Abstract
Insulators are defined as a class of regulatory elements that delimit independent transcriptional domains within eukaryotic genomes. According to previous data, an interaction (pairing) between some Drosophila insulators can support distant activation of a promoter by an enhancer. Here, we have demonstrated that pairs of well-studied insulators such as scs–scs, scs’–scs’, 1A2–1A2 and Wari–Wari support distant activation of the white promoter by the yeast GAL4 activator in an orientation-dependent manner. The same is true for the efficiency of the enhancer that stimulates white expression in the eyes. In all insulator pairs tested, stimulation of the white gene was stronger when insulators were inserted between the eye enhancer or GAL4 and the white promoter in opposite orientations relative to each other. As shown previously, Zw5, Su(Hw) and dCTCF proteins are required for the functioning of different insulators that do not interact with each other. Here, strong functional interactions have been revealed between DNA fragments containing binding sites for either Zw5 or Su(Hw) or dCTCF protein but not between heterologous binding sites [Zw5–Su(Hw), dCTCF–Su(Hw), or dCTCF–Zw5]. These results suggest that insulator proteins can support selective interactions between distant regulatory elements.
The term ‘insulators’ refers to the class of DNA sequence elements that contribute to organization of independent gene function domains by restricting the enhancer and silencer functions. Insulators have two distinctive properties. First, insulators block the enhancer and silencer functions in a position-dependent manner, producing this effect when inserted between these regulatory elements and a promoter but not when located upstream or downstream of them (1–6). Insulators do not inactivate enhancers, silencers or promoters, which indicates that insulators interfere with signaling between these classes of regulatory elements (3,7,8). Second, insulators protect gene expression from positive and negative effects of chromatin surrounding the gene (9–11) and confer the capacity for position-independent transcription on transgenes stably integrated into the genome (12–16).
The Drosophila genome contains many sequences with an insulator function (17–25). The first insulators to be identified were scs and scs’ located at the boundaries of two heat shock 70 genes (4,13,26). Multiple sequences within scs and scs’ are required for their insulator function (5,27–31). Two proteins, Zw5 and BEAF, bind to scs and scs’, respectively, and partially account for their insulator properties (28,30,32,33).
The best characterized insulator consisting of reiterated binding sites for the Su(Hw) protein was found in the gypsy retrotransposon regulatory region (2,3). The Su(Hw) protein associates with hundreds of non-gypsy regions that do not contain clustered Su(Hw) binding sites, with the vast majority of them carrying a single copy of the corresponding sequence (34–38).
Binding sites for a Drosophila homolog of vertebrate insulator protein CTCF were recently identified in several insulators (Mcp, Fab-8, etc.) separating regulatory domains of the bithorax complex (39–42). In vertebrates, almost all known insulator elements were shown to interact with CTCF, a DNA-binding protein that contains 11 zinc fingers (25). It was shown that dCTCF is required for the enhancer-blocking activity of the Fab-8 insulator (39,40).
The first experimental evidence for the functional interaction between insulators came from the fact that insertion of two gypsy insulators between an enhancer (or silencer) and a promoter allowed the enhancer (silencer) to bypass the insulators and activate transcription (43–45). The same was also reported for several other insulators (46–51). Recently, we identified an insulator, named Wari, from the 3′-side of the white gene (52). Although Wari contains no binding sites for known insulator proteins, it can equally well interact with another copy of Wari and with unrelated Su(Hw)-dependent insulators, gypsy or 1A2.
On the other hand, pairs of scs or scs’ insulators proved to have a higher enhancer-blocking activity than either of the insulators in a single copy (53,54). To explain different behavior of insulators in tandems, it was suggested that only some of them are capable to tandem interaction resulting in mutual neutralization of their enhancer-blocking activity. However, an alternative explanation arose from the recent observation that two Mcp insulators placed between the enhancers and promoters allowed effective bypass only when they were inserted in opposite orientations relative to each other (49).
Indeed, we have demonstrated here that paired scs or scs’ insulators can functionally interact with each other, supporting distant activation of the white transcription by the eye enhancer and the yeast GAL4 activator. All insulator pairs tested (scs–scs, scs’–scs’, 1A2–1A2 and Wari–Wari) display orientation-dependent pairing, which may account for the fact that functional interactions between the pairs of many other insulators have not yet been revealed. We also found that DNA fragments containing binding sites for either Zw5, or Su(Hw), or dCTCF alone can support long-distance white activation by GAL4 only upon interaction with another copy of the same fragment, being incapable of interacting in heterologous pairs.
MATERIALS AND METHODS
Plasmid constructions
The constructs were made on the basis of the CaSpeR vector (55). The 5-kb BamHI−BglII fragment (56) containing the coding region (yc) was subcloned into CaSpeR2 (yc-C2). The 3-kb SalI−BamHI fragment containing the yellow regulatory region (yr) was subcloned into pGEM7 cleaved by BamHI–XhoI (yr plasmid). The eye enhancer (Ee) flanked by frt sites was then inserted into the yr plasmid cleaved by BglII at –1874 relative to the yellow transcription start site (yr-frt(Ee). The pCaSpew15(+RI) plasmid was constructed by inserting an additional EcoRI site at +3291 bp of mini-white gene in the pCaSpew15 plasmid. An insulator located at the 3′-side of the mini-white gene (Wari insulator) was deleted from pCaSpew15(+RI) by digestion with EcoRI to produce the pCaSpeRΔ700 plasmid. The yellow gene coding region, a BamHI–Eco47III fragment from the yc-C2 plasmid, was subcloned into pCaSpeRΔ700 digested with BamHI and Eco47III to produce yc-C2Δ plasmid.
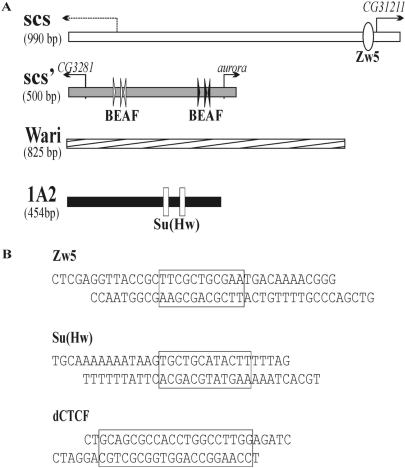
The scs’ insulator corresponded to an ∼500-bp fragment, numbered 1–501 in the scs’ GenBank sequence (accession number X63732). This fragment contains high- and low-affinity BEAF binding sites and two promoters of the CG3281 and aurora genes (28,31,57). The scs insulator corresponded to a 990-bp PvuII–PvuII fragment numbered 510–1503 bp in the GenBank scs sequence (accession number X63731). This fragment has an enhancer-blocking activity similar to that of the full-length scs and contains Zw5 binding sites (27,30) and two promoters (58,59). The 825-bp sequence containing the white-abutting resident insulator (Wari) is numbered 2 684 773–2 683 995 bp (accession no. NC_004354.3) (52). This fragment was PCR-amplified with 5′-cgcaaggagtagccgacatatat-3′ and 5′-ctttggagtacgaaatgcgtcg-3′ primers. The 454-bp sequence of the 1A2 insulator (1A2) numbered 255 315–255 768 bp (accession no. NC_004354.3) was PCR-amplified with 5′-ggagtactactaccaggc-3′ and 5′-caagaacatttccgatatg-3′ primers.
The sequences of the Su(Hw), dCTCF and Zw5 binding sites are shown in Figure 1B. The plasmid containing four reiterated Su(Hw)-binding sites (S×4) was made by tetramerization of the third Su(Hw) binding site, as described (66). The synthetic dCTCF-binding region (C×4) was made by multiplication of the dCTCF binding site from Fab-8, as described (51). The synthetic Zw5-binding region (Z×8) was created by concatamerization of oligonucleotides containing the 32-bp binding site of the natural scs insulator (30). Two pairs of single-stranded 37-bp oligonucleotides (corresponding to the sense and antisense strands) were synthesized so as to contain overhangs for either XhoI or SalI. The sequences of the oligonucleotides were 5′-ctcgaggttaccgcttcgctgcgaatgacaaaacggg-3′ (sence) and 5′-gtcgacccgttttgtcattcgcagcgaagc ggtaacc-3′ (antisense). The desired concatamers (Z×8, C×4, S×4) were isolated, purified and cloned into the pBluSK plasmid. The resulting DNA fragment was verified by sequencing and inserted between two lox or two frt sites. The DNA fragment containing the dCTCF and Su(Hw) binding sites (Cx4Sx4) was made by cloning the S×4 blunt-end fragment into Cx4–pSK cleaved by Eco32I.
Figure 1.
(A) Schemes of the scs, scs’, Wari and 1A2 insulators. The scs insulator contains a cryptic promoter (59) and the promoter of the CG31211 gene. The scs’ insulator contains promoters of the CG3281 and aurora genes. Arrows indicate gene promoters. The Zw5 binding site within scs is shown as a white oval. Positions of the CGATA motifs within scs’ are shown as arrowheads, with the arrow indicating the direction of the motif (5′-CGATA-3′). Clusters of three CGATA motifs form low-affinity (white arrows) and high-affinity (black arrows) binding sites for the BEAF protein (28). Su(Hw) binding sites in the 1A2 insulator are shown as white rectangles. (B) Sequences of the oligonucleotides used to produce the Zw5, Su(Hw) and dCTCF synthetic binding regions. The core binding sites are boxed.
All constructs were made according to two general schemes. In the first scheme, a fragment X (scs or Wari or Zw5 binding sites) was inserted in the direct or reverse orientation into the yr-frt(Ee) plasmid cleaved by Eco47III at –893 relative to the yellow transcription start site. As a result, the frt-flanked eye enhancer in these constructs was placed between the enhancers required for yellow expression in the wing and body, respectively.
In the second scheme, a fragment X (scs, scs’, Wari, 1A2, Z×8, S×4, C×4, C×4S×4) flanked by frt sites (frt(X)) was inserted in the direct or reverse orientation into the G4-Δyr plasmid cleaved by KpnI at –343 relative to the yellow transcription start site (G4-Δyr-frt(X)). In these constructs, the yellow enhancers were deleted.
A fragment X (scs, scs’, Wari, 1A2, Z×8, S×4, CTCF×4) flanked by lox sites (lox(X)) was cloned into yc-C2Δ (yc-C2Δ- lox(X)) or into yc-C2 (yc-C2-lox(X)) at +4964 relative to the yellow transcription start site between the yellow and white genes. Next, yr-frt(Ee)-X or G4-Δyr-frt(X) fragments were cloned into the corresponding yc-C2Δ- lox(X) or into yc-C2-lox(X) plasmids cleaved by XbaI–BamHI.
Generation and analysis of transgenic lines
The construct and P25.7wc plasmid were injected into yacw1118 preblastoderms (60). The resultant flies were crossed with yacw1118 flies, and the transgenic progeny were identified by their eye color. Chromosome localization of various transgene insertions was determined by crossing the transformants with the yacw1118 balancer stock carrying dominant markers, In(2RL),CyO for chromosome 2 and In(3LR)TM3,Sb for chromosome 3.
Lines with DNA fragment excisions were obtained by crossing the flies bearing the transposons with the Flp (w1118; S2CyO, hsFLP, ISA/Sco; +) or Cre (yw; Cyo, P[w+,cre]/Sco; +) recombinase-expressing lines (61,62). Cre recombinase induces 100% excisions in the next generation. A high level of FLP recombinase (almost 90% efficiency) was produced by daily heat-shock treatment for 2 h during the first 3 days after hatching. All excisions were confirmed by PCR analysis with the pairs of primers flanking the insertion site located at –343 relative to the yellow transcription start site (5′-tagatcaaataaagtcccta-3′ and 5′-gtttggtatgatttttggccttc-3′), and the insertion site between the yellow and white genes (5′-ttttcttgagcggaaaaagcgga-3′ and 5′-atctacattctccaaaaaagggt-3′). Details of the crosses used for genetic analysis and excision of functional elements are available upon request.
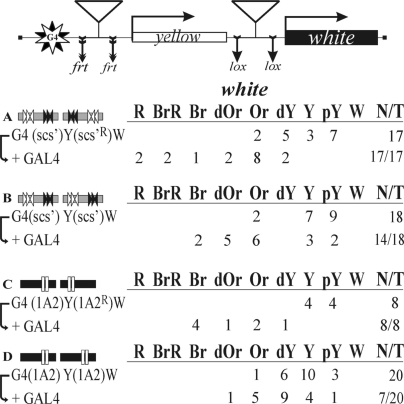
To induce GAL4 expression, we used the modified yw1118; P[w−, tubGAL4]117/TM3,Sb line (Bloomington Center #5138), in which the marker mini-white gene was deleted as described (49).
The white (w) phenotype was estimated from eye pigmentation in adult flies. Wild-type white expression determined the bright red eye color (R); in the absence of white expression, the eyes were white (W). Intermediate levels of white expression (in increasing order) were reflected in the eye color ranging from pale yellow (pY) to yellow (Y), dark yellow (dY), orange (Or), dark orange (dOr), and, finally, brown (Br) or brownish red (BrR).
RESULTS
Insulator bypass depends on the relative orientation of two scs insulators inserted between the eye enhancer and the white promoter
The scs insulator (Figure 1) was mapped in the 990-bp region including two promoters and a binding site for the Zw5 protein involved in enhancer blocking (27,30,63). Previously, it was found that the scs insulator activity was increased when two copies of this element were inserted in a tandem arrangement (53,54). In these experiments, however, two scs copies were inserted in the same direction between closely spaced enhancers and promoters, and the enhancer–promoter interaction across the insulators could be prevented due to spatial constrains. If so, increasing the distance between insulators and placing them in opposite orientations relative to each other might facilitate insulator bypass.
To check this assumption, we chose the regulatory region of the white gene that has been extensively used in insulator studies (4,13,14,34,44,52). The white gene determines eye pigmentation and is regulated by a specific enhancer (64). In our constructs, the yellow gene was inserted between the eye enhancer and the mini-white gene, with the eye enhancer being flanked by frt sites (Figure 2). Parentheses in construct designations indicate the elements flanked by frt or lox sites for in vivo excision by crossing, as described in Materials and methods section; such excisions are denoted by (Δ) in the primary (expression) data tables. Comparing eye pigmentation in flies from the transgenic line before and after the deletion of the eye enhancer, we could estimate its contribution to white expression.
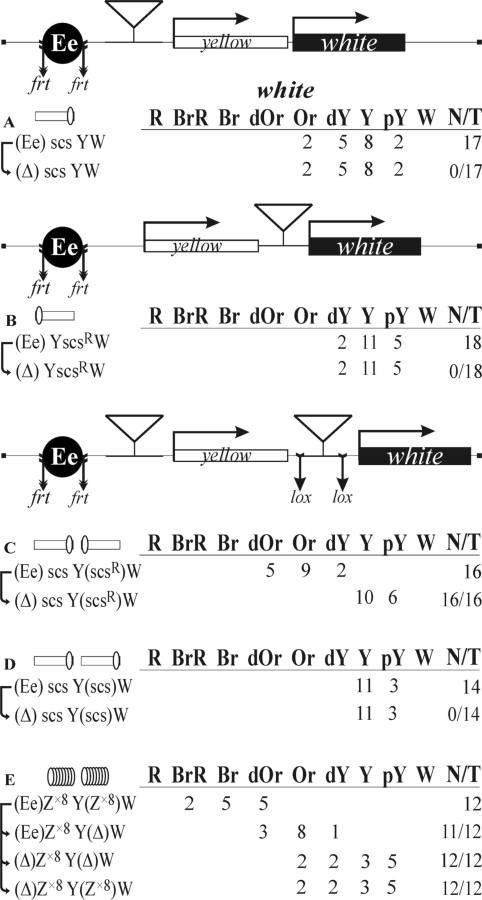
Figure 2.
Insulator bypass depends on the relative orientation of two scs insulators inserted between the eye enhancer and the white promoter. (A and B) Experimental evidence that one copy of scs in both orientations can effectively insulate the eye enhancer. (C and D) Experimental evidence that scs insulators are capable of functional interaction. (E) Tests for the functional interaction between Zw5 binding sites. Reductive schemes of the transgenic constructs are shown (not to scale). The yellow and white genes are shown as boxes, with arrows indicating the direction of their transcription. The eye enhancer (Ee) is shown as a black circle. Downward arrows indicate target sites for Flp recombinase (frt) or Cre recombinase (lox). The same sites in construct names are denoted by parentheses. The scs insulator is shown as a white box, with a white oval indicating the Zw5 binding site. The superscript index ‘R’ indicates that the corresponding element is inserted in the reverse orientation in the construct. The ‘white’ column shows the numbers of transgenic lines with different levels of white expression. The wild-type white expression determined the bright red eye color (R); in the absence of white expression, the eyes were white (W). Intermediate levels of pigmentation, with the eye color ranging from pale yellow (pY) through yellow (Y), dark yellow (dY), orange (Or), dark orange (dOr) and brown (Br) to brownish red (BrR), reflect the increasing levels of white expression. N is the number of lines in which flies acquired a new white (w) phenotype upon deletion (Δ) of the specified DNA fragment (the eye enhancer or scs); T is the total number of lines examined for each particular construct.
To assess the enhancer-blocking activity of the scs insulator, we made constructs in which the scs insulator was inserted either in the direct orientation, according to its position in genome (scs) near the eye enhancer (Figure 2A), or in the reverse orientation (scsR) near the white promoter (Figure 2B). We obtained a total of 35 transgenic lines carrying these constructs. The eye color in these flies ranged from pale yellow to orange, and the deletion of the eye enhancer did not affect eye pigmentation. These results show that a single copy of scs completely blocked the eye enhancer.
Next, we examined the activity of the eye enhancer in transgenic flies carrying two copies of the scs insulator inserted near the eye enhancer and the white promoter in either opposite or the same orientation relative to each other (Figure 2C and D). In the case of the construct carrying the scs insulators inserted in opposite orientations (Figure 2C), we obtained 16 transgenic lines in which flies had eye pigmentation ranging from dark yellow to dark orange. The deletion of the eye enhancer resulted in a noticeable reduction of eye pigmentation in all transgenic lines, indicating that the eye enhancer was capable of stimulating white expression. When the scs insulators were placed in the same orientation (Figure 2D), eye pigmentation ranged from pale yellow to yellow in all 14 transgenic lines obtained, with the deletion of the eye enhancer having no effect on white expression. Therefore, the pair of scs insulators completely blocked the enhancer. These results indicate that relative orientation of the two scs copies is critical for the ability of the eye enhancer to stimulate the white promoter across the insulator pair.
The scs insulator contains a binding site for the Zw5 protein that is necessary for its enhancer-blocking activity (30,63). It was shown that four Zw5 binding sites could partially block the eye enhancer (30). Thus, it is possible that Zw5 participates in pairing between two scs insulators. To test this possibility, we prepared oligos containing eight binding sites for Zw5 (Z×8), which insulated the eye enhancer better than oligos with four Zw5 binding sites (data not shown). In the construct (Figure 2E), we inserted the Z×8 oligo (proximal) near the eye enhancer flanked by frt sites and Z×8 flanked by lox sites (distal) near the white promoter. Eye pigmentation was compared in transgenic lines before and after the deletion of the distal Z×8 oligo flanked by lox sites. This deletion resulted in a considerable reduction of eye pigmentation. In derivative transgenic lines with the deleted enhancer, eye pigmentation in flies with and without the distal Z×8 oligo was the same. These results support the model that the interaction between protein complexes bound to the Zw5 binding sites promotes white activation by the eye enhancer.
Pairing of scs insulators or Zw5 binding sites facilitates long-distance stimulation of the white promoter by the GAL4 activator
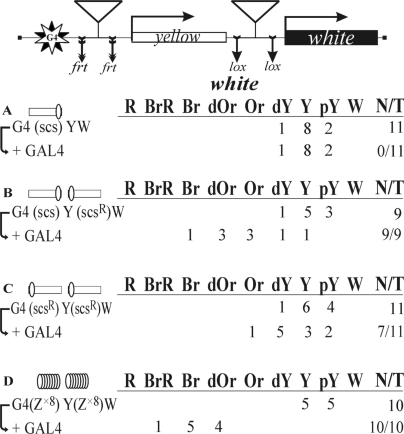
To confirm that the scs insulators can functionally interact at a distance, we used the GAL4/white model system based on the finding that the GAL4 activator cannot stimulate the white promoter across the yellow gene (49). To test whether the interaction between the scs insulators can facilitate white stimulation by GAL4, we inserted ten GAL4 binding sites (designated G4) at the 5′ side of the yellow gene. As a result, the distance between the mini-white gene and the GAL4 binding sites reached almost 5 kb. To express the GAL4 protein, we used a transgenic line carrying the GAL4 gene under control of the ubiquitous tubulin promoter (49).
In the control construct, a single copy of scs was inserted near the GAL4 binding sites (Figure 3A). In all 11 transgenic lines tested, GAL4 failed to stimulate white activation. Next, we checked if the interaction between two scs insulators inserted in opposite orientations would facilitate white activation by GAL4 (Figure 3B). These transgenic lines showed strong white activation by GAL4, indicating that the pair of scs insulators in such an arrangement supported communication between the GAL4 activator and the promoter complex. We then inserted two scs insulators in the same orientation (Figure 3C) and observed a relatively weak white activation by GAL4, compared to that in transgenic lines carrying the scs insulators inserted in opposite orientations. This is evidence that the scs insulators functionally interact in an orientation-dependent manner.
Figure 3.
Testing the functional interaction between scs insulators or Zw5 binding sites in the GAL4/white model system. The GAL4 binding sites (indicated as G4) are at a distance of ∼5 kb from the white promoter. A reductive scheme of transgenic construct used to examine the functional interaction between the insulators is presented in the upper part of the figure. ‘+GAL4’ indicates that eye phenotypes in transgenic lines were examined after the induction of GAL4 expression. N is the number of lines in which flies acquired a new w phenotype upon induction of GAL4. For other designations, see Figure 2.
Finally, we checked whether Zw5 binding sites could support white activation by GAL4. Once again, we used the oligos containing eight Zw5 binding sites (Z×8), which were inserted near the GAL4 binding sites and the white promoter (Figure 3D). In all 10 transgenic lines tested, GAL4 strongly stimulated white transcription, confirming our previous observation that Zw5 binding sites can functionally interact with each other.
Taken together, these results show that scs insulators can functionally interact in the orientation dependent manner and that the Zw5 protein may contribute to their pairing.
Functional interaction between two Wari insulators depends on their relative orientation
As shown in our previous study (52), pairing between two copies of the Wari insulator is required for the effective blocking of the enhancers. Hence, the question arose as to whether the relative orientation of the Wari insulators is of significance for their functional interaction.
At first, we examined the interaction between the Wari insulators in the eye enhancer/white promoter model system. The first copy of Wari was inserted in the direct orientation near the eye enhancer that was flanked by the frt sites. The second copy, flanked by the lox sites, was inserted near the white promoter in either the opposite (Figure 4A) or the same orientation (Figure 4B). To improve the enhancer-blocking activity of these insulators, the endogenous Wari located at the 3′-end of the white gene was left intact. Thus, the resulting constructs contained three copies of the Wari insulator.
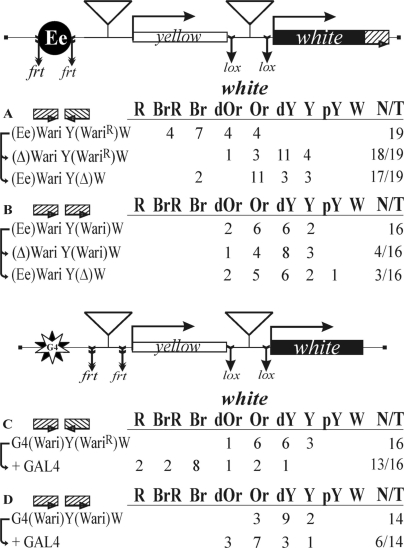
Figure 4.
Testing the functional interaction between Wari insulators (A and B) in the eye enhancer/white model and (C and D) in the GAL4/white model. The Wari insulator is shown as a hatched box. For other designations, see Figures 2 and 3.
When Wari insulators were placed in opposite orientations relative to each other (Figure 4A), we observed high levels of eye pigmentation, which decreased considerably upon deletion of one Wari insulator or the eye enhancer. Thus, the functional interaction between Wari insulators allowed the eye enhancer to stimulate white expression more effectively. In contrast, transgenic flies carrying two copies of Wari insulators in the same orientation (Figure 4B) had relatively weak eye pigmentation. The deletion of either eye enhancer or Wari changed the eye pigmentation only slightly and in a minor part of corresponding transgenic lines. These results show that the relative orientation of Wari insulators is important for eye enhancer/white promoter communication.
Next, we examined the interaction between Wari insulators in the GAL4/white assay. The insulators flanked by either lox or frt sites were inserted near the GAL4 binding sites and the white promoter in either the opposite or the same orientation (Figure 4C and D). In this case, the Wari insulator was removed from the 3′-side of the mini-white gene. When they were placed in opposite orientations, GAL4 strongly activated white expression (Figure 4C), whereas insulators in the same orientation allowed only weak stimulation of white expression by GAL4 (Figure 4D). Thus, the relative orientation of the interacting Wari insulators determines the efficiency of white stimulation by GAL4.
The pairing between two scs’ or 1A2 insulators supports long-distance white activation by GAL4
To determine whether orientation-dependent pairing is a common property of Drosophila insulators, we tested two other well-studied endogenous Drosophila insulators, 1A2 (34,35) and scs’ (4,13,28,32), in the GAL4/white assay.
The scs’ insulators were inserted either in opposite orientations (Figure 5A) or in the same orientation (Figure 5B). In both cases, the scs’ insulators markedly enhanced white activation by GAL4, confirming their ability to interact with each other. Once again, the relative orientation of the scs’ insulators proved to influence the level of white stimulation by GAL4.
Figure 5.
Testing the functional interaction between (A and B) two scs’ or (C and D) two 1A2 insulators. The scs’ insulator is shown as a gray box with the black and white arrows indicating binding sites for the BEAF protein. The 1A2 insulator is shown as a black box with white rectangles indicating Su(Hw) binding sites. For other designations, see Figures 2 and 3.
In addition, two similar constructs were made with the 1A2 insulators inserted in either opposite or the same orientation (Figure 5C and D). We observed that white activation by GAL4 depended on the relative orientation of the 1A2 insulators. When the insulators were in opposite orientations, GAL4 strongly stimulated white expression (Figure 5C); when their orientation was the same, only relatively weak stimulation was observed (Figure 5D). Thus, in the model of white activation by GAL4, the 1A2 insulators appear to interact in an orientation-dependent manner.
Su(Hw), Zw5 and dCTCF binding sites are capable of selective pairing only with their copies
In several previous studies, no functional interactions between different insulators were observed (53,54). Likewise, our experiments with the eye enhancer/white and GAL4/white model systems also did not reveal any functional interactions between heterologous insulators such as gypsy [with 12 binding sites of Su(Hw)], scs (with one binding site for Zw5) and Fab-8 (with two binding sites for dCTCF) (data not shown).
However, the gypsy, scs and Fab-8 insulators have a complex structure and may contain binding sites for additional proteins involved in the insulator activity. Hence, we decided to examine functional interactions between the oligos containing binding sites for Zw5, Su(Hw) and dCTCF proteins. Recently, we found that the functional interaction between two DNA fragments containing four consensus binding sites for the dCTCF protein (C×4) supported long-distance activation of white by the GAL4 activator (51) (Figure 6A). In this study, a functional interaction was revealed between DNA fragments Z×8 containing eight binding sites for Zw5 (Figures 3D and 6B).
Figure 6.
Testing the functional interaction between DNA fragments containing binding sites for different insulator proteins, dCTCF (black ovals), Zw5 (white ovals), Su(Hw) (white rectangles) or composite DNA fragments containing dCTCF and Su(Hw) binding sites (black ovals and white rectangles). For designations, see Figures 2 and 3. The results with G4(Cx4)Y(Cx4)W were taken from Kyrchanova et al. (51).
Previously, it was shown that four Su(Hw) binding sites function as a strong insulator (65), and we made the oligos containing four copies of the third Su(Hw) binding site (S×4) from the gypsy insulator (66). Here, we found that the functional interaction between the S×4 DNA fragments can facilitate white activation by GAL4 (Figure 6C). This is evidence that all three insulator proteins can support long-distance interactions in the GAL4/white model system.
Next, we analyzed functional interactions between DNA fragments containing binding sites for different proteins: Z×8 and C×4 (Figure 6D), C×4 and S×4 (Figure 6E), and Z×8 and S×4 (Figure 6F). No white activation by GAL4 was observed in any of these variants, indicating that insulator proteins could selectively support interactions within the genome.
Finally, we tested if composite DNA fragments containing four consecutive binding sites for each of the dCTCF and Su(Hw) proteins (S×4C×4) could functionally interact in an orientation dependent manner. Such fragments were inserted in the GAL4/white model system either in opposite orientations (Figure 6G) or in the same orientation (Figure 6H) relative to each other. Strong white activation by GAL4 was observed only when the DNA fragments were inserted in opposite orientations. This result showed that the relative orientation of the composite DNA fragments containing binding sites for two different insulator proteins determines the ability of the GAL4 activator to stimulate white expression.
DISCUSSION
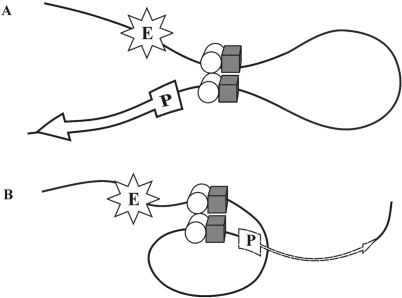
The results of this and previous studies (43–46,49–52) confirm that most of well-studied insulators can functionally interact in pairs, but the functional effect of this interaction depend on their relative orientation. A probable explanation to this orientation-dependent effect is that there are at least two insulator-bound proteins involved in specific protein–protein interactions. If so, the pairing of insulators, depending on their relative orientation, may lead to the formation of two loop configurations (Figure 7). In our model system, when the insulators are in opposite orientations relative to each other, the configuration of the loop formed upon their pairing is favorable for communication between regulatory elements located outside the loop, as these elements are brought in close proximity to each other (Figure 7A). Such a loop configuration can provide for the observed strong white stimulation by GAL4 or effective bypass of the insulators by the eye enhancer. In contrast, pairing between two insulators located in the same orientation leads to the formation of the loop that spatially separates regulatory elements (Figure 7B), with the consequent weakening of white activation by the eye enhancer or GAL4.
Figure 7.
Two models of pairing between the insulators. Presumptive proteins responsible for insulator pairing are shown as a white cylinder and a gray cube. Solid and dotted arrows indicate high and low levels of transcription, respectively. Other designations: (P) promoter, (E) enhancer.
It appears that most of Drosophila insulators contain binding sites for more than one insulator protein. For example, the scs insulator is assembled from a discrete number of functionally redundant DNA elements (27), and it is likely that Zw5 is only one of several proteins that are responsible for the activity of this insulator (27,30,63). The enhancer-blocking activity of the 1A2 insulator depends on the presence of not only two Su(Hw) binding sites but also of certain additional sequences, which indicates that at least one more transcriptional factor, in addition to Su(Hw), is necessary for its functioning (34,35,67). A direct test of other genome regions containing one or several endogenous Su(Hw) binding sites in the transgene assay shows that most of them effectively block enhancers, suggesting that additional proteins bound to non-gypsy regions contribute to the insulator function of Su(Hw) (36,37,67,68).
It is of interest that, as shown previously, the functional interaction between gypsy insulators, each containing 12 binding sites for the Su(Hw) protein alone, is less sensitive to their relative orientation (53,54). Here, we observed that the relative orientation of the scs’ insulators had only a slight effect on white stimulation by GAL4. Note that the weak scs’ insulator probably contains binding sites for only one protein, BEAF (8,32,33). On the other hand, stimulation of white by GAL4 in experiments with composite DNA fragments containing binding sites for two insulator proteins, dCTCF and Su(Hw), displayed striking dependence on their relative orientation. These results are in agreement with the proposed model that the binding of at least two different insulator proteins is essential for effective orientation-dependent interaction between insulators. However, more information about proteins bound to insulators is required to construct the model comprehensively explaining the phenomenon of insulator pairing.
Previously (53,54), no interaction between unrelated insulators was revealed. The results of this study show that DNA fragments containing binding sites for either of three different insulator proteins—Zw5, Su(Hw) and dCTCF—can effectively support long-distance interactions in pairs, with no functional interaction being observed between heterologous DNA fragments containing binding sites for different insulator proteins. Thus, insulator proteins can ensure selective long-distance interactions in chromosomes. For example, the interaction between gypsy insulators can support activation of the yellow promoter by enhancers separated by many megabases (69). In mammals, the interaction of the imprinting control region on chromosome 7 with the Wsb1/Nf1 locus on chromosome 11 depends on the presence of the CTCF protein (70).
Interestingly, although no functional interaction is observed between binding sites for Su(Hw) and dCTCF, both these proteins interact with CP190, the protein required for the enhancer-blocking activity of dCTCF- and Su(Hw)-dependent insulators (42,71,72). CP190 contains the BTB/POZ domain involved in homodimerization and the additional domain that interacts in vitro with the Mod(mdg4) protein, another component of the Su(Hw) insulator complex (73). Thus, the presence of the same protein in two different insulator complexes does not ensure the functional interaction between them. Further extensive studies are required to elucidate the proteins and their domains that are involved in selective long-distance interactions.
FUNDING
The Ministry of Science and Education of the Russian Federation (02.512.11.2252); the Molecular and Cellular Biology Program of the Russian Academy of Sciences; the International Research Scholar Award from the Howard Hughes Medical Institute (to P.G.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to N.A. Gorgoluk for his help in preparing the article.
REFERENCES
- 1.Gyurkovics H, Gausz J, Kummer J, Karch F. A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J. 1990;9:2579–2585. doi: 10.1002/j.1460-2075.1990.tb07439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holdridge C, Dorsett D. Repression of hsp70 heat shock gene transcription by the suppressor of hairy-wing protein of Drosophila melanogaster. Mol. Cell Biol. 1991;11:1894–1900. doi: 10.1128/mcb.11.4.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geyer PK, Corces VG. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 1992;6:1865–1873. doi: 10.1101/gad.6.10.1865. [DOI] [PubMed] [Google Scholar]
- 4.Kellum R, Schedl P. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol. Cell Biol. 1992;12:2424–2431. doi: 10.1128/mcb.12.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sigrist CJ, Pirrotta V. Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics. 1997;147:209–221. doi: 10.1093/genetics/147.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mallin DR, Myung JS, Patton JS, Geyer PK. Polycomb group repression is blocked by the Drosophila Suppressor of Hairy-wing [su(Hw)] insulator. Genetics. 1998;148:331–339. doi: 10.1093/genetics/148.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai HN, Levine M. Modulation of enhancer−promoter interactions by insulators in the Drosophila embryo. Nature. 1995;376:533–536. doi: 10.1038/376533a0. [DOI] [PubMed] [Google Scholar]
- 8.Scott KS, Geyer PK. Effects of the su(Hw) insulator protein on the expression of the divergently transcribed Drosophila yolk protein genes. EMBO J. 1995;14:6258–6267. doi: 10.1002/j.1460-2075.1995.tb00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell. 2002;109:551–562. doi: 10.1016/s0092-8674(02)00756-0. [DOI] [PubMed] [Google Scholar]
- 10.Mutskov VJ, Farrell CM, Wade PA, Wolffe AP, Felsenfeld G. The barrier function of an insulator couples high histone acetylation levels with specific protection of promoter DNA from methylation. Genes Dev. 2002;16:1540–1554. doi: 10.1101/gad.988502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oki M, Kamakaka RT. Blockers and barriers to transcription: competing activities? Curr. Opin. Cell Biol. 2002;14:299–304. doi: 10.1016/s0955-0674(02)00327-7. [DOI] [PubMed] [Google Scholar]
- 12.Bonifer C, Vidal M, Grosveld F, Sippel AE. Tissue specific and position independent expression of the complete gene domain for chicken lysozyme in transgenic mice. EMBO J. 1990;9:2843–2848. doi: 10.1002/j.1460-2075.1990.tb07473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- 14.Roseman RR, Pirrotta V, Geyer PK. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 1993;12:435–442. doi: 10.1002/j.1460-2075.1993.tb05675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roseman RR, Johnson EA, Rodesch CK, Bjerke M, Nagoshi RN, Geyer PK. A P element containing suppressor of hairy-wing binding regions has novel properties for mutagenesis in Drosophila melanogaster. Genetics. 1995;141:1061–1074. doi: 10.1093/genetics/141.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burgess-Beusse B, Farrell C, Gaszner M, Litt, M. Mutskov V, Recillas-Targa F, Simpson M, West A, Felsenfeld G. The insulation of genes from external enhancers and silencing chromatin. Proc. Natl Acad. Sci. 2002;99:16433–16437. doi: 10.1073/pnas.162342499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun FL, Elgin SC. Putting boundaries on silence. Cell. 1999;99:459–462. doi: 10.1016/s0092-8674(00)81534-2. [DOI] [PubMed] [Google Scholar]
- 18.Kuhn EJ, Geyer PK. Genomic insulators: connecting properties to mechanism. Curr. Opin. Cell Biol. 2003;15:259–265. doi: 10.1016/s0955-0674(03)00039-5. [DOI] [PubMed] [Google Scholar]
- 19.Capelson M, Corces VG. Boundary elements and nuclear organization. Biol. Cell. 2004;96:617–629. doi: 10.1016/j.biolcel.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 20.Brasset E, Vaury C. Insulators are fundamental components of the eukaryotic genomes. Heredity. 2005;94:571–576. doi: 10.1038/sj.hdy.6800669. [DOI] [PubMed] [Google Scholar]
- 21.Zhao H, Dean A. Organizing the genome: enhancers and insulators. Biochem. Cell Biol. 2005;83:516–524. doi: 10.1139/o05-054. [DOI] [PubMed] [Google Scholar]
- 22.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 23.Maksimenko OG, Chetverina DA, Georgiev PG. Insulators of higher eukaryotes: properties, mechanisms of action, and role in transcription regulation. Genetika. 2006;42:1029–1044. [PubMed] [Google Scholar]
- 24.Valenzuela L, Kamakaka RT. Chromatin insulators. Annu. Rev. Genet. 2006;40:107–138. doi: 10.1146/annurev.genet.39.073003.113546. [DOI] [PubMed] [Google Scholar]
- 25.Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr. Opin. Genet. Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Udvardy A, Maine E, Schedl P. The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J. Mol. Biol. 1985;185:341–358. doi: 10.1016/0022-2836(85)90408-5. [DOI] [PubMed] [Google Scholar]
- 27.Vazquez J, Schedl P. Sequences required for enhancer blocking activity of scs are located within two nuclease-hypersensitive regions. EMBO J. 1994;13:5984–5993. doi: 10.1002/j.1460-2075.1994.tb06944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao K, Hart CM, Laemmli UK. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell. 1995;81:879–889. doi: 10.1016/0092-8674(95)90008-x. [DOI] [PubMed] [Google Scholar]
- 29.Dunaway M, Hwang JY, Xiong M, Yuen HL. The activity of the scs and scs’ insulator elements is not dependent on chromosomal context. Mol. Cell Biol. 1997;17:182–189. doi: 10.1128/mcb.17.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaszner M, Vazquez J, Schedl P. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev. 1999;13:2098–2107. doi: 10.1101/gad.13.16.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuhn EJ, Hart CM, Geyer PK. Studies of the role of the Drosophila scs and scs’ insulators in defining boundaries of a chromosome puff. Mol. Cell Biol. 2004;24:1470–1480. doi: 10.1128/MCB.24.4.1470-1480.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hart CM, Zhao K, Laemmli UK. The scs’ boundary element: Characterization of boundary element-associated factors. Mol. Cell Biol. 1997;17:999–1009. doi: 10.1128/mcb.17.2.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cuvier O, Hart CM, Laemmli UK. Identification of a class of chromatin boundary elements. Mol. Cell Biol. 1998;18:7478–7486. doi: 10.1128/mcb.18.12.7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Golovnin A, Birukova I, Romanova O, Silicheva M, Parshikov A, Savitskaya E, Pirrotta V, Georgiev P. An endogenous Su(Hw) insulator separates the yellow gene from the Achaete-scute gene complex in Drosophila. Development. 2003;130:3249–3258. doi: 10.1242/dev.00543. [DOI] [PubMed] [Google Scholar]
- 35.Parnell TJ, Viering MM, Skjesol A, Helou C, Kuhn EJ, Geyer PK. An endogenous suppressor of hairy-wing insulator separates regulatory domains in Drosophila. Proc. Natl Acad. Sci. 2003;100:13436–13441. doi: 10.1073/pnas.2333111100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parnell TJ, Kuhn EJ, Gilmor B, Helou C, Wold M, Geyer PK. Identification of genomic sites that bind the Drosophila suppressor of hairy-wing insulator protein. Mol. Cell Biol. 2006;26:5983–5993. doi: 10.1128/MCB.00698-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramos E, Ghosh D, Baxter E, Corces VG. Genomic organization of gypsy chromatin insulators in Drosophila melanogaster. Genetics. 2006;172:2337–2349. doi: 10.1534/genetics.105.054742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adryan B, Woerfel G, Birch-Machin I, Gao S, Quick M, Meadows L, Russell S, White R. Genomic mapping of Suppressor of Hairy-wing binding sites in Drosophila. Genome Biol. 2007;8:R167. doi: 10.1186/gb-2007-8-8-r167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith ST, Munhall A, Grewe B, Bartkuhn M, Arnold R, et al. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 2005;6:165–170. doi: 10.1038/sj.embor.7400334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ciavatta D, Rogers S, Magnuson T. Drosophila CTCF is required for Fab-8 enhancer blocking activity in S2 cells. J. Mol. Biol. 2007;373:233–239. doi: 10.1016/j.jmb.2007.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holohan EE, Kwong C, Adryan B, Herold M, Renkawitz R, Russell S, White R. CTCF genomic binding sites in Drosophila and the organization of the bithorax complex. PLoS Genet. 2007;3:e112. doi: 10.1371/journal.pgen.0030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mohan M, Bartkuhn M, Herold M, Philippen A, Heinl N, Bardenhagen I, Leers J, White RAH, Renkawitz-Pohl R, Saumweber H, et al. The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J. 2007;26:4203–4214. doi: 10.1038/sj.emboj.7601851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cai HN, Shen P. Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science. 2001;291:493–495. doi: 10.1126/science.291.5503.493. [DOI] [PubMed] [Google Scholar]
- 44.Muravyova E, Golovnin A, Gracheva E, Parshikov A, Belenkaya T, Pirrotta V, Georgiev P. Loss of insulator activity by paired Su(Hw) chromatin insulators. Science. 2001;291:495–498. doi: 10.1126/science.291.5503.495. [DOI] [PubMed] [Google Scholar]
- 45.Comet I, Savitskaya E, Schuettengruber B, Negre N, Lavrov S, Parshikov A, Juge F, Gracheva E, Georgiev P, Cavalli G. PRE-mediated bypass of two Su(Hw) insulators targets PcG proteins to a downstream promoter. Dev. Cell. 2006;11:1–8. doi: 10.1016/j.devcel.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 46.Conte C, Dastugue B, Vaury C. Coupling of enhancer and insulator properties identified in two retrotransposons modulates their mutagenic impact on nearby genes. Mol. Cell Biol. 2002;22:1767–1777. doi: 10.1128/MCB.22.6.1767-1777.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melnikova L, Juge F, Gruzdeva N, Mazur A, Cavalli G, Georgiev P. Interaction between the GAGA factor and Mod(mdg4) proteins promotes insulator bypass in Drosophila. Proc. Natl Acad. Sci. 2004;101:14806–14811. doi: 10.1073/pnas.0403959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gruzdeva N, Kyrchanova O, Parshikov A, Kullyev A, Georgiev P. The Mcp element from the bithorax complex contains an insulator that is capable of pairwise interactions and can facilitate enhancer–promoter communication. Mol. Cell Biol. 2005;25:3682–3689. doi: 10.1128/MCB.25.9.3682-3689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kyrchanova O, Toshchakov S, Parshikov A, Georgiev P. Study of the functional interaction between Mcp insulators from the Drosophila bithorax complex: Effects of insulator pairing on enhancer–promoter communication. Mol. Cell Biol. 2007;27:3035–3043. doi: 10.1128/MCB.02203-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodin S, Kyrchanova O, Pomerantseva E, Parshikov A, Georgiev P. New properties of Drosophila Fab-7 insulator. Genetics. 2007;177:113–121. doi: 10.1534/genetics.107.075887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kyrchanova O, Toshchakov S, Podstreshnaya Y, Parshikov A, Georgiev P. Functional interaction between the Fab-7 and Fab-8 boundaries and the upstream promoter region in the Drosophila Abd-B gene. Mol. Cell Biol. 2008;28:4188–4195. doi: 10.1128/MCB.00229-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chetverina D, Savitskaya E, Maksimenko O, Melnikova L, Zaytseva O, Parshikov A, Galkin AV, Georgiev P. Red flag on the white reporter: a versatile insulator abuts the white gene in Drosophila and is omnipresent in mini-white constructs. Nucleic Acids Res. 2008;36:926–937. doi: 10.1093/nar/gkm992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuhn EJ, Viering MM, Rhodes KM, Geyer PK. A test of insulator interactions in Drosophila. EMBO J. 2003;22:2463–2471. doi: 10.1093/emboj/cdg241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Majumder P, Cai HN. The functional analysis of insulator interactions in the Drosophila embryo. Proc. Natl Acad. Sci. 2003;100:5223–5228. doi: 10.1073/pnas.0830190100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pirrotta V. Vectors for P-mediated transformation in Drosophila. Biotechnology. 1988;10:437–456. doi: 10.1016/b978-0-409-90042-2.50028-3. [DOI] [PubMed] [Google Scholar]
- 56.Geyer PK, Corces VG. Separate regulatory elements are responsible for the complex pattern of tissue-specific and developmental transcription of the yellow locus in Drosophila melanogaster. Genes Dev. 1987;1:996–1004. doi: 10.1101/gad.1.9.996. [DOI] [PubMed] [Google Scholar]
- 57.Glover DM, Leibowitz MH, McLean DA, Parry,H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 1995;81:95–105. doi: 10.1016/0092-8674(95)90374-7. [DOI] [PubMed] [Google Scholar]
- 58.Avramova Z, Tikhonov A. Are scs and scs’ “neutral” chromatin boundaries of the 87A7 locus in vivo? Trends Genet. 1999;15:138–139. doi: 10.1016/s0168-9525(99)01712-6. [DOI] [PubMed] [Google Scholar]
- 59.Hogga I, Karch F. Transcription through the iab-7 cis-regulatory domain of the bithorax complex interferes with maintenance of Polycomb-mediated silencing. Development. 2002;129:4915–4922. doi: 10.1242/dev.129.21.4915. [DOI] [PubMed] [Google Scholar]
- 60.Karess RE, Rubin GM. Analysis of P transposable element functions in Drosophila. Cell. 1984;38:135–146. doi: 10.1016/0092-8674(84)90534-8. [DOI] [PubMed] [Google Scholar]
- 61.Golic KG, Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- 62.Siegal ML, Hartl DL. Application of Cre/loxP in Drosophila. Site-specific recombination and transgene co-placement. Methods Mol. Biol. 2000;136:487–495. doi: 10.1385/1-59259-065-9:487. [DOI] [PubMed] [Google Scholar]
- 63.Blanton J, Gaszner M, Schedl P. Protein:protein interactions and pairing of boundary elements in vivo. Genes Dev. 2003;17:664–675. doi: 10.1101/gad.1052003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qian S, Varjavand B, Pirrotta V. Molecular analysis of the zeste–white interaction reveals a promoter-proximal element essential for distant enhancer-promoter communication. Genetics. 1992;131:79–90. doi: 10.1093/genetics/131.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scott KS, Taubman AD, Geyer PK. Enhancer blocking by the Drosophila gypsy insulator depends upon insulator anatomy and enhancer strength. Genetics. 1999;153:787–798. doi: 10.1093/genetics/153.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Golovnin A, Melnick E, Mazur A, Georgiev P. Drosophila Su(Hw) insulator can stimulate transcription of a weakened yellow promoter over a distance. Genetics. 2005;170:1133–1142. doi: 10.1534/genetics.104.034587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Soshnev AA, Li X, Wehling MD, Geyer PK. Context differences reveal insulator and activator functions of a Su(Hw) binding region. PLoS Genet. 2008;4:e10000159. doi: 10.1371/journal.pgen.1000159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuhn-Parnell EJ, Helou C, Marion DJ, Gilmore BL, Parnell TJ, Wold MS, Geyer PK. Investigation of the properties of non-gypsy suppressor of Hairy-wing-binding sites. Genetics. 2008;179:1263–1273. doi: 10.1534/genetics.108.087254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kravchenko E, Savitskaya E, Kravchuk O, Parshikov A, Georgiev P, Savitsky M. Pairing between gypsy insulators facilitates the enhancer action in trans throughout the Drosophila genome. Mol. Cell Biol. 2005;25:9283–9291. doi: 10.1128/MCB.25.21.9283-9291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 71.Pai C-Y, Lei EP, Ghosh D, Corces VG. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol. Cell. 2004;16:737–748. doi: 10.1016/j.molcel.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 72.Gerasimova TI, Lei EP, Bushey AM, Corces VG. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol. Cell. 2007;28:761–772. doi: 10.1016/j.molcel.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Golovnin A, Mazur A, Kopantseva M, Kurshakova M, Gulak PV, Gilmore B, Whitfield WGF, Geyer P, Pirrotta V, Georgiev P. Integrity of the Mod(mdg4)-67.2 BTB domain is critical to insulator function in Drosophila. Mol. Cell Biol. 2007;27:963–974. doi: 10.1128/MCB.00795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]