Abstract
Poly(ADP-ribose) (pADPr) is a polymer assembled from the enzymatic polymerization of the ADP-ribosyl moiety of NAD by poly(ADP-ribose) polymerases (PARPs). The dynamic turnover of pADPr within the cell is essential for a number of cellular processes including progression through the cell cycle, DNA repair and the maintenance of genomic integrity, and apoptosis. In spite of the considerable advances in the knowledge of the physiological conditions modulated by poly(ADP-ribosyl)ation reactions, and notwithstanding the fact that pADPr can play a role of mediator in a wide spectrum of biological processes, few pADPr binding proteins have been identified so far. In this study, refined in silico prediction of pADPr binding proteins and large-scale mass spectrometry-based proteome analysis of pADPr binding proteins were used to establish a comprehensive repertoire of pADPr-associated proteins. Visualization and modeling of these pADPr-associated proteins in networks not only reflect the widespread involvement of poly(ADP-ribosyl)ation in several pathways but also identify protein targets that could shed new light on the regulatory functions of pADPr in normal physiological conditions as well as after exposure to genotoxic stimuli.
INTRODUCTION
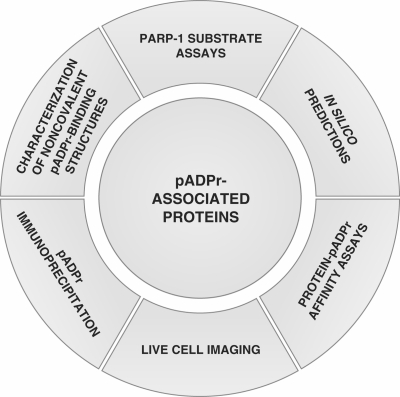
The activation of poly(ADP-ribose) polymerases (PARPs) has been the subject of numerous studies in which poly(ADP-ribose) (pADPr), a branched polymer assembled upon the catalytic transfer of ADP-ribose moieties from NAD, was initially regarded as a posttranslational modification. Indeed, the covalent attachment of pADPr chains to chromatin-associated proteins, such as histones has been known for decades (1). Since then, a growing body of work on pADPr metabolism across a broad range of model systems has identified new pADPr-associated proteins. Whether these proteins are covalently poly(ADP-ribosyl)ated, noncovalent pADPr binding proteins or exhibiting both properties, they can collectively be termed pADPr-associated proteins. The methods used so far to identify pADPr-associated proteins are summarized in Figure 1. Current strategies include various biochemical and biological validation approaches, bioinformatics analysis for the prediction of pADPr binding motifs or in vivo cell imaging. In spite of the increasing number of tools currently used to identify pADPr-associated proteins, very little literature addresses the question of poly(ADP-ribosyl)ation from a pADPr binding perspective. Until recently, most of the attention to pADPr-associated proteins has been focused on covalent poly(ADP-ribosyl)ation because of the drastic consequences generally observed on protein properties (2). However, several studies are now pointing to important noncovalent interactions between pADPr and various signaling proteins and an expanding number of proteins are now known to bind in a noncovalent manner to pADPr.
Figure 1.
Current experimental strategies for the identification of pADPr-associated proteins. Six types of experimental approaches have been used to date to identify poly(ADP-ribosyl)ated and pADPr binding proteins (pADPr-associated proteins). As illustrated clockwise starting from the top: (i) identification of in vitro and in vivo poly(ADP-ribosyl) transferase activity of PARP-1 onto acceptor proteins (covalent modifications) using immunological or radioactivity-based detection methods; (ii) identification of noncovalent pADPr binding regions within protein amino acid sequences based on similarity with a consensus pADPr binding motif; (iii) affinity-based assays for the identification, validation and quantitative evaluation of noncovalent pADPr binding [surface plasmon resonance (SPR), electrophoretic mobility shift assays (EMSA), peptide and protein polymer blot analysis, mono- and bidimensional gel electrophoresis coupled with polymer blot analysis followed by MS]; (iv) in vivo evaluation of recruitment and accumulation of pADPr binding proteins using micro irradiation-induced DNA damage assays in live cell experiments; (v) immunoprecipitation assays using specific anti-pADPr antibodies to pull-down pADPr-associated proteins; and (vi) the characterization of PARP-associated protein domains as functional noncovalent pADPr binding modules (e.g. the Macro domain A1pp or the zinc-finger domain PBZ).
Three protein motifs have been characterized to mediate noncovalent pADPr binding. Pleschke et al. (3) were the first to report a noncovalent pADPr binding motif in DNA damage checkpoint proteins. This characteristic pADPr binding motif, composed of interspersed basic and hydrophobic amino acid residues, suggested for the first time that pADPr can act as a mediator of protein–protein interactions. A second pADPr binding motif has been shown by Karras and colleagues (4) to reside within macro domains in the form of a conserved ligand binding pocket (5). The pADPr binding of macro domain-containing proteins is particularly interesting given that this module is mostly found in helicase proteins associated with DNA and/or RNA unwinding activity, an important function in DNA replication, DNA repair and DNA recombination processes. A recent study from Ahel and colleagues (6) has revealed a third structure (zinc-finger PBZ) found in DNA repair and checkpoint proteins, which also mediates specific pADPr binding.
These noncovalent pADPr motifs can be the object of pADPr-affinity evaluation. Indeed, Fahrer and colleagues (7) have published a quantitative method to assess the binding affinity of noncovalent pADPr binding proteins. They reported remarkably high noncovalent affinity to pADPr, this affinity being correlated to pADPr chain length, in consistency with the length-dependent behavior of pADPr in cell death (8), and the nanomolar affinity of macro domains.
Although some proteins bind pADPr through either a covalent or noncovalent bond, some proteins closely associated to PARPs may actually be modified by both mechanisms. Evidence is mounting that pADPr's functions extend far beyond that of a covalent posttranslational modification. pADPr can indeed be viewed as an effector molecule that modulate the properties of its acceptor proteins.
In view of the scarcity of information presently available and the potentially large number of pADPr-associated proteins, we conducted a proteome-wide investigation to identify candidates for pADPr binding and to determine if diversity exists in the roles of pADPr in cell function. First, in silico identification of consensus pADPr binding motifs was used to systematically identify putative pADPr binding proteins amongst a nonredundant human protein database. Second, synthetic peptides derived from the sequences of selected proteins that contained motifs with high homology to the predicted pADPr consensus binding site were tested for in vitro binding to pADPr. Positive binding sequences were aligned to refine the consensus pADPr binding motif into a more stringent pattern for increased confidence in pADPr binding predictions. Finally, in silico predictions were supported by protein–pADPr affinity assays coupled with large-scale mass spectrometry (MS)-based proteome analysis. Polymer blot analysis of two-dimensionally separated HeLa cell extracts revealed several proteins that were predicted to bind pADPr as well as pADPr-associated proteins identified by liquid-chromatography tandem MS (LC-MS/MS) that were immunoprecipitated from cell cultures exposed to extensive alkylation-induced DNA damage. Identified protein candidates were classified according to biological functions and their known interactions with pathways-related proteins so as to organize them into biologically meaningful clusters. Collectively, our results provide novel insights into the pADPr interactome and generate, for the first time, a large-scale MS-based proteomic resource for identifying pADPr binding candidates.
MATERIALS AND METHODS
In silico prediction of pADPr binding proteins
Putative pADPr binding proteins were first screened on the basis of the original consensus pADPr binding motif proposed by Pleschke et al. (3). The following original pattern value [AVILMFYW]1-X2-[KR]3-X4-[AVILMFYW]5-[AVILMFYW]6-[KR]7-[KR]8-[AVILMFYW]9-[AVILMFYW]10-[KR]11 was searched against the Homo sapiens entries of the Swiss-Prot database (20 070 human entries out of 392 667, as indexed on 23 July 2008) using the PattInProt search engine located on the NPS@ server (Network Protein Sequence @nalysis, http://npsa-pbil.ibcp.fr/) (9). A similarity level cut-off of 90% was applied. The refined pADPr binding motif [HKR]1-X2-X3-[AIQVY]4-[KR]5-[KR]6-[AILV]7-[FILPV]8 was screened using the same database but without mismatch (100% similarity).
Cell culture, siRNA transfections and induction of DNA damage
Human neuroblastoma SK-N-SH and HeLa cervical carcinoma cell lines were cultured (air/CO2, 19:1, 37°C) in DMEM medium supplemented with 10% fetal bovine serum (Hyclone-ThermoFisher Scientific, Ottawa, Canada). Penicillin (100 U/ml) and streptomycin (100 mg/ml) (Wisent, St-Bruno, Canada) were added to culture media. When growth of SK-N-SH cells reached ∼50% confluency, cells were transfected with 5 nM PARG siRNA [hPARG: AAGAUGAGAAUGGUGAGCGAAdTdT and hPARG control siRNA (mismatch): AAGAUGAGAAUCCUGAGCGAAdTdT] using HiPerfect reagent (Qiagen, Mississauga, Canada). Silencing was conducted over 6 days, passaging cells every 48 h to achieve maximum PARG knockdown. Alkylating DNA damage was performed using 100 μM N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) for 5 min.
Two-dimensional polymer blot analysis
Five hundred micrograms of total HeLa cell protein extracts were subjected to high-resolution two-dimensional electrophoresis essentially following the method developed by O’Farrell (10). Briefly, cells were lyzed in a lysis buffer containing 9 M urea, 2% NP-40 and 2% ampholines (1.6% pH 3–10 and 0.4% pH 5–7). The first dimension (isoelectric focusing) was performed on 3 × 125 mm tubes using the Model 225 tube gel casting stand and tube gel adapter (Bio-Rad, Mississauga, Canada) for the Protean II xi cell (Bio-Rad), during which proteins are separated according to their isoelectric points. The second dimension (SDS–PAGE) was performed using 20 × 20 cm gels with the Protean II xi cell. Two identical gels were prepared: one gel was silver stained (Vorum protocol) according to Mortz et al. (11) to visualize the separation pattern. The second gel was transferred onto 0.45 μm PVDF membrane. Polymer blot analysis and identification of pADPr binding proteins by MALDI-TOF MS was performed as described (12).
Peptide polymer blot analysis
Peptides corresponding to the predicted pADPr binding domain of selected proteins found by in silico analysis were synthesized using Fmoc solid-phase peptide synthesis with an Applied Biosystems 433A peptide synthesizer (Supplementary Table S4). The quality of peptide synthesis was controlled by MALDI-TOF MS analysis and the purity of peptide was evaluated using analytical HPLC. Peptides were dissolved in TBS-T (10 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.1% Tween-20). One microgram of each peptide was spotted onto a 21 mm × 50 mm nitrocellulose film-slide (Grace Bio-Labs, Bend, OR, USA), air-dried, rinsed three times with TBS-T and incubated for 1 h at room temperature with gentle agitation in TBS-T containing 250 nM 32P-labeled pADPr synthesized as described (12). It was then washed with TBS-T buffer until no radioactivity could be detected in the washes. The membrane was subsequently air-dried and subjected to autoradiography on an Instant Imager (Perkin Elmer, Woodbridge, Canada), which analyzes and quantifies the distribution of radioactivity on flat samples.
pADPr immunoprecipitation
SK-N-SH cells were seeded onto three 150 mm cell-culture dishes and grown up to 80–90% confluency. Cells were incubated with freshly prepared 100 µM MNNG for 5 min before extraction. All further steps were performed on ice or at 4°C. Two PBS washes were carried out prior to the extraction with 2 ml/plates of lysis buffer [20 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5% NP-40, Complete™ protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN, USA)]. Cell lysates were pooled, adjusted to 2 M NaCl and placed on ice for 15–20 min. After 30 s of gentle mixing, the cell extract was extensively dialyzed against ice-cold lysis buffer. Immunoprecipitation experiments were performed using Dynabeads™ magnetic beads covalently coupled with Protein G (Invitrogen, Burlington, Canada). The Dynabeads™ were washed two times with 1 ml of 0.1 M sodium acetate buffer, pH 5.0, coated with 10–15 μg of mouse monoclonal anti-pADPr antibody clone 10H (Tulip Biolabs, West Point, PA, USA) or equivalent amount of normal mouse IgGs (Calbiochem-EMD Biosciences, San Diego, CA, USA). The antibody-coupled Dynabeads™ were incubated for 1 h at room temperature with 1 ml of PBS containing 1% (w/v) BSA (Sigma-Aldrich, Oakville, Canada) to block nonspecific antibody binding sites. The beads were finally washed three times with 1 ml of lysis buffer and added to the pADPr-protein extract for 2 h incubation with gentle agitation. Samples were washed three times with two volumes of lysis buffer for 5 min. Protein complexes were eluted using 150 μl of 3× Laemmli sample buffer containing 5% β-mercaptoethanol and boiled for 5 min in a water bath. Proteins were resolved using 4–12% Criterion™ XT Bis–Tris gradient gel (Bio-Rad) and stained with Sypro Ruby (Bio-Rad) according to the manufacturer's instructions. Images were acquired using a CCD-based Chemi-Imager 4000 imaging system and AlphaEase software 3.3 (Alpha Innotech Corporation, San Leandro, CA, USA).
PARG activity assays
32P-labeled automodified PARP-1 was synthesized essentially as described by Ménard and Poirier (13) in a total reaction volume of 900 µl containing 100 mM Tris–HCl, pH 8.0, 10 mM MgCl2, 8 mM dithiothreitol, 10% (v/v) glycerol, 23 µg of calf thymus-activated DNA (Sigma-Aldrich), 1 mM NAD and 75 µCi of 32P-labeled NAD (GE Healthcare, Baie d'Urfée, Canada). Ethanol was added to this preparation dropwise to 10% (v/v) final concentration, with constant mixing, and the reaction mixture was incubated for 3 min at 30°C. The reaction was started by adding 20 U of PARP-1 purified up to the DNA-cellulose step as described by Zahradka and Ebisuzaki (14). After 30 min at 30°C, during which time the enzyme was modified by covalent linkage of pADPr chains to its automodification domain, 100 µl of 3 M sodium acetate, pH 5.2 and 700 µl of propan-2-ol were added as described by Brochu et al. (15). The reaction mixture was kept on ice for 30 min and then centrifuged at 10 000g for 10 min at 4°C. The pellet was washed twice with ice-cold 80% (v/v) ethanol, resuspended and incubated at 60°C for 2 h in 1 ml containing 1 M KOH and 50 mM EDTA to release the pADPr from automodified PARP-1 by alkali digestion. Protein-free pADPr was purified on dihydroxyboronyl Bio-Rex (DHBB) affinity resin as described in Shah et al. (16). PARG activity was measured by analyzing the production of ADP-ribose monomers from in vitro synthesized protein-free pADPr. PARG extracts used for immunoprecipitation experiments were incubated with 10 μM 32P-labeled pADPr for 20 min at 30°C. Aliquots were then spotted on PEI-F (polyethyleneimine F) cellulose thin layer chromatography (TLC) sheets (Macherey-Nagel, Bethlehem, PA, USA) and developed in 0.3 M LiCl and 0.9 M ethanoic (acetic) acid according to Ménard and Poirier (13).
Immunoblotting
Protein extracts eluted from Dynabeads™ were separated on SDS–PAGE and then transferred onto 0.2 μm nitrocellulose membrane (Bio-Rad). After incubating 1 h with blocking solution (PBS-T containing 5% nonfat milk), the membrane was probed overnight at room temperature with shaking, by primary antibodies to PARP-1 [clone C2-10 (1:5000)], pADPr [10H (1:2500) Tulip Biolabs, 96-10 (1:5000)], IQGAP1 [(1:1000) Cell Signaling], Apoptosis-inducing factor (AIF) [(1:5000) Sigma], Ku80 [(1:5000) Oncogene Research Products, San Diego, CA, USA], DNA-PK [(1:5000) Calbiochem], Adaptins α/β/γ [(1:1000/1:5000/1:5000) BD Biosciences, Mississauga, Canada]. After washing with PBS-T, species-specific horseradish peroxidase-conjugated secondary antibody was added for 1 h at room temperature. Signals were detected with Western Lightning™ Chemiluminescence reagent plus kit (Perkin Elmer).
LC-MS/MS analysis
SDS–PAGE protein lanes corresponding to control and anti-pADPr immunoprecipitated extracts were cut into 33 gel slices per lane using a disposable lane picker (The Gel Company, CA, USA). Gel slices were deposited into 96-well plates. In-gel protein digest was performed on a MassPrep™ liquid handling station (Waters, Mississauga, Canada) according to the manufacturer's specifications and using sequencing-grade modified trypsin (Promega, Madison, WI, USA). Peptide extracts were dried out using a SpeedVac™.
Peptide extracts were separated by online reversed-phase (RP) nanoscale capillary LC (nanoLC) and analyzed by electrospray MS (ES MS/MS). The experiments were performed on a Thermo Surveyor MS pump connected to a LTQ linear ion trap mass spectrometer (Thermo Electron, San Jose, CA, USA) equipped with a nanoelectrospray ion source (Thermo Electron, San Jose, CA, USA). Peptide separation took place within a PicoFrit column BioBasic C18, 10 cm × 0.075 mm internal diameter (New Objective, Woburn, MA, USA) with a linear gradient from 2% to 50% solvent B (acetonitrile, 0.1% formic acid) in 30 min, at 200 nl/min. Mass spectra were acquired using data-dependent acquisition mode (Xcalibure software, version 2.0). Each full-scan mass spectrum (400–2000 m/z) was followed by collision-induced dissociation of the seven most intense ions. The dynamic exclusion function was enabled (30 s exclusion), and the relative collisional fragmentation energy was set to 35%.
Interpretation of tandem MS spectra
All MS/MS samples were analyzed using Mascot (Matrix Science, London, UK; version 2.2.0). Mascot was set up to search against human Uniref_100 protein database assuming a digestion with trypsin. Fragment and parent ion mass tolerance were, respectively, of 0.5 Da and 2.0 Da. Iodoacetamide derivative of cysteine was specified as a fixed modification. Deamidation of asparagines and glutamine, acetylation of lysine and arginine and oxidation of methionine were specified as variable modifications. Two missed cleavages were allowed.
Criteria for protein identification
Scaffold (version 01_07_00; Proteome Software Inc., Portland, OR, USA) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at >80.0% probability as specified by the Peptide Prophet algorithm (17). Protein identifications were accepted if they could be established at >90.0% probability and contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (18). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Using these stringent identification parameters, the rate of false positive identifications is <1%.
Modeling of pADPr-associated protein networks
A graphical representation of the functional connections between pADPr-associated proteins was first computed using filtered Bibliosphere (Genomatix Software, Ann Arbor, MI, USA) interaction maps (co-citation functional level B2). Bibliosphere outputs a bibliometric-based clustering of related genes. Searching for biological processes enrichment of the pADPr binding network against the whole Gene Ontology (GO) hierarchy was performed using BiNGO v2.0 [19] and visualized with Cytoscape v2.5 [20]. GO annotation's P-values were obtained by hypergeometric statistical testing (cluster versus the whole-annotation bank) and corrected using the Benjamin and Hochberg false discovery rate included in the BiNGO software. GO database was downloaded on 1 July 2007. GoMiner [21] was used to reveal protein distributions within selected GO categories. For both in silico analysis, gene identification codes were converted or retrieved using the protein's Swiss-Prot accession numbers.
Evaluation of protein domain and family distribution
The Pfam (http://pfam.sanger.ac.uk/) domains were obtained by parsing the ‘swisspfam’ (version Pfam 22.0, July 2007, 9318 families) database using in-house Ruby scripts (version 1.8.6) which output Microsoft Excel-compatible spreadsheets suitable for further analysis. Datasets were generated from the both the original and refined pADPr binding motifs, and the protein dataset that contains all the proteins identified by LC-MS/MS from pADPr immunoprecipitation experiments.
RESULTS
In silico identification of noncovalent pADPr binding proteins
There are currently three protein modules known to bind pADPr. However, they differ greatly in their abundance within the human proteome. The macro domain that has been demonstrated to bind pADPr (4,5) (A1pp) could only be found in 27 human proteins using the InterPRO (22) protein database. Human proteins with the conserved zinc-finger motif PBZ are even rarer, since there are only four human proteins reported to match this stringent motif. On the other hand, 862 human putative pADPr binding proteins were extracted from the Swiss-Prot database following a similarity search based on the noncovalent pADPr binding motif defined by Pleschke et al. (3) found within DNA damage checkpoint proteins (Supplementary Table S1). This pADPr binding sequence is based on the consensus motif hxbxhhbbhhb (where h stands for hydrophobic residues, b for basic residues and x for any amino acid residues). However, the level of conservation of the proposed pADPr binding motif is low but still characterized by preferred amino acid residues and some more variable positions. This is a rather broad pADPr binding signature that can be found in a much larger diversity of proteins than the macro domain and zinc-finger motifs. Consequently, we searched for human proteins with a high similarity level but without any strict requirement for specific amino acids at a given position within the pADPr binding sequence, except for the two positively charged lysine or arginine residues in the center of the motif which seem to be the less tolerant positions. Following these criteria, we screened the Swiss-Prot protein library using the PattInProt search engine (9) with a similarity level threshold of 90% which allows the detection of degenerated sequences. The resulting dataset of more than 800 proteins (Supplementary Table S1) represents an impressive number of proteins. Although it probably displays a high rate of false positives, this in silico screening test is considered valuable because it greatly increase the likelihood of detecting a pADPr binding protein.
Refinement of the original pADPr binding motif
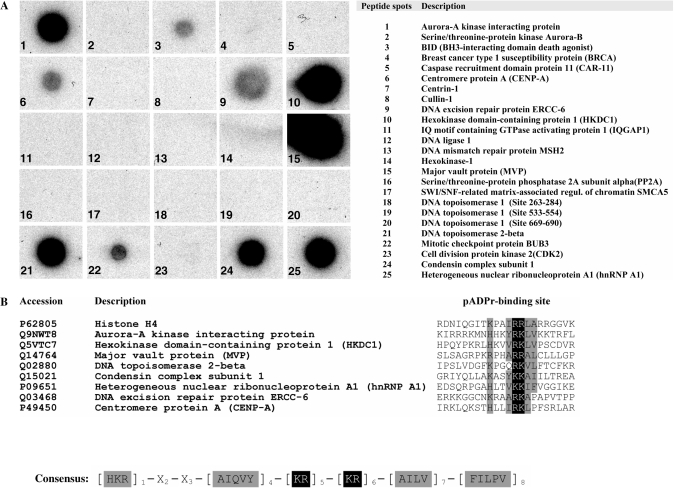
We first wanted to increase the number of experimentally validated pADPr binding sites so as to better define a consensus pADPr binding pattern and get a more representative view of the amino acid frequency at any given position. To achieve this goal, we selected some putative pADPr binding proteins suspected to be involved in pADPr-associated biological processes and synthesized peptides that corresponded to the region most similar to the consensus pattern. Table 1 presents the comprehensive list of pADPr binding regions that have been tested using synthetic peptides in polymer blot assays. For the present study, 24 new peptides were synthesized and assayed for pADPr binding. The hnRNP A1 peptide was used as a positive control since it has been validated as a strong and specific pADPr binding peptide (12). About one-third of the new peptides (9 out of 24) were characterized as pADPr binding peptides (Figure 2A). Most of the validated pADPr binding peptides share a restricted set of amino acids at specified positions of the pADPr binding consensus sequence (Figure 2B) from which we can derive a refined consensus motif. This modified consensus sequence with a much more limited set of amino acids at conserved positions, [HKR]1-X2-X3-[AIQVY]4-[KR]5-[KR]6-[AILV]7-[FILPV]8, represents a more stringent version of the original consensus sequence proposed by Pleschke and collaborators (3). In contrast to the more relaxed consensus, the two positively charged amino acids residues [KR]5-[KR]6 are strictly followed by either A, I, L or V ([AILV]7) which are classified as residues with alkyl side chains. These strong pADPr binding regions do not require a conserved hydrophobic residue at position 3, but the positively charged amino acid residues at position 1 is conserved. This refined pattern of validated pADPr binding sites is conserved in histone H4 (23,24) and hnRNP A1, whose pADPr binding motifs have been validated experimentally as strong binding sites (12).
Table 1.
Comprehensive protein sequence alignment of predicted pADPr binding motifs screened by polymer blot analysis
 |
The alignment includes known and putative pADPr binding proteins based on the pADPr binding motif (hxbxhhbbhhb)(3). The central amino acid couple K/R is shown on a black background. Proximal residues that fit the consensus are indicated under a gray background. Swiss-Prot accession codes, sites mapping and experimental pADPr binding validation are indicated for each peptide.
Figure 2.
Polymer blot analysis. (A) Synthetic peptides corresponding to putative pADPr binding proteins as revealed by in silico prediction based on the consensus sequence proposed by Pleschke et al. (3) were dot-blotted onto nitrocellulose-coated glass slides, screened with 32P-pADPr and autoradiographied (see Supplementary Table S1 for the complete list of predictions). Amino acid sequences used for peptide synthesis are provided in Supplementary Table S4. (B) A refined pADPr binding consensus is generated from the sequence alignment of validated pADPr binding peptides. Experimentally verified pADPr binding regions from pADPr dot blot analysis were aligned to derive a refined pADPr binding motif and to computationally screen protein sequence database with the goal of achieving higher reliability and minimizing false positive identifications (see Supplementary Table S2 for a listing of proteins that match the refined pADPr binding consensus).
The refined pattern was searched without mismatch tolerance against the Swiss-Prot database to extract a sub-dataset of pADPr binding proteins with high similarity to the pADPr binding sites that were validated in our study (Supplementary Table S2). Interestingly, the 526 unique proteins in this dataset are much better connected to DNA and chromatin functions than the original dataset. Unique proteins to the dataset generated with the refined pADPr binding motif include: DNA excision repair proteins ERCC-2 and ERCC-6, DNA polymerase subunit alpha B, DNA primase small subunit, DNA replication licensing factors MCM3, MCM5, MCM7, DNA topoisomerase 2-beta, mitochondrial DNA topoisomerase I, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (SMCA5), Centromere protein T (CENP-T), the DNA damage checkpoint response protein HUS 1, several histone metabolism-associated proteins, such as the histone acetyltransferases MYST3, MYST4, histone acetyltransferases p300 (25) and PCAF (26), or the newly identified pADPr binding protein DEK (27). Because several of these proteins are closely related to known pADPr-modulated pathways, the refined consensus appears to have a much higher specificity than its more relaxed counterpart. This refined motif lowers the screening effort to a more manageable level and should be considered as a first step in an attempt to identify pADPr binding proteins.
Identification of noncovalent pADPr binding proteins by polymer blot analysis following two-dimensional electrophoresis
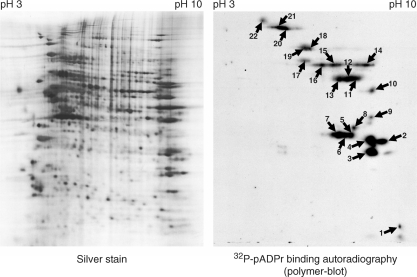
As in silico predictions are still limited in their ability to identify all pADPr binding proteins, it mostly serves a rapid approach to identify putative pADPr binding targets. Until they are more accurate, in silico-identified pADPr binding proteins will need to be validated experimentally. Thanks to the development of high-throughput proteomics technologies, proteome-wide identification of pADPr binding proteins is now manageable. Half a milligram of HeLa cell protein extract was resolved into discrete spots by two-dimensional electrophoresis and transferred onto PVDF membrane. pADPr binding proteins were identified by matrix-assisted laser desorption ionization (MALDI) MS after incubation with 32P-labeled pADPr and autoradiography (Figure 3). We selected tube-gel two-dimensional electrophoresis instead of the more recent immobilized pH gradient (IPG) two-dimensional isoelectric focusing (IEF) because of the increased sample loading capacity and because reproducibility was not as important as it would have been in a differential proteome analysis. Most of the pADPr binding proteins are found at high pH values (positively charged) as we would expect for proteins involved in DNA/RNA transactions or specifically involved in the binding of a negatively charged polyanion, such as pADPr. Known pADPr binding proteins, such as hnRNPs (12) still represent many of the abundant pADPr binding proteins identified by this method (Table 2). DNA-binding proteins, such as members of the zinc-finger protein family, are also found in several spots. In addition, the presence of the major vault protein (MVP) is interesting since its pADPr binding region, targeted in the peptide-binding assay, is the strongest of all tested (Figure 2A). Other proteins identified by MALDI MS also contain a pADPr binding site that match the refined consensus motif, including DNA replication licensing factor MCM5, Cullin 1 and Cullin 4A. Cell-cycle regulators CDK1 and CDK2 are also noteworthy identifications as well as Caspase-10. The identification of electrophoretically transferred, two-dimensionally separated proteins by MS suffers important limitations since high molecular weight proteins are particularly difficult to analyze this way and multiple co-migrating proteins in single spot can lead to ambiguous identifications.
Figure 3.
Two-dimensional polymer blot analysis. HeLa cell extracts were separated by tube-gel isoelectric focusing, resolved by SDS–PAGE and transferred onto PVDF membrane. The left panel shows the bidimensional protein separation as revealed by silver staining. The right-hand panel shows a corresponding two-dimensional-gel transferred onto a PVDF membrane for polymer blot analysis. Incubation with 32P-pADPr followed by autoradiography reveals a binding-protein pattern. Numbered spots were excised for identification by peptide mass fingerprinting (see Table 2 for complete spot identifications).
Table 2.
Identification of pADPr binding proteins by MALDI-TOF MS
| Spot number | Swiss-Prot accession | Protein description | Protein function |
|---|---|---|---|
| 1 | P10155 | 60 kDa SS-A/Ro ribonucleoprotein | RNA transcription |
| Q9Y6M9 | NADH dehydrogenase 1 beta subcomplex subunit 9 | Mitochondrial electron transport | |
| O76080 | AN1-type zinc-finger protein 5 | May be involved in transcriptional regulation | |
| 2 | P09651 | Heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) | RNA transcription |
| 3 | P17098 | Zinc-finger protein 8 | May be involved in transcriptional regulation |
| P56202 | Cathepsin W | Cysteine-type peptidase activity | |
| P23193 | Transcription elongation factor A protein 1 | RNA transcription | |
| P06493 | Cell-division control protein 2 homolog (CDK1) | Cell-cycle progression and RNA transcription | |
| P24941 | Cell-division protein kinase 2 (CDK2) | Cell-cycle progression and RNA transcription | |
| 4 | P09651 | Heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) | RNA transcription |
| P17038 | Zinc-finger protein 43 | May be involved in transcriptional regulation | |
| 5 | P22626 | Heterogeneous nuclear ribonucleoproteins A2/B1 | RNA transcription |
| 6 | P51523 | Zinc-finger protein 84 | May be involved in transcriptional regulation |
| P17024 | Zinc-finger protein 20 | May be involved in transcriptional regulation | |
| P50458 | LIM/homeobox protein Lhx2 | RNA transcription | |
| O00257 | E3 SUMO-protein ligase CBX4 (Polycomb 2 homolog) | RNA transcription | |
| P22626 | Heterogeneous nuclear ribonucleoproteins A2/B1 | RNA transcription | |
| 7 | P22626 | Heterogeneous nuclear ribonucleoproteins A2/B1 | RNA transcription |
| 8 | Q9UK10 | Zinc-finger protein 225 | May be involved in transcriptional regulation |
| P33992 | DNA replication licensing factor MCM5 | DNA replication | |
| O14628 | Zinc-finger protein 195 | May be involved in transcriptional regulation | |
| P51523 | Zinc-finger protein 84 | May be involved in transcriptional regulation | |
| 9 | No identification | ||
| 10 | P04720 | Elongation factor 1-alpha 1 | Translational elongation |
| 11 | P06213 | Insulin receptor | Insulin receptor signaling pathway |
| P20592 | Interferon-induced GTP-binding protein Mx2 | GTPase activity | |
| P11171 | Protein 4.1 | structural constituent of cytoskeleton | |
| Q02928 | Cytochrome P450 4A11 | Fatty acid metabolic process | |
| P31948 | Stress-induced-phosphoprotein 1 | Response to stress | |
| P50458 | LIM/homeobox protein Lhx2 | RNA transcription | |
| 12 | P31948 | Stress-induced-phosphoprotein 1 | Response to stress |
| 13 | Q01742 | Fibroblast growth factor receptor 2 | Fibroblast growth factor receptor activity |
| Q92851 | Caspase-10 [Precursor] | Induction of apoptosis | |
| P09651 | Heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) | RNA transcription | |
| 14 | Q9NZL3 | Zinc-finger protein 224 | May be involved in transcriptional regulation |
| 15 | No identification | ||
| 16 | No identification | ||
| 17 | No identification | ||
| 18 | No identification | ||
| 19 | P18206 | Vinculin | Cell adhesion |
| P26232 | Catenin alpha-2 | Cell adhesion | |
| Q13616 | Cullin-1 | G1/S transition of mitotic cell cycle | |
| Q14764 | Major vault protein | Vaults structure | |
| O14983 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | Calcium ion transport | |
| 20 | No identification | ||
| 21 | No identification | ||
| 22 | No identification |
HeLa cell extracts were separated by two-dimensional gel electrophoresis and the spots corresponding to pADPr binding proteins were revealed by polymer blot analysis. See Figure 3 for spot references.
Immunoprecipitation of pADPr-associated proteins following alkylation-induced DNA damage
The large-scale validation and identification of pADPr binding proteins mostly relied on polymer blot analysis projects based on one- or two-dimensional electrophoresis, both techniques requiring heavy denaturation steps for protein separation. An alternative approach to explore the nature of the interaction between cellular proteins and pADPr under more physiological conditions and with increased dynamic range is to use specific anti-pADPr antibodies to immunoprecipitate pADPr-associated proteins and pADPr-containing protein complexes. However, even if pADPr synthesis can be achieved by several PARPs, basal levels of pADPr in unstimulated cells are extremely low and probably insufficient to allow the identification of pADPr binding proteins in the absence of PARP-1 activation. Therefore, we used genotoxic stress to trigger activation of PARPs and thereby increase pADPr levels by 10- to 500-fold (2), up to concentrations appropriate for affinity purification. Nonetheless, because of the transient nature of pADPr following PARPs activation in response to DNA strand breaks, we needed a model in which pADPr levels would be sustained. Moreover, we needed a protein extraction procedure that would extract nuclear matrix-associated proteins since this structure represents a major site of pADPr metabolism. Indeed, Cardenas-Corona and colleagues (28) have shown that most of the pADPr is associated with the nuclear matrix and possibly tightly embedded within it since DNAse treatments are inefficient to release pADPr. In order to circumvent these two difficulties, pADPr hydrolysis was first significantly reduced by knocking down endogenous PARG in SK-N-SH cells, one of the most extensively studied neuroblastoma cell line used as a model for cytotoxicity, radiobiology and DNA damage response. With respect to the second concern, we performed high salt protein extractions, a procedure that has shown optimal results for the release of chromatin-bound pADPr. Extracts dialyzed against a nondenaturing immunoprecipitation buffer were used to pull-down pADPr and pADPr-associated proteins.
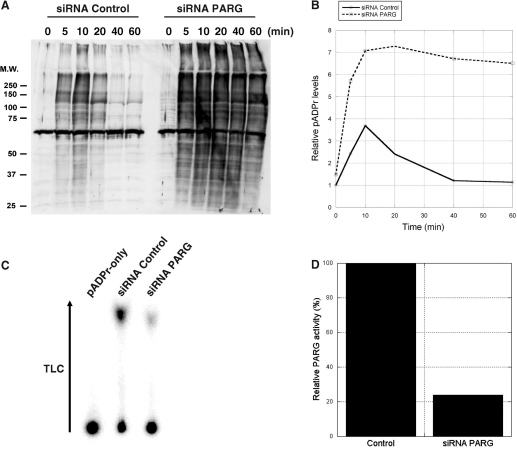
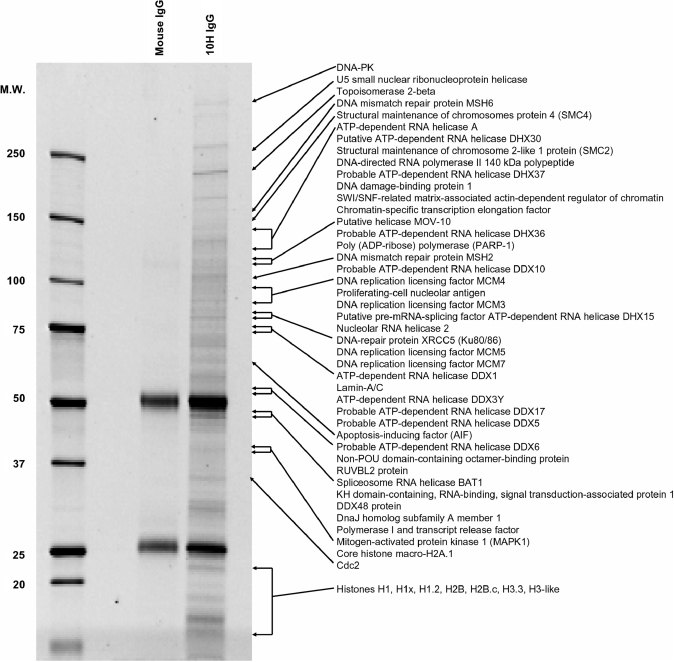
Figure 4A and B show that impaired pADPr turnover in PARG siRNA-treated cells results in sustained levels of pADPr. In contrast, pADPr levels in untreated cells do not increase as much and start decreasing after 10 min. Long-term PARG siRNA treatment (6 days) was necessary to reduce endogenous PARG levels by 80% as estimated from TLC PARG activity assays (Figure 4C and D), a maximum in PARG expression knockdown also reported by Cohausz et al. (29) in a pADPr -metabolizing enzymes study on alkylation-induced cell death. We coupled anti-pADPr mouse monoclonal antibodies (clone 10H) to magnetic Protein-G coated Dynabeads® to selectively pull-down pADPr-associated proteins. Normal mouse IgGs were used as a control reagent for immunoprecipitations using mouse monoclonal antibodies. Immunoprecipitated proteins were stained using SYPRO Ruby fluorescent stain after 1D SDS–PAGE (Figure 5). 10H-immunoprecipitated proteins are distributed across a wide range of molecular weights with minimal nonspecific binding from the control IgGs.
Figure 4.
pADPr levels are increased and sustained in SK-N-SH cells treated with PARG siRNA following alkylation-induced DNA damage. (A) Time course western blot analysis of pADPr accumulation in SK-N-SH cells following 100 μM MNNG treatment in both control and PARG siRNA treated cells. Crude protein extracts were loaded and subjected to SDS–PAGE and immunoblotting. Blots were revealed with anti-pADPr polyclonal antibody 96-10 as described in Materials and methods section. (B) Western blot quantification of pADPr levels from control (full line) and PARG siRNA-treated cells (dashed line) following 100 μM MNNG treatment. (C) Evaluation of PARG knockdown using PARG TLC activity assays. Silencing was conducted over 6 days, passaging cells every 48 h to achieve maximum PARG knockdown. Whole-cell extracts were prepared from cultured cells that had been treated with either control or PARG siRNA. PARG activities were detected by TLC analysis of reaction mixtures containing these cell extracts and 32P-labeled pADPr as a substrate. Substrate remained at the origin of the TLC plate, while ADP-ribose products migrated towards the top of the TLC plate following development by 0.3 M LiCl and 0.9 M ethanoic (acetic) acid. (D) Relative PARG residual activity in SK-N-SH cells after 6-day silencing as determined from the TLC quantification of PARG reaction products.
Figure 5.
SDS–PAGE analysis of pADPr-associated proteins from MNNG-treated and PARG-silenced SK-N-SH cells after immunoprecipitation with anti-pADPr antibodies. pADPr-associated proteins were immunoprecipitated using anti-pADPr mouse monoclonal antibody clone 10H bound to Protein G coated magnetic beads. Immunoprecipitates were resolved by 4–12% SDS–PAGE and stained with SYPRO Ruby fluorescent dye. Normal mouse IgGs were used to assess nonspecific binding. Selected proteins identified by LC-MS/MS, mostly involved in DNA/RNA transactions, are shown (see Supplementary Table S3 for complete protein listing).
LC-MS/MS identification of pADPr-associated proteins from SK-N-SH immunoprecipitates
Immunoprecipitated proteins resolved by SDS–PAGE were manually excised from the gel and identified using LC-MS/MS. More than 300 specific proteins were identified with stringent protein identification probabilities criteria as described in Materials and methods section (Supplementary Table S3). Hallmark proteins of the DNA damage response and repair pathways are collectively over-represented in the pADPr-associated protein dataset (Figure 5), with the notable identification of PARP-1, DNA-PK, XRCC5 (Ku80/86), MSH2, MSH6 and the DNA damage-binding protein 1 (DDB1/Xeroderma pigmentosum group E-complementing protein). There are also several important proteins involved in DNA replication and transcription including DNA replication licensing factors MCM3/MCM4/MCM5/MCM7, and DNA topoisomerase II beta whose corresponding pADPr binding region showed strong interaction with pADPr in polymer blot assays (Figure 2A). The minichromosome maintenance (MCM) proteins are DNA helicases involved in the maintenance of genomic stability in pathways that encompass DNA replication and repair by catalyzing the transient opening of DNA duplexes (30). Other helicases, such as the RNA helicases (DDX, DHX, DEAD/H families) are also remarkably abundant in the pADPr-associated protein dataset.
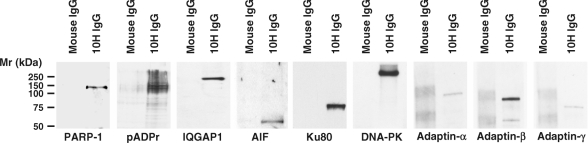
The number of observed peptides by LC-MS/MS for a given protein is semi-quantitative as it often reflects the relative abundance of the protein in the analyzed sample. Predominant proteins were estimated from their number of unique peptides. Supplementary Table S3, which lists all the proteins identified by LC-MS/MS in immunoprecipitates, presents the proteins in order of decreasing number of peptides. Proteins identified with high sequence coverage like PARP-1, IQGAP1, DNA-PK or Ku80 (XRCC5) were validated by western blot (Figure 6). In addition, Figure 6 shows that pADPr is selectively enriched in the pull-down extracts. AIF, which had been identified with only two unique peptides, also appeared with high specificity in western blot, thus validating the MS data. The presence of vesicle-associated adaptins isoforms α, β and γ in pADPr immunoprecipitates was also validated by western blot analysis (Figure 6). Unexpectedly, many vesicular proteins involved in the control of endosomal dynamics, such as the coatomer subunit alpha (COPA) and clathrin-associated adaptor protein AP-2, are also part of the pADPr-associated dataset. It is however noteworthy that adaptor proteins AP-1 and AP-2 as well as golgi-associated proteins are predicted to be pADPr binding proteins based on in silico analysis (Supplementary Table S2). These results provide additional support for a link between poly(ADP-ribosyl)ation and trafficking of endosomal vesicles (31–33).
Figure 6.
Validation of selected pADPr-associated proteins identified by LC-MS/MS using western blot analysis of pADPr immunoprecipitates. The specificity of the pADPr immunoprecipitation using anti-pADPr 10H monoclonal antibodies was evaluated by immunoblot analysis as described in Materials and methods section. The same proteins were not precipitated by normal mouse IgGs, confirming the specificity of the pull-down.
In silico predictions and experimental discoveries are obviously complementary approaches for the exploration of the pADPr interactome. The LC-MS/MS identification of the splicing factor ASF/SF2 (Supplementary Table S3), a recently reported pADPr binding protein (34), in the protein dataset generated from pADPr immunoprecipitates illustrate this complementarity since no consensus motifs were able to predict its pADPr binding.
Modeling of pADPr-associated protein networks
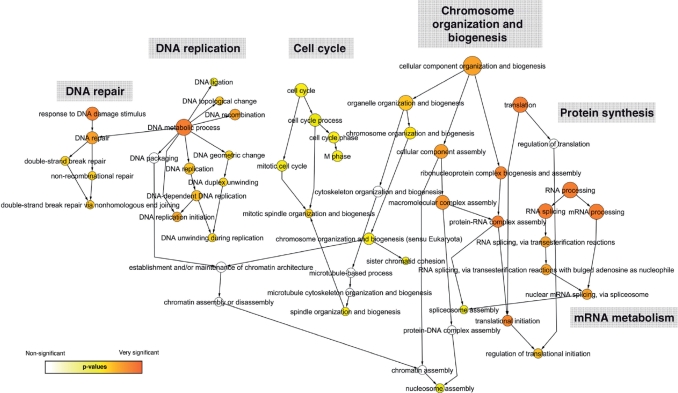
Unorganized datasets containing numerous proteins remain hard to analyze and the possibility to draw conclusions is hampered by the apparent complexity. We used systems biology data mining tools to elucidate the dynamic interplay of more than 300 putative pADPr-associated proteins in our dataset. Based on structured GOs annotations, we first determined which biological process categories were statistically over-represented in the immunoprecipitated pADPr-associated protein dataset. The statistical analysis was performed with a BiNGO (19) implementation in Cytoscape (20). BiNGO is a tool developed to highlight predominant functional themes in a dataset and to visualize them as an integrated molecular interaction network. Figure 7 shows the network distribution of predominant GO terms associated with pADPr-associated proteins in immunoprecipitates. Proteins involved in DNA/RNA transactions are clearly emerging from the protein population as we would expect. Statistically significant over-represented ontologies of pADPr-associated proteins are grouped into six categories that encompass the major PARP-1-dependent pathways (chromosome organization and biogenesis, DNA repair, DNA replication), pADPr-regulated functions (progression through cell cycle) and mRNA metabolism/protein synthesis for which several studies suggest a link with poly(ADP-ribosyl)ation reactions (12,35–38).
Figure 7.
Graphical network representation of pADPr-associated proteins identified by LC-MS/MS in anti-pADPr immunoprecipitates. Functional categorization of pADPr-associated proteins was performed using GO annotations. Over-represented categories were statistically identified using BiNGO and visualized with Cytoscape. The size of the terms (circles) is representative of the proportional protein abundance in the dataset and the color shading indicates the degree of statistical significance (darker shades indicate stronger significance).
A literature mining tool called Bibliosphere (Genomatix Software Inc.) was also used to visualize the relations between proteins based on the degree of interconnection for their corresponding genes (Supplementary Figure S1). In the Bibliosphere generated network, related genes are clustered around ‘hubs’ that play central role in the biological pathways. In this representation, PARP-1 is displayed as the central molecule linking all the pathways. The functional categories revealed by the Bibliosphere network are similar to the results obtained with the GO classification tool. Biological processes, such as structural organization and function of the nucleus and chromatin, cellular response to DNA double-strand breaks, DNA mismatch repair, DNA replication and genome integrity, apoptosis and mRNA metabolism/protein synthesis are predominant and strongly support the classification obtained with BiNGO's molecular interaction graph (Figure 7). BiNGO and Bibliosphere are two independent tools for data integration and visualization that provided converging results. However, they can complement each other and deliver a more global view of the pADPr-associated biological network.
Comparative distribution of pADPr binding candidates in predicted and experimental datasets
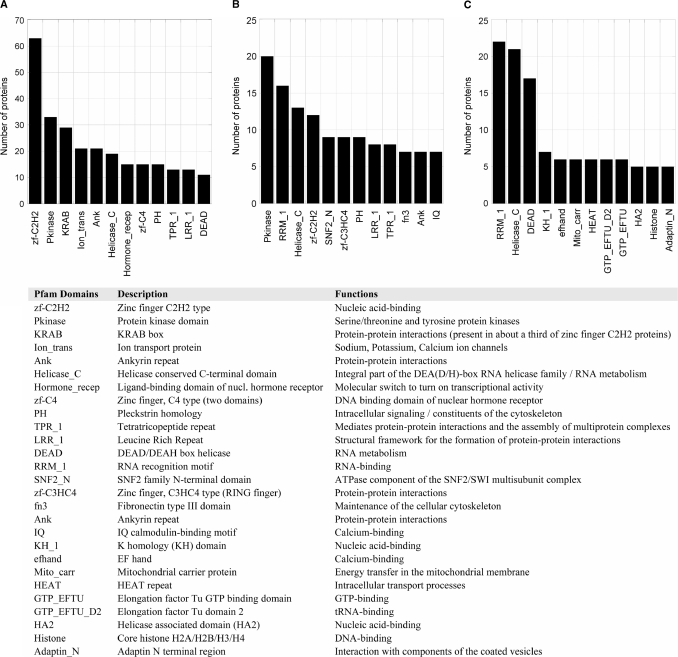
An estimation of the predominant protein domains found within the computationally predicted and experimentally verified pADPr binding protein datasets based on the Pfam protein families database (39) was performed (Figure 8). Clearly, proteins containing the classical zinc-finger motif (zf-C2H2) are over-represented in the in silico protein dataset obtained through searching a pattern with high similarity to the original motif (Figure 8A). However, the proteins revealed by our refined motif are enriched in additional pathways involving nucleic acids binding (Figure 8B). While zinc-finger proteins still represent an important category, RNA recognition motifs (RRM_1) now occur frequently as they do for the experimental dataset (Figure 8C). Interestingly, some members of the zf-C3HC4 (Ring finger) family are also found with this more stringent motif screen. Ring-finger proteins notably includes Rad 5/Rad 18 (40), BRCA1 (41) and RNF8 (42,43), a group of proteins known to mediate cooperation between ubiquitin-conjugating enzymes in DNA repair, and CHFR, a pADPr binding ubiquitin lyase involved in mitotic stress response (6,44). Another protein domain specifically abundant in the dataset generated with the refined pADPr binding motif is the SNF2_N domain (Figure 8B), which is found in proteins involved in a variety of processes including transcription regulation, DNA repair, DNA recombination and chromatin unwinding. Clearly, the in silico protein dataset generated with the refined pADPr binding motif is more closely related to the experimentally generated one. Compared with the dataset generated with the original consensus, both the in silico refined motif and the experimental dataset feature a high proportion of protein motifs related to RNA metabolism (RRM_1, Helicase_C, KH_1, DEAD, H2A) and chromatin-remodeling proteins. That strongly suggests that the refined pADPr binding motif dataset is representative of the actual biological diversity of pADPr binding proteins.
Figure 8.
Distribution of protein domains and families in computationally predicted and experimentally validated pADPr binding proteins. The graph shows the number of occurrences for the 12 most common Pfam domains in (A) putative pADPr binding proteins identified through a 90% similarity sequence search based on the original consensus pADPr binding motif (Supplementary Table S1), (B) a sub-dataset of putative pADPr binding proteins corresponding to the refined pADPr binding module derived from peptides polymer blot analysis (Supplementary Table S2) and (C) pADPr-associated proteins identified by MS in anti-pADPr immunoprecipitation assays in SK-N-SH cells following alkylation-induced DNA damage (Supplementary Table S3).
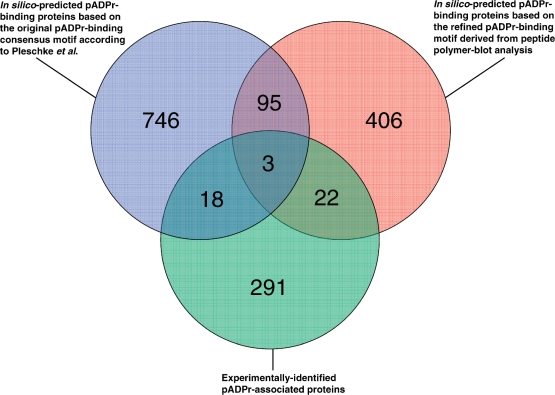
From the comparative evaluation of in silico-based prediction of pADPr binding proteins and pADPr-associated proteins found in pADPr immunoprecipitates, one can conclude that both the original and the refined consensus pADPr binding motif are appropriate for the prediction of pADPr binding proteins since both motifs are represented in about the same number of experimentally identified proteins sequences. This can be seen on the Venn diagram displaying the distribution of pADPr binding candidates among the three approaches selected for this study (Figure 9). At first look, it might seem that the number of experimentally validated proteins is low compared with the large number of predicted pADPr binding proteins. However, that statement should be put into context. First, in silico analysis only considers primary amino acid sequences; that is, although the use of a consensus sequence for database searching does improve the likelihood of detecting pADPr binding substrates, it does not take into account the structural determinants required for protein–pADPr interaction and hence leads to some false positives. Second, in silico predictions are performed over the entire human proteome without considering cell- or tissue-specific protein expression. Third, MS is not able to detect proteins of very low absolute or relative abundance whereas large dynamic range and low protein concentrations do not affect computer-based predictions. Finally, proteins that are part of stable pADPr-containing protein complexes or making strong interactions with proteins that are capable of pADPr binding proteins will not appear in predicted datasets but will likely be identified using experimental approaches. More rigorous studies will be required to improve prediction reliability for pADPr binding proteins and reach a lower rate of false positive identifications. Even if the protein datasets predicted by the two pADPr binding motifs both share a bias towards nucleic acid metabolism, only 98 proteins are common to both and their limited overlap makes them complementary. On the other side, 43 proteins identified empirically have a pADPr binding motif. This largely outnumbers any other studies that attempted to determine potential binding partners of pADPr. Moreover, we identified several other pADPr-associated proteins that can either be covalently poly(ADP-ribosyl)ated or noncovalent binders of pADPr without matching a typical pADPr binding motif [e.g. PARP-1, DNA-PK, XRCC5 (Ku86), AIF]. Collectively, these pADPr-associated proteins represent the most comprehensive and detailed information related to pADPr binding reported so far.
Figure 9.
Venn diagram illustrating the overlap between the three pADPr binding candidates datasets described in this study. Overlapping circles shows the distribution of in silico predicted proteins revealed through the use of the consensus pADPr binding motif proposed by Pleschke et al. (3) and the refined pADPr binding motif derived from sequence alignment of experimentally validated pADPr binding regions from peptide polymer blot analysis. The relationship between the two in silico prediction approaches and experimental identification of pADPr-associated proteins in pADPr immunoprecipitates is illustrated as a third intercepting circle.
DISCUSSION
The experimental demonstration of pADPr binding has traditionally been challenging because of the complexity of interactions between proteins and pADPr, and because of the transient nature of the pADPr-mediated biological response. With technological advances enabling biological questions to be addressed proteome-wide, the entire pADPr interactome is now amenable for scientific investigation. This is particularly useful in the context of poly(ADP-ribosyl)ation, since this phenomenon potentially influences many different cell functions through molecular interactions with numerous pADPr binding substrates (summarized in Table 3).
Table 3.
A summary of the major pADPr-regulated pathways
| Biological processes | pADPr binding and pADPr-associated proteins | Functions | References |
|---|---|---|---|
| DNA damage signaling and DNA repair | DNA-PK, XRCC1, XRCC5 (Ku86), XRCC6 (Ku70), MSH2, MSH6, DNA damage-binding protein 1 (Xeroderma pigmentosum group E-complementing protein), DNA excision repair protein ERCC-6, BRCA1, DNA repair and recombination protein RAD54-like, hnRNPs, Non-POU domain-containing octamer-binding protein (54 kDa nuclear RNA- and DNA-binding protein (PSF/p54(nrb))), DNA-repair protein complementing XP-C, alkylated DNA repair protein alkB homolog 4, XPA, p53, Mre11, ATM, p21, DNA ligase 3, Aprataxin PNK-like factor (APLF), DNA cross-link repair 1A protein (SMN1) | Detection and early response to DNA strand-breaks, DNA repair | (3,7,45–56) |
| Modification of chromatin structure | Condensin complex subunit 1 (hCAP-D2/Eg7), SMC2, SMC4, Chromobox protein homolog 1/5, Lamin-A/C, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5, Histones, Chromodomain-helicase-DNA-binding protein 1/5 | Compaction and decondensation of chromatin | (57–60) |
| Transcription | Telomerase reverse transcriptase, helicases, chromatin-specific transcription elongation factor FACT 140 kDa subunit, Transcriptional regulator ATRX, transcription intermediary factor 1-beta, probable global transcription activator SNF2L1, transcriptional activator protein Pur-alpha, transcription factor B1 (mitochondrial), activated RNA polymerase II transcription cofactor 4 variant, polymerase I and transcript release factor, Activating signal cointegrator 1 complex subunit 3, DNA-directed RNA polymerase II 140 kDa polypeptide, RUVBL2 protein, EBNA-2 co-activator variant, RHA helicase A, nucleolar RNA helicase 2, U5 snRNP-specific 116 kDa protein, U5 snRNP-specific 200 kDa protein, hnRNPs, DEAD-box RNA helicases p72 (DDX17), transcriptional repressor protein YY1, PARP-9/BAL1, PARP-14/BAL2, PARP-15/BAL3, HMG proteins, transcription factor SP1, Histone acetyltransferase p300, histone acetyltransferase PCAF | DNA-directed RNA synthesis | (24–26,61–71) |
| Replication | Helicases, MCM3, MCM4, MCM5, MCM7, DNA primase, DNA topoisomerase, DNA polymerase, replication factor C subunit 4, replication factor C subunit 5, telomerase reverse transcriptase | Synthesis of DNA strands from a parent molecule | (2,72) |
| RNA metabolism, RNA splicing and protein synthesis | Helicases, hnRNPs, SFQP, splicing factor 3B subunit 3, pre-mRNA-splicing factor SYF1, splicing factor 3B subunit 1, putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15, splicing factor, arginine/serine-rich 3, RNA-binding protein PNO1, SF2/ASF-1, pre-mRNA-splicing factor 19, regulator of nonsense transcripts 1, cytoplasmic FMR1-interacting protein 1, nuclease sensitive element-binding protein 1, spliceosome RNA helicase BAT1, mRNA turnover protein 4 homolog, elongation factor 1-alpha 1, elongation factor 2, polyadenylate-binding protein 1 | Pre-mRNA processing, transport, localization, mRNA stability | (12,35–38,73–78) |
| Cell death | AIF, hexokinase-1, HKDC1, cell-division cycle and apoptosis regulator protein 1, deleted in breast cancer gene 1 protein (DBC-1), BID, Caspase-10, prohibitin, Mitochondrial inner membrane protein (mitofilin), DEK, death-associated protein kinase 1/2/3, CAD | Regulation of cell survival, apoptosis | (24,79–85) |
| Cell-cycle and mitosis | Cell-division cycle and apoptosis regulator protein 1, Cullin-1, Cullin 4A, CDK1 (Cdc2), CDK2, Aurora-A kinase interacting protein, Cenp-A,Cenp-B, Cenp-T, spindle assembly checkpoint protein MAD1, NIMA-related protein kinase Nek10, protein regulator of cytokinesis 1, BUB3, centrosomal protein of 192 kDa, cell-division control protein 6 homolog, cell-division cycle-associated 7-like protein, cell-cycle checkpoint protein RAD17, nucleolar and spindle-associated protein 1, CHFR | Progression through cell-cycle, spindle assembly and structure, cell-cycle checkpoints | (6,86–93) |
Selected pADPr binding protein candidates revealed from in silico prediction-based or experimental analysis (bold) are proposed for each of the pathways.
The greatest advantage of a proteome-wide study like the one presented here lies in the acceleration of the pace at which pADPr binding candidates are discovered compared with traditional approaches. Although these candidates will require additional validation, their disclosure opens up considerable opportunity for new hypothesis-driven experiments.
Our understanding of poly(ADP-ribosyl)ation-related phenomena should grow more rapidly as studies target proteins with a putative pADPr binding motif, although motif prediction presents difficulties as matches are not strictly conserved. Pattern screening remains a valuable tool to identify interesting targets and their most probable pADPr binding region. This is especially true if one uses our refined pADPr binding motif as it generates a more restricted set of candidates than the original motif and better correlates with experimentally identified pADPr-associated proteins.
We believe that this work represents a valuable investigation into the pADPr interactome and reveals the potential of pADPr to impact a wide range of critical biological functions. By combining bioinformatics-based predictions, pADPr binding peptide blot assays, two-dimensional electrophoresis and large-scale LC-MS/MS identification of immunoprecipitated pADPr-associated proteins, this study represents the first large-scale proteomic identification of pADPr binding proteins and provides insights into the pathways that can be modulated by poly(ADP-ribosyl)ation.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
FUNDING
United States Public Health Services and National Institute of Neurological Disorders and Stroke (USPHS NINDS 039148); Canadian Institutes for Health Research (CIHR MOP-14052 and MOP-74648); scholarship from Fonds de la Recherche en Santé du Québec (FRSQ) (to J.P.G.); Strategic Training Program grant in genomics, proteomics and bioinformatics (CIHR STP-53894 to J.P.G.). Funding for open access charge: Canadian Institutes of Health Research (CIHR).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Christophe Combet (UMR 5086 CNRS, France) for help with PattInPROT and Pierre Gagné for critical reading of the article. We also thank Joanna M. Hunter for MALDI-TOF MS analysis and Louis M. Nicole for technical help with 2D-gels.
REFERENCES
- 1.Nishizuka Y, Ueda K, Honjo T, Hayaishi O. Enzymic adenosine diphosphate ribosylation of histone and poly adenosine diphosphate ribose synthesis in rat liver nuclei. J. Biol. Chem. 1968;243:3765–3767. [PubMed] [Google Scholar]
- 2.D’Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 3.Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 4.Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, Ladurner AG. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egloff MP, Malet H, Putics A, Heinonen M, Dutartre H, Frangeul A, Gruez A, Campanacci V, Cambillau C, Ziebuhr J, et al. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 2006;80:8493–8502. doi: 10.1128/JVI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 7.Fahrer J, Kranaster R, Altmeyer M, Marx A, Burkle A. Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Res. 2007;35:e143. doi: 10.1093/nar/gkm944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M, Klaus JA, Otsuka T, Zhang Z, Koehler RC, et al. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc. Natl Acad. Sci. USA. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: network protein sequence analysis. Trends Biochem. Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 10.O’Farrell PH. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 11.Mortz E, Krogh TN, Vorum H, Gorg A. Improved silver staining protocols for high sensitivity protein identification using matrix-assisted laser desorption/ionization-time of flight analysis. Proteomics. 2001;1:1359–1363. doi: 10.1002/1615-9861(200111)1:11<1359::AID-PROT1359>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 12.Gagne JP, Hunter JM, Labrecque B, Chabot B, Poirier GG. A proteomic approach to the identification of heterogeneous nuclear ribonucleoproteins as a new family of poly(ADP-ribose)-binding proteins. Biochem. J. 2003;371:331–340. doi: 10.1042/BJ20021675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menard L, Poirier GG. Rapid assay of poly(ADP-ribose) glycohydrolase. Biochem. Cell Biol. 1987;65:668–673. doi: 10.1139/o87-088. [DOI] [PubMed] [Google Scholar]
- 14.Zahradka P, Ebisuzaki K. Poly(ADP-ribose) polymerase is a zinc metalloenzyme. Eur. J. Biochem. 1984;142:503–509. doi: 10.1111/j.1432-1033.1984.tb08314.x. [DOI] [PubMed] [Google Scholar]
- 15.Brochu G, Duchaine C, Thibeault L, Lagueux J, Shah GM, Poirier GG. Mode of action of poly(ADP-ribose) glycohydrolase. Biochim. Biophys. Acta. 1994;1219:342–350. doi: 10.1016/0167-4781(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 16.Shah GM, Poirier D, Duchaine C, Brochu G, Desnoyers S, Lagueux J, Verreault A, Hoflack JC, Kirkland JB, Poirier GG. Methods for biochemical study of poly(ADP-ribose) metabolism in vitro and in vivo. Anal. Biochem. 1995;227:1–13. doi: 10.1006/abio.1995.1245. [DOI] [PubMed] [Google Scholar]
- 17.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 18.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 19.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 20.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, Sunshine M, Narasimhan S, Kane DW, Reinhold WC, Lababidi S, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, Copley R, et al. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224–D228. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panzeter PL, Zweifel B, Malanga M, Waser SH, Richard M, Althaus FR. Targeting of histone tails by poly(ADP-ribose) J. Biol. Chem. 1993;268:17662–17664. [PubMed] [Google Scholar]
- 24.Rouleau M, Aubin RA, Poirier GG. Poly(ADP-ribosyl)ated chromatin domains: access granted. J. Cell Sci. 2004;117:815–825. doi: 10.1242/jcs.01080. [DOI] [PubMed] [Google Scholar]
- 25.Hassa PO, Haenni SS, Buerki C, Meier NI, Lane WS, Owen H, Gersbach M, Imhof R, Hottiger MO. Acetylation of poly(ADP-ribose) polymerase-1 by p300/CREB-binding protein regulates coactivation of NF-kappaB-dependent transcription. J. Biol. Chem. 2005;280:40450–40464. doi: 10.1074/jbc.M507553200. [DOI] [PubMed] [Google Scholar]
- 26.Haenni SS, Hassa PO, Altmeyer M, Fey M, Imhof R, Hottiger MO. Identification of lysines 36 and 37 of PARP-2 as targets for acetylation and auto-ADP-ribosylation. Int. J. Biochem. Cell Biol. 2008;40:2274–2283. doi: 10.1016/j.biocel.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 27.Kappes F, Fahrer J, Khodadoust MS, Tabbert A, Strasser C, Mor-Vaknin N, Moreno-Villanueva M, Burkle A, Markovitz DM, Ferrando-May E. DEK is a poly(ADP-ribose) acceptor in apoptosis and mediates resistance to genotoxic stress. Mol. Cell. Biol. 2008;28:3245–3257. doi: 10.1128/MCB.01921-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cardenas-Corona ME, Jacobson EL, Jacobson MK. Endogenous polymers of ADP-ribose are associated with the nuclear matrix. J. Biol. Chem. 1987;262:14863–14866. [PubMed] [Google Scholar]
- 29.Cohausz O, Blenn C, Malanga M, Althaus FR. The roles of poly(ADP-ribose)-metabolizing enzymes in alkylation-induced cell death. Cell Mol. Life Sci. 2008;65:644–655. doi: 10.1007/s00018-008-7516-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forsburg SL. The MCM helicase: linking checkpoints to the replication fork. Biochem. Soc. Trans. 2008;36:114–119. doi: 10.1042/BST0360114. [DOI] [PubMed] [Google Scholar]
- 31.Chi NW, Lodish HF. Tankyrase is a golgi-associated mitogen-activated protein kinase substrate that interacts with IRAP in GLUT4 vesicles. J. Biol. Chem. 2000;275:38437–38444. doi: 10.1074/jbc.M007635200. [DOI] [PubMed] [Google Scholar]
- 32.Sbodio JI, Lodish HF, Chi NW. Tankyrase-2 oligomerizes with tankyrase-1 and binds to both TRF1 (telomere-repeat-binding factor 1) and IRAP (insulin-responsive aminopeptidase) Biochem. J. 2002;361:451–459. doi: 10.1042/0264-6021:3610451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeh TY, Sbodio JI, Tsun ZY, Luo B, Chi NW. Insulin-stimulated exocytosis of GLUT4 is enhanced by IRAP and its partner tankyrase. Biochem. J. 2007;402:279–290. doi: 10.1042/BJ20060793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malanga M, Czubaty A, Girstun A, Staron K, Althaus FR. Poly(ADP-ribose) binds to the splicing factor ASF/SF2 and regulates its phosphorylation by DNA topoisomerase I. J. Biol. Chem. 2008;283:19991–19998. doi: 10.1074/jbc.M709495200. [DOI] [PubMed] [Google Scholar]
- 35.Elkaim R, Thomassin H, Niedergang C, Egly JM, Kempf J, Mandel P. Adenosine diphosphate ribosyltransferase and protein acceptors associated with cytoplasmic free messenger ribonucleoprotein particles. Biochimie. 1983;65:653–659. doi: 10.1016/s0300-9084(84)80029-2. [DOI] [PubMed] [Google Scholar]
- 36.Thomassin H, Martins de Sa C, Scherrer K, Maniez C, Mandel P. Cytoplasmic poly(ADP-ribose) polymerase and poly(ADP-ribose) glycohydrolase in AEV-transformed chicken erythroblasts. Mol. Biol. Rep. 1988;13:35–44. doi: 10.1007/BF00805637. [DOI] [PubMed] [Google Scholar]
- 37.Jesser M, Chypre C, Hog F, Mandel P. Cytoplasmic poly(ADP-ribose)polymerase from mouse plasmacytoma free messenger ribonucleoprotein particles: purification and characterization. Biochem. Biophys. Res. Commun. 1993;195:558–564. doi: 10.1006/bbrc.1993.2082. [DOI] [PubMed] [Google Scholar]
- 38.Tanuma S, Endo H. Purification and characterization of an (ADP-ribose)n glycohydrolase from human erythrocytes. Eur. J. Biochem. 1990;191:57–63. doi: 10.1111/j.1432-1033.1990.tb19093.x. [DOI] [PubMed] [Google Scholar]
- 39.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ulrich HD, Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000;19:3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Starita LM, Parvin JD. The multiple nuclear functions of BRCA1: transcription, ubiquitination and DNA repair. Curr. Opin. Cell Biol. 2003;15:345–350. doi: 10.1016/s0955-0674(03)00042-5. [DOI] [PubMed] [Google Scholar]
- 42.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 44.Chaturvedi P, Sudakin V, Bobiak ML, Fisher PW, Mattern MR, Jablonski SA, Hurle MR, Zhu Y, Yen TJ, Zhou BB. Chfr regulates a mitotic stress pathway through its RING-finger domain with ubiquitin ligase activity. Cancer Res. 2002;62:1797–1801. [PubMed] [Google Scholar]
- 45.Haince JF, McDonald D, Rodrigue A, Dery U, Masson JY, Hendzel MJ, Poirier GG. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 2008;283:1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 46.Haince JF, Kozlov S, Dawson VL, Dawson TM, Hendzel MJ, Lavin MF, Poirier GG. Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. J. Biol. Chem. 2007;282:16441–16453. doi: 10.1074/jbc.M608406200. [DOI] [PubMed] [Google Scholar]
- 47.Malanga M, Althaus FR. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem. Cell Biol. 2005;83:354–364. doi: 10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- 48.Ruscetti T, Lehnert BE, Halbrook J, Le Trong H, Hoekstra MF, Chen DJ, Peterson SR. Stimulation of the DNA-dependent protein kinase by poly(ADP-ribose) polymerase. J. Biol. Chem. 1998;273:14461–14467. doi: 10.1074/jbc.273.23.14461. [DOI] [PubMed] [Google Scholar]
- 49.Marti TM, Kunz C, Fleck O. DNA mismatch repair and mutation avoidance pathways. J. Cell. Physiol. 2002;191:28–41. doi: 10.1002/jcp.10077. [DOI] [PubMed] [Google Scholar]
- 50.Wittschieben BO, Wood RD. DDB complexities. DNA Repair. 2003;2:1065–1069. doi: 10.1016/s1568-7864(03)00113-7. [DOI] [PubMed] [Google Scholar]
- 51.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 52.Carpenter B, MacKay C, Alnabulsi A, MacKay M, Telfer C, Melvin WT, Murray GI. The roles of heterogeneous nuclear ribonucleoproteins in tumour development and progression. Biochim. Biophys. Acta. 2006;1765:85–100. doi: 10.1016/j.bbcan.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 53.Bouchard VJ, Rouleau M, Poirier GG. PARP-1, a determinant of cell survival in response to DNA damage. Exp. Hematol. 2003;31:446–454. doi: 10.1016/s0301-472x(03)00083-3. [DOI] [PubMed] [Google Scholar]
- 54.Bladen CL, Udayakumar D, Takeda Y, Dynan WS. Identification of the polypyrimidine tract binding protein-associated splicing factor.p54(nrb) complex as a candidate DNA double-strand break rejoining factor. J. Biol. Chem. 2005;280:5205–5210. doi: 10.1074/jbc.M412758200. [DOI] [PubMed] [Google Scholar]
- 55.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rulten SL, Cortes-Ledesma F, Guo L, Iles NJ, Caldecott KW. APLF (C2orf13) is a novel component of poly(ADP-ribose) signaling in mammalian cells. Mol. Cell. Biol. 2008;28:4620–4628. doi: 10.1128/MCB.02243-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heale JT, Ball ARJr, Schmiesing JA, Kim JS, Kong X, Zhou S, Hudson DF, Earnshaw WC, Yokomori K. Condensin I interacts with the PARP-1-XRCC1 complex and functions in DNA single-strand break repair. Mol. Cell. 2006;21:837–848. doi: 10.1016/j.molcel.2006.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hirano T. At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- 59.Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008;22:832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Faraone-Mennella MR. Chromatin architecture and functions: the role(s) of poly(ADP-RIBOSE) polymerase and poly(ADPribosyl)ation of nuclear proteins. Biochem. Cell Biol. 2005;83:396–404. doi: 10.1139/o05-042. [DOI] [PubMed] [Google Scholar]
- 61.Heo K, Kim H, Choi SH, Choi J, Kim K, Gu J, Lieber MR, Yang AS, An W. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol. Cell. 2008;30:86–97. doi: 10.1016/j.molcel.2008.02.029. [DOI] [PubMed] [Google Scholar]
- 62.Wacker DA, Frizzell KM, Zhang T, Kraus WL. Regulation of chromatin structure and chromatin-dependent transcription by poly(ADP-ribose) polymerase-1: possible targets for drug-based therapies. Subcell. Biochem. 2007;41:45–69. doi: 10.1007/1-4020-5466-1_3. [DOI] [PubMed] [Google Scholar]
- 63.Ha HC, Hester LD, Snyder SH. Poly(ADP-ribose) polymerase-1 dependence of stress-induced transcription factors and associated gene expression in glia. Proc. Natl Acad. Sci. USA. 2002;99:3270–3275. doi: 10.1073/pnas.052712399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mattiussi S, Tempera I, Matusali G, Mearini G, Lenti L, Fratarcangeli S, Mosca L, D’Erme M, Mattia E. Inhibition of Poly(ADP-ribose)polymerase impairs Epstein Barr Virus lytic cycle progression. Infect. Agent Cancer. 2007;2:18. doi: 10.1186/1750-9378-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deng Z, Atanasiu C, Zhao K, Marmorstein R, Sbodio JI, Chi NW, Lieberman PM. Inhibition of Epstein-Barr virus OriP function by tankyrase, a telomere-associated poly-ADP ribose polymerase that binds and modifies EBNA1. J. Virol. 2005;79:4640–4650. doi: 10.1128/JVI.79.8.4640-4650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen Y, Schnetz MP, Irarrazabal CE, Shen RF, Williams CK, Burg MB, Ferraris JD. Proteomic identification of proteins associated with the osmoregulatory transcription factor TonEBP/OREBP: functional effects of Hsp90 and PARP-1. Am. J. Physiol. Renal Physiol. 2007;292:F981–F992. doi: 10.1152/ajprenal.00493.2005. [DOI] [PubMed] [Google Scholar]
- 67.Ko BC, Turck CW, Lee KW, Yang Y, Chung SS. Purification, identification, and characterization of an osmotic response element binding protein. Biochem. Biophys. Res. Commun. 2000;270:52–61. doi: 10.1006/bbrc.2000.2376. [DOI] [PubMed] [Google Scholar]
- 68.Kraus WL, Lis JT. PARP goes transcription. Cell. 2003;113:677–683. doi: 10.1016/s0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- 69.Aguiar RC, Takeyama K, He C, Kreinbrink K, Shipp MA. B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. J. Biol. Chem. 2005;280:33756–33765. doi: 10.1074/jbc.M505408200. [DOI] [PubMed] [Google Scholar]
- 70.Huang JY, Chen WH, Chang YL, Wang HT, Chuang WT, Lee SC. Modulation of nucleosome-binding activity of FACT by poly(ADP-ribosyl)ation. Nucleic Acids Res. 2006;34:2398–2407. doi: 10.1093/nar/gkl241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zaniolo K, Desnoyers S, Leclerc S, Guerin SL. Regulation of poly(ADP-ribose) polymerase-1 (PARP-1) gene expression through the post-translational modification of Sp1: a nuclear target protein of PARP-1. BMC Mol. Biol. 2007;8:96. doi: 10.1186/1471-2199-8-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Simbulan-Rosenthal CM, Rosenthal DS, Boulares AH, Hickey RJ, Malkas LH, Coll JM, Smulson ME. Regulation of the expression or recruitment of components of the DNA synthesome by poly(ADP-ribose) polymerase. Biochemistry. 1998;37:9363–9370. doi: 10.1021/bi9731089. [DOI] [PubMed] [Google Scholar]
- 73.Zhang S, Schlott B, Gorlach M, Grosse F. DNA-dependent protein kinase (DNA-PK) phosphorylates nuclear DNA helicase II/RNA helicase A and hnRNP proteins in an RNA-dependent manner. Nucleic Acids Res. 2004;32:1–10. doi: 10.1093/nar/gkg933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gagne JP, Bonicalzi ME, Gagne P, Ouellet ME, Hendzel MJ, Poirier GG. Poly(ADP-ribose) glycohydrolase is a component of the FMRP-associated messenger ribonucleoparticles. Biochem. J. 2005;392:499–509. doi: 10.1042/BJ20050792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fakan S, Leduc Y, Lamarre D, Brunet G, Poirier GG. Immunoelectron microscopical distribution of poly(ADP-ribose)polymerase in the mammalian cell nucleus. Exp. Cell Res. 1988;179:517–526. doi: 10.1016/0014-4827(88)90289-3. [DOI] [PubMed] [Google Scholar]
- 76.Tran EJ, Zhou Y, Corbett AH, Wente SR. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol. Cell. 2007;28:850–859. doi: 10.1016/j.molcel.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 77.Chypre C, Maniez C, Mandel P. Cytoplasmic poly(ADP-ribose) polymerase associated with free messenger ribonucleoprotein particles in rat brain. J. Neurochem. 1988;51:561–565. doi: 10.1111/j.1471-4159.1988.tb01075.x. [DOI] [PubMed] [Google Scholar]
- 78.Chypre C, Le Calvez C, Hog F, Revel MO, Jesser M, Mandel P. Phosphorylation of cytoplasmic poly (ADP-ribose) polymerase linked to free ribonucleoprotein particles by an associated protein kinase C. C. R. Acad. Sci. III. 1989;309:471–476. [PubMed] [Google Scholar]
- 79.Azoulay-Zohar H, Israelson A, Abu-Hamad S, Shoshan-Barmatz V. In self-defence: hexokinase promotes voltage-dependent anion channel closure and prevents mitochondria-mediated apoptotic cell death. Biochem. J. 2004;377:347–355. doi: 10.1042/BJ20031465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lai Y, Chen Y, Watkins SC, Nathaniel PD, Guo F, Kochanek PM, Jenkins LW, Szabo C, Clark RS. Identification of poly-ADP-ribosylated mitochondrial proteins after traumatic brain injury. J. Neurochem. 2008;104:1700–1711. doi: 10.1111/j.1471-4159.2007.05114.x. [DOI] [PubMed] [Google Scholar]
- 81.Landshamer S, Hoehn M, Barth N, Duvezin-Caubet S, Schwake G, Tobaben S, Kazhdan I, Becattini B, Zahler S, Vollmar A, et al. Bid-induced release of AIF from mitochondria causes immediate neuronal cell death. Cell Death Differ. 2008;15:1553–1563. doi: 10.1038/cdd.2008.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Le Bras M, Rouy I, Brenner C. The modulation of inter-organelle cross-talk to control apoptosis. Med. Chem. 2006;2:1–12. doi: 10.2174/157340606775197787. [DOI] [PubMed] [Google Scholar]
- 83.Mishra S, Murphy LC, Murphy LJ. The Prohibitins: emerging roles in diverse functions. J. Cell. Mol. Med. 2006;10:353–363. doi: 10.1111/j.1582-4934.2006.tb00404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sundararajan R, Chen G, Mukherjee C, White E. Caspase-dependent processing activates the proapoptotic activity of deleted in breast cancer-1 during tumor necrosis factor-alpha-mediated death signaling. Oncogene. 2005;24:4908–4920. doi: 10.1038/sj.onc.1208681. [DOI] [PubMed] [Google Scholar]
- 85.Yu SW, Andrabi SA, Wang H, Kim NS, Poirier GG, Dawson TM, Dawson VL. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc. Natl Acad. Sci. USA. 2006;103:18314–18319. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kanai M, Tong WM, Wang ZQ, Miwa M. Haploinsufficiency of poly(ADP-ribose) polymerase-1-mediated poly(ADP-ribosyl)ation for centrosome duplication. Biochem. Biophys. Res. Commun. 2007;359:426–430. doi: 10.1016/j.bbrc.2007.05.108. [DOI] [PubMed] [Google Scholar]
- 87.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 88.Fu J, Bian M, Jiang Q, Zhang C. Roles of Aurora kinases in mitosis and tumorigenesis. Mol. Cancer Res. 2007;5:1–10. doi: 10.1158/1541-7786.MCR-06-0208. [DOI] [PubMed] [Google Scholar]
- 89.Monaco L, Kolthur-Seetharam U, Loury R, Murcia JM, de Murcia G, Sassone-Corsi P. Inhibition of Aurora-B kinase activity by poly(ADP-ribosyl)ation in response to DNA damage. Proc. Natl Acad. Sci. USA. 2005;102:14244–14248. doi: 10.1073/pnas.0506252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Saxena A, Saffery R, Wong LH, Kalitsis P, Choo KH. Centromere proteins Cenpa, Cenpb, and Bub3 interact with poly(ADP-ribose) polymerase-1 protein and are poly(ADP-ribosyl)ated. J. Biol. Chem. 2002;277:26921–26926. doi: 10.1074/jbc.M200620200. [DOI] [PubMed] [Google Scholar]
- 91.Saxena A, Wong LH, Kalitsis P, Earle E, Shaffer LG, Choo KH. Poly(ADP-ribose) polymerase 2 localizes to mammalian active centromeres and interacts with PARP-1, Cenpa, Cenpb and Bub3, but not Cenpc. Hum. Mol. Genet. 2002;11:2319–2329. doi: 10.1093/hmg/11.19.2319. [DOI] [PubMed] [Google Scholar]
- 92.Chang P, Coughlin M, Mitchison TJ. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat. Cell Biol. 2005;7:1133–1139. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- 93.Chang P, Jacobson MK, Mitchison TJ. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–649. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.