Abstract
RNA-dependent RNA polymerases (RdRP) form an important class of enzymes that is responsible for genome replication and transcription in RNA viruses and involved in the regulation of RNA interference in plants and fungi. The RdRP kinetics have been extensively studied, but pausing, an important regulatory mechanism for RNA polymerases that has also been implicated in RNA recombination, has not been considered. Here, we report that RdRP experience a dramatic, long-lived decrease in its elongation rate when it is reinitiated following stalling. The rate decrease has an intriguingly weak temperature dependence, is independent of both the nucleotide concentration during stalling and the length of the RNA transcribed prior to stalling; however it is sensitive to RNA structure. This allows us to delineate the potential factors underlying this irreversible conversion of the elongation complex to a less active mode.
INTRODUCTION
Template-directed polymerization of nucleotides (NTPs) is an essential process in all living entities. Accordingly, enzymes catalyzing these processes operate in both cellular organisms and in viruses. In RNA viruses, RNA-dependent RNA polymerases (RdRPs) are the essential catalytic components of the polymerization machinery. RdRPs are also encoded by numerous cellular organisms, where they initiate or amplify the regulatory mechanisms known as RNA silencing (1). The structure and reaction mechanisms of viral RdRPs display similarity with many other nucleic acid polymerases, but nonetheless incorporate subtle differences. For instance, viral RdRPs adopt the ‘right hand-like’ conformation typical for numerous nucleic acid polymerases, but they display a distinct ‘closed-hand’ conformation, rather than the more common ‘open-hand’ structure (2,3).
Viral RdRPs are capable of carrying out two distinct reactions, replication and transcription. These reactions are completed in four steps: (i) template recognition and binding, (ii) initiation, (iii) elongation and (iv) termination. The binding of viral RdRPs to template RNA exhibits characteristically low binding constants (4,5), but binding may nonetheless be enhanced by specific nucleotide sequences and/or RNA secondary structures (6,7). During initiation and elongation, viral RdRPs perform a nucleotidyl transfer reaction to polymerize the complementary RNA strand (8). While RNA or protein primers may be required for initiation, most RdRPs initiate RNA synthesis de novo (3).
To date, the kinetic studies of viral RdRP mechanism have neglected the effect of viral RdRP stalling. Partial RNA products isolated from poliovirus- and tobacco mosaic virus-infected cells suggest that the RdRP indeed frequently stalls, leading to compromised processivity during RNA elongation in vivo (9). It has been suggested that viral RdRP pausing could be brought about by specific RNA sequences and secondary structures (9), and is likely a prerequisite for viral RNA recombination (10,11). Furthermore, rational drug design against viral RdRPs could benefit from the analysis of stalled viral RdRPs (12). Finally, while stalling of viral RdRPs has been utilized to separate the elongation and initiation stages of the replication and transcription reactions (13), the consequences of stalling on the elongation rates have not been examined.
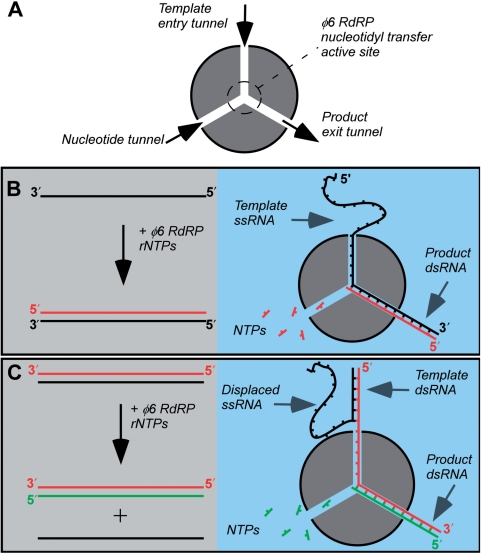
To quantitatively study the effect of stalling on the kinetics of viral RdRPs, we have used the RdRP from bacteriophage φ6 (φ6 RdRP) as a model system (Figure 1A). φ6 RdRP catalyzes primer-independent de novo RNA synthesis on single-stranded RNA (ssRNA) and double-stranded RNA (dsRNA) templates and elongates with high processivity (14). During replication, the single-stranded sense RNA strand [(+)RNA] serves as the template for conversion to a dsRNA genome (Figure 1B). In contrast, during transcription, the (−)RNA strand within the dsRNA genome serves as the template for a new (+)RNA strand, which leaves the original (+)RNA strand as a single-stranded by-product (Figure 1C). In both processes, φ6 RdRP initiates exclusively at the free 3′-end of the template strand, which enters the φ6 RdRP through the template tunnel leading from the polymerase surface to the active site in the center of the enzyme structure (Figure 1A). By contrast, internal initiation of the elongation complex is not thought to occur (Supplementary Figure S1) (15).
Figure 1.
Bacteriophage φ6 RdRP performs replication and transcription. (A) A schematic of the φ6 RdRP structure. A nucleotidyl transfer active site is linked to the enzyme surface via three tunnels: the template entry tunnel, the NTP tunnel, and the product exit tunnel. (B) During replication, a complementary antisense RNA strand [(−)RNA; red line] is polymerized onto a sense RNA template [(+)RNA; black line]. The 3′-end of the template strand accesses the enzyme's active site through the template tunnel. In the presence of NTPs, product dsRNA exits through the product tunnel. (C) During transcription, the (+)RNA strand is displaced while φ6 RdRP polymerizes a new (+)RNA strand (green line) onto the (−)RNA template. Here, the dsRNA genome is first unwound, and the (+)RNA strand is displaced at the entrance to the template tunnel, allowing only the (−)RNA strand to enter the template tunnel. The dsRNA product exits through the product tunnel. The polarities of the RNA strands are indicated in the schematics.
Here, we measure the rate of RNA elongation by φ6 RdRP and demonstrate that while it is possible to reinitiate the polymerase following stalling induced by nucleotide deprivation, the resulting elongation rate is drastically and irreversibly reduced. This was quantified by measuring the elongation rate of the reinitiated complex using gel electrophoresis. Neither the NTP concentration nor the length of the RNA synthesized prior to stalling had an effect on the rate reduction following reinitiation. We attribute the reduction in the elongation rate to a transition to a sub-optimal conformational state of the elongation complex, brought about by stalling, and demonstrate that the conversion to the sub-optimal conformational state depends on the structure of the RNA template. In addition to the potential biological importance of the stalling, in vitro studies of polymerases have used stalling to measure elongation rates (13,16–18). In light of our findings, it is important to design the experimental setup in a way that a possible reduction in the elongation rate due to stalling is taken into account.
MATERIALS AND METHODS
Purification of the recombinant RdRP from bacteriophage φ6
The NdeI–EcoRI restriction fragment from pEM2 plasmid (14) was transferred into a pET-28a(+) vector (Novagen, USA). The resultant plasmid pAA5 was propagated in Escherichia coli BL21(DE3) (19). φ6 RdRP was expressed as previously described (14), except for 25 μg/ml kanamycin. The cells were harvested and resuspended in 50 mM Tris–HCl (pH 8.0), 300 mM NaCl, 1 mM imidazole and disrupted. The supernatant was loaded onto a Ni-NTA affinity column (Qiagen, Valencia, CA, USA). After two successive washes with imidazole buffers (10 mM and 20 mM), φ6 RdRP was eluted in 50 mM Tris–HCl (pH 8.0), 300 mM NaCl, 250 mM imidazole. Further purification was done using HiTrap™ Heparin HP and Q HP columns (GE Healthcare, USA). φ6 RdRP was eluted with a linear NaCl gradient (from 0.1 M–1 M NaCl) in 50 mM Tris–HCl (pH 8.0), 0.1 mM EDTA. The purified protein was stored in the elution buffer (containing 300 mM NaCl) at 4°C.
Preparation of RNA molecules
ssRNAs were obtained via in vitro run-off transcription using PCR-amplified sections of the pBB10 plasmid (20) as described previously (primers listed in Table S1) (21). To favor terminal incorporation of cytidine into the 3′-end of the transcripts used as templates for φ6 RdRP, the transcription mixtures were supplemented with 20 mM CTP.
dsRNA molecules were obtained by hybridizing complementary ssRNAs in 0.5 × SSC (Promega, Madison, WI, USA) using a ‘gradual-cool’ temperature program (21). dsRNA template for transcription reactions was obtained by hybridizing three RNA molecules (4 kb RNA, 1.3 kb RNA and 4 kb ‘main’ RNA) in a molar ratio of 1:1:1. Alternatively, a molar ratio of 1:4 was used in hybridizations between longer and shorter complementary ssRNA molecules for calibration of the electrophoretic mobilities. All hybridized RNAs were purified as described in (21).
A kinetic study of elongation by reinitiation of stalled φ6 RdRP
The elongation rate of φ6 RdRP was typically assayed in a reaction mixture containing 80 nM RNA template, 2.6 μM φ6 RdRP, 50 mM HEPES pH 7.9, 20 mM ammonium acetate, 5 mM MgCl2, 2 mM MnCl2, 0.1 mM EDTA pH 8.0, 0.1% Triton X-100, 5% (v/v) Superase•In, and 2.5 mM of each nucleotide. A 10-fold lower enzyme to RNA template ratio was tested and shown to have no effect on the kinetics of the reinitiated reaction (data not shown). Prior to the reactions, RNA was heat-denatured by incubation at 65°C for 15 min, followed by fast cooling to 4°C. The ‘stalled’ reactions were initiated using only three NTPs (ATP, CTP, and GTP). The ‘unstalled’ reactions were incubated in the absence of NTPs for the same time period. After 15 min incubation at temperature T1, the temperatures of the stalled and unstalled reactions were changed to temperature T2, and left to equilibrate for 5 min. The stalled φ6ECs were reinitiated by adding UTP, and the unstalled reactions were initiated by the addition of all four NTPs. Aliquots were taken at different time points after the addition of missing nucleotides, mixed with EDTA to 45 mM final concentration, and placed on ice. The products were analyzed using 0.75% or 1.5% agarose gels for the transcription and replication reactions, respectively. The agarose gels were preloaded at 2 V/cm for 15 min, and the electrophoresis was carried out at 5 V/cm at 4°C. The reaction products were visualized with ethidium bromide staining. The electrophoretic mobilities of the RNA replication intermediates were compared to a 2-log DNA ladder calibrated using a series of RNA hybrids (Supplementary Data and Figure S2). For experiments including a heparin trap (13), 9 mg/ml heparin (Sigma-Aldrich, St. Louis, MO, USA) was added after the stalling reactions, and the reactions were incubated at 22°C for 5 min prior to the addition of UTP.
RESULTS
Reinitiated φ6EC replicates with a reduced rate
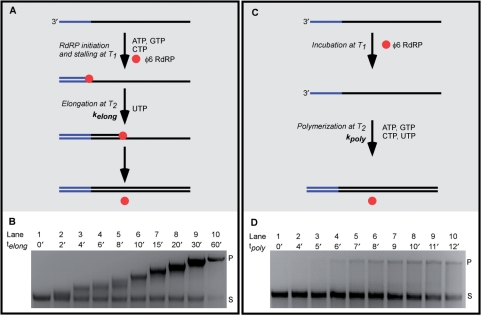
In replication, a φ6 RdRP elongation complex (φ6EC) can be stalled in vitro using a limited selection of NTPs and a template molecule in which the 3′-terminal region is devoid of one or more of the nucleotides (Figure 2A; for proof of stalling on short oligos, see Supplementary Data and Figure S3). Elongation can then be reinitiated by the addition of the missing NTP(s), yielding an entirely double-stranded product. To study the effect of stalling on the rate of φ6 RdRP replication, we selected a 4193 nt long replication template (4 kb ssRNA) in which the first occurrence of adenine was 50 nt from the 3′-end. The stalled φ6EC exhibited an electrophoretic mobility indistinguishable from that of free 4 kb ssRNA (Supplementary Figure S4). Following stalling and reinitiation by UTP addition, aliquots were collected at successive time points and analyzed on agarose gel (Figure 2B). After reinitiation, a fraction of the replication template retained the electrophoretic mobility of free 4 kb ssRNA (Supplementary Figure 2B, lanes 2–10), which corresponds to either free 4 kb ssRNA or to inactive stalled φ6EC. The electrophoretic mobility of successfully reinitiated φ6ECs decreased with time as φ6 RdRP progressed along the 4 kb ssRNA (Figure 2B, lanes 2–10). Notably, the band corresponding to the reinitiated φ6ECs stayed well-defined, suggesting that the stalled φ6ECs reinitiated in a synchronized manner. Termination of the replication reaction was detected by a stabilization of the electrophoretic mobility of the reinitiated φ6ECs (data not shown). This occurred between 30 min and 60 min after reinitiation, from which we deduce an overall kelong between 1.2 and 2.3 nt·s−1. (For the discussion of the measured rates please see Supplementary Data).
Figure 2.
A reinitiated φ6 RdRP elongation complex (φ6EC) and randomly-initiated φ6 RdRP replication show distinct electrophoretic profiles on agarose gels. (A) Schematic of stalling and reinitiation of φ6EC. In the presence of three NTPs (ATP, GTP, CTP), a φ6EC is stalled at the 50th nt from the 3′-end of the template at temperature T1. The sequence elongated prior to stalling is shown in blue. After UTP addition, the stalled φ6EC reinitiates and synthesizes the complementary strand at temperature T2. (B) Agarose gel of the elongation intermediates after reinitiation of the stalled φ6EC on 4 kb ssRNA template. Aliquots were taken at different times after reinitiation (telong). Letters S and P indicate 4 kb ssRNA and 4 kb dsRNA, respectively. (C) Schematic of a randomly-initiated φ6 RdRP replication. 4 kb ssRNA was incubated with φ6 RdRP, and all four NTPs were subsequently added simultaneously. (D) Agarose gel of aliquots of randomly-initiated φ6 RdRP replication taken at different polymerization times (tpoly).
A control experiment was carried out to measure the overall replication polymerization rate (kpoly) of unstalled φ6 RdRP under identical reaction conditions (Figure 2C). To measure kpoly, the 4 kb ssRNA was first incubated with φ6 RdRP for the same duration as above, but in the absence of NTPs. Subsequently, all four NTPs were simultaneously added and aliquots were taken at successive time points. At time zero, only free 4 kb ssRNA could be detected (Figure 2C, lane 1). In contrast to the reinitiated φ6ECs, at later times distinct bands could only be detected for the free 4 kb ssRNA and the final replication product (4 kb dsRNA) (Figure 2C, lanes 2–10), consistent with an unsynchronized population of φ6ECs in this experiment. The first appearance of 4 kb dsRNA product occurred 6 min after the addition of NTPs (Figure 2C, Lane 4), corresponding to a minimal kpoly of 12 nt s−1. Surprisingly, the observed kpoly is at least six-fold higher than kelong, despite the fact that kpoly is a composite of kelong and the rates of accompanying initial stages of replication (e.g. φ6 RdRP binding and initiation of φ6EC). This suggests that the randomly-initiated φ6EC elongates considerably faster than the reinitiated φ6EC.
An accurate determination of kelong and kpoly
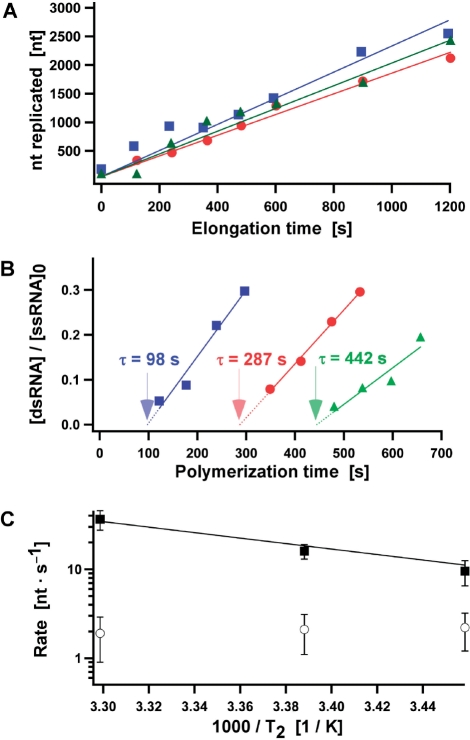
To support this initial observation, we obtained a more quantitative determination of kelong and kpoly in the case of reinitiated and randomly initiated φ6ECs, respectively. We converted the electrophoretic mobilities of the elongation intermediates of reinitiated φ6EC to a number of replicated nucleotides by using a calibration curve relating the two quantities (Supplementary Data and Figure S2). We converted only the earlier time points in Figure 2B, because the decreasing differences in the electrophoretic mobilities in the later reaction stages precluded an accurate determination of the number of replicated nucleotides. The results show that the number of replicated nucleotides after reinitiation increased linearly with time, indicating that the reinitiated φ6EC exhibited a constant kelong (Figure 3A, red points). kelong was deduced from the slope of the linear fit (Figure 3A, solid red line) and averaged 2 ± 1 nt s−1 (mean and standard deviation determined from six experiments).
Figure 3.
Stalling of φ6EC reduces the elongation rate (kelong) following reinitiation. (A) Progression of the reinitiated φ6EC along the 4 kb ssRNA at three different values of T2 (T2 = 16°C, green triangles; T2 = 22°C, red circles; T2 = 30°C, blue squares). Stalling of the φ6EC was carried out at T1 = 22°C. The number of replicated nucleotides (nt) was determined from the electrophoretic mobilities of elongation intermediates using the calibration curve in Figure S2C as described in the Supplementary Data. Solid lines are linear fits to the data. kelong are determined from the slopes of the corresponding linear fits. (B) The relative concentration of the replication products as a function of the polymerization time during randomly-initiated φ6 RdRP replication at three different values of T2 (T2 = 16°C, green triangles; T2 = 22°C, red circles; T2 = 30°C, blue squares). The incubation of 4 kb ssRNA and φ6 RdRP prior to the addition of the NTPs was carried at T1 = 22°C. Values of τ, representing the minimal sum of the initiation and elongation times during randomly-initiated φ6 RdRP replication, were determined by the x-intercept extrapolated from the linear fits and used to calculate the polymerization rates (kpoly) as described in the text. (C) Arrhenius plot of the elongation and polymerization rates for the reinitiated φ6EC (open circles), and for randomly-initiated φ6 RdRP replication (filled squares), respectively. The data obtained with random initiation was fitted to the Arrhenius equation (solid line).
Independently, we determined an improved estimate for kpoly for randomly-initiated φ6RdRP replication, by extrapolating the data to the minimum time τ necessary for the conversion of a single 4 kb ssRNA to its 4 kb dsRNA product, as previously reported (14). For example, to extract the value of τ from the data in Figure 2D, the normalized intensity of the band corresponding to 4 kb dsRNA was plotted as a function of polymerization time, and the experimental points were fitted to a straight line (Figure 3B, solid red line). The x-intercept of the linear fit yielded τ = 287 ± 42 s, which corresponds to kpoly = 15 ± 3 nt s−1 (mean and standard deviation deduced from three experiments). These more accurate values of kelong and kpoly thus establish that the elongation rate of randomly-initiated φ6EC exceeds that of reinitiated φ6EC by nearly an order of magnitude.
kelong of the reinitiated φ6EC is independent of temperature
To gain insight into the mechanisms underlying the observed rate reduction, we compared the temperature dependence of kelong and kpoly. To measure the temperature dependence of kelong, we first stalled φ6EC at temperature T1 for 15 min, equilibrated for 5 min at temperature T2, and reinitiated the φ6EC (Figure 2A). We first fixed T1 at 22°C and varied T2 (16°C, 22°C, or 30°C) (Supplementary Figure S5). Using calibration described above, we found that the number of replicated nucleotides by the reinitiated φ6ECs increased linearly with time for all three T2 (Figure 3A, green, red and blue solid lines correspond to T2 = 16°C, T2 = 22°C, and T2 = 30°C, respectively). Surprisingly, we found kelong to be independent of T2 (kelong of 2 ± 1 nt s−1 for all three T2). The experiment was then repeated at different T1 (16°C and 30°C) (Supplementary Figure S5). Here, too, we found that kelong remained constant during replication and was unaffected by changes in T1. Furthermore, the data reveals that kelong is independent of T1. In summary, we can conclude that over the range tested, kelong of the reinitiated φ6EC is independent of temperature.
For comparison, we applied the same temperature variation to the unstalled enzyme. As above, three different values of T1 and T2 were tested (16°C, 22°C, and 30°C for both T1 and T2) (Supplementary Figure S6). kpoly was determined by measuring τ as in Figure 2D. At fixed T1 = 22°C, we found that kpoly increased with increasing T2, from 9 ± 4 nt s−1 at T2 = 16°C, to 15 ± 3 nt s−1 at T2 = 22°C, and finally to 43 ± 14 nt s−1 at T2 = 30°C (mean and standard deviation deduced from three experiments). No detectable changes in the measured kpoly were observed when T1 was changed to 16°C or 30°C (Supplementary Figure S6). A comparison of kelong and kpoly is revealing: first, kpoly is consistently greater than kelong over the entire temperature range probed (Figure 3C): the rate reduction is not a particularity of experiments performed at room temperature; in addition, in contrast to the temperature-insensitive kelong, the polymerization rate kpoly of randomly-initiated φ6EC showed significant dependence on T2 (Figure 3C), from which we deduced an activation free energy for the rate-determining step of 24 kBT by fitting the data to an Arrhenius equation (solid line in Figure 3C).
kelong is insensitive to the nucleotide concentration during stalling and to the length of elongated RNA prior to stalling
To relate the observed differences between the kinetic profiles of reinitiated and randomly-initiated φ6ECs to specific regions of φ6RdRP structure, we investigated a number of factors that could affect the interactions within the φ6EC. These experiments were performed at a fixed T1 and T2 of 22°C.
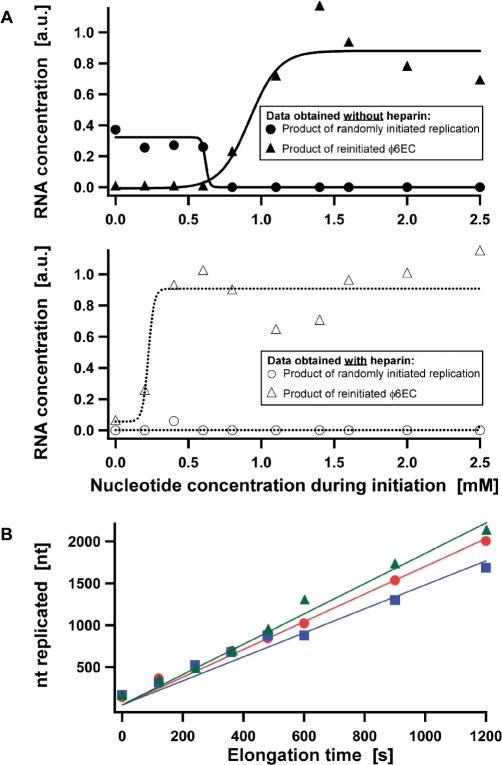
To determine whether the NTP concentration during stalling affected the rate of the reinitiated φ6ECs, we varied it from 0–2.5 mM per NTP. The stalled φ6ECs were reinitiated by the addition of a mixture of all four NTPs to bring the final concentration of each NTP after reinitiation to 2.5 mM, as above. Aliquots were collected at 4 min and 8 min after reinitiation and loaded on agarose gel (Supplementary Figure S7A). In the complete absence of NTPs during stalling, only the products of randomly-initiated φ6ECs were detected, as expected (Figure 4A). As the NTP concentration during the stalling stage was increased from 0.2 mM to 0.6 mM, this remained the case: only the products of randomly-initiated replication were present, and no detectable slow population of reinitiated φ6EC was observed (Figure 4A top, circles). However, once the NTP concentration exceeded 0.8 mM, the products of randomly-initiated replication were no longer observed; instead the slowly-migrating, distinct bands of elongation intermediates attributable to reinitiated φ6ECs appeared (Figure 4A top, triangles). Concentrations of reinitiated φ6ECs increased as the NTP concentration during stalling was further increased to 1.4 mM, but beyond 1.4 mM no further increase was detected. We note that little variation was observed between the electrophoretic mobilities of the reinitiated φ6EC, indicating similar kelong (Supplementary Figure S7A).
Figure 4.
Effects of the NTP concentration during the stalling stage and the length of dsRNA synthesized prior stalling on kelong of the reinitiated φ6EC. (A) φ6 RdRP stalling was carried out at different NTP concentrations, but the reinitiated φ6EC performed the reaction at a constant NTP concentration of 2.5 mM per NTP. The reinitiated φ6EC kinetics at each NTP concentration were studied in the absence and presence of heparin. Heparin was added after the stalling step, prior to reinitiation. Relative concentrations of the replication products and intermediates at 8 min after the addition of UTP were obtained by dividing the intensity of the corresponding band with the intensity of the 3 kb dsDNA band of the dsDNA ladder. In both plots, circles refer to the product of the randomly-initiated replication (4 kb dsRNA), whereas triangles correspond to the replication intermediates of the reinitiated φ6EC (Supplementary Figure S7). The experimental points were fitted to a sigmoidal curve. (B) The number of nucleotide replicated prior stalling was 3, 7, or 50 nts and the data is indicated with circles, squares, and triangles, respectively. Solid lines are linear fits to the data.
Finally, to absolutely confirm that the bands detected below 0.8 mM NTP concentration during stalling were indeed the products of randomly-initiated replication, the experiments were repeated in the presence of heparin following stalling (Supplementary Figure S7B). Heparin is known to inactivate free (i.e. unstalled) RNA polymerase (13), and can thus prevent random initiation after stalling. We indeed observed the products of randomly-initiated replication either entirely disappeared or were greatly reduced in the presence of heparin (Figure 4A bottom, circles). In contrast, the presence of heparin did not reduce the concentration of the reinitiated φ6ECs; however, it decreased the nucleotide concentration during stalling at which successfully reinitiated φ6ECs were detected (Figure 4A bottom, triangles). We can thus conclude that NTP concentration during the stalling stage affects the likelihood of forming stalled φ6ECs, leaving the subsequent dynamics as captured by kelong entirely unaffected.
The site at which the φ6EC stalls on the template strand determines the length of the dsRNA protruding from the φ6 RdRP product tunnel (Figure 1B). To investigate the effect of the dsRNA length on the kelong of reinitiated φ6EC, we synthesized replication templates with a stalling site at the third or seventh nucleotide. These replication templates were designed to stall φ6EC in the presence of three NTPs (i.e. ATP, CTP, and GTP) and reinitiate by the addition of UTP, as in the case of 4 kb ssRNA. On all these templates, the number of replicated nucleotide increased linearly with the elongation time (Figure 4B), as observed previously. Similarly, the observed kelong were measured to be 2 ± 1 nt s−1 in all instances (mean and standard deviation deduced from three experiments). Thus, we find the dynamics of the reinitiated φ6EC to be insensitive to the length of the elongated RNA prior stalling.
Kinetics of the reinitiated elongation complex during φ6 RdRP transcription
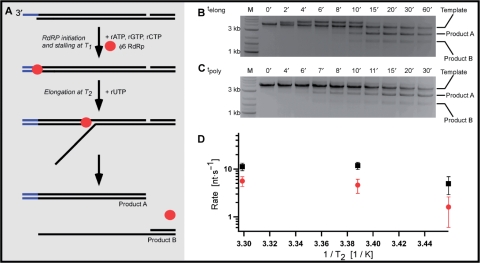
Different secondary structures of replication and transcription templates may interact differently with φ6 RdRP within the stalled φ6ECs (Figure 1), as RNA secondary structure elements located in viral RNAs have been shown to regulate RdRP polymerization (22,23). To test the role of template secondary structure on φ6EC kinetics after reinitiation, a 4 kb long transcription template was designed with a 50 nt long single-stranded sequence devoid of adenine at the 3′-end of the transcribed strand (Figure 5A). The design of the transcription template included a nick solely to facilitate separation of the template from the product on gel. After φ6EC stalling and introduction of the missing UTP, φ6EC can reinitiate yielding a branched elongation intermediates. At the end of transcription, a completely double-stranded 3 kb dsRNA (Product A) and partially double-stranded RNA (Product B) remained.
Figure 5.
Stalling affects the kelong during transcription less than during replication. (A) Schematic representation of φ6 RdRP transcription with stalling. The transcription template was synthesized by hybridizing three ssRNAs. The single-stranded sequence transcribed prior stalling is positioned at the 3′-end of the template strand. φ6 RdRP is stalled with three NTPs at temperature T1, and transcription reinitiated by the addition of UTP at temperature T2. When the reaction is complete, Products A and B result. (B) Agarose gel analysis of aliquots collected at different time points after the reinitiation of the stalled φ6EC. The transcription template, Product A and Product B bands are indicated. Transcription intermediates have the lowest electrophoretic mobility and can be observed at 2′, 4′, 6′, 8′ and 10′ after reinitiation. (C) Transcription by φ6 RdRP in the absence of stalling. Aliquots collected at different polymerization times were analyzed on agarose gel. (D) An Arrhenius plot of the experimentally-observed elongation (red circles) and polymerization rates (black squares) of the reinitiated φ6EC and randomly initiated φ6 RdRP transcription, respectively, during the transcription reaction.
Similarly to the above study of φ6RdRP replication, we measured kelong of the reinitiated φ6EC and kpoly of randomly initiated φ6EC during transcription. First, kelong on the transcription template after reinitiation was assayed at three different T2 (16°C, 22°C or 30°C). The elongation intermediates could be readily observed and displayed a decreased electrophoretic mobility compared to the free transcription template or the stalled φ6EC as a result of their higher molecular weight and branched structure (Supplementary Figure 5B). The electrophoretic mobility of Product A corresponds well to that of 2876 bp long dsRNA, and the electrophoretic mobility of Product B agrees with the value predicted by the previously constructed calibration curves of RNA hybrids (Supplementary Figure S2C). As in the replication reaction, an appreciable fraction of the transcription template binds φ6 RdRP, stalls the φ6EC, and enables the synchronized reinitiation of the stalled φ6EC after UTP addition, as judged from the well-defined bands corresponding to the elongation intermediates (Figure 5B, lane 2). In these transcription experiments, the more complex structure of the elongation intermediates prevented determination of kelong from the electrophoretic mobilities of the reaction intermediates. Rather, kelong was determined by dividing the length of the transcribed template by the typical time required to complete transcription. This time was approximated by measuring the mid-point between time t1, when the Product A band was first detected, and time t2, when the intensity of the elongation intermediate was observed to decrease. We report that kelong during transcription was comparable to kelong during replication at the lowest T2 (1.6 ± 1.0 nt s−1 at T2 = 16°C) but increased somewhat at higher T2 (4.6 ± 1.5 nt s−1 at T2 = 22°C) and 5.6 ± 1.3 nt s−1 at T2 = 30°C (red circles in Figure 5D) (mean and standard deviation deduced from three experiments).
We then investigated the kinetics of transcription by randomly-initiated φ6EC. In the transcription reaction with the unstalled φ6EC, the transcription products could readily be observed (Figure 5C). However, no elongation intermediates were detected, due to a lack of synchrony in the population of φ6ECs. We plotted the intensity of the Product A band as a function of the polymerization time and fitted the experimental points to a line. The τ values obtained were used to determine kpoly as above. At T1 = 22°C, the kpoly obtained for the randomly-initiated transcription was 5 ± 2 nt s−1 at T2 = 16°C, kpoly = 12 ± 2 nt s−1 at T2 = 22°C, and kpoly = 11 ± 2 nt s−1 at T2 = 30°C (black squares in Figure 5D). We thus observe that the values of kelong are again consistently slower than the values of kpoly of the randomly-initiated φ6 RdRP. However, the observed differences are less pronounced than in the case of the replication reaction. In addition, in transcription, the kelong and kpoly display similar temperature sensitivities, in contrast to the replication reaction, where considerable differences in the temperature trends were observed.
DISCUSSION
We have established that a reinitiated φ6EC displays a decreased kelong compared to a randomly-initiated φ6EC. The reduced kelong of the reinitiated φ6EC remained constant during the replication of at least the first 2.5 kb of the 4 kb ssRNA replication template (Figure 3A). Furthermore, our estimate of the time point at which replication of 4 kb ssRNA was complete (Figure 2B) yielded an overall elongation rate between 1.2 nt s−1 and 2.3 nt s−1, in a good agreement with the rate determined for the replication of the first 2.5 kb via fitting the electrophoretic mobilities of the replication intermediate (2 ± 1nt s−1). Thus, the reinitiated φ6EC elongates with a reduced rate throughout the template, from which we conclude that the conversion undergone by φ6EC during the stalling step is irreversible.
This irreversible conversion to the inhibited φ6EC state must be promoted during stalling, as the initial stages prior to φ6EC stalling proceeded similarly in both stalled and unstalled reactions. An upper limit for the time needed for the conversion to the inhibited φ6EC state could be deduced from the shortest time required to observe a synchronized population of reinitiated φ6ECs (2 min at T1 = 22°C; Supplementary Figure S8). Analysis of the temperature dependence of the reinitiated and randomly-initiated φ6EC showed considerably different behaviour for the two complexes (Figure 3C). The kinetics of a randomly-initiated φ6EC could be described well within an Arrhenius model and yielded activation energy of 24 kBT for polymerization. In contrast, the kinetics of a reinitiated φ6EC showed little temperature dependence of the elongation rates, implying that the conversion event during the stalling step may be insensitive to temperature. The irreversible conversion was similarly insensitive to both the NTP concentration during stalling and the length of the replicated template before stalling (Figure 4).
It is interesting to consider the possible nature of this irreversible conversion. A comparison of replication and transcription revealed that inhibition of the reinitiated φ6EC was appreciably greater with single- than double-stranded templates (Figures 3 and 5). This suggests that the template type has an effect on the conversion to the inhibited φ6EC state. This may be akin to the regulation of activity observed for other nucleic acid polymerases via either specific (e.g. RNA hairpin-protein) or nonspecific (e.g. electrostatic, hydrophobic) interactions (24–26). The interactions between φ6 RdRP and its template that are responsible for the conversion to the inhibited state do not appear to be related to a particular RNA secondary structure, as the template strands were denatured prior to stalling and were thus most likely present as an ensemble of different RNA folds stable under the applied reaction conditions. The fact that replication is affected more than transcription may suggest the nature of the nonspecific interactions involved (e.g. hydrophobic interactions promoted by the exposed bases of ssRNA). However, the fact that replication is affected more than transcription may also implicate other nonspecific interactions that occur with a higher probability in the case of ssRNA template simply as a consequence of its lower persistence length, which gives rise to a lower radius of gyration (27) and thus a higher local concentration of the template at the enzyme surface. Nonspecific RNA-φ6 RdRP interactions may cause the template RNA to interfere with the different tunnels in the enzyme and impair their proper functioning (e.g. they might limit exchange of free NTPs in the NTP channel, exit of the dsRNA in the product tunnel, or entry of the template strand in the template tunnel), resulting in a decrease in the overall elongation rates. Alternatively, RNA-φ6 RdRP interactions could trigger conformational changes within the φ6RdRP that result in suboptimal reaction rates.
It is evident that complex enzymes such as φ6 RdRPs are regulated through multiple mechanisms, and their sum total dictates both the enzyme rates. It is thus intriguing that even a simple stalling event can lead to considerable changes in enzyme dynamics. It remains to be seen whether the reduction in elongation rate after stalling is a general property of viral RdRPs. The arrest of the elongation complex could be biologically relevant during the viral life cycle, in which RNA replication and transcription are highly regulated (28). In addition, it could affect the frequency of RNA recombination and thus the adaptation of RNA viruses to their environment (10,11). More technically, many experiments have applied stalling protocols to measure elongation rates of polymerases (13,16–18). As shown here for φ6 RdRP, these measurements may provide inaccurate values, in the absence of confirmation that the enzyme is unaffected by stalling. For example, when T7 RNA polymerase (T7 RNAP) was stalled and subsequently reinitiated, the resulting elongation rate was as low as 2 nt s−1 as judged from a study by Ferrari and co-workers (17). By contrast, unstalled T7 RNAP exhibited elongation rates that range from 40–400 nt s−1 (29–31). Finally, polymerases may not be the only enzymes whose activity can be regulated via stalling. For instance, Kowalczykowski and co-workers recently showed that RecBCD helicase switches lead motors in response to the stalling at a specific DNA sequence, similarly resulting in a rate reduction of motor translocation (32).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Nanoned, The Netherlands Organization for Scientific Research, and the European Science Foundation (grants to N.H.D.); the Finish Center of Excellence Program 2006-2011 (1213467 to D.H.B.), the Academy of Finland and Nanotechnology (700036 to D.H.B.); Helsinki Graduate School of Biotechnology and Molecular Biology (funding of A.P.A.). Funding for open access charge: The Netherlands Organization for Scientific Research.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank W. Kamping and R. Tarkiainen for their experimental work and R. Tuma for fruitful discussions.
REFERENCES
- 1.Cerutti H, Casas-Mollano JA. On the origin and functions of RNA-mediated silencing: From protists to man. Curr. Genet. 2006;50:81–99. doi: 10.1007/s00294-006-0078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferrer-Orta C, Arias A, Escarmis C, Verdaguer N. A comparison of viral RNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 2006;16:27–34. doi: 10.1016/j.sbi.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Ortin J, Parra F. Structure and function of RNA replication. Annu. Rev. Microbiol. 2006;60:305–326. doi: 10.1146/annurev.micro.60.080805.142248. [DOI] [PubMed] [Google Scholar]
- 4.Arnold JJ, Gohara DW, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3Dpol): Pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mn2+ Biochemistry. 2004;43:5138–5148. doi: 10.1021/bi035213q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cramer J, Jaeger J, Restle T. Biochemical and pre-steady-state kinetic characterization of the hepatitis C virus RNA polymerase (NS5BΔ21, HC-J4) Biochemistry. 2006;45:3610–3619. doi: 10.1021/bi051483s. [DOI] [PubMed] [Google Scholar]
- 6.Thal MA, Wasik BR, Posto J, Hardy RW. Template requirements for recognition and copying by Sindbis virus RNA-dependent RNA polymerase. Virology. 2007;358:221–232. doi: 10.1016/j.virol.2006.08.022. [DOI] [PubMed] [Google Scholar]
- 7.Yang HY, Gottlieb P, Wei H, Bamford DH, Makeyev EV. Temperature requirements for initiation of RNA-dependent RNA polymerization. Virology. 2003;314:706–715. doi: 10.1016/s0042-6822(03)00460-4. [DOI] [PubMed] [Google Scholar]
- 8.van Dijk AA, Makeyev EV, Bamford DH. Initiation of viral RNA-dependent RNA polymerization. J. Gen. Virol. 2004;85:1077–1093. doi: 10.1099/vir.0.19731-0. [DOI] [PubMed] [Google Scholar]
- 9.Neufeld KL, Richards OC, Ehrenfeld E. Purification, characterization, and comparison of poliovirus RNA polymerase from native and recombinant sources. J. Biol. Chem. 1991;266:24–24. [PubMed] [Google Scholar]
- 10.Mindich L. Packaging, replication and recombination of the segmented genomes of bacteriophage φ6 and its relatives. Virus Res. 2004;101:83–92. doi: 10.1016/j.virusres.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 11.Worobey M, Holmes EC. Evolutionary aspects of recombination in RNA viruses. J. Gen. Virol. 1999;80:2535–2543. doi: 10.1099/0022-1317-80-10-2535. [DOI] [PubMed] [Google Scholar]
- 12.Koch U, Narjes F. Recent progress in the development of inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. Curr. Top. Med. Chem. 2007;7:1302–1329. doi: 10.2174/156802607781212211. [DOI] [PubMed] [Google Scholar]
- 13.Sun JH, Kao CC. RNA synthesis by the BMV RNA-dependent RNA polymerase: transition from initiation to elongation. Virology. 1997;233:63–73. doi: 10.1006/viro.1997.8583. [DOI] [PubMed] [Google Scholar]
- 14.Makeyev EV, Bamford DH. Replicase activity of purified recombinant protein P2 of double-stranded RNA bacteriophage φ6. EMBO J. 2000;19:124–133. doi: 10.1093/emboj/19.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DL. A mechanism for initiating RNA-dependent RNA polymerization. Nature. 2001;410:235–240. doi: 10.1038/35065653. [DOI] [PubMed] [Google Scholar]
- 16.Davenport RJ, Wuite G. JL, Landick R, Bustamante C. Single-molecule study of transcriptional pausing and arrest by E. coli RNA polymerase. Science. 2000;287:2497–2500. doi: 10.1126/science.287.5462.2497. [DOI] [PubMed] [Google Scholar]
- 17.Ferrari R, Rivetti C, Dieci G. Transcription reinitiation properties of bacteriophage T7 RNA polymerase. Biochem. Biophys. Res. Commun. 2004;315:376–380. doi: 10.1016/j.bbrc.2004.01.071. [DOI] [PubMed] [Google Scholar]
- 18.Tolic-Norrelykke SF, Engh AM, Landick R, Gelles J. Diversity in the rates of transcript elongation by single RNA polymerase molecules. J. Biol. Chem. 2004;279:3292–3299. doi: 10.1074/jbc.M310290200. [DOI] [PubMed] [Google Scholar]
- 19.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 20.Petrushenko ZM, Lai CH, Rai R, Rybenkov VV. DNA reshaping by MukB – Right-handed knotting, left-handed supercoiling. J. Biol. Chem. 2006;281:4606–4615. doi: 10.1074/jbc.M504754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vilfan ID, Kamping W, van den Hout M, Candelli A, Hage S, Dekker NH. An RNA toolbox for single-molecule force spectroscopy studies. Nucleic Acids Res. 2007;35:6625–6639. doi: 10.1093/nar/gkm585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deiman B, Kortlever RM, Pleij C.WA. The role of the pseudoknot at the 3′ end of turnip yellow mosaic virus RNA in minus-strand synthesis by the viral RNA-dependent RNA polymerase. J. Virol. 1997;71:5990–5996. doi: 10.1128/jvi.71.8.5990-5996.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osman TAM, Hemenway CL, Buck KW. Role of the 3′ tRNA-like structure in TMV minus-strand RNA synthesis by the viral RNA-dependent RNA polymerase in vitro. J. Virol. 2000;74:11–11. doi: 10.1128/jvi.74.24.11671-11680.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Artsimovitch I, Landick R. Interaction of a nascent RNA structure with RNA polymerase is required for hairpin-dependent transcriptional pausing but not for transcript release. Genes Dev. 1998;12:3110–3122. doi: 10.1101/gad.12.19.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rivetti C, Guthold M, Bustamante C. Wrapping of DNA around the E.coli RNA polymerase open promoter complex. EMBO J. 1999;18:4464–4475. doi: 10.1093/emboj/18.16.4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toulokhonov I, Landick R. The flap domain is required for pause RNA hairpin inhibition of catalysis by RNA polymerase and can modulate intrinsic termination. Mol. Cell. 2003;12:1125–1136. doi: 10.1016/s1097-2765(03)00439-8. [DOI] [PubMed] [Google Scholar]
- 27.Sedlak M. In: Physical chemistry of polyelectrolytes. Radeva T, editor. New York: CRC Press; 2001. pp. 1–58. [Google Scholar]
- 28.Plumet S, Duprex WP, Gerlier D. Dynamics of viral RNA synthesis during Measles virus infection. J. Biol. Chem. 2005;79:6900–6908. doi: 10.1128/JVI.79.11.6900-6908.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim JH, Larson RG. Single-molecule analysis of 1D diffusion and transcription elongation of T7 RNA polymerase along individual stretched DNA molecules. Nucleic Acids Res. 2007;35:3848–3858. doi: 10.1093/nar/gkm332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skinner GM, Baumann CG, Quinn DM, Molloy JE, Hoggett JG. Promoter binding, initiation, and elongation by bacteriophage T7 RNA polymerase - A single-molecule view of the transcription cycle. J. Biol. Chem. 2004;279:3239–3244. doi: 10.1074/jbc.M310471200. [DOI] [PubMed] [Google Scholar]
- 31.Uptain SM, Kane CM, Chamberlin MJ. Basic mechanisms of transcript elongation and its regulation. Annu. Rev. Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 32.Spies M, Amitani I, Baskin RJ, Kowalczykowski SC. RecBCD enzyme switches lead motor subunits in response to χ recognition. Cell. 2007;131:694–705. doi: 10.1016/j.cell.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.