Abstract
We describe a method to identify and recover minor human immunodeficiency virus type 1 (HIV-1) sequence variants from a complex population. The original heteroduplex tracking assay (HTA) was modified by incorporating a biotin tag into the probe to allow for direct sequence determination of the query strand. We used this approach to recover sequences from minor HIV-1 variants in the V3 region of the env gene, and to identify minor drug-resistant variants in pro. The biotin-HTA targeting of the V3 region of env allowed us to detect minor V3 variants, of which 45% were classified as CXCR4-using viruses. In addition, the biotin-protease HTA was able to detect mixtures of wild-type sequence and drug-resistance mutations in four subjects that were not detected by bulk sequence analysis. The biotin-HTA is a robust assay that first separates genetic variants then allows direct sequence analysis of major and minor variants.
INTRODUCTION
Genetically unstable organisms present a special challenge to therapeutic intervention. Genetic instability coupled with large population sizes leads to population diversity that can harbor advantageous mutations in the face of changing selective pressures. Our knowledge about genetic complexity is always limited by our ability to sample minor variants within the population. One example of this phenomenon is the human immunodeficiency virus type 1 (HIV-1), which maintains genetic diversity in its population that can impact evolution of escape from immune selection, drug resistance and changes in target cell specificity (1).
Several approaches are available for sampling genetic complexity. Bulk sequence analysis of the total population suffers from limited sensitivity, and cannot reliably detect variants that comprise less than ∼25% of the population (2, 3). A strategy of cloning a PCR product generated from a complex population followed by sequencing individual clones is limited by the number of clones analyzed, and the accurate detection of minor variants in the population requires a large sampling of clones (3, 4). Allele-specific PCR can detect variants in the 0.1–1% range, and although there is no information about linkage to other sequence variation (5, 6), further analysis of the allele-specific product can generate some linkage information (7). More recent pyrosequencing approaches offer deep sequencing capability, although the high error rate necessitates oversampling of sequences which reduces sensitivity, and bioinformatics approaches are necessary to handle the large data output (8).
An alternative approach to dissecting genetic diversity is the heteroduplex tracking assay (HTA). The HTA is a gel-based assay that separates viral variants based on sequence differences, and can resolve variants that comprise as little as 1–3% of the total viral population (9–11). In this assay, the desired genomic region is amplified by PCR, and a radioactively labeled probe is annealed to the PCR products. Heteroduplexes are generated between the probe and PCR products, and any insertions, deletions or clustered mutations will result in a bend or kink in the DNA helix, conferring an altered migration through a nondenaturing polyacrylamide gel. However, this approach is limited in that it is not linked to direct sequence analysis.
In this report, we describe a modification of the HTA strategy that couples the ability to separate genetic variants in a gel-based assay with direct sequence analysis of the separated variants. The ability to directly sequence the query strand of each heteroduplex in the gel was accomplished by adding a biotinylated-nucleotide tag to the radiolabeled probe strand. Using the biotinylated probe, we were able to purify the labeled heteroduplex and subsequently separate the query strand from the probe. The purified query strand was then subjected to PCR amplification and conventional sequence analysis. We used this approach to identify sequences from minor variants in the V3 region of the HIV-1 env gene that predict changes in target cell tropism, and to examine the heterogeneity of the pro gene population during the evolution of resistance to protease inhibitors.
MATERIALS AND METHODS
Plasma samples
All plasma samples were obtained as excess tissue samples and with Institutional Review Board approval. Samples for the V3 studies were from subjects in a ritonavir efficacy study (12), and were provided by Dale Kempf (Abbott Laboratories) unlinked to personal identifiers. Samples for the protease studies were entry time-point plasma samples obtained from subjects in the ACTG 359 clinical trial (13).
Plasmids and probes
The V3 Mut-1 probe plasmid was generated using the V3JR-FL plasmid pJN27 developed by Nelson et al. (14). A BamHI restriction site located upstream of the probe insert in pJN27 was destroyed, and the EcoRI site located at the start of the probe sequence was mutated to a new BamHI site using the Quikchange site-directed mutagenesis strategy. The 5′-GATC-3′ overhang after BamHI cleavage permits sequential labeling of the antisense strand in a fill-in reaction with biotin-labeled dGTP and 35S-dATP. To limit PCR amplification of the probe sequence from the isolated heteroduplex, the upstream V3 PCR primer binding site in the pJN27 plasmid was mutated, reducing the efficiency of amplification by 1000-fold. The final sequence of the V3 Mut-1 probe at the upstream V3 boundary was 5′-GATCCGGCTTGAATCTGTAGAAATTAATTGTACACGACAAAA…-3′. The final probe sequence at the downstream V3 boundary in the probe is 5′-…CATTGTAACATTAGTAGAGCAAAAAAGCCGAATTAATTCTGCA-3′. Introduced nucleotide changes are indicated by bolded and underlined letters, and plasmid sequence included in the V3 probe is shown in italics.
The pR-EBm protease probe plasmid was generated from the multiple-site-specific HTA probe pR-EB previously described by Resch et al. (15). The site-directed mutagenesis procedure described above was used to generate the pR-EBm probe. The probe was modified for this study by introducing base mismatches in the upstream PCR primer binding site to limit amplification from the probe sequence from the isolated heteroduplex. Additionally, a BamHI site was introduced at the 3′ end to accommodate biotin- and radiolabeling of the sense strand. The 5′ probe sequence is 5′-TATGGATAACTAAAGGAAGCTCTATTAGATACTGGCT …-3′, and the 3′ probe sequence is 5′-… TCAGATTGGTTGCACTTTAAATTTTCCATCG(biotin-dGTP)(S35-dATP)-3′. Introduced nucleotide changes are indicated by bolded, underlined letters, and plasmid sequence included in the probe is shown in italics.
Probe labeling
The V3 Mut-1 probe plasmid (10 μg) was digested with BamHI (60 μl total volume) followed by a fill-in reaction with 4 nmol Biotin-11-dGTP (PerkinElmer), 0.05 mCi [α-35S]-dATP (1250 Ci/mmol; PerkinElmer), 20 U of the Klenow fragment of DNA polymerase I and 0.6 μl of 1M DTT. The reaction was incubated for 15 min at room temperature followed by the addition of 1.4 µl 0.5 M EDTA and heat inactivation for 15 min at 80°C. The excess nucleotides were then removed using the Qiagen PCR Purification kit (Qiagen), and the V3 Mut-1 probe was released from the vector by PstI digestion. The pR-EBm probe plasmid was labeled using the same procedure, except that after purification the pR-EBm probe was released from the vector by NdeI digestion.
RNA Isolation, RT-PCR and HTA
Procedures for viral RNA isolation from plasma, RT-PCR, and the HTA were conducted as previously described (14, 15). Briefly, viral RNA was isolated from blood plasma (140 μl) using the QIAmp Viral RNA kit (Qiagen). Reverse transcription and PCR amplification of the 140 bp V3 amplicon were conducted with 5 μl of purified RNA (from 50 μl column elution volume) and the primers HIVV3F [Hxb2 7142-7171 (5′-GAATCTGTAGAAATTAATTGTACAAGACCC-3′)] and HIVV3R [Hxb2 7239-7211 (5′-CCATTTTGCTCTACTAATGTTACAATGT-3′)] by using the Qiagen One-Step RT-PCR kit (Qiagen) as per manufacturer's instructions. Amplification of a 247-bp region of pro was conducted using the procedure stated above and the primers PRAMPUP [Hxb2 2305-2334 (5′-AACTAAAGGAAGCTCTATTAGATACAGGAG-3′)] and PRAMPDW [Hxb2 2551-2525 (5′-GGAAAATTTAAAGTGCAACCAATCTGA-3′)]. Heteroduplex annealing reactions consisted of 1 µl of 10× annealing buffer (14, 16), 0.1 μg biotinylated and radioactively-labeled probe and 8 μl of unpurified PCR product. The reactions were denatured and the DNA was annealed at room temperature to allow heteroduplex formation. The heteroduplexes were separated by native polyacrylamide gel electrophoresis (as described in refs 14 and 15), and the separated heteroduplexes were visualized by autoradiography. The relative abundance of each detected heteroduplex was determined by exposing the HTA gel to a phosphorimager screen, and the percent abundance of each variant was calculated using ImageQuant software (Molecular Dynamics).
Band extraction, DNA purification and sequence analysis
The dried HTA gels were aligned with the exposed autoradiography film, and the desired labeled bands were excised from the gel. The gel fragment was transferred to a sterile 1.5 ml microcentrifuge tube. Crush and Soak buffer (120 μl; 500 mM NH4OAc, 0.1% SDS, 0.1 mM EDTA) was then added, and the tubes were incubated at 37°C overnight. After incubation, the tubes were centrifuged for 2 min at 14 000 r.p.m. to pellet any gel fragments, and the supernatants were transferred to a 96-well PCR plate. Streptavidin-coated Dynabeads® (Invitrogen) (5 μl per reaction) were washed twice then resuspended in the same volume in 2× B&W buffer [10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 2 M NaCl]. The bead suspension was then aliquoted into a clean 96-well PCR plate (5 μl per well). The plate containing the beads was placed on the magnet for 1 min and the supernatant was removed. The plate was removed from the magnet, and the beads were resuspended in 80 μl of 2× B&W buffer. An equal volume (80 μl) of the gel-extracted heteroduplex product was then added to the beads. The plate was then incubated for 17 min at room temperature in a MixMate shaker (Eppendorf) at 700 r.p.m. to prevent the beads from settling. After the incubation, the plate was placed on the magnet for 2 min, and the supernatant was removed and discarded. The beads were washed three times in 1× B&W buffer (20 μl), and then washed twice in 1× SSC (pH 7.0; 50 μl). After the last wash, the beads were resuspended in 20 μl of freshly prepared 0.15 M NaOH and incubated for 5 min at room temperature to denature the heteroduplexes. The plate was then placed on the magnet for 1 min, and the supernatant containing the denatured query strand was transferred to a clean 96-well PCR plate. The supernatant was neutralized by adding 2.4 μl of 10× TE (pH 7.5) and 2.4 μl of 1.25 M acetic acid.
The purified query strand DNA was then amplified with the Platinum Taq DNA Polymerase High Fidelity PCR kit (Invitrogen) as per the manufacturer's instructions, using 2 μl DNA and the PCR primers described above. The V3 sequencing primers are V3SeqF [Hxb2 7147-7176 (5′-TGTAGAAATTAATTGTACAAGACCCAACAA-3′)] and V3SeqR [Hxb2 7234-7205 (5′-TTTGCTCTACTAATGTTACAATGTGCTTGT-3′)]. The protease PCR primers described above were used to sequence the pro PCR products. Bulk sequencing was also conducted on the original PCR products amplified from viral RNA. GenBank accession numbers FJ347037-FJ347099.
RESULTS
Biotinylated probes and HTA allow the identification and recovery of minor HIV-1 variants from a complex population
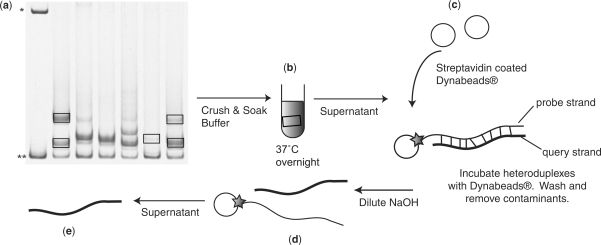
The HTA is capable of separating diverse genetic variants in a gel-based format (9–11,14–16). We have modified the original HTA method to permit direct sequence determination of the query strand of the target heteroduplex and adapted the approach to a 96-well plate format. To accomplish direct sequencing of the query strand we incorporated a biotin tag into the probe strand (see Materials and methods section) to allow for the purification of the labeled heteroduplex followed by the separation of the query and probe strands (Figure 1).
Figure 1.
The biotin-HTA strategy. (a) The region of interest is PCR amplified from the HIV-1 genome and mixed with a biotinylated probe to generate heteroduplexes between the probe and PCR product. The heteroduplexes are then separated by polyacrylamide gel electrophoresis based on DNA bending. The single-stranded probe band is denoted by the asterisk, and the double-stranded probe band is denoted by double asterisks. The labeled heteroduplexes are excised from the dried polyacrylamide gel. (b) The gel fragments are soaked overnight to recover heteroduplex DNA from the gel fragments. (c) The supernatant containing the biotin-tagged heteroduplexes is then incubated with streptavidin-coated magnetic Dynabeads®. The biotin-tagged heteroduplexes bind to the streptavidin-coated magnetic Dynabeads®, and contaminants are removed by washing. (d) The query DNA strand in the heteroduplex is eluted from the probe DNA with dilute NaOH, which allows the probe strand to remain attached to the magnetic beads. (e) The purified query strand is then used as template in another round of PCR amplification, and the PCR product is sequenced using standard technology.
In this approach a query PCR product is generated, annealed to a radiolabeled/biotin-labeled probe, and the heteroduplexes are resolved by native polyacrylamide gel electrophoresis. Gel slices containing the labeled heteroduplexes are then excised from dried polyacrylamide gel using X-ray film that had been exposed to the gel as a guide. The excised gel slices are rehydrated and soaked overnight to recover heteroduplex DNA from the gel fragments. The biotin-tagged heteroduplexes are then purified away from the gel fragments and any unlabeled co-migrating DNA (i.e. heteroduplexes formed between the unlabeled PCR products) by binding the labeled heteroduplexes to streptavidin-coated magnetic Dynabeads®, followed by washing. The query DNA strand in the heteroduplex is then eluted from the probe DNA with dilute NaOH, which allows the probe strand to remain attached to the magnetic beads and thus removed from the solution. The purified query DNA strand is then used as the template in another round of PCR amplification, and the PCR product is sequenced using standard technology. Below we describe the application of this approach to the sequence-based identification of HIV-1 variants that enter cells using the CXCR4 coreceptor, and of variants that encode drug resistance in subjects failing therapy with a protease inhibitor, in each case resolving these variants from genotypic mixtures.
Minor X4 variants are identified and recovered using a biotinylated-V3 HTA probe
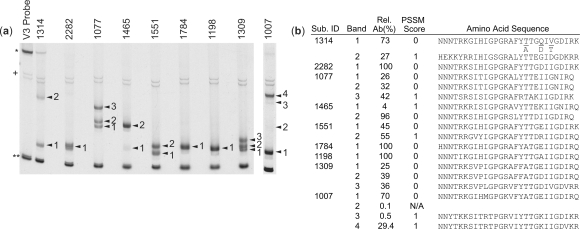
We analyzed the V3 region of env for 34 HIV-1-infected subjects with CD4+ T cell counts <150 cells/µl to test the ability of our biotin-HTA method to detect and recover minor V3 variants in complex viral populations. Figure 2a shows the separation of genotypic mixtures of the V3 region of the env gene in the HTA gel (14, 17). We were able to obtain V3 sequence from 63 of the 69 visible heteroduplexes in the polyacrylamide gels, and phosphorimager analysis was used to define the relative fraction of each variant. For these subjects the average number of variants was two variants per subject, and the number of variants ranged from one (12 subjects) up to five (one subject).
Figure 2.
Identification and recovery of minor V3 variants using a biotinylated-V3 HTA. (a) Biotin-V3 HTA gel for nine representative subjects. The asterisk indicates the single-stranded probe band, double asterisks indicate the double-stranded probe band, the plus sign indicates background bands and arrowheads indicate shifted heteroduplex bands. (b) The numbered bands were extracted, purified and sequenced to determine the V3 sequence of each band. It is important to note that the V3 PCR primers we used to amplify the V3 products extend into the first four and the last three amino acids of the V3 sequence, and the V3 sequences provided in Figure 2 and GenBank (Acc. # FJ347037-FJ347099) do not include these amino acids. However, to accurately predict coreceptor usage using PSSM we added the JRFL consensus amino acid sequence to the beginning (CTRP) and end (AHC) of each V3 sequence. The relative abundance (Rel. Ab.) and PSSM score (0 = R5-like sequence, 1 = ×4-like sequence, N/A = not applicable) for each sequence are listed in (b).
We used the modified 11/25 rule (18, 19) and the position-specific scoring matrix (PSSM) (20) to predict the coreceptor usage of these V3 sequences based on the viral genotype. A total of 63 V3 sequences were generated for these 34 subjects, of which 48 corresponded to an R5-using genotype and 15 corresponded to an X4-using genotype (Figure 2 and data not shown). The biotin-V3 HTA method recorded the presence of minor R5 or X4 V3 variants (defined as <30% of the total population) in 18 of the 34 subjects, and a total of 26 bands with a relative abundance of <30% were excised and sequenced. We obtained sequence for 20 of the bands while we failed to obtain sequence for six bands. The relative abundance of the six variants for which sequence was not obtained ranged from 0.1% to 13% of the total population, with the failure to obtain usable sequence attributable to either very low abundance or near co-migration. Of the 20 minor V3 sequences we recovered, nine were classified as X4-using viruses. We were able to detect two of these minor X4 sequences as mixtures in bulk sequence analysis, while the other seven minor X4 variants were not detected by bulk sequence analysis due to low relative abundance. All of the V3 variants that were extracted from the gel and PCR amplified migrated to the same position in the gel as the original band amplified from viral RNA (data not shown). One limitation of this procedure is that major bands in the gel trail upwards, and PCR products of minor variants that migrate right above a major variant in the gel may contain a small amount of the major variant. However, in most cases (55 out of 69 variants) the V3 sequence obtained from the PCR products had clean peaks, and the major variant was not detected in the sequence analysis (data not shown). For the remaining 14 variants that had multiple peaks present in the sequence analysis, and also displayed a large amount of the contaminating major variant, purified samples were re-amplified and purified again to obtain the target sequence. Using this enrichment method, we were able to determine the sequence of eight additional variants. Overall, we were able to obtain clear sequence for ∼90% of all bands detected and ∼75% of all detected bands representing <30% of the population, and the use of biotin-HTA resolved all mixtures seen with bulk sequence analysis.
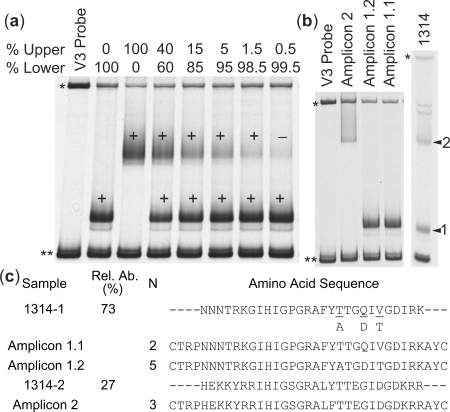
In order to further examine the sensitivity and specificity of the biotin-HTA, several control experiments were conducted. First, we mixed together two V3 PCR products at varying ratios and then separated the mixtures using the biotin-V3 HTA. The bands were then excised from the gel, purified and sequenced. In this control experiment, we were able to obtain sequence from a band that represented as little as 1.5% of the total population (Figure 3a). Sequence was not obtained from the band representing 0.5% of the population; however, we did not re-amplify the product in this case which can allow increased sensitivity of detection, as described above. To address specificity, single genome amplification (SGA) (21) of the env gene was conducted on blood plasma from subject 1314 in order to compare the relative abundance of variants determined using biotin-HTA to sampling of the viral population by sequencing individual viral templates. We generated and sequenced 10 SGA amplicons and compared the V3 regions to the sequences identified using the biotin-V3 HTA. The 10 amplicons represented two migration patterns in the V3-HTA, with seven of the amplicons having one migration rate (shown as amplicons 1.1 and 1.2 in Figure 3b), and three of the amplicons having the other migration rate (labeled amplicon 2 in Figure 3b). This closely approximated the ratio of 73% and 27% for these two bands as determined by V3-HTA (Figure 3c), and this result is consistent with a similar control done previously with a V1/V2-HTA probe (16). The altered migration of heteroduplexes in the HTA is based on sequence differences between the strands, especially clustered mutations and insertion/deletions, with variable sensitivity to detect isolated point mutations. This feature can be seen in the sequence differences between the bands that were resolved by the biotin V3-HTA (amplicons 1 and 2, Figure 3c), but the co-migration of sequences that differed by only a few nucleotides (amplicons 1.1 and 1.2, Figure 3c). In this case the level of variability in the isolated HTA band was revealed as a mixed peak in the sequence chromatogram when that band was isolated and sequenced (data not shown).
Figure 3.
Examination of the sensitivity of the biotin-V3 HTA. (a) Mixtures of two V3 PCR products were separated by the biotin-V3 HTA. The bands were cut from the gel, purified and sequenced. The plus sign indicates bands for which sequence was obtained, and the minus signindicates a band for which the sequence was not obtained. The asterisk indicates the single-stranded probe band, and double asterisks indicate the double-stranded probe band. (b and c) SGA of the env gene was conducted on Subject 1314. The migration of three different SGA-V3 PCR products and the original 1314 V3 PCR product are shown in (b). The V3 amino acid sequence of the extracted bands and the SGA amplicons, the relative abundance (Rel. Ab.) and number of SGA amplicons (N) are listed in (c).
Minor protease drug-resistant variants are detected using a biotinylated-protease HTA probe
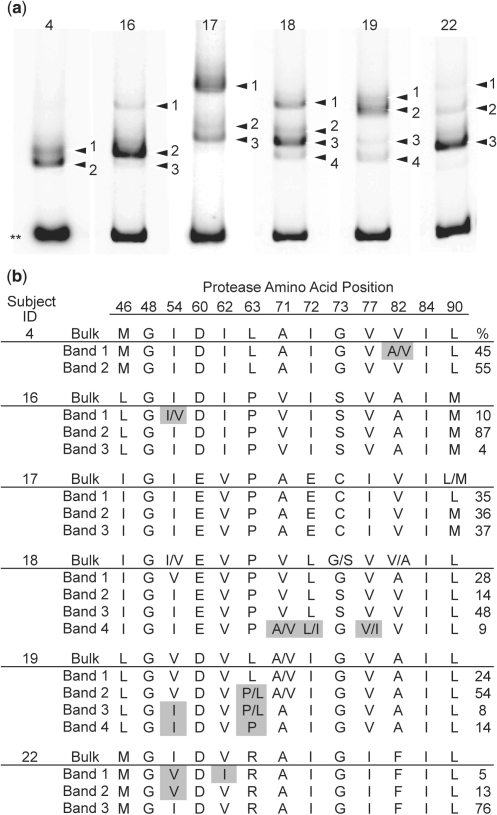
We have applied the biotin-HTA to separate HIV-1 pro variants, encoding the viral protease, and to sequence these variants to detect mutations that confer resistance to protease inhibitors. The biotin-protease HTA is a modification of the previously described multiple site-specific HTA (MSS-HTA) (15) (see Materials and methods section) with the probe designed to query mutations at protease codon positions 46, 48, 54, 82, 84 and 90. We used both our own bulk sequence analysis and the biotin-protease HTA to screen 19 subjects who had previously failed therapy that included the protease inhibitor IDV. A total of 10 subjects had no primary resistance mutations by either method, although some had viral genotypes that included polymorphic mutations that can become enriched with therapy (22) (data not shown) (Stanford Database, http://hivdb.stanford.edu/). The remaining nine subjects all exhibited primary resistance mutations (Stanford database, http://hivdb.stanford.edu/). For three of these subjects we detected a homogeneous population by both methods (data not shown). The results of the biotin-HTA analysis and the bulk sequence analysis for the remaining six subjects are shown in Figure 4.
Figure 4.
Detection of minor protease inhibitor resistance mutations using a biotinylated-protease HTA. (a) Protease-specific MSS-HTA gel for six representative subjects. Double asterisks indicate the double-stranded probe band and arrowheads indicate shifted heteroduplex bands. (b) Numbered bands were extracted and sequenced to identify the predicted amino acid at the positions indicated. For each subject, the bulk PCR sequence is listed above the band sequences. Mixtures are indicated where identified by sequencing. The relative abundance of each band is listed in the right-most column as a percentage of the total. The amino acids inferred from the bulk sequence for Subject 4 represent the wild-type consensus. Amino acid positions which the probe was designed to query are in bold (46, 48, 54, 82, 84, 90). Shaded amino acids represent positions at which heterogeneity was identified by HTA but not by bulk sequence analysis.
Bulk sequence analysis was able to identify mutations in variants that comprised as little as 28% of the population (Figure 4, subject 18, band 1), but also missed mutations that existed in variants making up as much as 45% of the population (Figure 4, subject 4, band 1), perhaps due to poor incorporation of the chain terminating nucleotide at that position. In contrast, using the biotin-HTA approach we were able to identify primary resistance mutations at the query positions in three subjects (subject 4, band 1; subject 16, band 1; and subject 22, bands 1 and 2) that were not detected in bulk sequencing (Figure 4). Exclusive linkage between resistance mutations I54V and V82A was established in one subject (Figure 4, subject 18, band 1). The least abundant variant we identified comprised 4% of the total population (Figure 4b, subject 16, band 3), although this variant differed by only one synonymous change from the bulk sequence (data not shown). Resistance mutations were sometimes identified as mixtures in excised bands (Figure 4, subject 4, band 1; subject 16, band 1; subject 18, band 4). This can result from either trailing of products from a major lower band, or, in the case of positions that the probe was not designed to query, the inability of the probe to distinguish between variants at that position (Figure 4, subject 18, band 4; subject 19, bands 1–3). Despite existing as mixtures in these cases, resistance mutations were enriched, and therefore detected, in electrophoretically separated variants when they were otherwise not detected by bulk sequence analysis. One subject had a resistance mutation (subject 15, V82A; data not shown) that was detected in a separate clinically approved bulk sequence analysis protocol but not in the HTA analysis, while three other subjects displayed resistance mutations that were detected by HTA but not by the clinically approved analysis (Figure 4, subject 4, V82A; subject 16 and 18, I54V).
DISCUSSION
The biotin-HTA allows for the detection and direct sequence analysis of multiple variants in a complex population. Using the biotin-HTA targeting the V3 region of env, we were able to detect and directly determine the sequence of 20 minor (<30% abundance) R5 and X4 viral variants, of which 45% were classified as X4-using viruses. In addition, we used a biotin-protease HTA to detect and sequence specific drug resistance mutations in pro in 19 subjects failing treatment with an HIV-1 protease inhibitor. Fifteen of the 19 subjects had matching genotypes for the clinical screening data and the HTA analysis, while three subjects displayed resistance mutations that were detected by HTA but not by a clinically approved bulk sequence analysis.
An RNA heteroduplex generator-tracking assay (RNA-HTA) has been developed that uses a degradable RNA probe to allow direct sequence analysis of the query strand, which was tested by examining HIV-1 variants with drug-resistance mutations in the protease coding domain (23). Using this method, variants that comprised 1% of the total population were identified and subjected to sequence analysis. One drawback of the RNA-HTA method is that unlabeled DNA–DNA heteroduplexes generated from variants within the PCR product can co-migrate with the labeled RNA–DNA heteroduplexes, going undetected in the gel but giving rise to sequence mixtures during the subsequent amplification and sequence analysis. Our biotin-HTA method introduces the ability to purify specifically the biotin/radiolabeled probe: PCR product heteroduplexes from unlabeled PCR product heteroduplexes that might migrate to the same location in the polyacrylamide gel. Similar to what Kapoor et al. (23) saw in their studies, we also find that major variants trail upward in the gel and can be detected in the PCR products extracted from upper bands. In the case that an unreadable sequence is detected using the biotin-HTA procedure, it is possible to simply re-amplify the now enriched mixture and run another biotin-HTA followed by sequence analysis.
In a control experiment we were able to obtain sequence information from a band that comprised 1.5% of the signal, and as seen previously (16), the HTA provides sampling that is similar to that obtained by other methods but has the advantage of being able to query many more templates than these other methods (Figure 3). During the analysis of the subject samples, we were able to obtain the sequence of a variant that represented 0.5% of the total population (subject 1007, Figure 2b) by running the extracted V3 PCR product on another biotin-HTA and re-amplifying the purified product, thus allowing for further enrichment of the minor variant. A useful comparison of the potential utility of the HTA is as follows. If one were to sequence 300 clones (assuming they are derived from independent templates in the PCR amplification), there is a 95% chance of detecting variants present at a 1% level. In this case detection of presence is more robust than the estimate of relative abundance; also, the amount of work essentially increases linearly as the number of clones analyzed increases. HTA is able to detect signals in the 1% range in the gel-based detection system. However, sampling quality can be significantly better since this approach is not limited by the number of templates sampled, i.e. sampling 300 templates is as easy as sampling 3000 templates. This allows the HTA approach to measure more accurately the relative abundance of even minor species in those circumstances where larger numbers of templates are available to be sampled, and to validate their abundance by comparing repeat amplifications.
Different HIV-1 entry inhibitors are being explored as possible drugs to block HIV-1 infection and to treat current infections. Several CCR5-inhibitors are approved or in development for the treatment of HIV-1 infection, including maraviroc (Pfizer) (24, 25) and vicriviroc (Schering Plough) (26, 27). CCR5-inhibitors work by binding the CCR5 coreceptor to block the entry of HIV-1. CXCR4-using viruses are naturally resistant to these CCR5-binding drugs and therefore this class of inhibitor is not indicated for subjects with X4 variants. In addition, coreceptor switching has been documented in several subjects using CCR5-inhibitors, although the level of CXCR4-emerging viruses was lower than expected (25, 28). In the subjects where CXCR4-using viruses emerged after treatment with a CCR5 inhibitor, phylogenetic analysis showed that these variants were a result of the outgrowth of a minor CXCR4-using variant that was present prior to the initiation of drug therapy (28). The biotin-V3 HTA that we have developed can detect and genotype minor V3 variants in the range of 1.5% of the total HIV-1 population, and could be used as an initial genotypic screen for CXCR4-using variants in people considering CCR5-inhibitor drug therapy.
Additionally, genotypic testing for the presence of resistance mutations is the standard-of-care for people initiating and failing therapy (Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents, U.S. D.H.H.S., http://www.aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGL.pdf). The biotin-protease HTA provides an example where complex mixtures of sequences can be examined with greater sensitivity to detect resistance mutations. The biotin-HTA methods used here for the detection and direct sequence analysis of minor HIV-1 variants should be applicable to other complex viral and non-viral populations where increased sensitivity of detection of minor genetic variants is important.
FUNDING
National Institutes of Health (R37-AI44667 to R.S., T32-AI07001 to G.S. and T32-GM07092 to W.I.). Funding for open access charge: NIH (R37-AI44667).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Rushina Cholera for assistance in doing these experiments. We also thank Dale Kempf from Abbott and the ACTG 359 study team for making samples available for this study.
REFERENCES
- 1.Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 2.Larder BA, Kohli A, Kellam P, Kemp SD, Kronick M, Henfrey RD. Quantitative detection of HIV-1 drug resistance mutations by automated DNA sequencing. Nature. 1993;365:671–673. doi: 10.1038/365671a0. [DOI] [PubMed] [Google Scholar]
- 3.Palmer S, Kearney M, Maldarelli F, Halvas EK, Bixby CJ, Bazmi H, Rock D, Falloon J, Davey RTJr, Dewar RL, et al. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 2005;43:406–413. doi: 10.1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu SL, Rodrigo AG, Shankarappa R, Learn GH, Hsu L, Davidov O, Zhao LP, Mullins JI. HIV quasispecies and resampling. Science. 1996;273:415–416. doi: 10.1126/science.273.5274.415. [DOI] [PubMed] [Google Scholar]
- 5.Halvas EK, Aldrovandi GM, Balfe P, Beck IA, Boltz VF, Coffin JM, Frenkel LM, Hazelwood JD, Johnson VA, Kearney M, et al. Blinded, multicenter comparison of methods to detect a drug-resistant mutant of human immunodeficiency virus type 1 at low frequency. J. Clin. Microbiol. 2006;44:2612–2614. doi: 10.1128/JCM.00449-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paredes R, Marconi VC, Campbell TB, Kuritzkes DR. Systematic evaluation of allele-specific real-time PCR for the detection of minor HIV-1 variants with pol and env resistance mutations. J. Virol. Methods. 2007;146:136–146. doi: 10.1016/j.jviromet.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson JA, Li JF, Wei X, Lipscomb J, Bennett D, Brant A, Cong ME, Spira T, Shafer RW, Heneine W. Simple PCR assays improve the sensitivity of HIV-1 subtype B drug resistance testing and allow linking of resistance mutations. PLoS ONE. 2007;2:e638. doi: 10.1371/journal.pone.0000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delwart EL, Gordon CJ. Tracking changes in HIV-1 envelope quasispecies using DNA heteroduplex analysis. Methods. 1997;12:348–354. doi: 10.1006/meth.1997.0489. [DOI] [PubMed] [Google Scholar]
- 10.Delwart EL, Sheppard HW, Walker BD, Goudsmit J, Mullins JI. Human immunodeficiency virus type 1 evolution in vivo tracked by DNA heteroduplex mobility assays. J. Virol. 1994;68:6672–6683. doi: 10.1128/jvi.68.10.6672-6683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delwart EL, Shpaer EG, Louwagie J, McCutchan FE, Grez M, Rubsamen-Waigmann H, Mullins JI. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 12.Cameron DW, Heath-Chiozzi M, Danner S, Cohen C, Kravcik S, Maurath C, Sun E, Henry D, Rode R, Potthoff A, et al. Randomised placebo-controlled trial of ritonavir in advanced HIV-1 disease. The Advanced HIV Disease Ritonavir Study Group. Lancet. 1998;351:543–549. doi: 10.1016/s0140-6736(97)04161-5. [DOI] [PubMed] [Google Scholar]
- 13.Gulick RM, Hu XJ, Fiscus SA, Fletcher CV, Haubrich R, Cheng H, Acosta E, Lagakos SW, Swanstrom R, Freimuth W, et al. Randomized study of saquinavir with ritonavir or nelfinavir together with delavirdine, adefovir, or both in human immunodeficiency virus-infected adults with virologic failure on indinavir: AIDS Clinical Trials Group Study 359. J. Infect. Dis. 2000;182:1375–1384. doi: 10.1086/315867. [DOI] [PubMed] [Google Scholar]
- 14.Nelson JA, Fiscus SA, Swanstrom R. Evolutionary variants of the human immunodeficiency virus type 1 V3 region characterized by using a heteroduplex tracking assay. J. Virol. 1997;71:8750–8758. doi: 10.1128/jvi.71.11.8750-8758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Resch W, Parkin N, Stuelke EL, Watkins T, Swanstrom R. A multiple-site-specific heteroduplex tracking assay as a tool for the study of viral population dynamics. Proc. Natl Acad. Sci. USA. 2001;98:176–181. doi: 10.1073/pnas.011511298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitrinos KM, Hoffman NG, Nelson JA, Swanstrom R. Turnover of env variable region 1 and 2 genotypes in subjects with late-stage human immunodeficiency virus type 1 infection. J. Virol. 2003;77:6811–6822. doi: 10.1128/JVI.77.12.6811-6822.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson JA, Baribaud F, Edwards T, Swanstrom R. Patterns of changes in human immunodeficiency virus type 1 V3 sequence populations late in infection. J. Virol. 2000;74:8494–8501. doi: 10.1128/jvi.74.18.8494-8501.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffman NG, Seillier-Moiseiwitsch F, Ahn J, Walker JM, Swanstrom R. Variability in the human immunodeficiency virus type 1 gp120 Env protein linked to phenotype-associated changes in the V3 loop. J. Virol. 2002;76:3852–3864. doi: 10.1128/JVI.76.8.3852-3864.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Resch W, Hoffman N, Swanstrom R. Improved success of phenotype prediction of the human immunodeficiency virus type 1 from envelope variable loop 3 sequence using neural networks. Virology. 2001;288:51–62. doi: 10.1006/viro.2001.1087. [DOI] [PubMed] [Google Scholar]
- 20.Jensen MA, Li FS, van′t Wout AB, Nickle DC, Shriner D, He HX, McLaughlin S, Shankarappa R, Margolick JB, Mullins JI. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J. Virol. 2003;77:13376–13388. doi: 10.1128/JVI.77.24.13376-13388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, Derdeyn CA, Farmer P, Hunter E, Allen S, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shafer RW. Rationale and uses of a public HIV drug-resistance database. J. Infect. Dis. 2006;194(Suppl 1):S51–S58. doi: 10.1086/505356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kapoor A, Jones M, Shafer RW, Rhee SY, Kazanjian P, Delwart EL. Sequencing-based detection of low-frequency human immunodeficiency virus type 1 drug-resistant mutants by an RNA/DNA heteroduplex generator-tracking assay. J. Virol. 2004;78:7112–7123. doi: 10.1128/JVI.78.13.7112-7123.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dorr P, Westby M, Dobbs S, Griffin P, Irvine B, Macartney M, Mori J, Rickett G, Smith-Burchnell C, Napier C, et al. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob. Agents Chemother. 2005;49:4721–4732. doi: 10.1128/AAC.49.11.4721-4732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fatkenheuer G, Pozniak AL, Johnson MA, Plettenberg A, Staszewski S, Hoepelman AI, Saag MS, Goebel FD, Rockstroh JK, Dezube BJ, et al. Efficacy of short-term monotherapy with maraviroc, a new CCR5 antagonist, in patients infected with HIV-1. Nat. Med. 2005;11:1170–1172. doi: 10.1038/nm1319. [DOI] [PubMed] [Google Scholar]
- 26.Gulick RM, Su Z, Flexner C, Hughes MD, Skolnik PR, Wilkin TJ, Gross R, Krambrink A, Coakley E, Greaves WL, et al. Phase 2 study of the safety and efficacy of vicriviroc, a CCR5 inhibitor, in HIV-1-Infected, treatment-experienced patients: AIDS clinical trials group 5211. J. Infect. Dis. 2007;196:304–312. doi: 10.1086/518797. [DOI] [PubMed] [Google Scholar]
- 27.Strizki JM, Tremblay C, Xu S, Wojcik L, Wagner N, Gonsiorek W, Hipkin RW, Chou CC, Pugliese-Sivo C, Xiao Y, et al. Discovery and characterization of vicriviroc (SCH 417690), a CCR5 antagonist with potent activity against human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 2005;49:4911–4919. doi: 10.1128/AAC.49.12.4911-4919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westby M, Lewis M, Whitcomb J, Youle M, Pozniak AL, James IT, Jenkins TM, Perros M, van der Ryst E. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J. Virol. 2006;80:4909–4920. doi: 10.1128/JVI.80.10.4909-4920.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]