Abstract
The Arabidopsis RNA-binding protein AtGRP8 undergoes negative autoregulation at the post-transcriptional level. An elevated AtGRP8 protein level promotes the use of a cryptic 5′ splice site to generate an alternatively spliced transcript, as_AtGRP8, retaining the 5′ half of the intron with a premature termination codon. In mutants defective in nonsense-mediated decay (NMD) abundance of as_AtGRP8 but not its pre-mRNA is elevated, indicating that as_AtGRP8 is a direct NMD target, thus limiting the production of functional AtGRP8 protein. In addition to its own pre-mRNA, AtGRP8 negatively regulates the AtGRP7 transcript through promoting the formation of the equivalent alternatively spliced as_AtGRP7 transcript, leading to a decrease in AtGRP7 abundance. Recombinant AtGRP8 binds to its own and the AtGRP7 pre-mRNA, suggesting that this interaction is relevant for the splicing decision in vivo. AtGRP7 itself is part of a negative autoregulatory circuit that influences circadian oscillations of its own and the AtGRP8 transcript through alternative splicing linked to NMD. Thus, we identify an interlocked feedback loop through which two RNA-binding proteins autoregulate and reciprocally crossregulate by coupling unproductive splicing to NMD. A high degree of evolutionary sequence conservation in the introns retained in as_AtGRP8 or as_AtGRP7 points to an important function of these sequences.
INTRODUCTION
Post-transcriptional regulation has come into focus as an important mechanism to control gene expression in higher plants (1–4). It occurs at multiple levels including pre-mRNA maturation, mRNA transport, translation and breakdown. The major players are RNA-binding proteins that influence the fate of an mRNA molecule either directly by binding to defined RNA sequences and structural elements or indirectly through protein–protein interaction (5).
One important protein domain known to interact with RNA molecules is the RNA recognition motif (RRM) (6). It is composed of a four-stranded antiparallel β-sheet with two α-helices. The highly conserved octapeptide RNP1 and hexapeptide RNP2 sequence motifs are located in the β3 and β1 sheets and contain conserved aromatic residues making contacts to the RNA substrate. A systematic survey disclosed 196 RRM-containing proteins in the genome of Arabidopsis thaliana (5). Among those, the 16 kDa AtGRP8 (A. thaliana glycine-rich RNA-binding protein 8) protein combines a single N-terminal RRM with a C-terminal region enriched in glycine repeats with some interspersed serine, tyrosine and arginine residues (7–9). It is also known as GR-RBP8, GRP8 or CCR1 (cold and circadian regulated 1) (5,10,11). Both AtGRP8 and AtGRP7 encoding an orthologous RNA-binding protein that shares 77% sequence identity undergo circadian oscillations with a peak at the end of the daily light phase (9,11). Notably, AtGRP8 is subject to negative regulation by AtGRP7, as in transgenic plants ectopically overexpressing AtGRP7 under control of the CaMV (Cauliflower Mosaic Virus) 35S RNA promoter, AtGRP8 oscillations are strongly depressed (9). This regulation occurs at the post-transcriptional level through reduction of constitutive splicing and stimulation of alternative splicing at a cryptic intronic 5′ splice site of the AtGRP8 pre-mRNA, leading to an alternative splice variant (as_AtGRP8) with a premature termination codon (PTC) in the retained part of the intron.
AtGRP7 uses the same mechanism for negative autoregulation: rising protein levels promote alternative splicing by binding to its own pre-mRNA, and the alternatively spliced transcript (as_AtGRP7) is degraded via a pathway involving the nonsense-mediated decay (NMD) components AtUPF1 (UP FRAMESHIFT PROTEIN 1) and AtUPF3 (12,13). Thus, AtGRP7 is part of a negative feedback loop through which it influences its own oscillation at the post-transcriptional level. This feedback loop is thought to operate as a slave oscillator downstream of the circadian clock, transducing temporal information within the cell by regulating other transcripts (13,14).
We have recently found that AtGRP8 acts in concert with AtGRP7 to influence the transition to flowering in A. thaliana (15). Apart from this, little is known about AtGRP8 function within the cell.
Here we show that AtGRP8 forms a feedback loop that is interlocked with the AtGRP7 feedback loop. Recombinant AtGRP8 interacts with its own mRNA in vitro and promotes negative autoregulation in vivo by causing alternative splicing. The parts of the intron that are retained in the unproductively spliced transcripts show an exon-like evolutionary conservation, pointing to a functional role. As the alternatively spliced AtGRP8 and AtGRP7 transcripts are bona fide NMD targets, it appears that the interlocked AtGRP7/AtGRP8 feedback loops harness autogenous alternative splicing-activated decay via the NMD pathway to fine-tune the expression of their components.
MATERIALS AND METHODS
AtGRP8 overexpression in transgenic plants
The protein-coding region of AtGRP8 was amplified by PCR from the cDNA with the upstream primer 5′ GGCCATGGCTGAAGTTGAGT 3′ and the downstream primer 5′ CCGGATCCTTTACCAGCCGCCACCAC 3′ covering the translation start and stop (bold) and comprising engineered NcoI and BamHI sites (underlined), respectively. The PCR product was inserted between the CaMV 35S RNA promoter with the duplicated enhancer fused to the Tobacco Mosaic Virus omega translational enhancer and the CaMV polyadenylation signal (9). To express AtGRP8-RQ, the Arg47Gln mutation was introduced by PCR-mutagenesis (see below), sequenced and reinserted into the original plasmid by BglII/SacI digestion. The entire casettes were inserted into the binary vector pHPT1 that was obtained by replacing the promoter-less β-glucuronidase gene of pGPTV-HPT by the pUC19 polylinker (16). Arabidopsis thaliana L. Columbia plants were transformed by vacuum infiltration (17).
Plant growth
Seeds were germinated on one-half strength MS plates (18) containing 0.5% sucrose and the appropriate antibiotic and grown in 16 h light/8 h dark cycles at a constant temperature of 20°C. After 2 weeks, resistant plants were transferred to one-half strength MS plates without antibiotics.
Recombinant GST-AtGRP8 and GST-AtGRP8-RQ
Recombinant GST-AtGRP8 protein was constructed by inserting the AtGRP8 coding region into NotI-EcoRI-cut pGEX-6P1 vector (GE Healthcare, Freiburg, Germany).
To generate the mutant variant GST-AtGRP8-RQ, Arg47 in GST-AtGRP8 was exchanged for glutamine by PCR with Phusion Polymerase (Finnzymes, Espoo, Finland) using the overlapping primers RQfor CGAGAGTGGAAGATCCCAAGGAT-TCGGATTCGTCA and RQrev TGACGAATCCGAATCCTTGGGATCTTCCACTCTCG. Silent mutations were introduced into neighbouring amino acids creating a diagnostic StyI site (underlined). The mutation was verified by sequencing. The recombinant proteins were expressed in E. coli BL21 DE3. Affinity purification by chromatography on Glutathione Sepharose (GE Healthcare) and concentration of the eluate by centrifugation through Centricon® 30 filter devices (Millipore, Billerica, MA, USA) were done as described (12,13).
RNA-binding assay
Synthetic oligoribonucleotides (ORN) were purchased from Biomers (Ulm, Germany). RNA bandshifts with recombinant AtGRP8 and the ORNs labelled at the 5′ end with γ-[32P] ATP were performed as previously described (12). To determine binding affinities, 50 fmol of labeled ORN-binding substrates were incubated with increasing amounts of recombinant protein, respectively, in the presence of 1 µg tRNA. Bound and free RNA were quantified and Kd values calculated based on three independent experiments as described (12).
RNA analysis
Isolation of total RNA from Arabidopsis plants and hybridization of RNA gel blots with the gene-specific AtGRP7 and AtGRP8 probes were performed as described (8,19). Semiquantitative RT–PCR on retrotranscribed total RNA was done as described (15). Primers are listed in Table S2.
Immunoblot analysis
Protein extraction from Arabidopsis plants and incubation of protein gel blots with antipeptide antibodies raised against AtGRP8 and AtGRP7, followed by chemiluminescence detection were done as described (12).
Secondary structure prediction
Evolutionary conserved RNA secondary structure elements were determined with the RNA-Decoder program (20,21). RNA-Decoder takes as input a fixed alignment of evolutionarily related RNA sequences and an evolutionary tree relating them and predicts as output the conserved secondary structures supported by the evolutionary patterns detected in the input alignment. A set of nine pre-mRNA sequences comprising AtGRP8 orthologs with the same gene structure comprising a 5′ UTR, two protein-coding exons of phase 0, a single intron and a 3′ UTR (Table S1) were assembled. To generate the input alignment, we first compiled separate alignments for 5′ UTRs, exon 1, intron, exon 2 and 3′ UTRs using ClustalW (Version 1.83) (22), which we then combined manually into longer alignments. To maximize the alignment quality, the alignments of the pre-mRNA sequences of exons 1 and 2 were based on the ClustalW alignments of the corresponding encoded amino-acid sequences. As the resulting full-length pre-mRNA alignment (1407 nt) was too long to analyze in a single chunk, we generated an mRNA input alignment with UTRs (1055 nt) to investigate conserved secondary structures in or near the UTRs and a pre-mRNA input alignment without UTRs (1093 nt) to investigate conserved secondary structures in or near the intron.
RESULTS AND DISCUSSION
AtGRP8 negatively autoregulates its own pre-mRNA
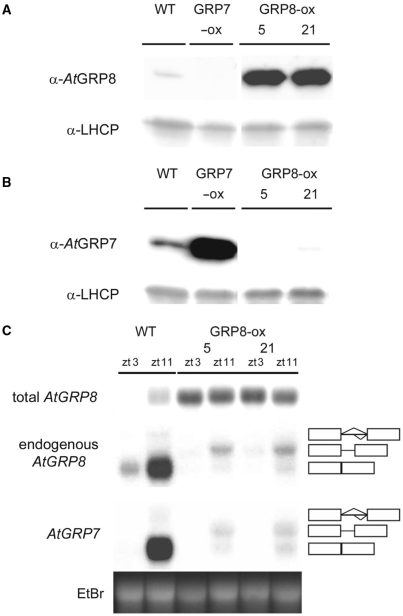
To begin to investigate the molecular properties of the predicted RNA-binding protein AtGRP8, we generated transgenic plants ectopically overexpressing the AtGRP8 coding region under control of the CaMV promoter with the duplicated enhancer (AtGRP8-ox plants). Immunoblot analysis using a specific AtGRP8 antibody identified transgenic lines with strongly elevated AtGRP8 protein levels (Figure 1A). Compared to WT plants, the total AtGRP8 transcript level was strongly elevated in AtGRP8-ox plants harvested at zt3 (zeitgeber time 3, that is 3 h after lights on), the circadian minimum of AtGRP8 oscillations, and zt11, the circadian maximum, due to the expression of the transgene (Figure 1C). The endogenous AtGRP8 transcript forms are selectively detected with a gene-specific probe derived from the 5′ UTR that is not contained in the overexpression construct (Figure 1C). In WT plants, the endogenous AtGRP8 mRNA and a small amount of its pre-mRNA containing the 283-nt intron were present. In the AtGRP8-ox plants, almost no mature endogenous AtGRP8 mRNA was detectable. Instead, an intermediate size transcript appears at a low level, corresponding to the alternatively spliced AtGRP8 transcript (as_AtGRP8) that is generated through the use of a cryptic 5′ splice site within the intron. These data indicate that AtGRP8 exerts negative autoregulation on its own pre-mRNA.
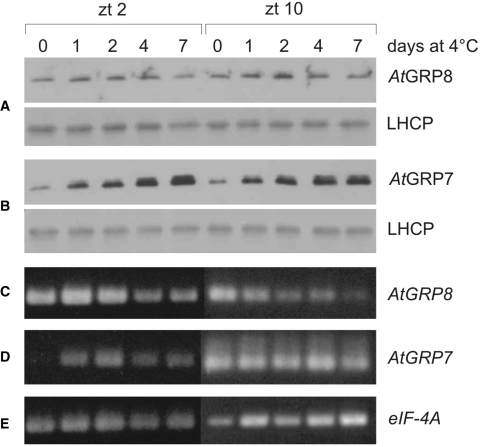
Figure 1.
Influence of ectopic AtGRP8 overexpression on the endogenous AtGRP7 and AtGRP8. (A) The immunoblot with total protein of WT plants, the AtGRP8-ox lines 5 and 21 and an AtGRP7-ox line, harvested at zt11, was probed with the AtGRP8 antibody (top) and an antibody against LHCP (light harvesting chlorophyll-binding protein) as loading control (bottom). The absence of crossreaction with overexpressed AtGRP7 protein in AtGRP7-ox plants demonstrates the specificity of the antibody. (B) A duplicate blot was probed with the AtGRP7 antibody (top) and the LHCP antibody (bottom). (C) WT and AtGRP8-ox plants were harvested at zt3 and zt11. The RNA gel blot was hybridized with the AtGRP8 cDNA to determine the total AtGRP8 transcript level. The stripped blot was rehybridized with the gene-specific probe to monitor the endogenous AtGRP8 transcript and subsequently with the gene-specific AtGRP7 probe. The position of the pre-mRNA, as_AtGRP8 and as_AtGRP7 retaining the first half of the intron and the mature mRNA are indicated. Boxes represent exons, lines represent the first and second half of the intron, respectively. The ethidium-bromide stained gel shows equal loading.
In WT plants, as_AtGRP8 is hardly detectable. Nevertheless, corresponding cDNAs have been isolated from cDNA libraries (DS, unpublished results). In response to the elevated AtGRP8 protein level, a shift to a cryptic 5′ splice site occurs and as_AtGRP8 accumulates to a low level at the expense of the mature mRNA in AtGRP8-ox plants. Because the retained part of the intron contains in-frame stop codons, no full-length AtGRP8 protein can be produced from the as_AtGRP8 transcript. Thus, like AtGRP7, AtGRP8 negatively autoregulates, and the molecular underpinnings of the AtGRP8 feedback loop are similar to the way AtGRP7 influences its own circadian oscillations (12,13).
AtGRP8 promotes alternative splicing of the AtGRP7 pre-mRNA
To test whether AtGRP8 would also influence AtGRP7 transcript abundance, the AtGRP7 transcript level was compared in WT and AtGRP8-ox plants (Figure 1C). In WT plants, the mature AtGRP7 mRNA and a small amount of the pre-mRNA were detected at the circadian maximum. When AtGRP8 was expressed at high levels, a low amount of the as_AtGRP7 transcript appeared at the expense of the mature mRNA. Accordingly, AtGRP7 protein was barely detectable in the AtGRP8-ox plants (Figure 1B).
AtGRP8 RNA and protein steady-state abundance previously have been shown to be under negative control by AtGRP7 (12,13). Consistent with this, AtGRP8 RNA and protein levels are elevated in the atgrp7-1 T-DNA insertion mutant due to relief of repression (15). Our data now show that not only does AtGRP7 influence AtGRP8 abundance, but also AtGRP8 in turn regulates AtGRP7. Similarly, another Arabidopsis splicing regulator, the Ser/Arg-rich (SR) protein atRSZ33 crossregulates alternative splicing of a conserved long intron in the atRSp31 gene, equivalent to the effect on its own pre-mRNA (23,24). The alternative atRSp31 form accumulates to high levels and thus is stable in contrast to as_AtGRP8.
Recombinant AtGRP8 binds to its own pre-mRNA
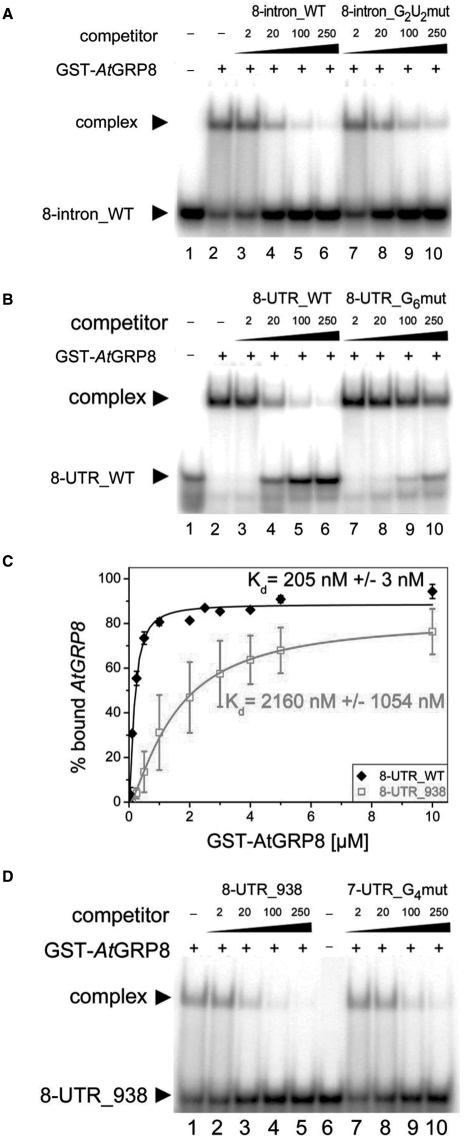
As AtGRP7 autoregulation involves AtGRP7 binding to its own 3′ UTR and intron, it is conceivable that AtGRP8 might similarly interact with its own pre-mRNA. Therefore, we performed RNA-bandshift assays to test the AtGRP7-binding sites determined previously (12) for interaction with recombinant AtGRP8 (Table 1). For the intronic binding site 8-intron_WT GST-AtGRP8 formed a retarded complex which was competed more efficiently by the ORN 8-intron_WT than by the mutated counterpart 8-intron_G2U2mut with exchanges of two G and two U residues (Figure 2A). For the 3′ UTR, a retarded complex was observed for 8-UTR_WT that was more efficiently outcompeted by unlabeled ORN 8-UTR_WT than by the mutated counterpart 8-UTR_G6mut with mutation of six G residues (Figure 2B). Sequence alignment of the AtGRP7-binding sites upon the AtGRP8 intron and 3′ UTR had uncovered a second motif with homology to the AtGRP7-binding site within the AtGRP8 3′ UTR (data not shown). However, determination of the Kd values revealed that AtGRP8 has a 10-fold lower affinity for the corresponding ORN 8-UTR_938 (Table 1) compared to 8-UTR_WT (Figure 2C). Furthermore, the complex was outcompeted by an excess of unlabeled 8-UTR_938 to the same degree as by a negative control ORN 7-UTR_G4mut (Table 1) (Figure 2D). Based on the low affinity and lack of specificity, 8-UTR_938 was excluded as binding site. Taken together, our data show that AtGRP8 binds to its own pre-mRNA and recognizes sequences in the second half of the intron and the 3′ UTR that also comprise targets for AtGRP7.
Table 1.
Sequences of oligoribonucloetides used as RNA-binding substrates
| Name | Sequence |
|---|---|
| 8-UTR_WT | GUUUUUGGUUUAGAUUUGGUUUUGUGU |
| 8-UTR_G6mut | GUUUUUAAUUUAAAUUUAAUUUUAUGU |
| 8-UTR_938 | CGUUUGGUUUACUUUUUUGAUGAAACA |
| 8-intron_WT | CUUCCACGAUUGUUUUUGCUGAUGUGU |
| 8-intron_G2U2mut | CUUCCACGAUUAUCCUUACUGAUGUGU |
| 7-UTR_WT | AUUUUGUUCUGGUUCUGCUUUAGAUUUGAUCU |
| 7-UTR_G4mut | AUUUUAUUCUAAUUCUGCUUUAGAUUUAAUCU |
| 7-intron_WT | GUUCAGUUUUGUUGGAUUGUUUUGCUGAUCUG |
| 7-intron_G6mut | GUUCAAUUUUAUUAAAUUAUUUUACUGAUCUG |
Sequences of the AtGRP8 3′ UTR (8-UTR_WT and 8-UTR_938) and intron (8-intron_WT) ORN and the corresponding mutated 8-UTR_G6mut and 8-intron_G2U2mut with six G residues exchanged for A or two G and two U residues exchanged for A or C (in bold), respectively, as well as sequence of AtGRP7 3′ UTR (7-UTR_WT) and intron (7-intron_WT) ORN and the corresponding mutated 7-UTR_G4mut and 7-intron_G6mut with four or six G residues exchanged for A (in bold), respectively, are shown.
Figure 2.
Binding of recombinant AtGRP8 to the AtGRP8 transcript. (A) GST-AtGRP8 protein was incubated with labelled 8-intron_WT ORN in the presence of 5 µg of tRNA and 2, 20, 100 and 250 pmol of unlabelled 8-intron_WT ORN (lanes 3–6) or 8-intron_G2U2mut (lanes 7-10), respectively. Lane 1, free ORN. (B) GST-AtGRP8 protein was incubated with labelled 8-UTR_WT ORN in the presence of 5 µg of tRNA and 2, 20, 100 and 250 pmol of unlabelled 8-UTR_WT ORN (lanes 3–6) or 8-UTR_G6mut (lanes 7-10), respectively. Lane 1, free ORN. (C) To compare binding affinities, 50 fmol of labelled 8-UTR_WT ORN or 8-UTR_938 were incubated with 0.01, 0.05, 0.1, 0.25, 0.5, 1, 2, 2.5, 5 and 10 µM or 0.1, 0.25, 0.5, 1, 2, 3, 4, 5 and 10 µM of GST-AtGRP8, respectively. All reactions contained 1 µg tRNA. Bound and free RNA were quantified and Kd values calculated based on the mean of three independent experiments as described (12). (D) GST-AtGRP8 protein was incubated with labelled 8-UTR_938 ORN in the presence of 5 µg of tRNA and 2, 20, 100 and 250 pmol of unlabelled 8-UTR_938 ORN (lanes 2–5) or 7-UTR_ G4mut (lanes 7–10), respectively. Lane 6, free ORN.
To assess the secondary structure of these putative binding sites in the context of the entire pre-mRNA, AtGRP8 was searched for secondary structure elements conserved in plant orthologs with RNA-Decoder (20,21). Among the numerous existing secondary structure prediction programs, RNA-Decoder is unique in that it explicitly takes the known protein-coding regions of an input alignment into account. This feature is important when searching partly protein-coding sequences such as pre-mRNAs for conserved secondary structure elements, as the evolutionary pattern due to amino-acid conservation needs to be carefully distinguished from the evolutionary pattern due to secondary structure conservation (20,21).
In order to detect conserved secondary structures in the AtGRP8 pre-mRNA, we assembled a set of nine pre-mRNA sequences comprising AtGRP8 as well as eight AtGRP8 orthologs from the Brassicaceae mustard and oilseed rape, the other dicotyledoneous plants tobacco and Pelargonium, and the monocotyledoneous plants rice and maize (Table S1). As input tree to RNA-Decoder, we used the tree predicted by ClustalW (22) for the nine encoded protein sequences. The predicted secondary structures for the AtGRP8 and, for comparison, the AtGRP7 pre-mRNAs are shown in Figure S1. Interestingly, the predicted AtGRP8-binding sequences map to conserved regions, pointing to an important role of the secondary structure for the function of these motifs. 8-intron_WT and 8-UTR_WT are predicted to be single-stranded. In contrast, 8-UTR_938 is predicted to be partially double-stranded and thus may be less accessible to interacting proteins, in line with the very low binding affinity (Figure 2C).
Recombinant AtGRP8 binds to the AtGRP7 pre-mRNA
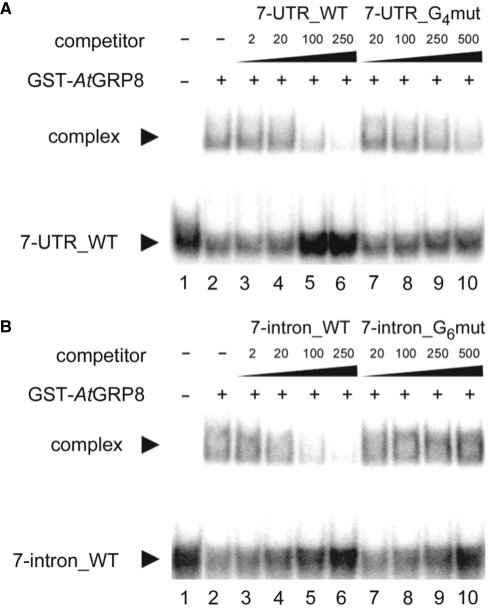
The data shown in Figure 1 indicate that AtGRP8 negatively influences AtGRP7 pre-mRNA splicing in the same way as AtGRP7 autoregulates. As the shift to the alternative splice form relies on AtGRP7 binding to its own pre-mRNA, we tested whether recombinant AtGRP8 would bind in vitro to the AtGRP7 target sites within the AtGRP7 pre-mRNA (Table 1). Complex formation was observed with 7-UTR_WT that was completely lost upon addition of 250 pmol of unlabeled 7-UTR_WT, but much less reduced with 500 pmol of the mutated 7-UTR_G4mut (Figure 3A). Also for the ORN spanning the AtGRP7-binding site within the second half of the intron a strong interaction with GST-AtGRP8 was found which was abolished by 250 pmol of unlabeled 7-intron_WT but not affected by 500 pmol of 7-intron G6mut (Figure 3B). This shows that AtGRP8 also binds to the AtGRP7 intron and 3′ UTR with a certain specificity. Thus, AtGRP7 and AtGRP8 may recognize overlapping or identical motifs within both pre-mRNAs. To compare the binding affinities, Kd values were determined for all interactions (Table 2). For the four binding sites, the Kd values were in the same order of magnitude as those previously determined for GST-AtGRP7 protein (12). Therefore, presently we cannot infer which of the possible interactions between AtGRP7 or AtGRP8 and the respective target sites may prevail in vivo.
Figure 3.
Binding of recombinant AtGRP8 to the AtGRP7 transcript. (A) GST-AtGRP8 protein was incubated with labelled 7-UTR_WT ORN in the presence of 5 μg of tRNA and 2, 20, 100 and 250 pmol of unlabelled 7-UTR_WT ORN (lanes 3–6) or 20, 100, 250 and 500 pmol of unlabelled 7-UTR_G4mut (lanes 7–10), respectively. Lane 1, free ORN. (B) GST-AtGRP8 protein was incubated with labelled 7-intron_WT ORN in the presence of 5 μg of tRNA and 2, 20, 100 and 250 pmol of unlabelled 7-intron_WT ORN (lanes 3–6) or 20, 100, 250 and 500 pmol of unlabelled 7-intron_G6mut ORN (lanes 7–10), respectively. Lane 1, free ORN.
Table 2.
Kd values for recombinant GST-AtGRP8-RQ and GST-AtGRP8
| ORN | Protein | Kd [M] | Kd RQ/Kd WT |
|---|---|---|---|
| 8-UTR_WT | GST-AtGRP8-WT | 2.05 ± 0.03 × 10–7 | |
| GST-AtGRP8-RQ | 1.31 ± 0.35 × 10–6 | 6.4 | |
| 7-UTR_WT | GST-AtGRP8-WT | 3.18 ± 2.27 × 10–7 | |
| GST-AtGRP8-RQ | 3.22 ± 0.97 × 10–6 | 10.1 | |
| 8-intron_WT | GST-AtGRP8-WT | 1.64 ± 0.65 × 10–6 | |
| GST-AtGRP8-RQ | 2.29 ± 0.36 × 10–5 | 14.0 | |
| 7-intron_WT | GST-AtGRP8-WT | 4.26 ± 0.52 × 10–7 | |
| GST-AtGRP8-RQ | 3.59 ± 2.02 × 10–6 | 8.4 |
Kd values were determined as described in (12). The ratio between the Kd value for the mutated protein and the WT protein is indicated.
Mutation of AtGRP8 RNP1 Arg47 impairs RNA-binding activity in vitro
Binding of AtGRP8 to its own and the AtGRP7 pre-mRNA in vitro suggests that this interaction may initiate alternative splicing and down-regulation of endogenous AtGRP7 and AtGRP8 in vivo. To investigate whether this in vitro binding activity correlated with in vivo function, a mutation was introduced into the RRM of recombinant GST-AtGRP8. The conserved arginine (R) 47 predicted to lie at the beginning of the β3 strand that is part of the RNA-binding platform was exchanged for glutamine (Q) (25–27). Indeed, we have shown that recombinant GST-AtGRP7-RQ with an analogous mutation of R49 has a 6-fold lower affinity for its target sites than WT GST-AtGRP7 protein while folding of the protein is not affected (12).
The AtGRP8-RQ mutant protein was expressed as GST-fusion and tested for binding to the AtGRP7 and AtGRP8 target sites. Only weak interaction was observed with Kd values about one order of magnitude higher for GST-AtGRP8-RQ compared to GST-AtGRP8 (Table 2). Competition assays revealed that 8-UTR_WT competed much better for binding of GST-AtGRP8-RQ than 8-UTR_G6mut and 7-UTR_WT competed much better for binding of GST-AtGRP8-RQ than 7-UTR_G4mut (data not shown). Thus, mutation of the conserved R47 reduces the binding affinity rather than the specificity of the interaction.
The AtGRP8 R47Q mutation impairs but does not abolish promotion of alternative AtGRP8 and AtGRP7 splicing
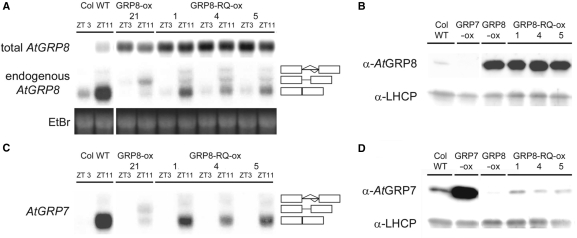
To test the impact of the RQ mutation on AtGRP8 activity in vivo, plants constitutively overexpressing the mutated AtGRP8-RQ protein were generated (AtGRP8-RQ-ox) and compared to plants overexpressing WT AtGRP8 protein (AtGRP8-ox). Transgenic plants were identified with a total AtGRP8 transcript level constitutively elevated at zt3 and zt11 (Figure 4A, top). An immunoblot analysis confirmed expression of intact mutated protein, as total AtGRP8 protein was elevated to a level similar to that in AtGRP8-ox plants overexpressing WT AtGRP8 protein (Figure 4B). However, in contrast to AtGRP8-ox plants, the mature endogenous AtGRP8 mRNA was still detected in addition to as_AtGRP8 (Figure 4A, middle). Thus, an elevated AtGRP8-RQ level only partially shifts the splice site in favor of the cryptic 5′ splice site with concomitant reduction in as_AtGRP8 abundance, as observed in AtGRP8-ox plants. We infer from this that the weakened interaction of AtGRP8-RQ with its own pre-mRNA impairs but does not abolish negative autoregulation of the AtGRP8 pre-mRNA.
Figure 4.
Molecular characterization of transgenic AtGRP8-RQ ox lines. (A) WT, AtGRP8-ox and AtGRP8-RQ-ox plants were harvested at zt3 and zt11. The RNA gel blot was hybridized with the AtGRP8 cDNA to determine the total transcript level (top) and with the gene-specific probe to monitor the endogenous AtGRP8 transcript (middle). The position of the pre-mRNA, as_AtGRP8 retaining the first half of the intron and the mRNA are indicated. Boxes represent exons, lines represent the first and the second half of the intron, respectively. The ethidium-bromide stained gel shows equal loading (bottom). (B) The immunoblot with total protein of WT plants, the AtGRP8-ox lines 5 and 21, and the AtGRP8-RQ-ox lines 1, 4 and 5, harvested at zt11, was probed with the AtGRP8 antibody (top) and an antibody against LHCP as loading control (bottom). (C) The RNA gel blot shown in (A) was stripped and rehybridized with the AtGRP7 probe. (D) The immunoblot with the same protein extracts as shown in (B) was probed with the AtGRP7 antibody (top) and the LHCP antibody (bottom).
Overexpression of WT AtGRP8 led to an accumulation of as_AtGRP7 and an almost complete loss of the mature mRNA (Figure 4C). In contrast, no alternatively spliced AtGRP7 transcript was detected in AtGRP8-RQ-ox plants. Thus, the R47Q mutation reduces the effect AtGRP8 protein has on splice site selection. The level of mature AtGRP7 mRNA was only weakly reduced compared to WT plants, consistent with the idea that the residual binding activity of AtGRP8-RQ still causes some production of as_AtGRP7 at the expense of the AtGRP7 mRNA. However, the alternatively spliced transcript is hardly detectable due to its short half life (13). Accordingly, almost no AtGRP7 protein was detected in AtGRP8-ox plants, whereas a small amount of AtGRP7 protein was detectable in AtGRP8-RQ-ox lines (Figure 4D). Altogether, the interaction of AtGRP8 with the AtGRP7 and AtGRP8 pre-mRNAs in vivo is relevant for the negative autoregulation and crossregulation. Presumably, binding of AtGRP8 and AtGRP7 triggers additional factors that act in concert to regulate the choice of splice sites (12).
Distinct cold response of AtGRP8 and AtGRP7
The reciprocal regulation between AtGRP7 and AtGRP8 uncovered here may suggest that the proteins are able to fully substitute for each other. Therefore we compared their expression patterns. According to publically available microarray data, AtGRP7 and AtGRP8 are expressed at a similar level across plant tissues except for a lower AtGRP8 abundance in siliques and later stages of seed development (Figure S2) (28). Moreover, the circadian maximum of AtGRP8 transcript oscillation is only slightly advanced relative to the AtGRP7 peak (29). Both AtGRP7 and AtGRP8 transcripts have been described as being upregulated by cold (11,30). We investigated steady-state protein levels under these conditions using specific anti-peptide antibodies. Two-week-old Col plants were transferred to 4°C at light onset and harvested without cold treatment and after 1, 2, 4 and 7 days, respectively. AtGRP8 protein abundance rose marginally at best after transfer to 4°C, whereas AtGRP7 protein abundance steadily increased up to day 7 (Figure 5A, B). The AtGRP8 transcript level was only weakly increased at the circadian minimum (zt2) after 1 day of cold treatment but declined afterwards (Figure 5C). Also at the circadian maximum (zt10), AtGRP8 transcript levels declined during extended exposure to cold. In contrast, a strong increase of the AtGRP7 transcript level was observed at the circadian minimum (zt2). At the circadian maximum (zt10), the level in plants transferred to 4°C was not elevated beyond that of control plants. These data indicate that AtGRP7 and AtGRP8 differ in their response to cold and thus do not act entirely redundant, as their expression pattern is not fully congruent.
Figure 5.
Differential regulation by cold of AtGRP8 and AtGRP7. (A) Col plants grown for 2 weeks in 16 h light/8 h dark cycles at 20°C were transferred to 16-h light/8-h dark cycles at 4°C and harvested on day 0, 1, 2, 4 and 7 at zt2 and zt10, respectively. (B) Immunoblots with total protein extracts from the same plants were probed with the AtGRP8 (A, top) and AtGRP7 (B, top) antibodies and an LHCP antibody (A and B, bottom) as loading control. (C) Semiquantitative RT-PCR of AtGRP8. (D) Semiquantitative RT-PCR of AtGRP7. (E) Semiquantitative RT-PCR of eIF-4A as constitutive control. The exponential range was determined by comparing the signal with increasing number of cycles. The absence of genomic DNA was confirmed with nonretrotranscribed RNA.
To determine whether the exposure to low temperatures may influence the alternative splicing, the pattern of the alternative splice forms and mature mRNAs in 2-week-old plants exposed to cold for 1, 2 or 5 days was compared to that in untreated plants (Figure S3). as_AtGRP7 accumulated in parallel with the RNA. While the AtGRP8 mRNA only weakly and transiently increased, a stronger upregulation was observed for as_AtGRP8. Thus, only for AtGRP8 a slight change in the ratio of the alternative splice forms is observed in the cold. Previous studies have demonstrated changes in the alternative splicing pattern of Arabidopsis SR genes upon cold treatment which may give rise to proteins with different domains and, consequently, changes in splicing of downstream targets (31,32).
as_AtGRP8 and as_AtGRP7 are direct targets of the NMD pathway
Degradation of as_AtGRP8 and as_AtGRP7 is dependent on AtUPF1 and AtUPF3, key components of the NMD pathway of mRNA surveillance that ensures clearance of PTC-containing mRNAs from the cellular transcriptome (12). Recently, NMD has emerged as a widespread regulatory mechanism of physiological gene expression (33–35). In the Arabidopsis lba (low β-amylase) mutant which has a point mutation in the UPF1 gene a suite of transcripts show higher steady-state abundance, as expected for NMD substrates (36). But additionally several transcripts are down-regulated with a high proportion of sugar-inducible mRNAs, implicating AtUPF1 in sugar signalling (36).
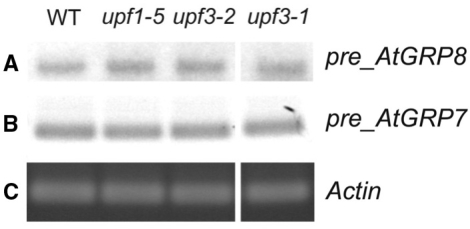
By analogy, the elevated as_AtGRP8 and as_AtGRP7 levels in upf1 and upf3 mutants (12) may be an indirect consequence of the reduced AtUPF1 or AtUPF3 levels. To distinguish such an indirect effect of AtUPF1 and AtUPF3 on transcription from a direct effect on transcript abundance, we assayed the pre-mRNA levels by RT–PCR. AtGRP8 pre-mRNA steady-state abundance was not changed in the upf1 and upf3 mutants (Figure 6A) and also the AtGRP7 pre-mRNA level was indistinguishable from WT (Figure 6B) in contrast to the strongly elevated as_AtGRP7 and as_AtGRP8 levels found in upf1-5, upf3-1 and upf3-2 (12). This indicates that the AtUPF1- and AtUPF3-dependent reduction of as_AtGRP8 and as_AtGRP7 is due to post-transcriptional destabilization rather than an indirect consequence of transcriptional inhibition. In higher plants, an increasing number of PTC-containing transcripts have been found to be stabilized when UPF1 and/or UPF3 functions are impaired (36–39). It has not been investigated whether some of them may be influenced either through an NMD-independent function of AtUPF1 and AtUPF3 or as a consequence of a cognate transcription factor undergoing NMD. In humans, in a survey of potential NMD targets more than 5% of the genes detected on Affymetrix GeneChips were affected by the presence or absence of UPF1 (40). For 15 out of 16 selected transcripts the pre-mRNA level was also changed, however, suggesting that the vast majority of those transcripts are affected indirectly through altered transcription rather than being bona fide NMD targets (40).
Figure 6.
Effect of upf1 and upf3 mutations on steady-state abundance of the AtGRP8 and AtGRP7 pre-mRNAs. RNA from the upf1-5, upf3-1 and upf3-2 mutants and WT harvested at zt10 was reverse-transcribed. PCR amplification of the AtGRP8 (A) and AtGRP7 pre-mRNA (B) was performed using specific primers and 24 cycles. The gel with the PCR products was blotted and hybridized with the AtGRP8 cDNA (A) or AtGRP7 cDNA (B). Amplification with ACTIN primers served as control (C).
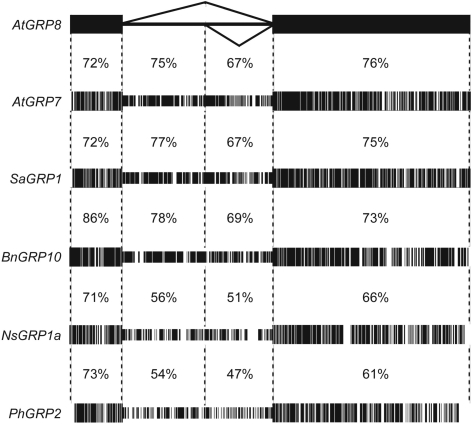
If the unproductive AtGRP8 and AtGRP7 splicing elicited by elevated levels of AtGRP7 and AtGRP8 protein indeed is functionally relevant, this may be reflected at the level of sequence conservation. The gene structure of the small glycine-rich RNA-binding proteins with a single RRM is well conserved among different plant species, harboring a single intron of similar size between RNP2 and RNP1, and a predicted cryptic 5′ splice site (8,19). Notably, the retained upstream part of the intron exhibits a higher degree of conservation than the downstream part spliced out in the as_AtGRP7 and as_AtGRP8 variants (Figure 7). A comparison of AtGRP8 with AtGRP7 and orthologs from mustard, oilseed rape, tobacco and Pelargonium shows that the degree of conservation reaches that of the surrounding exons (Figure 7, Table S1). The open reading frames contain an in frame termination codon within the first half of the intron and code for predicted 5 kDa polypeptides comprising only the RNP2 moiety of the RRM. As intact RNP1 is important for high-affinity RNA binding of AtGRP7 (12) and AtGRP8 (Table 2), these truncated polypeptides presumably do not interact with the RNA target sites in a productive manner but theoretically, they could interfere with AtGRP7 and AtGRP8 function. So far, however, these polypeptides have not been detected in Arabidopsis WT plants, the upf mutants or in Sinapis alba (data not shown). Thus, the conservation likely is not due to protein coding and therefore implies a regulatory function, e.g. to convey degradation when retained.
Figure 7.
Conservation of the retained part of the GRP introns. The nucleotide sequence corresponding to AtGRP8 exon I, exon II and the intron was aligned with the corresponding sequence of AtGRP7, SaGRP1 (8), BnGRP10 (46), NsGRP1a (47) and PhGRP2 (48).
A similar scenario has been observed for the ribosomal protein L12 in Caenorhabditis elegans (41). Elevated L12 levels promote the formation of an alternatively spliced transcript with a PTC due to removal of a partial intron. Thus, this autoregulatory circuit also couples unproductive splicing with destruction via the NMD pathway and, like for AtGRP7 and AtGRP8, the retained part of the intron displays a striking sequence conservation in worms (41).
Notably, the AtGRP7 transcript oscillates with one of the highest amplitudes observed for circadian genes in Arabidopsis (19). Following transcriptional activation by the circadian clock during the day and a steep rise in mRNA level, the AtGRP7 mRNA needs to decay during the declining phase below a level attained by time-of-day dependent transcriptional control alone in order to prevent damping (19,35). One way to accomplish high-amplitude mRNA oscillations would be by severely decreasing mRNA half life during the course of the day (42). However, such a scenario would also impact non-clock functions of AtGRP7 and AtGRP8, as in addition to their regulation by the circadian clock AtGRP8 and in particular AtGRP7 respond to numerous external factors (28). Transcriptional activation by external regulatory cues presumably would not bring about sufficiently high levels of a short- lived mRNA during a considerable part of the day to allow increase in protein steady-state abundance. Thus, AS-NMD elicited by the proteins themselves may be an efficient way to adjust mRNA and consequently protein levels.
Utilization of NMD for negative autoregulation has been widely observed for proteins involved in splicing regulation. The mammalian SR protein SC35 downregulates its expression through alternative splicing, resulting in an unstable transcript (43). However, in contrast to as_AtGRP7 and as_AtGRP8, the alternative SC35 transcripts retain full coding capacity. Crossregulation recently has been described for the polypyrimidine tract-binding protein (PTB), a global repressor of alternative splicing in nonneuronal cells, and its neurally expressed paralogue nPTB. PTB induces skipping of its own exon 11, producing an NMD substrate, and also induces exon-skipping in nPTB, resulting in a switch from PTB to nPTB expression during the development of neurons that impacts neural-specific splicing patterns of downstream targets. nPTB is able to autoregulate at the level of exon skipping when PTB is absent (44). Spellman showed that nPTB is able to repress PTB exon 11, however whether this occurs in vivo has not yet been determined (45).
For AtGRP7 and AtGRP8 we have found autoregulation and reciprocal crossregulation, showing that the newly identified AtGRP8 feedback loop is interconnected with the AtGRP7 feedback loop previously identified as a circadian slave oscillator (9). The connection between the two regulatory circuits may serve to integrate input by diverse stimuli, and the crossregulation may fine tune and balance the expression of both proteins.
In the future it will be important to determine common target transcripts of AtGRP7 and AtGRP8 and to determine the relative influence both proteins have on such downstream transcripts.
CONCLUSION
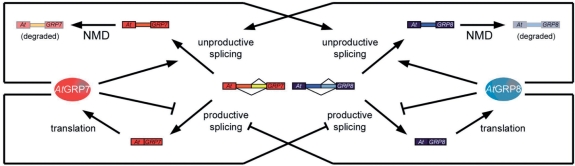
We show here that the clock-controlled RNA-binding protein AtGRP8 forms an interlocked post-transcriptional negative feedback loop with the AtGRP7 autoregulatory circuit. Both proteins negatively autoregulate and reciprocally crossregulate by binding to their pre-mRNAs and promoting unproductive splicing coupled to degradation via the NMD pathway (Figure 8). Thus, we extend the examples of quantitative post-transcriptional control by AS-NMD to autoregulatory loops in output pathways of the circadian clock.
Figure 8.
Model of the interlocked AtGRP7 and AtGRP8 feedback loops. Increasing AtGRP7 and AtGRP8 protein (depicted as ellipses) levels promote use of the cryptic intronic 5′ splice sites, leading to unproductively spliced as_AtGRP7 and as_AtGRP8 transcripts that are degraded via the NMD pathway.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Research Council (SFB 613 and STA 653/2 to D.S.); NSERC (Discovery Grant to I.M.M.). J.C.S. is a fellow of the German National Academic Foundation. Funding for open access charge: German Research Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Kristina Neudorf and Elisabeth Detring for expert technical assistance and Martina Lummer and Dr Christian Heintzen for critical comments on the manuscript. The upf1 and upf3 mutants were kindly provided by Dr Brendan Davies.
REFERENCES
- 1.Cheng Y, Chen X. Posttranscriptional control of plant development. Curr. Opin. Plant Biol. 2004;7:20–25. doi: 10.1016/j.pbi.2003.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Staiger D. RNA-binding proteins and circadian rhythms in Arabidopsis thaliana. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2001;356:1755–1759. doi: 10.1098/rstb.2001.0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reddy AS. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 2007;58:267–294. doi: 10.1146/annurev.arplant.58.032806.103754. [DOI] [PubMed] [Google Scholar]
- 4.Lorkovic ZJ, Wieczorek Kirk DA, Lambermon MH, Filipowicz W. Pre-mRNA splicing in higher plants. Trends Plant Sci. 2000;5:160–167. doi: 10.1016/s1360-1385(00)01595-8. [DOI] [PubMed] [Google Scholar]
- 5.Lorkovic ZJ, Barta A. Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 2002;30:623–635. doi: 10.1093/nar/30.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 7.van Nocker S, Vierstra RD. Two cDNAs from Arabidopsis thaliana encode putative RNA binding proteins containing glycine-rich domains. Plant Mol. Biol. 1993;21:695–699. doi: 10.1007/BF00014552. [DOI] [PubMed] [Google Scholar]
- 8.Heintzen C, Melzer S, Fischer R, Kappeler S, Apel K, Staiger D. A light- and temperature-entrained circadian clock controls expression of transcripts encoding nuclear proteins with homology to RNA-binding proteins in meristematic tissue. Plant J. 1994;5:799–813. doi: 10.1046/j.1365-313x.1994.5060799.x. [DOI] [PubMed] [Google Scholar]
- 9.Heintzen C, Nater M, Apel K, Staiger D. AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 1997;94:8515–8520. doi: 10.1073/pnas.94.16.8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu ZQ, Guo M, Jeong BR, Tian F, Elthon TE, Cerny RL, Staiger D, Alfano JR. A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature. 2007;447:284–288. doi: 10.1038/nature05737. [DOI] [PubMed] [Google Scholar]
- 11.Carpenter CD, Kreps JA, Simon AE. Genes encoding glycine-rich Arabidopsis thaliana proteins with RNA-binding motifs are influenced by cold treatment and an endogenous circadian rhythm. Plant Physiol. 1994;104:1015–1025. doi: 10.1104/pp.104.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schöning JC, Streitner C, Page DR, Hennig S, Uchida K, Wolf E, Furuya M, Staiger D. Autoregulation of the circadian slave oscillator component AtGRP7 and regulation of its targets is impaired by a single RNA recognition motif point mutation. Plant J. 2007;52:1119–1130. doi: 10.1111/j.1365-313X.2007.03302.x. [DOI] [PubMed] [Google Scholar]
- 13.Staiger D, Zecca L, Wieczorek Kirk DA, Apel K, Eckstein L. The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J. 2003;33:361–371. doi: 10.1046/j.1365-313x.2003.01629.x. [DOI] [PubMed] [Google Scholar]
- 14.Rudolf F, Wehrle F, Staiger D. Slave to the rhythm. The Biochemist. 2004;26:11–13. [Google Scholar]
- 15.Streitner C, Danisman S, Wehrle F, Schöning JC, Alfano JR, Staiger D. The small glycine-rich RNA-binding protein AtGRP7 promotes floral transition in Arabidopsis thaliana. Plant J. 2008;56:239–250. doi: 10.1111/j.1365-313X.2008.03591.x. [DOI] [PubMed] [Google Scholar]
- 16.Becker D, Kemper E, Schell J, Masterson R. New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 1992;20:1195–1197. doi: 10.1007/BF00028908. [DOI] [PubMed] [Google Scholar]
- 17.Bechthold N, Ellis J, Pelletier G. In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. Science de la vie/Life Sci. 1993;316:1194–1199. [Google Scholar]
- 18.Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962;15:473–497. [Google Scholar]
- 19.Staiger D, Apel K. Circadian clock-regulated expression of an RNA-binding protein in Arabidopsis: characterisation of a minimal promoter element. Mol. Gen. Genet. 1999;261:811–819. doi: 10.1007/s004380050025. [DOI] [PubMed] [Google Scholar]
- 20.Pedersen JS, Meyer IM, Forsberg R, Simmonds P, Hein J. A comparative method for finding and folding RNA secondary structures within protein-coding regions. Nucleic Acids Res. 2004;32:4925–4936. doi: 10.1093/nar/gkh839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pedersen JS, Forsberg R, Meyer IM, Hein J. An evolutionary model for protein-coding regions with conserved RNA structure. Mol. Biol. Evol. 2004;21:1913–1922. doi: 10.1093/molbev/msh199. [DOI] [PubMed] [Google Scholar]
- 22.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalyna M, Lopato S, Barta A. Ectopic expression of atRSZ33 reveals its function in splicing and causes pleiotropic changes in development. Mol. Biol. Cell. 2003;14:3565–3577. doi: 10.1091/mbc.E03-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalyna M, Lopato S, Voronin V, Barta A. Evolutionary conservation and regulation of particular alternative splicing events in plant SR proteins. Nucleic Acids Res. 2006;34:4395–4405. doi: 10.1093/nar/gkl570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding J, Hayashi MK, Zhang Y, Manche L, Krainer AR, Xu RM. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999;13:1102–1115. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jessen TH, Oubridge C, Teo C.-H, Pritchard C, Nagai K. Identification of molecular contacts between the U1 A small nuclear ribonucleoprotein and U1 RNA. EMBO J. 1991;10:3447–3456. doi: 10.1002/j.1460-2075.1991.tb04909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagai K, Oubridge C, Jessen TH, Li J, Evans PR. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature. 1990;348:515–520. doi: 10.1038/348515a0. [DOI] [PubMed] [Google Scholar]
- 28.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis Microarray Database and Analysis Toolbox. Plant Physiol. 2004;136:2621–6232. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 30.Kwak KJ, Kim YO, Kang H. Characterization of transgenic Arabidopsis plants overexpressing GR-RBP4 under high salinity, dehydration, or cold stress. J. Exp. Bot. 2005;56:3007–3016. doi: 10.1093/jxb/eri298. [DOI] [PubMed] [Google Scholar]
- 31.Lazar G, Goodman H. The Arabidopsis splicing factor SR1 is regulated by alternative splicing. Plant Mol. Biol., 2000:571–581.. doi: 10.1023/a:1006394207479. [DOI] [PubMed] [Google Scholar]
- 32.Palusa SG, Ali GS, Reddy AS. Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant J. 2007;49:1091–1107. doi: 10.1111/j.1365-313X.2006.03020.x. [DOI] [PubMed] [Google Scholar]
- 33.Rehwinkel J, Raes J, Izaurralde E. Nonsense-mediated mRNA decay: Target genes and functional diversification of effectors. Trends Biochem. Sci. 2006;31:639–646. doi: 10.1016/j.tibs.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 34.Guan Q, Zheng W, Tang S, Liu X, Zinkel RA, Tsui KW, Yandell BS, Culbertson MR. Impact of nonsense-mediated mRNA decay on the global expression profile of budding yeast. PLoS Genet. 2006;2:e203. doi: 10.1371/journal.pgen.0020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGlincy NJ, Smith CW. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem. Sci. 2008;33:385–393. doi: 10.1016/j.tibs.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 36.Yoine M, Ohto MA, Onai K, Mita S, Nakamura K. The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in Arabidopsis. Plant J. 2006;47:49–62. doi: 10.1111/j.1365-313X.2006.02771.x. [DOI] [PubMed] [Google Scholar]
- 37.Hori K, Watanabe Y. UPF3 suppresses aberrant spliced mRNA in Arabidopsis. Plant J. 2005;43:530–540. doi: 10.1111/j.1365-313X.2005.02473.x. [DOI] [PubMed] [Google Scholar]
- 38.Arciga-Reyes L, Wootton L, Kieffer M, Davies B. UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 2006;47:480–489. doi: 10.1111/j.1365-313X.2006.02802.x. [DOI] [PubMed] [Google Scholar]
- 39.Wu J, Kang J.-H, Hettenhausen C, Baldwin IT. Nonsense-mediated mRNA decay (NMD) silences the accumulation of aberrant trypsin proteinase inhibitor mRNA in Nicotiana attenuata. Plant J. 2007;51:693–706. doi: 10.1111/j.1365-313X.2007.03173.x. [DOI] [PubMed] [Google Scholar]
- 40.Viegas MH, Gehring NH, Breit S, Hentze MW, Kulozik AE. The abundance of RNPS1, a protein component of the exon junction complex, can determine the variability in efficiency of the Nonsense Mediated Decay pathway. Nucleic Acids Res. 2007;35:4542–4551. doi: 10.1093/nar/gkm461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitrovich QM, Anderson P. Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes Dev. 2000;14:2173–2184. doi: 10.1101/gad.819900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.So WV, Rosbash M. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 1997;16:7146–7155. doi: 10.1093/emboj/16.23.7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sureau A, Gattoni R, Dooghe Y, Stevenin J, Soret J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001;20:1785–1796. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M., Jr., Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spellman R, Llorian M, Smith CW. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol. Cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bergeron D, Beauseigle D, Bellemare G. Sequence and expression of a gene encoding a protein with RNA-binding and glycine-rich domains in Brassica napus. Biochim. Biophys. Acta. 1993;1216:123–125. doi: 10.1016/0167-4781(93)90047-h. [DOI] [PubMed] [Google Scholar]
- 47.Hirose T, Sugita M, Sugiura M. cDNA structure, expression and nucleic acid-binding properties of three RNA-binding proteins in tobacco: occurrence of tissue-specific alternative splicing. Nucleic Acids Res. 1993;21:3981–3987. doi: 10.1093/nar/21.17.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clark DG, Richards C, Brown KM. Characterization of circadian-regulated mRNAs encoding glycine-rich RNA-binding proteins in Pelargonium hortorum. Physiol. Plant. 1999;106:409–414. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.