 2007), and the generality of the effect is demonstrated palpably here. Evidently, the importance of a broad distribution, in permitting a large population size, or as a buffer against habitat loss, is such that it transcends biological differences among taxonomic groups, as well as differences in the environments and the threatening processes among regions. Other than geographical range size, the most commonly significant predictors in the taxon-specific models were adult body mass, HPD and latitude. Body mass is strongly correlated with risk-promoting aspects of biology such as a slow life history and with external threatening processes such as hunting intensity. Moreover, body mass interacts with other risk-promoting factors such that larger mammal species seem to be most sensitive to slow life history, low population density and other factors (Cardillo et al. 2005). Human population may be a good general proxy for threat intensity (Cardillo et al. 2004), although it will be more strongly correlated with some types of threatening processes (e.g. hunting) than others (e.g. invasive species). Latitude may also be a general proxy for a range of environmental or anthropogenic factors that influence extinction risk: for example, mammals of larger body size tend to be found at higher latitudes, while temperate latitudes are often most heavily modified by human activity.
There were no other predictors that could qualify as general, prescriptive indicators of high extinction risk across mammal taxa or regions. There are three probable reasons for this. First, mammal clades differ greatly in life-history and ecological strategies, and therefore in the responses of species populations to the threatening processes. Broadly speaking, life-history speed (and hence rates of maximum population increase) in mammals scales with body size (Bielby et al. 2007). Thus, in the MAM for rodents, the group with the smallest mean body mass, none of the variation in extinction risk is attributable to differences in life history, but in the ungulates (for which mean body mass is much larger) life history is of overriding importance. This is consistent with a previous finding (Cardillo et al. 2005) that biological traits tend to be important predictors of extinction risk in larger species, but are less important in smaller species (bats appear to be an exception, but they tend to have slower life history relative to body size than other mammals; Jones & Purvis 1997).
Second, the predominant threatening processes experienced by mammal populations vary among taxa and regions (table S2 in the electronic supplementary material). Different threat types most strongly affect species with different biological traits (Isaac & Cowlishaw 2004; Price & Gittleman 2007), and this may partly explain the variability in our models. However, there is no clear correspondence between the proportion of variance in extinction risk explained by different predictor types (figure 1) and the most prevalent threatening processes listed in the Red List (table S2 in the electronic supplementary material), whether across clades or across regions. This may reflect the lack of a consistent, quantitative method of assigning threatening processes to mammal species under the Red List scheme.
Third, a more procedural reason for the variability in extinction risk models may simply be the effect of our patchy knowledge of the biology of mammal species. In the compilation of data as extensive as the one we have used (PanTheria), a large amount of missing data is inevitable. It is also an unavoidable fact that we know far more about some groups of mammals (in particular, primates and carnivores) than others (especially rodents), so that the missing data are non-randomly distributed with respect to phylogeny. In the extreme case, variables that are very poorly represented for a given taxon (e.g. population density for bats) cannot appear in the final model for that taxon. In general, however, there is no close correspondence between data completeness and the composition of the final models for each taxon (table S3 in the electronic supplementary material), although few of the predictor variables present in the final models are represented by less than half of the species in the relevant taxon. Higher frequencies of missing values within taxa may result in models that are less robust to highly influential observations. However, our model-fitting procedure provided insurance against this by systematically identifying influential contrasts at each step of the model-fitting process, and assessing whether their removal made any qualitative difference.
Techniques for filling in missing values, such as multiple imputation (MI), are widely advocated by statisticians (Little & Rubin 2002), and have indeed been used in a recent comparative extinction risk study (Fisher et al. 2003). However, we are unaware of any MI algorithm capable of dealing correctly with the strongly hierarchical phylogenetic structure inherent in a comparative biological dataset. The result is that biological variables imputed using currently available MI algorithms suffer a loss of phylogenetic signal (M. Cardillo & A. Purvis 2008, unpublished data). We therefore believe that our heuristic approach to finding the best models from incomplete datasets remains the most powerful approach until a phylogenetically explicit MI method is developed. Nonetheless, we note that a dataset for primates with missing values estimated using MI (M. Cardillo & A. Purvis 2008, unpublished data) yielded an extinction risk model qualitatively identical to the primate model presented here. This offers some encouragement that our results may not be strongly biased by missing values.
5. Conclusions
The comparative approach to identifying patterns and correlates of extinction risk has contributed a great deal to our understanding of the ecological processes underlying species declines (Fisher & Owens 2004). Based on our results for mammals, we concur with Fisher & Owens (2004) that models of relatively narrow taxonomic scope are likely to be more informative for practical conservation than more broadly focused models. How can comparative models of extinction risk help inform conservation policy? One way is by identifying species and areas most sensitive to human impacts on the basis of their biology (Cardillo et al. 2004, 2006). This is an especially important exercise for species in areas where natural habitats are still largely intact, but likely to come under increasing pressure over the coming decades. For example, over the next 15 years, a large proportion of the tropical forest of the Amazon Basin is expected to be destroyed or heavily degraded under proposed development schemes (Laurance et al. 2001; Soares-Filho et al. 2006), and the potential for mammal extinctions is high (Grelle 2005). In such areas, comparative models could help prioritize species for preventative conservation efforts, if species are ranked by their inherent vulnerability to decline (i.e. predicted extinction risk), rather than their current threat status. Many mammal species are still too poorly known to assign to an IUCN Red List category, and it may even be possible to use comparative models to aid in assigning species to provisional Red List categories.
Acknowledgments
We thank R. Beck, O. Bininda-Emonds, E. Boakes, C. Carbone, T. Clary, C. Connolly, M. Cutts, J. Davies, J. Foster, R. Grenyer, M. Habib, V. Kanchaite, R. Liu, M. Miyamoto, J. O'Dell, D. Orme, C. Plaster, S. Price, E. Rigby, J. Rist, W. Sechrest, M. Tambutti, A. Teacher and R. Vos for contributing to the construction of the databases. We also thank Emilia Martins and Navjot Sodhi for comments on an earlier draft. This work was funded by the grants from NERC (UK) to G.M.M. and A.P. (NER/A/S/2001/00581 and NE/B503492/1), from NSF (USA) to J.L.G. (DEB/0129009), an NERC fellowship to M.C. (NE/C517992/1) and a Columbia University Earth Institute fellowship to K.E.J.
Supplementary Material
Details of the datasets and threatening processes for mammals
References
- Balmford A. Extinction filters and current resilience: the significance of past selection pressures for conservation biology. Trends Ecol. Evol. 1996;11:193–196. doi: 10.1016/0169-5347(96)10026-4. doi:10.1016/0169-5347(96)10026-4 [DOI] [PubMed] [Google Scholar]
- Bielby J, Mace G.M, Bininda-Emonds O.R.P, Cardillo M, Gittleman J.L, Jones K.E, Orme C.D.L, Purvis A. The fast-slow continuum in mammalian life history: an empirical reevaluation. Am. Nat. 2007;169:748–757. doi: 10.1086/516847. doi:10.1086/516847 [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds O.R.P, et al. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. doi:10.1038/nature05634 [DOI] [PubMed] [Google Scholar]
- Cardillo M, Purvis A, Sechrest W, Gittleman J.L, Bielby J, Mace G.M. Human population density and extinction risk in the world's carnivores. PLoS Biol. 2004;2:909–914. doi: 10.1371/journal.pbio.0020197. doi:10.1371/journal.pbio.0020197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardillo M, Mace G.M, Jones K.E, Bielby J, Bininda-Emonds O.R.P, Sechrest W, Orme C.D.L, Purvis A. Multiple causes of high extinction risk in large mammal species. Science. 2005;309:1239–1241. doi: 10.1126/science.1116030. doi:10.1126/science.1116030 [DOI] [PubMed] [Google Scholar]
- Cardillo M, Mace G.M, Gittleman J.L, Purvis A. Latent extinction risk and the future battlegrounds of mammal conservation. Proc. Natl Acad. Sci. USA. 2006;103:4157–4161. doi: 10.1073/pnas.0510541103. doi:10.1073/pnas.0510541103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CIESIN 2000 Gridded population of the world. See http://sedac.ciesin.columbia.edu/plue/gpw
- Eisenhauer J.G. Regression through the origin. Teach. Stat. 2003;25:76–80. doi:10.1111/1467-9639.00136 [Google Scholar]
- ESRI 2002 ArcMap Version 9, ESRI.
- Fisher D.O, Owens I.P.F. The comparative method in conservation biology. Trends Ecol. Evol. 2004;19:391–398. doi: 10.1016/j.tree.2004.05.004. doi:10.1016/j.tree.2004.05.004 [DOI] [PubMed] [Google Scholar]
- Fisher D.O, Blomberg S.P, Owens I.P.F. Extrinsic versus intrinsic factors in the decline and extinction of Australian marsupials. Proc. R. Soc. B. 2003;270:1801–1808. doi: 10.1098/rspb.2003.2447. doi:10.1098/rspb.2003.2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland T, Harvey P.H, Ives A.R. Procedures for the analysis of comparative data using phylogenetically independent contrasts. Syst. Biol. 1992;41:18–32. doi:10.2307/2992503 [Google Scholar]
- Grelle C.E.V. Predicting extinction of mammals in the Brazilian Amazon. Oryx. 2005;39:347–350. doi:10.1017/S0030605305000700 [Google Scholar]
- Grenyer R, et al. Global distribution and conservation of rare and threatened vertebrates. Nature. 2006;444:93–96. doi: 10.1038/nature05237. doi:10.1038/nature05237 [DOI] [PubMed] [Google Scholar]
- Isaac N.J.B, Cowlishaw G. How species respond to multiple extinction threats. Proc. R. Soc. B. 2004;271:1135–1141. doi: 10.1098/rspb.2004.2724. doi:10.1098/rspb.2004.2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IUCN. IUCN; Gland, Switzerland: 2004. 2004 IUCN red list of threatened species.www.redlist.org [Google Scholar]
- Johnson C.N, Delean S, Balmford A. Phylogeny and the selectivity of extinction in Australian marsupials. Anim. Conserv. 2002;5:135–142. doi:10.1017/S1367943002002196 [Google Scholar]
- Jones K.E, Purvis A. An optimum body size for mammals? Comparative evidence from bats. Funct. Ecol. 1997;11:751–756. doi:10.1046/j.1365-2435.1997.00149.x [Google Scholar]
- Jones K.E, Purvis A, Gittleman J.L. Biological correlates of extinction risk in bats. Am. Nat. 2003;161:601–614. doi: 10.1086/368289. doi:10.1086/368289 [DOI] [PubMed] [Google Scholar]
- Laurance W.F, Cochrane M.A, Bergen S, Fearnside P.M, Delamonica P, Barber C, D'Angelo S, Fernandes T. ENVIRONMENT: the future of the Brazilian Amazon. Science. 2001;291:438–439. doi: 10.1126/science.291.5503.438. doi:10.1126/science.291.5503.438 [DOI] [PubMed] [Google Scholar]
- Little R.J.A, Rubin D.B. Wiley; Hoboken, NJ: 2002. Statistical analysis with missing data. [Google Scholar]
- McKinney M.L. Extinction vulnerability and selectivity: combining ecological and paleontological views. Annu. Rev. Ecol. Syst. 1997;28:495–516. doi:10.1146/annurev.ecolsys.28.1.495 [Google Scholar]
- O'Grady J.J, Burgman M.A, Keith D.A, Master L.L, Andelman S.J, Brook B.W, Hammerson G.A, Regan T, Frankham R. Correlations among extinction risks assessed by different systems of threatened species categorization. Conserv. Biol. 2004;18:1624–1635. doi:10.1111/j.1523-1739.2004.00109.x [Google Scholar]
- Owen-Smith N. Demography of a large herbivore, the greater kudu Tragelaphus strepsiceros, in relation to rainfall. J. Anim. Ecol. 1990;59:893–913. doi:10.2307/5021 [Google Scholar]
- Owen-Smith N, Mason D.R, Ogutu J.O. Correlates of survival rates for 10 African ungulate populations: density, rainfall and predation. J. Anim. Ecol. 2005;74:774–788. doi:10.1111/j.1365-2656.2005.00974.x [Google Scholar]
- Price S.A, Gittleman J.L. Hunting to extinction: biology and regional economy influence extinction risk and the impact of hunting in artiodactyls. Proc. R. Soc. B. 2007;274:1845–1851. doi: 10.1098/rspb.2007.0505. doi:10.1098/rspb.2007.0505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis A, Garland T. Polytomies in comparative analyses of continuous characters. Syst. Biol. 1993;42:569–575. doi:10.2307/2992489 [Google Scholar]
- Purvis A, Gittleman J.L, Cowlishaw G, Mace G.M. Predicting extinction risk in declining species. Proc. R. Soc. B. 2000a;267:1947–1952. doi: 10.1098/rspb.2000.1234. doi:10.1098/rspb.2000.1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis A, Jones K.E, Mace G.M. Extinction. Bioessays. 2000b;22:1123–1133. doi: 10.1002/1521-1878(200012)22:12<1123::AID-BIES10>3.0.CO;2-C. doi:10.1002/1521-1878(200012)22:12<1123::AID-BIES10>3.0.CO;2-C [DOI] [PubMed] [Google Scholar]
- Purvis A, Cardillo M, Grenyer R, Collen B. Correlates of extinction risk: phylogeny, biology, threat and scale. In: Purvis A, Gittleman J.L, Brooks T, editors. Phylogeny and conservation. Cambridge University Press; Cambridge, UK: 2005. pp. 295–316. [Google Scholar]
- R Development Core Team. R Development Core Team; Vienna, Austria: 2007. R: a language and environment for statistical computing.http://www.R-project.org [Google Scholar]
- Reynolds J.D. Life histories and extinction risk. In: Blackburn T.M, Gaston K.J, editors. Macroecology: concepts and consequences. Blackwell; Oxford, UK: 2003. pp. 195–217. [Google Scholar]
- Reynolds J.D, Dulvy N.K, Goodwin N.B, Hutchings J.A. Biology of extinction risk in marine fishes. Proc. R. Soc. B. 2005;272:2337–2344. doi: 10.1098/rspb.2005.3281. doi:10.1098/rspb.2005.3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues A.S.L, Pilgrim J.D, Lamoreux J.F, Hoffmann M, Brooks T.M. The value of the IUCN Red List for conservation. Trends Ecol. Evol. 2006;21:71–76. doi: 10.1016/j.tree.2005.10.010. doi:10.1016/j.tree.2005.10.010 [DOI] [PubMed] [Google Scholar]
- Sechrest, W. 2003 Global diversity, endemism, and conservation of mammals. PhD thesis, University of Virginia, Charlottesville.
- Soares-Filho B.S, et al. Modelling conservation in the Amazon basin. Nature. 2006;440:520–523. doi: 10.1038/nature04389. doi:10.1038/nature04389 [DOI] [PubMed] [Google Scholar]
- UNEP 2003 GRID databases See http://www.grid.unep.ch/data/index.php
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details of the datasets and threatening processes for mammals