Abstract
To investigate nucleic acid base pairing and stacking via atom-specific mutagenesis and crystallography, we have synthesized for the first time the 6-Se-deoxyguanosine phosphoramidite and incorporated it into DNAs via solid-phase synthesis with a coupling yield over 97%. We found that the UV absorption of the Se-DNAs red-shifts over 100 nm to 360 nm (ε = 2.3 × 104 M−1 cm−1), the Se-DNAs are yellow colored, and this Se modification is relatively stable in water and at elevated temperature. Moreover, we successfully crystallized a ternary complex of the Se-G-DNA, RNA and RNase H. The crystal structure determination and analysis reveal that the overall structures of the native and Se-modified nucleic acid duplexes are very similar, the selenium atom participates in a Se-mediated hydrogen bond (Se … H–N), and the SeG and C form a base pair similar to the natural G–C pair though the Se-modification causes the base-pair to shift (approximately 0.3 Å). Our biophysical and structural studies provide new insights into the nucleic acid flexibility, duplex recognition and stability. Furthermore, this novel selenium modification of nucleic acids can be used to investigate chemogenetics and structure of nucleic acids and their protein complexes.
INTRODUCTION
Selenium derivatization of proteins via selenomethionine has revolutionized protein X-ray crystallography via multiwavelength anomalous dispersion (MAD), and two thirds of new crystal structures of proteins have been determined via this strategy recently (1,2). Indirect derivatization of nucleic acids with the Se-labeled proteins for structure determination of five ribozymes, such as the hepatitis δ virus ribozyme and the flexizyme (3,4), was also reported. Inspired by these advances, our research group pioneered and developed covalent incorporation of selenium into DNAs and RNAs (5–8) for structure determination via MAD or single-wavelength anomalous dispersion (SAD) phasing. This novel research area has attracted many attentions and research activities in chemical synthesis, biochemistry and structural biology (9–15).
Besides structural study, we are exploring chemogenetic investigation of nucleic acid function by the atom-specific substitution of oxygen (atomic radius, 0.73 Å) with selenium (1.16 Å, from the same elemental Family VIA in the periodic table) as an atomic probe (12,15). As the genetic information storage, replication, and transcription are achieved via base-pairing, stacking interaction and size-and-shape impact of the nucleobase pairs, extensive research has been focused on studying the recognition and stability of nucleobase pairs and double-stranded structures (16–20). Interestingly, our recent study via replacement of thymidine 4-oxygen with selenium in DNA (SeT) has revealed that DNA is flexible and able to accommodate a large atom. In addition, we have discovered that the thymidine 4-selenium atom forms a hydrogen bond (Se … H–N) with the adenosine 6-amino group in DNA duplex (12), and the Se-nucleobase-derivatized DNA has an X-ray crystal structure virtually identical to the corresponding native DNA structure. This selenium substitution, which leads to the Se-mediated hydrogen bond, the Se-nucleobase stacking interaction and the Se-modified duplex, provides a unique opportunity to obtain new insights into the base-paring and stacking interactions, and the duplex recognition and stability.
Despite the synthesis of the 2′-deoxy-6-selenoguanosine and its derivatives several decades ago (21,22), synthesis of nucleic acids containing the Se-guanine remained a challenge, even though 6-S-purines have been introduced into nucleic acids (23,24). To explore the Se derivatization of guanine for structure and function studies, we report here the synthesis of a 6-Se-2′-deoxyguanosine phosphoramidite, its incorporation into oligonucleotides, and Se-DNA duplex stability investigation via UV-melting study. We also report here the observation of the colored DNAs containing the 6-Se-guanine and the Se-G DNA stability in water and at an elevated temperature. Moreover, we crystallized a ternary complex of the Se-G-DNA, RNA and RNase H, and determined its X-ray crystal structure. This is the first structure determination of a protein-nucleic acid complex on the basis of Se-derivatized nucleic acids and MAD phasing. We also report here that the overall nucleic acid structures of the native and modified duplexes are very similar, that the selenium atom forms a Se-hydrogen bond (Se … H–N), and the SeG and C form a base pair similarly to the native G–C pair, though the 6-selenium modification results in the base-pair shift for approximately 0.3 Å. This Se-base-pair shift leads to a reduction in the base-stacking interaction, which explains the decrease in UV-melting temperatures of the modified duplexes, comparing to the corresponding native ones. Our exciting and novel discoveries will open a new research avenue in structure and function studies of nucleic acids as well as their protein complexes.
MATERIALS AND METHODS
Synthesis of 6-Se-deoxyguanosine phosphoramidite (3)
N2-[2-(4-tert-butylphenoxy)acetyl]-6-(2-cyanoethyl)seleno-5′-O-(4,4′-dimethoxytriphenylmethyl)-2′-deoxyguanosine (2). 2,4,6-(Triisopropylbenzene)sulfonyl chloride (300 mg, 1 mmol, 1.5 eq., TIBS–Cl) dissolved in CH2Cl2 (1 ml) was added to the solution of 1 (500 mg, 0.66 mmol), 4-dimethylaminopyridine (15 mg, 0.12 mmol, DMAP), and triethylamine (0.18 ml, 1.32 mmol, 2 eq., TEA) in CH2Cl2 (2 ml) under argon. This reaction was stirred at room temperature for 15 min (monitored by silica gel TLC to confirm the completion; 5% MeOH in CH2Cl2) and then injected to a solution of the sodium selenide (NCCH2CH2SeNa). The selenide solution (25) was prepared by injecting the NaBH4 suspension (150 mg in 4 ml of EtOH) into a flask containing di-(2-cyanoethyl) diselenide (700 mg, 2.64 mmol, 8 eq.) dissolved in ethanol (15 ml) on an ice-bath and under argon. After the selenium incorporation reaction was completed in an hour (monitored by TLC, 5% MeOH in CH2Cl2, product Rf = 0.43), water (10 ml) was added to the reaction flask. The solution was extracted with CH2Cl2 (3 × 20 ml). The combined organic layer was dried over MgSO4 (s), filtered and evaporated under reduced pressure. The crude product was then dissolved in methylene chloride (5 ml) and purified on a silica gel column equilibrated with methylene chloride. The column was eluded with a step-wise gradient of methanol-methylene chloride mixtures (CH2Cl2, 0.5%, 1.0%, 2.0% MeOH in CH2Cl2, 300 ml each) to afford product 2 as a white foam (460 mg, 80% yield over two reactions). Intermediate: 5′-O-(4,4′-Dimethoxytriphenylmethyl)-N2-[2-(4-tert-butylphenoxy)acetyl]-6-O-[2,4,6-(triisopropylbenzene)sulfonyl]-2′-deoxyguanosine. 1H-NMR (CD2Cl2) δ: 1.28–1.32 (m, 18H, 6× CH3-ipr), 1.36 (s, 9H, 3× CH3-tBu), 2.59–2.65 and 2.76–2.83 (2× m, J2′−1′ = 6.4 Hz, 2H, H-2′), 2.99 (h, J = 6.8 Hz, 1H, CH-ipr), 3.16 (br, 1H, OH), 3.35 and 3.47 (2× dd, J5′−4′ = 4.0 and J5′−5′ = 10.4 Hz, 2H, H-5′), 3.78 (s, 6H, 2× OCH3), 4.21–4.31 (m, 3H, 2× CH-ipr and H-4′), 4.64 (s, 2H, CH2–O), 4.85–4.88 (m, 1H, H-3′), 6.60 (t, J1′-2′ = 6.4 Hz, 1H, H-1′), 6.76–7.50 (m, 19H, CH-arom), 8.16 (s, 1H, H-8), 8.66 (br, 1H, NH); 13C-NMR (CD2Cl2) δ: 23.24 and 24.30 (CH3-ipr), 29.93 and 34.36 (CH-ipr), 31.20 (CH3-tBu), 34.07 (C-tBu), 40.36 (C-2′), 55.16 (OMe), 64.23 (C-5′), 68.11 (CH2-O), 72.31 (C-3′), 84.64 (C-1′), 86.44 (C-arom), 86.83 (C-4′), 113.05, 114.41, 124.06, 126.54, 126.81, 127.75, 128.04, 129.92, 130.08 (CH-arom), 120.57 (C-5), 131.18, 135.55, 135.88, 144.76, 145.18, 150.31, 150.76, 154.52, 154.68, 154.92, 158.63, 158.67, 166.40 (C=O). HRMS (ESI–TOF): molecular formula, C58H67N5O10S; [M + H]+: 1026.4678 (calc. 1026.4681). Compound 2: 1H-NMR (CD2Cl2) δ: 1.35 (s, 9H, 3× CH3-tBu), 2.47 (br, 1H, OH), 2.56–2.62 and 2.80–2.88 (2× m, J2′−1′ = 6.4 Hz, 2H, H-2′), 3.16 (t, J = 7.2 Hz, 2H, SeCH2CH2CN), 3.34 and 3.47 (2× dd, J5′-4′ = 4.0 and J5′−5′ = 10.0 Hz, 2H, H-5′), 3.58 (t, J = 7.2 Hz, 2H, Se–CH2–CH2–CN), 3.79 (s, 6H, 2× OCH3), 4.18–4.21 (m, 1H, H-4′), 4.68 (s, 2H, CH2–O), 4.80–4.84 (m, 1H, H-3′), 6.49 (t, J1′−2′ = 6.4 Hz, 1H, H-1′), 6.78–7.43 (m, 17H, CH-arom), 8.10 (s, 1H, H-8), 8.96 (br, 1H, NH); 13C-NMR (CD2Cl2) δ: 18.92 and 19.01 (SeCH2CH2CN), 30.94 (CH3–tBu), 33.83 (C–tBu), 40.04 (C-2′), 54.95 (OMe), 63.80 (C-5′), 67.76 (CH2–O), 72.00 (C-3′), 84.11 (C-1′), 86.17 (C-arom), 86.27 (C-4′), 112.79, 114.07, 126.39, 126.57, 127.56, 127.76, 129.73 (CH-arom), 118.93 (CN), 131.49 (C-5), 135.38, 135.42, 144.54, 144.99, 154.63, 158.39 (C-arom), 148.37 (C-4), 150.80 (C-2), 158.01 (C-6), 165.55 (C=O). HRMS (ESI–TOF): molecular formula, C46H48N6O7Se; [M + Na]+: 899.2651 (calc. 899.2642).
N2-[2-(4-tert-butylphenoxy)-acetyl]-6-(2-cyanoethyl)-seleno-5′-O-(4,4′-dimethoxytriphenylmethyl)-2′-deoxy-guanosine 3′-O-(2-cyanoethyl)-N,N-diisopropylamino phosphoramidite (3). Compound 2 (250 mg, 0.29 mmol) and 5-(benzylthio)-1H-tetrazole (27 mg, 0.15 mmol) were dried on a high vacuum overnight. Under argon, dry methylene chloride (1 ml) was added into the flask to dissolve them, followed by injection of 2-cyanoethyl N,N,N,N-tetraisopropylphosphorodiamidite (103 mg, 0.34 mmol, 1.2 eq.). The solution was stirred under argon at room temperature for 30 min. Reaction completion was indicated by TLC [CH2Cl2/EtOAc (7:3), product (Rf = 0.58 and 0.65): a mixture of two diastereomers]. The reaction was quenched with NaHCO3 (3 ml, sat.), stirred for 5 min, and extracted with CH2Cl2 (3 × 5 ml). The combined organic layer was dried over MgSO4 (s), filtered, and evaporated under reduced pressure. The crude products were purified on a silica gel column equilibrated with CH2Cl2, and eluted with CH2Cl2/EtOAc (7:3). After dissolving in CH2Cl2 (1 ml), these products were precipitated from pentane (200 ml) to yield the titled compound (3) as a white powder (230 mg, 75% yield). 1H-NMR (CD2Cl2, two sets of signals from a mixture of two diastereoisomers) δ: 1.15–1.24 (m, 24H, 8× CH3-ipr), 1.36 (s, 18H, 6× CH3–tBu), 2.51 and 2.65 (2 × t, J = 6.4 Hz, 4H, 2× OCH2CH2CN), 2.68–2.78 and 2.88–2.95 (2× m, J2′-1′ = 6.4 Hz, 4H, 2× H-2′), 3.21 and 3.22 (2 × t, J = 7.2 Hz, 4H, 2× SeCH2CH2CN), 3.35–3.50 (m, 4H, 2× H-5′), 3.58 (t, J = 7.2 Hz, 4H, 2× SeCH2CH2CN), 3.63–3.91 (m, 8H, 4× CH-ipr, 2× O–CH2–CH2–CN), 3.80 and 3.82 (2× s, 12H, 4× OCH3), 4.29–4.37 (m, 2H, 2× H-4′), 4.71 and 4.72 (2× s, 4H, 2× CH2–O), 4.78–4.88 (m, 2H, 2× H-3′), 6.44 (t, J1′-2′ = 6.4 Hz, 2H, 2× H-1′), 6.78–7.45 (m, 34H, CH-arom), 8.13 and 8.14 (2× s, 2H, 2× H-8), 8.91 and 8.92 (2× br, 2H, 2× NH); 13C-NMR (CD2Cl2) δ: 19.14 (SeCH2CH2CN), 19.43 and 19.45 (SeCH2CH2CN), 20.20, 20.27 and 20.36, 20.43 (OCH2CH2CN), 24.30, 24.34, 24.37, 24.41 (CH3-ipr), 31.21 (CH3-tBu), 34.08 (C-tBu), 39.44, 39.49 and 39.54, 39.57 (C-2′), 43.22, 43.34 (CH-ipr), 55.18 and 55.21 (OMe), 58.15, 58.34 and 58.29, 58.48 (OCH2CH2CN), 63.35 and 63.69 (C-5′), 68.08 (CH2–O), 73.38, 73.55 and 73.85, 74.03 (C-3′), 84.48 and 84.52 (C-1′), 85.85, 85.91 and 86.09, 86.13 (C-4′), 86.55 (C-arom), 112.59, 114.33, 126.61, 126.80, 126.83, 127.79, 128.01, 128.08, 129.99, 130.02, 130.06 (CH-arom), 117.66 and 117.82 (OCH2CH2CN), 119.29 (SeCH2CH2CN), 131.76 and 131.77 (C-5), 135.60, 135.65, 144.77, 145.12, 145.16, 155.00, 155.02, 158.64, 158.66 (C-arom), 148.72 and 148.76 (C-4), 151.13 and 151.16 (C-2), 158.29 and 158.35 (C-6), 165.76 (C=O); 31P-NMR (CD2Cl2, using H3PO4 as the standard) δ: 149.2. HRMS (ESI–TOF): molecular formula, C55H65N8O8PSe; [M + H]+: 1077.3902 (calc. 1077.3901).
Synthesis, purification and analysis of the Se-G DNAs
All syntheses were carried out in a 1-μmol scale and with the DMTr-on. The 6-Se-G derivatized oligonucleotides were prepared using the ultramild CE phosphoramidites (dA, dC and dG). Concentration of the Se-modified phosphoramidite (3) in acetonitrile was the same as those of the conventional phosphoramidites (0.1 M). The phosphoramidite coupling reaction was carried out using BTT activator (0.325 M). After the synthesis, the Se-oligonucleotides were cleaved from the solid support and fully deprotected overnight at room temperature with potassium carbonate (1 ml, 0.05 M in anhydrous methanol). The supernatant was evenly divided into two 2-ml Eppendorf tubes, followed by the addition of water (1 ml per tube) and triethylammonium acetate buffer [0.5 ml per tube, 2 M triethylammonium acetate (TEAAc), pH 7.1]. After filtration with 0.45 μ filter, the Se-DNA oligonucleotides were purified by reversed-phase high performance liquid chromatography (RP-HPLC) twice, with DMTr-on and DMTr-off.
The purification was carried out on Welchrom XB-C18 column (21.1 × 250 mm, 10 µ) with buffer A (10 mM TEAAc, pH 7.1) and buffer B (10 mM TEAAc, pH 7.1, in 50% acetonitrile). A flow rate of 6 ml/min and a gradient (starting from buffer A) were used with buffer B increased by 4.5% (for DMTr-on) or 2% (for DMTr-off) every minute over 20 min. By monitoring under 260 nm and 360 nm, the Se-DNAs were collected and lyophilized. For the 5′-DMTr deprotection, the lyophilized Se-DNAs with DMTr-on were treated with trichloroacetic acid solution (final concentration: 0.3% w/w) for 3 min, followed by the addition of TEAAc buffer (2 M) to adjust the pH to ≈7. Similarly, the Se-DNAs with DMTr-off were purified by HPLC again, followed by lyophilization. The purified Se-DNAs with DMTr-off were re-dissolved in water and analyzed by HPLC, UV and MS to confirm the high quality and integrity. Similarly, Se-DNA HPLC analysis was performed on a Welchrom XB-C18 column (4.6 × 250 mm, 5 μ) using the same buffer system. A flow rate of 1.0 ml/min and a gradient (starting from buffer A) of reaching 30% buffer B in 20 min were used.
UV absorption, thermo-stability, and duplex melting studies of the Se-G DNAs
To determine the extinction coefficient of DNA SeG by comparing with the native nucleotide, we synthesized and purified GG, SeGG and SeGSeG dimers, and their UV spectra were studied. By taking the advantage of HPLC separation and UV analysis, we have developed a useful HPLC–UV approach to allow separation and accurately measure and calculate the extinction coefficients of the base-modified nucleotides. The HPLC conditions are the same as those in the oligonucleotides analysis. To study the Se-DNA thermo-stability, 6-Se-G-DNAs were heated at 60°C for 1 h in the buffer of 100 mM NaH2PO4-Na2HPO4 (pH 7.6), followed by UV and HPLC analysis.
We measured the melting temperature of the Se-DNA-derivatized duplexes along with those of the native duplexes. Prior to acquisition of the melting curves, duplexes were annealed by heating to 70°C for 2 min, followed by slowly cooling to 5°C and keeping at the temperature for 3 h. Denaturation curves were acquired at 260 nm and 1 cm path length at heating or cooling rates of 0.5°C/min, using a UV-Vis spectrophotometer equipped with a six-sample thermo-stated cell block and a temperature controller. The experiments were performed using the samples (DNA duplexes, 1.0 μM) dissolved in the buffer of 50 mM NaCl, 10 mM NaH2PO4-Na2HPO4 (pH 6.5), 0.1 mM EDTA and 10 mM MgCl2.
RESULTS AND DISCUSSION
Synthesis of 6-Se-2′-deoxyguanosine phosphoramidite (3)
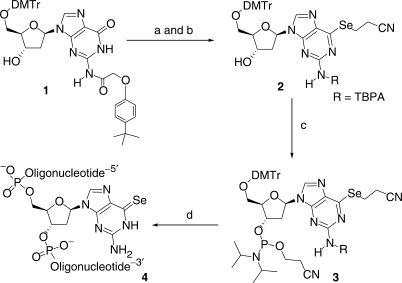
Our development of the 2-cyanoethyl-seleno protection and deprotection for the 4-Se-thymidine DNA synthesis (12) encouraged us to protect the 6-Se-functionality on deoxyguanosine with the same protecting group. In addition, the 2-cyanoethyl protecting group can be removed under ultramild conditions (0.05 M K2CO3 in methanol). Since strong basic conditions (such as NH3 treatment) can cause deselenization, we decided to use (4-tert-butylphenoxy)acetyl (TBPA) as the protecting group (26) for the 2-NH2 of this 6-Se-deoxyguanosine phosphoramidite (3), which can also be removed under the ultramild condition. Our synthesis (Scheme 1) started from the partially protected deoxyguanosine derivative (1). To avoid protection of the 3′-hydroxyl group, we have developed a condition that allows selective sulfonylation at the 6-position of deoxyguanosine in the presence of the free 3′-OH group, using 2,4,6-triisopropylbenzenesulfonyl chloride (27). Without purification of the activated intermediate, the protected 6-Se-deoxyguanosine derivative (2) was obtained (80% yield over two steps) by the substitution of the activating group at the 6-position with sodium 2-cyanoethylselenide, which was generated by the reduction of di-(2-cyanoethyl) diselenide with NaBH4 (25). The 6-Se-deoxyguanosine derivative (2) was converted to the corresponding phosphoramidite (3) in a satisfactory yield.
Scheme 1.
Synthesis of the 6-(2-cyanoethyl)seleno guanosine phosphoramidite (3) and oligonucleotides containing the 6-Se-G (4). Reagents and conditions: (a) TIBS, DMAP, TEA, CH2Cl2, room temperature; (b) diselenide, NaBH4/EtOH, −5°C, 80% yield in two steps; (c) phosphoramidite, BTT, CH2Cl2, 75% yield; (d) solid-phase synthesis. TIBS = 2,4,6-triisopropylbenzene-1-sulfonyl chloride, DMAP = 4-dimethylaminopyridine, CH2Cl2 = dichloromethane, TEA = triethylamine, EtOH = ethanol, NaBH4 = sodium borohydride, diselenide = (NCCH2CH2Se)2, phosphoramidite = 2-cyanoethyl tetraisopropyl-phosphorodiamidite, BTT = 5-(benzylthio)-1H-tetrazole.
Synthesis of the 6-Se-G DNAs
Strong basic conditions (such as NH3 treatment) for nucleobase deprotection cause the deselenization of 6-Se-G (SeG), thus the ultramild protecting group [(4-tert-butylphenoxy)acetyl] (26) is used for the 2-NH2 of 3. The 6-Se-G derivatized oligonucleotides were prepared using the ultramild CE phosphoramidites (dA, dC and dG; 28), 3, and BTT activator (29). These ultramild protecting groups can be removed under the ultramild deprotection condition (the K2CO3 treatment; 12). When the oligonucleotides contain many dG residues, phenoxyacetic anhydride (Pac2O), instead of acetic anhydride, is used in the capping step to avoid the dG acetylation, which is difficult to remove under the K2CO3 treatment.
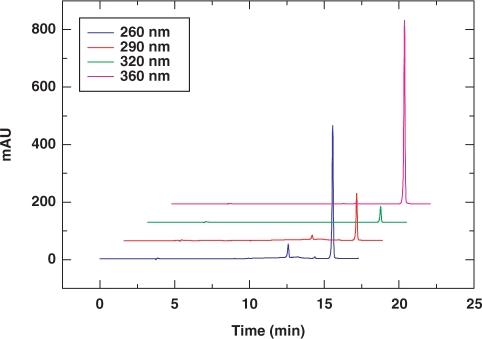
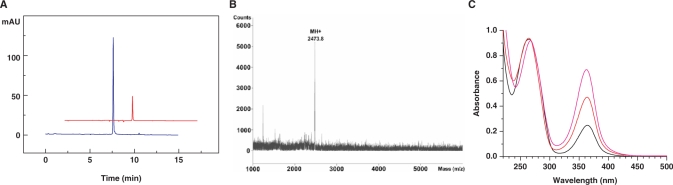
To measure the coupling efficiency of the 6-Se-G phosphoramidite (3), we synthesized 5′-DMTr-SeGG dinucleotide, analyzed it by RP-HPLC (Figure 1), and compared it with the native 5′-DMTr-GG synthesis and analysis (Supplementary Data), which indicated a high coupling yield (over 97%). Typical HPLC, UV and MS analyses of the SeG-DNAs are shown in Figure 2. More MS data of the synthesized SeG-DNAs are shown in Table 1. The purified Se-DNAs are yellow colored, which is the first observation of the colored DNAs containing the 6-Se-deoxyguanosine. In addition, our synthesis and analysis indicated that the 6-Se-G functionality of the Se-DNAs is relatively stable under aqueous conditions and air.
Figure 1.
RP-HPLC analysis of the deprotected and crude 5′-DMTr-SeGG dimer. The crude dimer was monitored at four different wavelengths: blue 260 nm, red 290 nm, green 320 nm, and pink 360 nm. HPLC conditions: Welchrom C18-XB column (4.6 × 250 mm, 5 μ), 25°C, 1 ml/min, gradient from buffer A to 30% buffer B in 20 min.
Figure 2.
HPLC, MS and UV analyses of the SeG-DNAs. (A) RP-HPLC analysis of 5′-d(GAATCA-SeG-GTGTC)-3′ [monitored at 260 nm (blue) and 360 nm (red)]. The sample was analyzed on a Welchrom XB-C18 column (4.6 × 250 mm, 5 μ) at a flow of 1.0 ml/min and with a linear gradient of 5 to 50% B in 10 min, with a retention time of 7.6 min. Buffer A: 10mM TEAAc (pH 7.1); B: 60% acetonitrile in 10 mM TEAAc (pH 7.1). (B) MS analysis of 5′-d(GT-SeG-TACAC)-3′.Molecular formula: C78H99N30O45P7Se; [M+H]+: 2473.8 (calcd: 2473.6). (C)UV spectra of the SeG-DNAs containing one SeG (ATG-SeG-TGCAC, black), two SeGs (ATG-SeG-T-SeG-CAC, red), and three SeGs (AT-SeG-SeG-T-SeG-CAC, pink).
Table 1.
MALDI-TOF MS data of the SeG-modified oligonucleotides
| Entry | Se-oligonucleotides | Measured (calcd.) m/z |
|---|---|---|
| a | 5′-T-SeG-T-3′ | [M + H]+: 973 (973) |
| C30H40N9O19P2Se: FW 971.6 | ||
| b | 5′-TT-SeG-T-3′ | [M + H]+: 1244 (1244) |
| C40H52N11O24P3Se: FW 1242.8 | ||
| c | 5′-ATG-SeG-TGCTC-3′ | [M + H]+: 2793 (2794) |
| C88H112N32O53P8Se: FW 2792.8 | ||
| d | 5′-ATG-SeG-T-SeG–CTC-3′ | [M + H]+: 2858 (2857) |
| C88H112N32O52P8Se2: FW 2855.7 | ||
| e | 5′-AT-SeG-SeG-T-SeG–CTC-3′ | [M + H]+: 2920 (2920) |
| C88H112N32O51P8Se3: FW 2918.7 | ||
| f | 5′-GT-SeG-TACAC-3′ | [M + H]+: 2474 (2474) |
| C78H99N30O45P7Se: FW: 2472.6 | ||
| g | 5′-G-SeG-GTACAC-3′ | [M + H]+: 2498 (2499) |
| C78H98N33O44P7Se: FW 2497.6 | ||
| h | 5′-GC-SeG-TATACGC-3′ | [M + H]+: 3092 (3092) |
| C97H123N38O57P9Se: FW: 3091.0 | ||
| i | 5′-GCG-SeG-ATACGC-3′ | [M + H]+: 3116 (3116) |
| C97H122O56N41P9Se: FW: 3115.4 |
Determination of extinction coefficient of DNA SeG (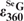 ) and UV spectroscopic studies
) and UV spectroscopic studies
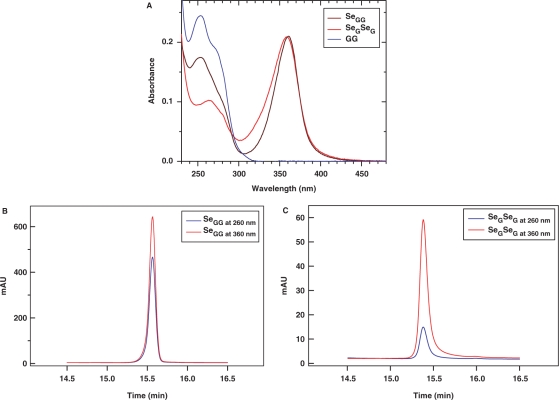
To determine the extinction coefficient of DNA SeG by comparing with the native nucleotide, we synthesized and purified GG, SeGG and SeGSeG dimers, and their UV spectra are presented in Figure 3. By taking the advantage of HPLC separation and UV analysis, we have developed this useful HPLC–UV approach to accurately measure and calculate the extinction coefficients of the base-modified nucleotides. Our experimental results indicate that SeG in DNA absorbs at both 254 nm and 360 nm (λmax = 267 nm and 360 nm) while native G (λmax = 254 nm) does not absorb at 360 nm (Figure 3A). The SeG λmax values of SeGG (λmax = 359 nm) and SeGSeG (λmax = 361 nm) are virtually identical; the average λmax of SeG is 360 nm. Since the extinction coefficient of G at 260 nm (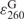 = 1.22 × 104 M−1cm−1) is known (30), we performed HPLC analysis of SeGG and SeGSeG under both 260 nm and 360 nm (Figure 3B and C), and their peak areas were quantified, respectively. First, the absorption ratio at 260 nm and 360 nm of SeGSeG (
= 1.22 × 104 M−1cm−1) is known (30), we performed HPLC analysis of SeGG and SeGSeG under both 260 nm and 360 nm (Figure 3B and C), and their peak areas were quantified, respectively. First, the absorption ratio at 260 nm and 360 nm of SeGSeG (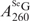 /
/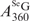 ) was calculated and determined as α value. In Figure 3B, the α value is used to calculate the 260-nm absorption contribution from SeG of the SeGG. The net 260-nm absorption from G of the SeGG (
) was calculated and determined as α value. In Figure 3B, the α value is used to calculate the 260-nm absorption contribution from SeG of the SeGG. The net 260-nm absorption from G of the SeGG (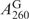 ) is obtained by subtraction of the SeG 260-nm contribution from the total SeGG absorption at 260 nm (Figure 3B). Thus, we deduced Equation (3) from Equation (1) and (2) presenting the SeGG. Since
) is obtained by subtraction of the SeG 260-nm contribution from the total SeGG absorption at 260 nm (Figure 3B). Thus, we deduced Equation (3) from Equation (1) and (2) presenting the SeGG. Since 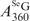 can be directly measured and
can be directly measured and 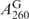 can be accurately calculated from this SeGG in Figure 3B, we determined
can be accurately calculated from this SeGG in Figure 3B, we determined 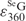 as 2.3 × 104 M−1cm−1. Similarly, from Equation (2) and (4) presenting the SeGSeG, we deduced Equation (5) and calculated the ratio of
as 2.3 × 104 M−1cm−1. Similarly, from Equation (2) and (4) presenting the SeGSeG, we deduced Equation (5) and calculated the ratio of 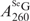 /
/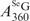 in Figure 3C, thereby accurately determining
in Figure 3C, thereby accurately determining 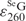 as 5.3 × 103 M−1 cm−1.
as 5.3 × 103 M−1 cm−1.
| 1 |
| 2 |
| 3 |
| 4 |
| 5 |
Excitingly, we observed that invisible DNA turns into colored DNA via the single atom replacement with selenium, while natural DNAs are colorless. Comparing with the native deoxyguanosine nucleotide (UV λmax = 254 nm, ε = 1.22 × 104 M−1 cm−1; 30), the UV spectrum of the 6-Se-deoxyguanosine nucleotide (λmax = 360 nm, ε = 2.3 × 104 M−1 cm−1) reveals a higher absorption and a large red-shift over 100 nm, thereby leading to the appearance of yellow color. This Se-nucleotide visualization is probably due to the ease of the delocalization of the selenium electrons on the nucleobase, requiring less energy for the electron excitation, thereby resulting in the large UV red-shift. In contrast, the 6-S-deoxyguanosine nucleotide (6-S-dG, λmax = 339 nm; 24) shows a smaller red-shift from the deoxyguanosine nucleotide and remains colorless.
Figure 3.
Calculation of 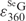 via UV and HPLC analyses. (A) UV absorption spectra of GG dimer (blue line), SeGG dimer (black line), and SeGSeG dimer (red line); (B) RP-HPLC analysis of SeGG dimer at 260 nm (blue line) and 360 nm (red line); (C) RP-HPLC analysis of SeGSeG dimer at 260 nm (blue line) and 360 nm (red line).
via UV and HPLC analyses. (A) UV absorption spectra of GG dimer (blue line), SeGG dimer (black line), and SeGSeG dimer (red line); (B) RP-HPLC analysis of SeGG dimer at 260 nm (blue line) and 360 nm (red line); (C) RP-HPLC analysis of SeGSeG dimer at 260 nm (blue line) and 360 nm (red line).
Thermostability studies of the 6-Se-G SeNAs
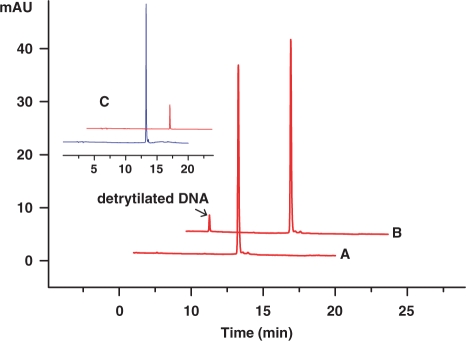
The DNA containing 6-Se-G [5′-DMTr-d(GAATCA-SeG-GTGTC)-3′] was heated at 60°C for 1 h in the buffer of 100 mM NaH2PO4-Na2HPO4 (pH 7.6) (Figure 4). Besides a small amount of the DNA detritylation (≈5%), deselenization of the Se-G-DNA was insignificant (≤1%, determined by the Se-DNA total decrease), which shows that the 6-Se-G functionality is relatively stable in aqueous solution at the elevated temperature. Since stability in the air is required in most crystallization experiments, we have monitored the Se-derivatized DNAs for weeks by HPLC, and found insignificant deselenization.
Figure 4.
Thermostability studies of the 6-Se-G-DNA. The sample [5′-DMT-d(GAATCA-SeG-GTGTC)-3′] was dissolved in a 100 mM phosphate buffer (pH 7.6) and analyzed by HPLC at 360 nm. (A) before heating; (B) after heating at 60°C for 1 h. (C) HPLC analysis of the Se-G-DNA, monitored at both 267 nm and 360 nm, before heating.
UV-melting study of the Se-G DNA duplexes
The UV-melting temperatures were measured (Tm, Table 2) to examine the impact of the SeG residue incorporation on the thermodynamic stability of DNA duplexes. Two typical melting tempeature curves of the native and modified duplexes are shown in Figure 5. It has been observed in the literature (31) that the 6-S-G modification can slightly destablize nucleic aicd duplex and cause drop in the UV-melting temperature (Tm) up to ≈3°C per S-modification. Since selenium atomic size is larger than sulfur atomic size, we expected that the 6-Se substitution can cause more decrease in the duplex stability and melting temperature as well. As predicted, these Se-DNA duplexes with single, double or triple Se-G-modifications indeed show significant decrease in melting temperatures (up to 11°C per modification on DNA with one Se atom, Table 2).
Table 2.
UV Melting temperatures of the SeG-modified oligonucleotides
| Entry | DNA pairs | Se-DNA Tm (native) °C |
|---|---|---|
| a | 5′-CGTACC TACAGTT-SeG-T-3′ | 51.1 ± 0.2 (55.0 ± 0.1) |
| 3′-GCATGGATGTCAA—C-A-5′ | ||
| b | 5′-Py-A-SeG–A-ACTGTAGGTACG | 55.2 ± 0.1 (58.3 ± 0.1) |
| 3′-T—C-BrU-TGACATCCATGC-5′ | ||
| c | 5′-TACTAAC-SeG-TAGTA-3′ | 47.9 ± 0.1 (54.0 ± 0.3) |
| d | 5′-GAATCC-SeG–CTGTC-3′ | 42.0 ± 0.1 (53.0 ± 0.1) |
| 3′-CTT AGG–C-GACAG-5′ | ||
| e | 5′-GAATCT-SeG–CTGTC-3′ | 40.0 ± 0.2 (48.0 ± 0.2) |
| 3′-CTT AGA–C-GACAG-5′ | ||
| f | 5′-GAATCA-SeG-GTGTC-3′ | 38.4 ± 0.2 (47.0 ± 0.1) |
| 3′-CTT AGT—C-CACAG-5′ | ||
| g | 5′-GC-SeG-TATACGC-3′ | 28.5 ± 0.3 (38.0 ± 0.2) |
| h | 5′-ATG-SeG-TGCTC-3′ | 32.3 ± 0.3 (42.5 ± 0.3) |
| 3′-TAC—C-ACGAG-5′ | ||
| i | 5′-ATG-SeG-T-SeG–CTC-3′ | 17.0 ± 0.5 (42.5 ± 0.3) |
| 3′-TAC—C-A—C-GAG-5′ | ||
| j | 5′-AT-SeG-SeG-T-SeG–CTC-3′ | 9.6 ± 1.0 (42.5 ± 0.3) |
| 3′-TA—C—C-A—C-GAG-5′ |
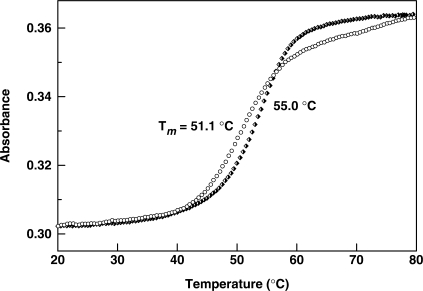
Figure 5.
Normalized melting temperature curves of the non- and Se-modified DNA duplexes. The Se-DNA duplex: 5′-CGTACCTACAGTT-SeG-T-3′ and 5′-ACAACTGTAGGTACG-3′ (Open circle, Tm = 51.1°C); the corresponding native DNA duplex: (filled diamond, 55.0°C).
Interestingly, we also observed that Tms of the Se-DNA duplexes in Entry a and b of Table 2, where the Se-modifications are close to the 5′ or 3′ terminus, are just 3–4°C lower than that of the corresponding native complex. In addition, a small change in Tm was also observed when the Se-modification was introduced to a region containing a bulge or a flipping nucleotide. For instance, the self-complementary sequence in Entry c forms two A-bulges in the duplex, where these two A-nucleotides flip out in the crystal structure (32). Tm of this duplex with the Se-modification close to these two As drops only 2-3°C per Se-modification. These results suggest that the degree of the Se-G-duplex destabilization is dependent on the modification location, sequence and secondary structure of nucleic acids. Since the large size of selenium atom, which requires more space surrounding the modification site, is the major factor of the duplex instability, less thermo-destabilization is observed when the Se-modification is introduced to a position close to secondary structures that are more dynamic and flexible. This unique effect of the selenium modification can be taken advantage of in identifying and studying secondary structures of nucleic acids, such as DNAzymes and ribozymes, and nucleic acid-protein complexes (33–36). For structural studies of nucleic acids and their protein complexes, it is better to place this modification close to the termini of DNA or RNA, pyrimidines, or the internal bulges or loops. It is also a good idea to put the modification in RNA or DNA loops.
Structure of Se-DNA/RNA in a protein complex
In order to further study the Se-G-modification and Se-derivatized duplex of nucleic acids, we attempted to crystallize a Se-DNA/RNA/RNase H ternary complex and determine its X-ray crystal structure. Furthermore, we attempted to demonstrate the proof of principle on the structural determination of protein-nucleic acid complexes via Se-derivatized nucleic acids and phasing. By using the RNase H complex as a model system, which was determined previously (37,38), we successfully demonstrated the structure determination of a protein–nucleic acid complex on the basis of the nucleic acid Se-derivatization and MAD phasing. Besides facilitation of the structure determination, our selenium-modification study has revealed new insights into RNase H catalysis. The crystallization, structure refinement and determination results, and the enzyme mechanism study will be published elsewhere.
In this report, we focus here on the study of the Se-G-modification and its structural impact on base pairing and stacking interaction. It is easier to synthesize, derivatize, and purify DNAs than RNAs, thus the DNA portion of the DNA/RNA hybrid (5′-ATGTCG-p-3′/5′-UCGACA-3′; one-base overhang at both 5′-ends) was derivatized with the selenium functionality on two Gs (G3 and G6). The plasmid expressing Bacillus halodurans RNase H (D132N mutant; 37,38) was a kind gift from Yang's laboratory at the National Institute of Health. The 3D crystal structure of the Se-DNA/RNA/RNase H complex (PDB ID: 2R7Y) was successfully determined on the basis of the DNA Se-derivatization and phase information obtained from selenium scattering.
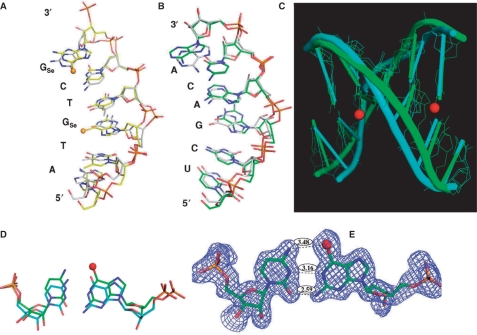
Both the native and Se-DNA-derivatized complexes were crystallized in C2 space group with similar unit cell dimensions. Our study reveals that the protein structures of both the native (2.70 Å resolution, PDB ID: 2G8U; 37,38) and modified (1.80 Å resolution, PDB ID: 2R7Y) complexes are virtually identical, and that the nucleic acid global structures of the native and Se-modified duplexes are very similar (Figure 6) though the nucleobases shift locally (Figure 6A–D). Probably due to flexibility of the over-hung ends, more structural differences are observed at the DNA and RNA termini. The distance between 1-NH of G3 in the DNA sequence and the N3 of C5 in the RNA sequence, and the distance between exo-2-NH2 of G3 and exo-2-O of C5 are 3.16 Å and 2.59 Å, respectively (the corresponding H-bond lengths of the native G–C pair: 2.99 Å and 2.95 Å). These distances indicate the retention of the two native hydrogen bonds of the G3–C5 base pair. Since the Se atomic radius is 0.43 Å larger than that of O and hydrogen bond length is normally 2.7–3.2 Å, the distance (3.48 Å) between the G3 exo-6-Se and C5 exo-4-NH2 (the native H-bond length: 2.99) indicates a selenium-mediated H-bond (Figure 6E). Thus, the SeG3–C5 base pair consists of three hydrogen bonds (exo-6-Se/exo-4-NH2, 1-NH/N3, and exo-2-NH2/exo-2-O). Interestingly, the hydrogen bond length (2.59 Å) between G3 exo-2-NH2 and C5 exo-2-O is 0.36 Å shorter than the corresponding native bond (2.95 Å).
Figure 6.
The superimposed global and local structures of the 6-Se-G-modified (2R7Y) and native (2G8U) DNA/RNA duplexes (5′-ATGTCG-p-3′/5′-UCGACA-3′) of the nucleic acid–protein complex; the balls represent selenium atoms in the Se-derivatized DNA (5′-AT-SeG-TC-SeG-p-3′). (A) The structure of the Se-DNA sequence (2R7Y, in yellow) is superimposed over the corresponding native (2G8U, in grey); (B) The structure of the RNA sequence (2R7Y, in green) is superimposed over the corresponding native (2G8U, in grey); (C) The duplex structure of the Se-DNA/RNA hybrid (2R7Y, in green) is superimposed over the corresponding native (2G8U, in cyan); (D) The comparison of the Se-modified (in green) and native (in cyan) G3/C5 base-pair structures; (E) The Se-G3/C5 base pair (2R7Y) with the experimental electron density shows three hydrogen bonds (exo-6-Se/exo-4-NH2, 1-NH/N3, and exo-2-NH2/exo-2-O) with bond lengths in 3.48 Å, 3.16 Å and 2.59 Å, respectively.
The bond length comparisons between the native and Se-mediated H-bonds are summarized in Table 3. Furthermore, the bond length (3.48 Å) of the Se … H–N hydrogen bond (exo-6-Se/exo-4-NH2) in the SeG3-C5 are very close to the bond length (3.35 Å) of the Se … H–N hydrogen bond (exo-4-Se/exo-6-NH2) previously discovered within the SeT-A base pair (12). Similarly, the SeG6 and C2 also form three H-bonds and behave in the same way (Table 3). Therefore, we have demonstrated that the SeG and C form a base pair that is similar to the natural G–C pair. Consistently, our UV-melting results agree with our structure study. The crystal structure indicates that in order to accommodate the large Se atom and to form the Se … H–N bond within the base pair and duplex, each SeG–C base pair unwinds slightly via minor adjustment of the sugar pucker (Figure 6D and E) and shifts ∼0.3 Å distance within the major groove (Figure 6D), when comparing to the corresponding native G–C pair. Our crystal structure results explain the decrease in the melting temperature after the sulfur or selenium modification, which causes the base-pair shift and reduces stacking interaction, thereby destabilizing the duplex structure. Our results also explain why larger selenium atom destabilizes the duplexes more than smaller sulfur atom (31). In addition, the results are consistent with our observation of the duplex slight destabilization when the Se-modifications are introduced to the positions close to the 5′ or 3′ terminus, bulge loops or flipping nucleotides, which are more dynamic, flexible and capable of accommodating a larger atom.
Table 3.
Hydrogen bond lengths of Se-mediated H-bonds comparing to the native ones
| Base pair | H-bond | Se-modified base pair | Native base pair |
|---|---|---|---|
| bond length (Å) | bond length (Å) | ||
| 6-Se-G3/C5 in 2R7Y | exo-6-Se/exo-4-NH2 | 3.48 | 2.99 |
| 1-NH/N3 | 3.16 | 2.99 | |
| exo-2-NH2/exo-2-O | 2.59 | 2.95 | |
| 6-Se-G6/C2 in 2R7Y | exo-6-Se/exo-4-NH2 | 3.43 | 3.07 |
| 1-NH/N3 | 3.19 | 3.15 | |
| exo-2-NH2/exo-2-O | 2.75 | 3.15 | |
| 4-Se-T/A in 2NSK | exo-4-Se/exo-6-NH2 | 3.35 | 2.87 |
| 3-NH/N1 | 3.02 | 2.78 |
CONCLUSIONS
In summary, we have synthesized the first 6-Se-deoxyguanosine phosphoramidite and incorporated it into oligonucleotides, via solid-phase synthesis under ultramild conditions, in nearly quantitative yield. We also discovered that the Se-G-containing DNAs are yellow and with strong UV absorption at 360 nm (ε = 2.3 × 104 M−1 cm−1). In addition, this Se-functionality is stable in aqueous solution and at the elevated temperature. Moreover, we crystallized a ternary complex of the Se-G-DNA, RNA and RNase H, and revealed that the global nucleic acid structures of the native and Se-modified duplexes are very similar, the SeG and C form a base pair similar to the native G–C pair, and the selenium atom participates in a Se-mediated hydrogen bond (Se … H–N). The G–C base pair accommodates the large selenium atom by shifting approximately 0.3 Å. As this base-pair shift reduces the stacking interaction, our structure work provides a clear picture on why the Se-modification or S-modification causes decrease in the UV-melting temperature. Our study also points out where to better place Se atom by incorporating it to the positions close to the termini or flexible secondary structures.
The Se-modified nucleic acids have been used for the first time in derivatization and structural determination of a protein–nucleic acid complex via Se-nucleic acid and MAD phasing. Our studies shed new light on the nucleic acid stability, flexibility, and duplex recognition governed by stacking interaction, base-pairing, and size-and-shape impact of the base pairs. In addition, this novel Se-modification of nucleic acids can be used to carry out chemogenetic and spectroscopic investigation of nucleic acids and their protein complexes, and to perform the macromolecular structure study via crystallography by MAD or SAD phasing. Besides the applications in structure and function studies, the Se-G DNAs (yellow color) may also have great potentials in construction of colored DNA nanoscale devices and structures (39) as well as in nucleic acid diagnosis (40).
Supplementary Data
Supplementary Data are available at NAR Online.
FUNDING
The GCC Distinguished Cancer Clinicians and Scientists Award, and the National Science Foundation (MCB-0517092 and CHE-0750235). Funding for open access charge: National Science Foundation (MCB-0517092).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Wei Yang at National Institute of Health for the kind gift of RNase H-expressing plasmid, and Drs Anand Saxena and Michael Becker at NSLS beamline X12C and X25 for their help in the data collection.
REFERENCES:
- 1.Hendrickson WA, Pahler A, Smith JL, Satow Y, Merritt EA, Phizackerley RP. Crystal structure of core streptavidin determined from multiwavelength anomalous diffraction of synchrotron radiation. Proc. Natl Acad. Sci. USA. 1989;86:2190–2194. doi: 10.1073/pnas.86.7.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hendrickson WA. Synchrotron crystallography. Trends Biochem. Sci. 2000;25:637–643. doi: 10.1016/s0968-0004(00)01721-7. [DOI] [PubMed] [Google Scholar]
- 3.Ferre-D AR, Zhou KH, Doudna JA. Crystal structure of a hepatitis δ virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 4.Xiao H, Murakami H, Suga H, Ferre-D AR. Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme. Nature. 2008;454:358–361. doi: 10.1038/nature07033. [DOI] [PubMed] [Google Scholar]
- 5.Carrasco N, Ginsburg D, Du Q, Huang Z. Synthesis of selenium-derivatized nucleosides and oligonucleotides for X-ray crystallography. Nucleos. Nucleot. Nucl. 2001;20:1723–1734. doi: 10.1081/NCN-100105907. [DOI] [PubMed] [Google Scholar]
- 6.Du Q, Carrasco N, Teplova M, Wilds CJ, Egli M, Huang Z. Internal derivatization of oligonucleotides with selenium for X-ray crystallography using MAD. J. Am. Chem. Soc. 2002;124:24–25. doi: 10.1021/ja0171097. [DOI] [PubMed] [Google Scholar]
- 7.Caton-Williams J, Huang Z. Biochemistry of selenium-derivatized naturally occurring and unnatural nucleic acids. Chem. Biodivers. 2008;5:396–407. doi: 10.1002/cbdv.200890040. [DOI] [PubMed] [Google Scholar]
- 8.Sheng J, Huang Z. Selenium Derivatization of Nucleic Acids for Phase Determination in Nucleic Acid X-ray Crystallography. Int. J. Mol. Sci. 2008;9:258–271. doi: 10.3390/ijms9030258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teplova M, Wilds CJ, Wawrzak Z, Tereshko V, Du Q, Carrasco N, Huang Z, Egli M. Covalent incorporation of selenium into oligonucleotides for X-ray crystal structure determination via MAD: proof of principle. Biochimie. 2002;84:849–858. doi: 10.1016/s0300-9084(02)01440-2. [DOI] [PubMed] [Google Scholar]
- 10.Moroder H, Kreutz C, Lang K, Serganov A, Micura R. Synthesis, oxidation behavior, crystallization and structure of 2′-methylseleno guanosine containing RNAs. J. Am. Chem. Soc. 2006;128:9909–9918. doi: 10.1021/ja0621400. [DOI] [PubMed] [Google Scholar]
- 11.Egli M, Pallan PS, Pattanayek R, Wilds CJ, Lubini P, Minasov G, Dobler M, Leumann CJ, Eschenmoser A. Crystal structure of homo-DNA and nature's choice of pentose over hexose in the genetic system. J. Am. Chem. Soc. 2006;128:10847–10856. doi: 10.1021/ja062548x. [DOI] [PubMed] [Google Scholar]
- 12.Salon J, Sheng J, Jiang J, Chen G, Caton-Williams J, Huang Z. Oxygen replacement with selenium at the midine 4-position for the Se base pairing and crystal structure studies. J. Am. Chem. Soc. 2007;129:4862–4863. doi: 10.1021/ja0680919. [DOI] [PubMed] [Google Scholar]
- 13.Jiang J, Sheng J, Carrasco N, Huang Z. Selenium derivatization of nucleic acids for crystallography. Nucleic Acids Res. 2007;35:477–485. doi: 10.1093/nar/gkl1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watts JK, Johnston BD, Jayakanthan K, Wahba AS, Pinto BM, Damha MJ. Synthesis and biophysical characterization of oligonucleotides containing a 4′-selenonucleotide. J. Am. Chem. Soc. 2008;130:8578–8579. doi: 10.1021/ja802205u. [DOI] [PubMed] [Google Scholar]
- 15.Caton-Williams J, Huang Z. Synthesis and DNA-polymerase incorporation of colored 4-selenothymidine triphosphate for polymerase recognition and DNA visualization. Angew. Chem. Int. Ed. Engl. 2008;47:1723–1725. doi: 10.1002/anie.200705213. [DOI] [PubMed] [Google Scholar]
- 16.Piccirilli JA, Krauch T, Moroney SE, Benner SA. Enzymatic incorporation of a new base pair into DNA and RNA extends the genetic alphabet. Nature. 1990;343:33–37. doi: 10.1038/343033a0. [DOI] [PubMed] [Google Scholar]
- 17.Moran S, Ren RX, Kool ET. A thymidine triphosphate shape analog lacking Watson-Crick pairing ability is replicated with high sequence selectivity. Proc. Natl Acad. Sci. USA. 1997;94:10506–10511. doi: 10.1073/pnas.94.20.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaput JC, Sinha S, Switzer C. 5-propynyluracil.diaminopurine: an efficient base-pair for non-enzymatic transcription of DNA. Chem. Commun. (Camb) 2002:1568–1569. doi: 10.1039/b204535d. [DOI] [PubMed] [Google Scholar]
- 19.Zimmermann N, Meggers E, Schultz PG. A novel silver(i)-mediated DNA base pair J. Am. Chem. Soc. 2002;124:13684–13685. doi: 10.1021/ja0279951. [DOI] [PubMed] [Google Scholar]
- 20.Leconte AM, Hwang GT, Matsuda S, Capek P, Hari Y, Romesberg FE. Discovery, characterization, and optimization of an unnatural base pair for expansion of the genetic alphabet. J. Am. Chem. Soc. 2008;130:2336–2343. doi: 10.1021/ja078223d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mautner HG, Chu SH, Jaffe JJ, Sartorelli AC. The Synthesis and Antineoplastic Properties of Selenoguanine, Selenocytosine and Related Compounds. J. Med. Chem. 1963;6:36–39. doi: 10.1021/jm00337a008. [DOI] [PubMed] [Google Scholar]
- 22.Milne GH, Townsend LB. Synthesis and antitumor activity of alpha-and beta-2′-deoxy-6-selenoguanosine and certain related derivatives. J.Med. Chem. 1974;17:263–268. doi: 10.1021/jm00249a002. [DOI] [PubMed] [Google Scholar]
- 23.Coleman RS, Arthur JC, McCary JL. Covalent cross-linking of duplex DNA using 4-thio-2′-deoxyuridine as a readily modifiable platform for introduction of reactive functionality into oligonucleotides. Tetrahedron. 1997;53:11191–11202. doi: 10.1093/nar/25.23.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadokura M, Wada T, Seio K, Sekine M. Synthesis of 4-thiouridine, 6-thioinosine, and 6-thioguanosine 3′,5′-O-bisphosphates as donor molecules for RNA ligation and their application to the synthesis of photoactivatable TMG-capped U1 snRNA fragments. J. Org. Chem. 2000;65:5104–5113. doi: 10.1021/jo991432z. [DOI] [PubMed] [Google Scholar]
- 25.Logan G, Igunbor C, Chen G.-X, Davis H, Simon A, Salon J, Huang Z. A Novel and Simple Strategy for Incorporation, Protection, and Deprotection of Selenium Functionality. Synlett. 2006;10:1554–1558. [Google Scholar]
- 26.Boal JH, Wilk A, Harindranath N, Max EE, Kempe T, Beaucage SL. Cleavage of oligodeoxyribonucleotides from controlled-pore glass supports and their rapid deprotection by gaseous amines. Nucleic Acids Res. 1996;24:3115–3117. doi: 10.1093/nar/24.15.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palin R, Grovea S.JA, Prossera AB, Zhang M.-Q. Mono-6-(O-2,4,6-triisopropylbenzenesulfonyl)-γ-cyclodextrin, a novel intermediate for the synthesis of mono-functionalised γ-cyclodextrins. Tetrahedron Lett. 2001;42:8897–8899. [Google Scholar]
- 28.Vu H, McCollum C, Jacobson K, Theisen P, Vinayak R, Spiess E, Andrus A. Fast oligonucleotide deprotection phosphoramidite chemistry for DNA synthesis. Tetrahedron Lett. 1990;31:7269–7272. [Google Scholar]
- 29.Welz R, Müller S. 5-(Benzylmercapto)-1H-tetrazole as activator for 2′-O-TBDMS phosphoramidite building blocks in RNA synthesis. Tetrahedron Lett. 2002;43:795–797. [Google Scholar]
- 30.Cavaluzzi MJ, Borer PN. Revised UV extinction coefficients for nucleoside-5′-monophosphates and unpaired DNA and RNA. Nucleic Acids Res. 2004;32:e13. doi: 10.1093/nar/gnh015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bohon J, de los Santos CR. Effect of 6-thioguanine on the stability of duplex DNA. Nucleic Acids Res. 2005;33:2880–2886. doi: 10.1093/nar/gki572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berglund JA, Rosbash M, Schultz SC. Crystal structure of a model branchpoint-U2 snRNA duplex containing bulged adenosines. RNA. 2001;7:682–691. doi: 10.1017/s1355838201002187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salehi-Ashtiani K, Luptak A, Litovchick A, Szostak JW. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science. 2006;303:1788–1792. doi: 10.1126/science.1129308. [DOI] [PubMed] [Google Scholar]
- 34.Silverman SK. Catalytic DNA (deoxyribozymes) for synthetic applications-current abilities and future prospects. Chem. Commun. (Camb.) 2008;30:3467–85. doi: 10.1039/b807292m. [DOI] [PubMed] [Google Scholar]
- 35.Morikawa K, Shirakawa M. Three-dimensional structural views of damaged-DNA recognition: T4 endonuclease V, E. coli Vsr protein, and human nucleotide excision repair factor XPA. Mutat. Res. 2000;460:257–275. doi: 10.1016/s0921-8777(00)00031-8. [DOI] [PubMed] [Google Scholar]
- 36.Benoff B, Yang H, Lawson CL, Parkinson G, Liu J, Blatter E, Ebright YW, Berman HM, Ebright RH. Structural basis of transcription activation: the CAP-alpha CTD-DNA complex. Science. 2002;297:1562–1566. doi: 10.1126/science.1076376. [DOI] [PubMed] [Google Scholar]
- 37.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 38.Nowotny M, Yang W. Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. Embo J. 2006;25:1924–1933. doi: 10.1038/sj.emboj.7601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding B, Seeman NC. Operation of a DNA robot arm inserted into a 2D DNA crystalline substrate. Science. 2006;314:1583–1585. doi: 10.1126/science.1131372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JG, Cheong KH, Huh N, Kim S, Choi JW, Ko C. Microchip-based one step DNA extraction and real-time PCR in one chamber for rapid pathogen identification. Lab Chip. 2006;6:886–895. doi: 10.1039/b515876a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.