Abstract
Short interfering RNA (siRNA) may down-regulate many unintended genes whose transcripts possess complementarity to the siRNA seed region, which contains 7 nt. The capability of siRNA to induce this off-target effect was highly correlated with the calculated melting temperature or standard free-energy change for formation of protein-free seed duplex, indicating that thermodynamic stability of seed duplex formed between the seed and target is one of the major factor in determining the degree of off-target effects. Furthermore, unlike intended gene silencing (RNA interference), off-target effect was completely abolished by introduction of a G:U pair into the seed duplex, and this loss in activity was completely recovered by a second mutation regenerating Watson–Crick pairing, indicating that seed duplex Watson–Crick pairing is also essential for off-target gene silencing. The off-target effect was more sensitive to siRNA concentration compared to intended gene silencing, which requires a near perfect sequence match between the siRNA guide strand and target mRNA.
INTRODUCTION
A growing body of evidence from large-scale knockdown experiments (1–5) suggests that short interfering RNA (siRNA) could generate off-target effects through a mechanism similar to that of target silencing by microRNAs (miRNAs) (6–8), which influence the expression levels of many transcripts in a tissue-specific manner (9). The 3′ UTRs of off-target transcripts or miRNA targets are complementary to the guide strand (GS) seed region, nucleotide positions 2–8 (Supplementary Figure S1) (3–6). Within cells, the GS binds to Ago to form an RNA-induced silencing complex (RISC) (10,11). In RISC, the seed nucleotides are presumed to be present on the surface of Ago in a quasi-helical form to serve as the entry or nucleation site for mRNA (12,13). However, little is known about the molecular basis that determines the efficiency of seed-dependent off-target gene silencing. To clarify this point, using a reporter system and microarray profiling, we examined the relationship between stability of the protein-free seed duplex and the capability of siRNA to induce off-target effects.
MATERIALS AND METHODS
Cell culture
Human HeLa cells were cultured and subjected to gene silencing as described previously (14,15). Cells were plated into an individual well of 24-well culture plate at 1 × 105 cells/ml (1 ml/well) 24 h prior transfection. Transfection was carried out using Lipofectamine 2000 (Invitrogen). Sequences of siRNAs chemically synthesized (Proligo) are listed in Supplementary Table S1.
Plasmid construction and gene-silencing assay
All plasmids constructed are derivatives of psiCHECK-1 (Promega). Chemically synthesized 75-bp-long ds oligonucleotides, each including three tandem repeats of an identical 23 bp (Supplementary Table S2) and cohesive XhoI/EcoRI ends, were inserted into the corresponding restriction enzyme sites of psiCHECK-1 to generate psiCHECK-cm and psiCHECK-sm (Figure 1A and B). Inserted cm or sm targets, each 21 nt in length, were expressed as part of the 3′ UTR of Renilla luc mRNA in transfected cells. The completely matched target (cm) matches the siRNA GS completely, whereas the seed-matched (sm) target consists of two parts. 3′ terminal 8 nt of the target are complementary in sequence to the 5′ end (nucleotide position 1) and the seed (positions 2–8) of the corresponding siRNA GS, while the remaining 13 nt are totally unhomologous to the GS (Supplementary Table S2).
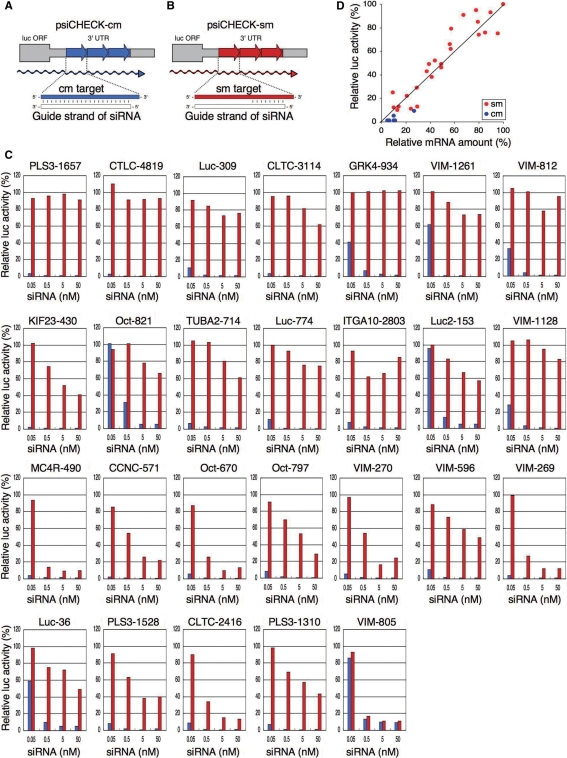
Figure 1.
Off-target gene silencing assay using reporter plasmids. Structures of (A) psiCHECK-cm and (B) psiCHECK-sm (the luc transcript of which, respectively, presents three tandem repeats of cm and sm in the 3′ UTR, shown with thick arrows). (C) Gene-silencing activities of 26 siRNAs were examined in HeLa cells as a function of siRNA concentration. In most cases, luc mRNA with cm was effectively inactivated. In contrast, inactivation of mRNA with sm varied significantly, depending on the siRNA concentration used for transfection. Sequences of siRNA, cm targets and sm targets are shown in Supplementary Tables S1 and S2. (D) Simultaneous reduction in mRNA amount and luc activity in psiCHECK-transfected cells. Ordinate represents relative luc activity (%) in different psiCHECK-sm and -cm transfected cells. Abscissa represents relative amount of luc mRNA (%) in the corresponding psiCHECK-sm and -cm transfected cells. Results strongly suggest that the reduction of luc activity (protein activity) is due mainly to gene-silencing-dependent degradation of the corresponding mRNA.
HeLa cells in a well of 24-well culture plate were transfected simultaneously with one of psiCHECK derivatives constructed above (10 ng), pGL3-Control (Promega, 1 µg) or phLuc-Control (0.5 µg) (14) and siRNA (0.05–50 nM). Cells were harvested 24 h after transfection and relative luc activity (Renilla luc activity/firefly luc activity) was determined using the Dual-Luciferase Reporter Assay System (Promega). pGL3-Control encoding firefly luc also served as a control for relative luc assay for siRNAs against endogenous genes. Used siRNAs against mammalian endogenous genes are: siVIM [human vimentin]-269, -270, -596, -805, -812, -1128 and -1261; siOct [mouse Oct4]-670, -797 and -821; siGRK4 [human G protein-coupled receptor kinase 4]-934; siPLS3 [human plastin3, T-isoform]-1310, -1528 and -1657; siCTLC [human clathrin heavy chain]-2416, -3114 and -4819; siKIF23 [human kinesin family member 23]-430; siTUBA2 [human tubulin alpha]-714; siITGA10 [human integrin alpha2]-2803; siMC4R [human melanocortin 4 receptor]-490; siCCNC [human cyclin C]-571. Gene silencing by siRNAs against firefly luc (siLuc-36, -309, -774 and 2-153) was carried out using phLuc-Control, which encodes luc different from firefly luc. siGY-441, an siRNA for GFP knockdown, used as an siRNA control.
Calculation of thermodymanic parameters
Standard Gibbs free energy change (ΔG) and Tm were calculated according to the nearest neighbor model (16) and the thermodynamic values for RNA–RNA (17). Dissociation constant (Kd) for the seed duplex was determined using the following formula, ΔG = –RTln(1/Kd), where T was 298.15 K. The calculation formula for Tm is as follows: Tm = {(1000 × ΔH)/[A + ΔS + ln(Ct/4)]} −273.15 + 16.6log[Na+]. ΔH (kcal/mol), sum of nearest neighbor enthalpy change. A, helix initiation constant (–10.8). ΔS, sum of nearest neighbor entropy change (17). R, gas constant (1.987 cal/deg/mol). Ct, total molecular concentration of strand (100 nM). [Na+] was fixed at 100 mM.
Microarray analysis
HeLa cells (1 × 105 cells/ml) were inoculated into an individual well culture plate 24 h prior to transfection. Cells were transfected with 50 nM siRNA. Total RNA (3 µg) was purified using RNeasy Kit (Qiagen) 24 h after transfection and hybridized to Human Genome U133 Plus 2.0 GeneChip (Affymetrix) containing about 47 400 human transcripts according to the manufacturer's protocol. RNA from mock-transfected cells, which treated with transfection reagent in the absence of siRNA, was used as a control. The transcript expression value was calculated using Microarray Suite 5.0 (MAS5) (18) with quantile normalization (19); those transcripts with strong enough hybridization signals to be called present (P) were used in this study. To identify transcripts that were downregulated on the array, the cumulative distribution of expression changes for those messages with the site versus those with no canonical site were compared. The statistical significance of their dissimilarity was quantified based on the P-value using Wilcoxon's rank-sum test (6).
Motif analysis of 3′ UTR and CDS
We mapped the probe sequences, which were taken from the annotation table provided on the Affymetrix Web site (http://www.affymetrix.com), to the RefSeq human mRNA sequences (release 24) to identify the target transcripts. We found 67 220 annotations for human transcripts to be corresponded to 54 675 of the probe sets. In microarray analyses using siVIM-270 and siVIM-812, the number of transcripts called P by MAS5 was estimated at 19 856. Of 19 856, 16 783 that represented RefSeq entries (25%) with the 3′ UTR were considered here. In the experiments using siCLTC-2416 and siCLTC-4819, 17 321 of 20 673 transcripts defined as P were analyzed. Seven nucleotide sequences matching the seed region (nucleotides 2–8) of GS were assigned as extracted 3′ UTR or CDS sequences.
Quantitative RT–PCR (qRT–PCR)
qRT–PCR was carried out using an aliquot of total RNA analyzed using the microarray. RNA was reverse-transcribed using the SuperScript First-Strand Synthesis System for RT–PCR (Invitrogen). The mixture of resultant cDNA and SYBR Green PCR Master Mix (Applied Biosystems) were incubated at 95°C for 10 min before the PCR reaction. The levels of PCR products were monitored with ABI PRISM 7000 sequence detection system and analyzed with ABI PRISM 7000 SDS software (Applied Biosystems). Each reaction ran in triplicate. The expression level of each sample was first normalized to the amount of β-actin and then to the mock transfection control. Used primer sets are listed in Supplementary Table S3.
RESULTS AND DISCUSSION
Variation in the efficiency of off-target effect according to the seed sequence
To determine the relationship between siRNA sequences and off-target effects, we introduced 26 pairs of cm and sm target sequences into an expression reporter plasmid, psiCHECK (Figure 1A and B) (14) and examined the change in luc activity in transfected HeLa cells as a function of siRNA concentration (Figure 1C). psiCHECK encodes the Renilla luc gene. Three tandem repeats of cm or sm target sequences were introduced into the region corresponding to the 3′ UTR of the luc mRNA to generate psiCHECK-cm or psiCHECK-sm, respectively. The cm targets, 21 nt in length, were completely complementary to the GS of siRNAs (Figure 1A), which were arbitrarily chosen from the highly functional class I siRNA (15), and used for determining the efficiencies of intended gene silencing, basically RNA interference (RNAi). Class I siRNA possesses A or U residues at position 1, three to six A/U residues in nucleotide positions 2–7 and G/C at position 19 (15). The GS of any class I siRNA is expected to be incorporated into RISC very effectively (15,20,21), such that RISC formation may not be a rate-limiting step in class I-siRNA-dependent unintended gene silencing. The sm sequence possesses complete complementarity to the entire seed region (positions 2–8) but not to the remaining non-seed region, positions 9–21 (Figure 1B), and was used for determining the efficiency of seed-dependent unintended off-target effect. The sm and cm sequences, along with homology to the corresponding siRNA GS are shown in Supplementary Tables S1 and S2. Under our experimental conditions, the content of luc mRNA produced within cells was estimated at about 300 copies per ng of the total RNA, a value one-hundredth of that found for β-actin mRNA. The luc activities measured using different psiCHECK-sm and -cm constructs targeted by the corresponding siRNAs were almost proportional to the levels of mRNA (Figure 1D), indicating that at least under these conditions most, if not all, of the luc activity reduction due to not only RNAi but also off-target effect is attributable to siRNA-dependent luc mRNA degradation.
As anticipated, all 26 siRNAs examined exhibited high activity for intended gene silencing at 50 nM (Figure 1C). Even at 0.5 nM most, if not all, siRNAs used reduced the activity of the luc gene with the cm target to less than 20%. In contrast, the off-target gene silencing calculated using sm target was much less effective and more susceptible to changes in siRNA concentration, with the exception of siVIM-805. Virtually no off-target effects were induced by transfection with any of the 26 siRNAs at 0.05 nM. Only 5 of the 26 siRNAs applied at 0.5 nM reduced luc activity to less than 35%. More than five siRNAs did not bring about any appreciable off-target effects even when the siRNA concentration for transfection was increased to 50 nM. The same RISC is considered to be capable of causing both intended RNAi and unintended off-target gene silencing (22). Thus, above findings may indicate that variations in the efficiency of unintended off-target gene silencing are due to a difference in the interactions between the GS entrapped in RISC and mRNA, i.e. a difference in the efficiency of seed duplex formation. High complementarity in the non-seed region in addition to that in the seed region is necessary for intended gene silencing. A seed/non-seed swapping experiment (Supplementary Figure S2) has shown that unlike seed complementarity to target mRNA, complementarity to the non-seed region is incapable of inducing any gene silencing effects. Notably, siVIM-805 might be an exceptional siRNA that exerts intended gene silencing by a mechanism similar to that for off-target gene silencing (Figure 1C).
In our experiments, the base at the 5′ end (position 1) was always A or U, and complementary to its mRNA counterpart. However, A/U pairing at position 1 may not be important for unintended gene silencing. First, the 5′ end of the GS is considered to be embedded in a pocket of Ago in the RISC (12,13). Second, little or no contribution of A/U-pairing at position 1 to the seed activity or RISC formation has been confirmed by various experiments, including microarray analysis (15,23) (Supplementary Figure S1).
Apparent correlation between seed-dependent off-target effect and thermodynamic stability of the siRNA seed duplex
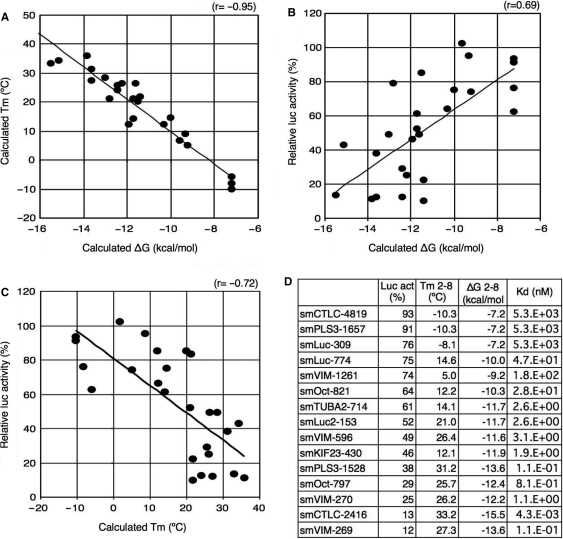
Melting temperature (Tm) and standard free energy change (ΔG) for the formation of the seed duplex may be good measures for the thermodynamic stability of the protein-free seed duplex. We first clarified their relationship. As shown in Figure 2A, the calculated Tm values of 26 seed duplexes in 100 mM NaCl, which varied from −10°C to 36°C, were negatively correlated with the calculated ΔG values, which ranged from −16 to −7 kcal/mol. The correlation coefficient was estimated at −0.95, indicating a very strong correlation. Therefore, a seed duplex with a high Tm is generally considered to be associated with a low ΔG value, and vice versa. Figure 2B demonstrates the positive correlation between relative luc activity compromised by the off-target effect and the calculated ΔG of the protein-free seed duplex. Shown in Figure 2C are the calculated Tm values of the protein-free seed duplex, which were negatively correlated with the compromised luc activity. Correlation coefficients of the former and the latter, respectively, were 0.69 and −0.72, indicating a close relationship between the seed-dependent off-target effect and the seed-duplex ΔG or Tm. The value for ΔG may be converted to the dissociation constant using the formula ΔG = –RTln(1/Kd) (24). Thereby, 15 Kd values were calculated and are listed in Figure 2D. The highest Kd value (CLTC-4819 seed-duplex with the highest ΔG, −7.2 kcal/mol) was found to be more than 106 times greater than the lowest one (CLTC-2416 seed-duplex with the lowest ΔG, −15.5 kcal/mol). This indicates a wide range of seed-duplex stability, which may account for the strong siRNA-concentration dependency of the off-target effect (Figure 1C). Thus, it may follow that the degree of off-target effects is primarily governed by the thermodynamic stability of duplex, 7 bp in length, which is formed between the GS seed region and its mRNA counterpart.
Figure 2.
Close relationship between efficiency of seed-dependent off-target gene silencing and the thermodynamic stability of the protein-free seed duplex. (A) The calculated Tm for the seed duplex decreases with increasing changes in standard free-energy (ΔG) for seed duplex formation (correlation coefficient, −0.95). (B) luc activity compromised by seed-dependent off-target gene silencing at 50 nM siRNA concentration was positively correlated with ΔG (correlation coefficient, 0.69). The luc activity data were collected from Figure 1C. (C) Correlation between seed-dependent gene silencing activity (luc activity) and the calculated Tm of the protein-free seed duplex. A set of luc activities compromised by seed-dependent gene silencing at 50 nM siRNA concentration were collected from Figure 1C. Tm value of the protein-free seed region (positions 2–8) was calculated using the nearest neighbor method. Relative luc activity and calculated Tm were correlated with each other and had a coefficient of −0.72. (D) List of calculated dissociation constants for protein-free seed duplexes. ΔG was converted to Kd using the formula ΔG = –RTln(1/Kd).
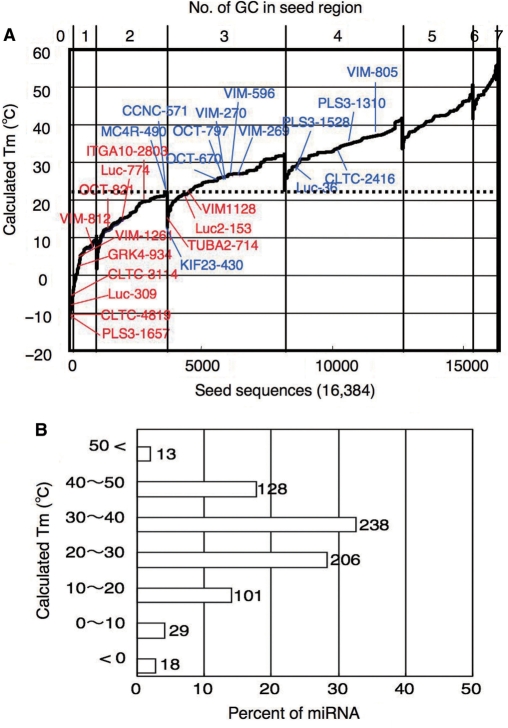
A considerable deviation was observed in luc activity measurements shown in Figure 2B and C. This may be due in part to differences in the non-seed sequence and/or its counterpart in target mRNA, since the target sequences that correspond to the non-seed region make an appreciable contribution to target recognition by miRNAs and/or siRNAs in microarray profiling (1,6,8) (Supplementary Figure S1). The reporter assay using eight different targets with common seed and different non-seed sequences showed that off-target effects are rather various, indicating possible involvement of siRNA non-seed region and/or its target counterpart in off-target effect (Supplementary Figure S3). However, correlation between seed-dependent gene silencing activity of siRNA used in this study (Figure 1C) and calculated Tm of protein-free seed duplex of them aligned in all possible 7-nt sequences (47 = 16 384) may suggest that 21.5°C serves as a kind of benchmark Tm (Figure 3A), which may discriminate almost off-target-free seed sequences from off-target-positive ones.
Figure 3.
Correlation between seed-dependent gene-silencing activity and calculated Tm of protein-free seed duplex. Gene-silencing activity was measured using relative luc activity in HeLa cells transfected with psiCHECK-sm and cognate siRNAs at 50 nM as shown in Figure 1C. Tm of the protein-free seed region (positions 2–8) was determined using nearest neighbor method (Figure 2C). (A) All possible 7-nt seed sequences (47 = 16 384) were ordered as a function of GC content and Tm values of their ds counterparts. Note that, because of its definition, class I siRNA cannot possess more than four GC in the seed region. Blue: combinations of target and siRNA, giving less than 50% relative luc activity. Red: combinations of target and siRNA with little or no off-target effect (luc activity >50%). Dotted line at 21.5°C may correspond to 50% luc activity reduction. (B) Tm distribution of 733 human microRNAs registered in miRbase. Ordinate represents calculated seed-duplex Tm. Abscissa represents percentage. Seventy-five percent of 565 siRNAs registered was estimated to be associated with Tm more than 21.5°C.
Genome-wide analysis of off-target gene silencing determined by the siRNA seed duplex stability
To further confirm the notion that off-target gene silencing is determined primarily by seed duplex stability, genome-wide expression profiling was carried out using four class I siRNAs targeting human vimentin (siVIMs) or clathrin heavy chain (siCLTCs) (Figure 4; Supplementary Figures S4 and S5). The seed duplexes generated by siVIM-270 and siCLTC-2416 have Tm values in 100 mM NaCl of 26.2°C and 33.2°C, respectively, while those generated by siVIM-812 and siCLTC-4819 are 8.8°C and −10.3°C, respectively. HeLa cells were transfected with 50 nM siRNA and the changes in the expression level of transcripts were analyzed after 24 h. The reporter assay described above predicted that siRNAs with high seed duplex Tm values (siVIM-270 and siCLTC-2416) may be good inducers, while those with low Tm values (siVIM-812 and siCLTC-4819) may be poor inducers of the off-target effect.
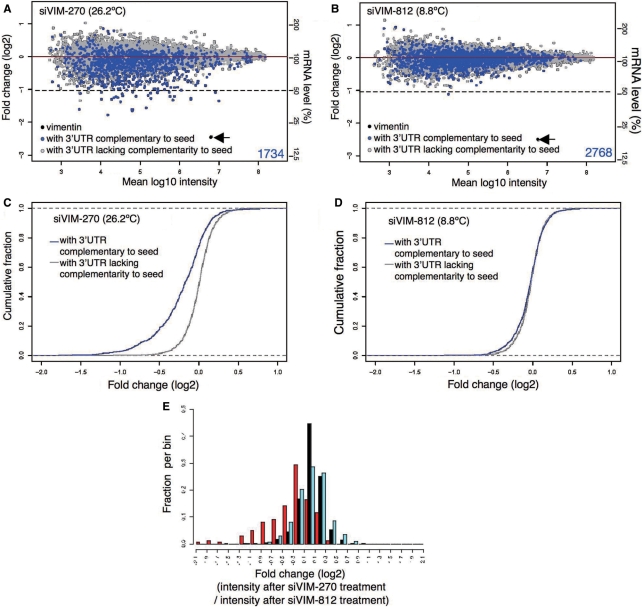
Figure 4.
Microarray-based off-target-effect profiling and data analysis. Profiles of transcript downregulation by (A) siVIM-270 and (B) siVIM-812. The cumulative distribution of transcripts from cells treated with (C) siVIM-270 and (D) siVIM-812, are shown as log2 of fold change to mock transfection. The blue line (C, D) indicates the cumulative fraction of transcripts with one or more sequence complementary to the siRNA seed in the 3′ UTR. The gray line (C, D) shows transcripts with no seed complementarity. Distributions of log fold change, defined as the log2 of expression in siVIM-270- or siVIM-812-transfected cells over that in mock-transfected cells, for mRNAs lacking seed complementarity are 0.221 ± 0.001 (C) and 0.016 ± 0.005 (D), respectively. (E) Comparison of microarray profiles of transcripts downregulated by siVIM-270 and siVIM-812. Only transcripts with 3′ UTR complementarity to two or more than two siRNA seed sequences were examined. Ordinate represents fraction. Abscissa represents signal intensity obtained after siVIM-270 treatment divided by that after siVIM-812 treatment. The blue and red bars denote the distribution of transcripts with 3′ UTR complementary to the VIM-812 and VIM-270 seed sequence, respectively. The black bar represents transcripts with seed complementarity to neither siVIM-270 nor siVIM-812.
As anticipated, all four siRNAs used effectively reduced the amount of vimentin and clathrin heavy chain mRNA, to less than 20% (arrows in Figure 4A and B; Supplementary Figure S5) as a result of intended RNAi effect. In contrast, the off-target-dependent reduction in the amount of mRNA depended significantly on the Tm of the seed-duplex. A high level of off-target effects was evident in the case of transfection with siVIM-270 (Figure 4C) and siCLTC-2416 (Supplementary Figure S4A), which were predicted as good off-target inducers. The expression levels of the transcripts with seed complementary sequences (positions 2–8 of siRNA guide strand) in 3′ UTRs were clearly reduced by both siRNAs in common, although the transcripts with complementarities in the other positions were not changed at least coincidentally (Supplementary Figure S1), in a manner consistent with the previous reports (3–6). Transfection with siVIM-812 (Figure 4D) and siCLTC-4819 (Supplementary Figure S4B) that were predicted as poor off-target inducers exhibited little off-target effects (Figure 4E, Supplementary Figure S4C). Results from microarray quantitative analysis were essentially identical to those obtained by qRT–PCR (Supplementary Figure S6). We conclude that the level of off-target gene silencing is determined by the thermodynamic stability of the seed duplex formed between the siRNA GS and the target mRNA.
Furthermore, in accordance with the recent papers described that not only 3′ UTR but also CDS is miRNA target (25,26), our (Supplementary Figure S1) and others’ (6) microarray analyses indicated that seed-dependent off-target silencing effects are also observed in transcripts containing seed complementary sequences in CDSs, although the effects were marginal. As in the case of 3′ UTR, the off-target effects by the transfection of siRNAs with low thermodynamic stabilities of seed duplexes (siVIM-812 and siCLTC-4819) were reduced in the transcripts with seed complementarity in CDSs (data not shown) compared to the results of siRNAs with high stabilities (siVIM-270 and siCLTC-2416; Supplementary Figure S1D and F). Thus, the thermodynamic stability of the seed duplex might be basic determinant of off-target silencing in both CDSs and 3′ UTRs, although the mechanism of marginal seed-dependent silencing effects in CDSs remains unknown.
The miRbase includes 733 human miRNA sequences (27). Our calculation indicated that 546 of the 733 were capable of providing seed duplexes of which Tm values are greater than 21.5°C (Figure 3B), suggesting that about 75% of miRbase miRNAs may exert relatively strong seed-dependent gene silencing.
Elimination of seed-dependent off-target gene silencing by G:U pairing
G:U pairing is not typical of Watson–Crick pairing, which consists of G:C and A:U pairs in the case of RNA (28). G:U pairs were frequently incorporated into RNA secondary structures, probably because the standard free-energy change for G:U pair formation is almost equivalent to that for A:U pair formation (28) (Supplementary Table S4). However, array analysis may suggest that G:U pairing causes a virtually complete elimination of the off-target effect (Figure 5) (8). To extend this point, G:U pairing mutations were introduced into the VIM-270 seed-duplex (Figure 6). Both target sequence and siRNA mutations were introduced by chemical oligonucleotide synthesis. The luc mRNA with a mutated target was expressed using psiCHECK. Note that all mutations producing G:U pairing at positions 2, 3 and 7, are target sequence mutations, which may not influence the efficiency of RISC formation (22). Figure 6 shows clearly that introduction of any G:U pairing to the seed duplex results in an almost complete loss of the off-target effect. The second mutation was introduced into the opposite strand to generate a phenotypic revertant of Watson–Crick pairing (Figure 6C–G). The off-target effect was almost completely recovered, irrespective of differences in location of the mutation within the seed duplex. In contrast to the off-target effect, G:U pairing afforded virtually no effect on intended gene silencing (Figure 6), indicating an apparent difference between the molecular mechanisms of intended RNAi and unintended off-target gene silencing.
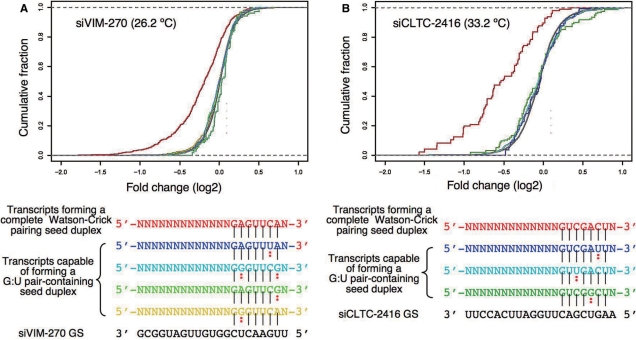
Figure 5.
Microarray analysis of the effect of G:U pairing in the seed duplex. Two siRNAs, (A) siVIM-270 and (B) siCLTC-2416, with high Tm values (26.2°C and 33.2°C, respectively) were used at 50 nM for transfection. Together, (A) and (B) indicate that transcripts in which the 3′ UTR is forming the seed duplex consisting only of Watson–Crick pairing are capable of exerting a significant off-target effect. Transcripts forming a G:U pair-containing seed duplex are associated with virtually no off-target effect. Nucleotide sequences of transcripts analyzed are shown in the lower margin. Red denotes transcripts forming a seed duplex consisting only of Watson–Crick base pairs [1043 transcripts in (A), 97 in (B)]. Other colors represent transcript groups that are capable of forming a G:U pair-containing seed duplex in the 3′ UTR. The numbers of transcripts are as follows: in (A), 1076 blue, 104 sky blue, 331 green and 931 yellow; in (B), 114 blue, 1226 sky blue and 157 green. Solid vertical line denotes Watson–Crick pairing. Dotted line denotes G:U pairing.
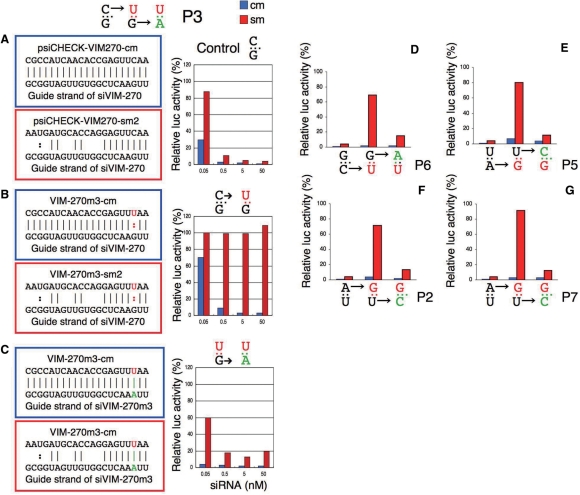
Figure 6.
Reporter plasmid analysis of the effect of G:U pairing in the seed duplex. Three copies of cm or sm targets were introduced into psiCHECK. Blue bars represent silencing of cm targets. Red bars refer to off-target gene (sm-target) silencing. Bases replaced by the first mutation are colored in red, while those replaced by the second mutation are colored in green. (A) Control experiment. The siVIM-270-dependent silencing of luc mRNA with VIM-270 completely matched (cm) or seed-matched (sm2) targets. Note the absence of any G:U pairing from the seed duplex. (B) The C residue opposite position 3 of the GS in the VIM-270-cm or VIM-270-sm2 targets was replaced with U to generate VIM-270m3-cm or VIM-270m3-sm2. This m3 mutation introduces a U:G pair at position 3 of both the cm and sm seed duplexes. Although cm-target silencing was not affected by the mutation, sm-target silencing (off-target effect) was almost completely abolished. (C) G→A and C→U substitutions were introduced at position 3 of siVIM-270 GS and its counterpart in the passenger strand (PS), respectively, as the second mutations to generate a siRNA mutant, siVIM-270m3. Thus, U:A pairing was introduced at ds position including nucleotide 3 of the GS to generate a phenotypic Watson–Crick pairing revertant. A significant recovery of off-target effect is evident. (D–G) Four additional examples of G:U pairing mutation and phenotypic reversion. Mutations were introduced into VIM270-cm, VIM270-sm or siVIM-270 to generate pairing mutants of the seed duplex at GS positions (D) 6, (E) 5, (F) 2 and (G) 7. In all cases examined, the first mutation (G:U pair introduction in the seed region) abolished nearly all off-target silencing activity, which was almost completely recovered by the second mutation, which regenerated Watson–Crick pairing.
As shown in Supplementary Table S4, the difference in ΔG values between the G:U pair-containing seed duplex and its cognate with an A:U pair is very small, if at all, indicating that a difference in ΔG, or thermodynamic stability of seed duplex, cannot account for a catastrophic change in off-target effects due to a G:U/A:U replacement. The substitution of any Watson–Crick pair by a G:U pair has been reported to result in structural perturbation, regardless of the sequence (28). Structural analysis of the Aquifex aeolicus Ago has shown that the phosphodiester backbone of five consecutive residues corresponding to position 2–6 of the GS, which adopts a helical pitch, has extensive surface and charge complementarity with the side chains in the Mid and PIWI domains of Ago (12,13). Thus, these structural constraints on the GS seed may make it difficult to form any G:U pairing between target mRNA and the GS seed binding to Ago in RISC.
CONCLUSION
We found that the seed-dependent off-target effect is highly correlated with the thermodynamic stability in the duplex formed between the seed region of the siRNA guide strand and its target mRNA. ΔG and Tm for the formation of the seed duplex, respectively, are positively and negatively correlated with seed-dependent gene silencing activity. The seed-dependent off-target gene silencing effect is almost completely eliminated with G:U pairing in the seed duplex. In this case, off-target effect is avoided probably because of the structural perturbation of seed duplex but not seed duplex stability.
Unlike the off-target effect, intended gene silencing, RNAi, was significantly tolerant to not only the presence of a G:U pair in the seed region (see e.g. Figure 6A and B), but also to changes in siRNA concentration (Figure 1C). We therefore consider that for RNAi, which requires a near perfect match between target mRNA and the siRNA GS, the first recognition of mRNA by the GS seed arm and subsequent target recognition by the non-seed region of GS are so highly cooperative that the second proof-reading process can easily surmount the G:U pairing problem in the initial mRNA nucleation process.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant-in-Aid for priority area and Genome Network Project from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to K.U.-T., partial). Funding for open access charge: Ministry of Education, Culture, Sports, Science and Technology.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotech. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 2.Scacheri PC, Rozenblatt-Rosen O, Caplen NJ, Wolfsberg TG, Umayam L, Lee JC, Hughes CV, Shanmugam KS, Bhattacharjee A, Meyerson M, et al. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc. Natl Acad. Sci., USA. 2004;101:1892–1897. doi: 10.1073/pnas.0308698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin X, Ruan X, Anderson MG, McDowell JA, Kroeger PE, Resik SW, Shen Y. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 2005;33:4527–4525. doi: 10.1093/nar/gki762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- 6.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 8.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Martinez J, Patkaniowska A, Urlaub H, Lührmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 11.Schwarz DS, Hutvágner G, Haley B, Zamore PD. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell. 2002;10:537–548. doi: 10.1016/s1097-2765(02)00651-2. [DOI] [PubMed] [Google Scholar]
- 12.Yuan Y-R, Pei Y, Ma JB, Kuryavyi V, Zhadina M, Meister G, Chen HY, Dauter Z, Tuschl T, Patel DJ. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma J-B, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ui-Tei K, Naito Y, Zenno S, Nishi K, Yamato K, Takahashi F, Juni A, Saigo K. Functional dissection of siRNA sequence by systematic DNA substitution: modified siRNA with a DNA seed arm is a powerful tool for mammalian gene silencing with significantly reduced off-target effect. Nucleic Acids Res. 2008;36:2136–2151. doi: 10.1093/nar/gkn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ui-Tei K, Naito Y, Takahashi F, Haraguchi T, Ohki-Hamazaki H, Juni A, Ueda R, Saigo K. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004;32:936–948. doi: 10.1093/nar/gkh247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panjkovich A, Melo F. Comparison of different melting temperature calculation methods for short DNA sequences. Bioinformatics. 2005;21:711–722. doi: 10.1093/bioinformatics/bti066. [DOI] [PubMed] [Google Scholar]
- 17.Freier AM, Kierzek R, Jaeger JA, Sugimoto N, Caruthers MH, Neilson T, Turner DH. Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl Acad. Sci. USA. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hubbell E, Liu WM, Mei R. Robust estimators for expression analysis. Bioinformatics. 2002;18:1585–1592. doi: 10.1093/bioinformatics/18.12.1585. [DOI] [PubMed] [Google Scholar]
- 19.Bolstad BM, Irizarry RA, Gautier L, Wu Z. Preprocessing high-density oligonucleotide arrays. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer; 2005. pp. 13–32. [Google Scholar]
- 20.Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 21.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 22.Filipowicz W. RNAi: The nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 23.Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2007;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turner DH. Thermodynamics of base pairing. Curr. Opin. Struct. Biol. 1996;6:299–304. doi: 10.1016/s0959-440x(96)80047-9. [DOI] [PubMed] [Google Scholar]
- 25.Duursma AM, Kedde M, Schrier M, Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14:872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 27.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varani G, McClain WH. The G·U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000;1:18–23. doi: 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.