Abstract
Retroelements are usually considered to be eukaryotic elements because of the large number and variety in eukaryotic genomes. By comparison, reverse transcriptases (RTs) are rare in bacteria, with only three characterized classes: retrons, group II introns and diversity-generating retroelements (DGRs). Here, we present the results of a bioinformatic survey that aims to define the landscape of RTs across eubacterial, archaeal and phage genomes. We identify and categorize 1021 RTs, of which the majority are group II introns (73%). Surprisingly, a plethora of novel RTs are found that do not belong to characterized classes. The RTs have 11 domain architectures and are classified into 20 groupings based on sequence similarity, phylogenetic analyses and open reading frame domain structures. Interestingly, group II introns are the only bacterial RTs to exhibit clear evidence for independent mobility, while five other groups have putative functions in defense against phage infection or promotion of phage infection. These examples suggest that additional beneficial functions will be discovered among uncharacterized RTs. The study lays the groundwork for experimental characterization of these highly diverse sequences and has implications for the evolution of retroelements.
INTRODUCTION
Reverse transcriptase (RT) was discovered in 1970 in two tumor-associated retroviruses, and subsequently in a variety of eukaryotic elements including long terminal repeat (LTR) and non-LTR retroelements, hepadnaviruses and telomerase (1–4). (In this article, we define a retroelement as any element containing a putative RT, regardless of its ability to retrotranspose.) Retroelements are abundant in eukaryotic genomes, and often constitute large portions of nuclear DNA, including ∼40% of the human genome and ∼60% in maize (5,6). In general, eukaryotic retroelements are thought to survive as selfish DNAs and to have deleterious effects on their hosts (7–9). It is not surprising, however, given their prevalence that beneficial roles have occasionally evolved. For example, telomerase and the HeT-A/TART retroelements function to protect chromosomal ends (10–12). Still, such adaptations remain the exception, and as a group the elements survive by spreading faster than they are lost. Thus, retroelements have expanded dramatically and are one of the major hallmarks of eukaryotic genomes.
In bacteria, reverse transcriptase was discovered considerably later, as a component of retrons (13,14). Characterized bacterial retroelements have now grown to three families, and include retrons, group II introns and diversity-generating retroelements (DGRs) (15–18). In contrast to eukaryotic elements, bacterial retroelements are not known to accumulate to high copy numbers within a genome, with the greatest number being 28 group II introns (∼1% of the genome) in the cyanobacterium Thermosynechococcus elongatus (19). Bacterial retroelements are also distinctive because they are structurally simple. Group II introns and retrons encode a single polypeptide, while some viral retroelements encode over a dozen polypeptides (20).
Group II introns are the best-characterized bacterial retroelements and the only type known to exhibit autonomous mobility. These retroelements consist of an RT encoded within a catalytic, self-splicing RNA structure. RT domains 1–7 constitute the palm and finger domains of the polymerase protein, and their sequences are generally alignable across RTs (21). In contrast, domains 0 and 2a are shared among only a subset of RT classes (22,23). Downstream of the RT domain is domain X, which corresponds to the thumb of the polymerase, and has a second function in facilitating splicing of its host intron (maturase activity) (24,25). Many group II open reading frames (ORFs) encode an endonuclease domain of the HNH family that is important for the mobility mechanism (26). Intron mobility is mediated by an ribonucleoprotein (RNP) consisting of intron lariat and two molecules of intron-encoded protein, which site-specifically recognizes and inserts intron sequence into a ∼20- to 35-bp DNA target (17). The mechanism, known as target-primed reverse transcription (TPRT), is well characterized and there are several variations (27,28).
The second type of bacterial RT, DGRs, does not appear to be mobile, but instead functions to diversify DNA sequences (18). For the phage BPP-1 of Bordetella pertussis, the retroelement consists of the RT, an RNA template (TR), an accessory protein (atd) and a target protein gene (mtd) that contains a C-terminal variable region (VR) (29). The role of the DGR is to produce diversity in the VR sequence of the phage tail protein, which is the region that contacts the bacterium during infection. In doing so, the DGR mediates tropism switching and allows the phage to infect cells with altered surface composition, as occurs when Bordetella cells alternate between pathogenic and free-living phases. RTs of DGRs appear to be more closely related to group II introns than retrons (30,31; our unpublished data).
Retrons, although discovered first, are the most unexplained of bacterial retroelements. They do not appear to be independently mobile, nor has a clear phenotype been associated with them. Retrons consist of an RT and overlapping multicopy single-stranded DNA/RNA (msRNA/msDNA) genes. The RNA transcript of the msRNA/msDNA sequence folds into a specific secondary structure to which the RT binds (32). The RT partially reverse transcribes the RNA to form a branched RNA–DNA molecule linked by a 2′–5′ bond. This msDNA accumulates to high levels in some cases, but it remains unknown what purpose the msDNA serves, for either the retron or the host cell (15,16,33).
An additional type of retroelement has been tentatively identified, but is not well characterized. We refer to these elements here as Abi (abortive phage infection) retroelements. The bacterium Lactococcus lactis encodes over 20 genetic systems that defend against phage infection. Each system blocks infection at one of many points in the phage replication cycle (34). Two of these systems, abiA and abiK, encode genes related to RT (35,36). Mutations within the RT motifs of abiK block abortive infection in vivo (35); however, AbiA and AbiK proteins have not been shown biochemically to have RT activity. An analogous property has been reported for orf570 of the P2-like phage P2-EC30, which is associated with resistance to phage T5 (37).
In the course of identifying group II introns in bacterial genomes, we found a number of RTs that appeared not to belong to group II introns, retrons or DGRs. Consequently, we undertook a comprehensive search and analysis of RTs in eubacteria, archaea, and phages to examine whether novel RT classes exist, and to determine how many types there might be. An independent study has previously identified eight novel lineages of retroelements by analyzing gene neighborhoods of the RTs (38). Here, we identify an additional 9 groupings and 38 unclassified RTs and present a comprehensive overview of RTs in bacteria.
METHODS
Compilation of the RT data set
RTs were identified in GenBank (July 2007) using PSI-BLAST (39) with a set of 25 RT queries (Supplementary Table S1). Fifteen of these RTs have been previously identified in searches designed to find distant RT relatives of group II introns, retrons and DGRs. For example, BLAST searches with AbiA and AbiK sequences produced weak hits having RT sequence motifs, and using these hits as queries in further cycles, a collection of putative RT ORFs was acquired. In PSI-BLAST, the first iteration identifies closely related sequences by standard BLASTP, while subsequent iterations construct a PSSM (position specific scoring matrix) based on the results of the previous cycle, and this sequence profile is then used as a query to identify more distant relatives. In practice, we find that ORFs acquired in the first search are sometimes absent in subsequent iterations, and that later iterations often result in only a small number of new sequences and can result in artifactual hits. Thus, we chose to use a large number of queries and a relatively small number of iterations to maximize the efficiency of the search. After each iteration, sequences with an e-value of <0.005 were retained. Iterations 2–4 recovered from 368 to 625 sequences, and pooling the results of all four iterations resulted in approximately 600–800 sequences. By using a variety of queries for PSI-BLAST, it was found that all group II intron queries resulted in highly similar RT sets (∼95% identical). Importantly, ORFs recovered from each query invariably included distantly related RTs (e.g. group II RTs identified Abi RTs and vice versa), and there was a large degree of overlap in the ORF collections regardless of the initial query. Equivalent searches were done for eubacteria, archaea, and phages and proteins smaller than 100 amino acids were discarded as probable fragments or nonspecific hits. The final collection of putative RTs was 1049 sequences, which included 11 RTs added during subsequent BLAST searches and analyses of novel groups.
Classification of sequences
For practical purposes, the 1049 RTs were divided into 10 sets, and each set was aligned with 32 reference sequences of group II introns, retrons, DGRs and Abi elements using CLUSTAL W (40). The reference sequences were chosen to represent the known phylogenetic breadth of each RT group (Supplementary Table S2). Initial phylogenetic analyses were done using the NJ algorithm with uncorrected distances as implemented in PAUP 4.0b10 (41). Sequences belonging to group II introns, retrons and DGRs were inferred by identifying the smallest monophyletic group including all reference sequences for a particular type of RT element. These groups were then aligned automatically, and refined manually in Bioedit v7.0.5.2 (42) in order to identify sequence hallmarks unique for each group (domain X for group II introns, regions X and Y for retrons, TR–VR repeats for DGRs). Other groupings were determined based on the identification of nearest neighbors using local BLAST and reciprocal BLAST, and examination for synapomorphies in the ORF regions outside the RT domains, i.e. appended domains and other shared sequences. For some RTs, additional phylogenetic analyses were done to confirm monophyly of classes and/or relationships to previously identified elements. This was done using the maximum likelihood criterion with the RtREV model (43) including an invariant sites parameter as implemented in RAxML (44). For these analyses, 100 bootstrap replicates were also completed. Due to limited number of alignable characters, this could only be done for a subset of the RTs. Specifically one tree was made for group II introns and the group II-like classes (213 amino acids with 112 RTs), a second tree of retrons (157 amino acids with 96 retrons) and a third one of DGRs (data not shown).
RESULTS AND DISCUSSION
Compilation and classification of bacterial RT genes
RT ORFs in eubacteria, archaea and phages were identified in GenBank using PSI-BLAST and a diverse set of 25 bacterial RT queries (see Methods section). The queries included ORFs representing group II introns, retrons, DGRs and Abi elements, as well as 15 RTs previously identified as unlikely to belong to these classes (Supplementary Table S1). For each query, four iterations of PSI-BLAST were done and the hits from all four searches were pooled. In preliminary searches, we found that this search strategy (i.e. relatively few iterations with a large number of queries) was the best method for retrieving the most comprehensive and diverse set of sequences (see Methods section for further details). The results of the 25 searches were pooled into a single nonredundant set for a total of 1049 putative RT ORFs (Supplementary Data S1).
In order to test the thoroughness of this strategy, we repeated the PSI-BLAST search at the end of the study using three newly identified RTs (gi|124009057, gi|91202364, gi|89895550) which are not closely related to any initial queries. The purpose was to find whether ‘outlier’ RTs would identify even more diverse RT sequences. However, these queries primarily retrieved the RTs found in the initial search, and no new groups were discovered. For this reason, as well as the overall redundancy of RTs found when using different queries, we believe that the search strategy approached saturation, and that our data set reflects the composition of RTs present in GenBank.
The collected RT sequences are highly diverse, with fewer than 100 amino acids alignable across the set, thus preventing robust phylogenetic analyses (below). Nevertheless, neighbor-joining (NJ) trees were useful in the initial steps of clustering related sequences and identifying the RTs of group II introns, retrons and DGRs. NJ trees were constructed in batches of approximately 100 unknown ORFs along with 30 reference sequences spanning the diversity of group II introns, retrons and DGRs (Supplementary Table S2). ORFs were putatively assigned to one of these classes based on the NJ trees, and further alignments within each subset allowed confirmation and refinement of the classifications based on sequence hallmarks. Identifications of group II introns were supported by the presence of a domain X motif (25), and also by comparison with our highly curated intron database [(45); www.fw.ucalgary.ca/group2introns/]. In some cases, group II introns were confirmed by generating intron RNA secondary structures. In this way, 742 ORFs were designated group II introns, although many are duplicate copies or fragments (below). Similarly, 113 ORFs were assigned as retron RTs and were supported by the sequence hallmarks X and Y (15). Thirty-six DGRs were verified by identification of potential TR–VR repeats adjacent to the RT (29).
Classification of Abi RTs is problematic because the three elements reported to have abortive infection properties (AbiA, AbiK and Abi-P2) are distantly related, with only three RT domains alignable between them (Figure 1). One possibility is that Abi elements constitute an extremely diverse but monophyletic lineage of RTs involved in phage abortive infection. Alternatively, multiple lineages of RTs may have evolved roles in abortive infection. In either case, one cannot confidently conclude which other RTs in the set might have abortive infection properties. Hence, we assign only three close relatives as belonging to the AbiK group and eight to the Abi-P2 group, while AbiA has no close relatives.
Figure 1.
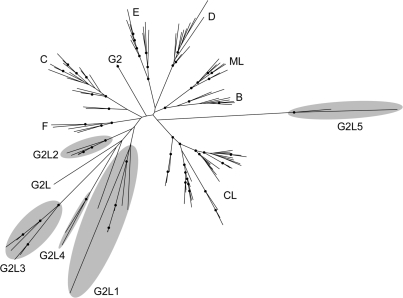
RTs identified in eubacterial, archaeal and phage genomes. RTs were classified based on limited phylogenetic analyses, BLAST scores and sequence examination for synapomorphies. The number of RTs per group is indicated in parentheses and by triangle size. Domain structures are depicted in black (alignable with group II introns), gray (uncertain alignment) and white (lack of alignability). Additional domains, including domains of unknown function (DUFs), are in light gray squares, with parentheses indicating that some members lack the domain. In this figure, the designation of an extension as a DUF indicates that the amino acid sequence is conserved within the group. Groups corresponding to lineages in Kojima and Kanehisa (38) are as follows: G2L1 [H1, H2, H3 in (38)]; G2L2, (H4); UG1 (F1); UG3 (G); UG5 (F2); UG6 (E); UG8 (G); UG9 (D); retrons (A); and DGRs (B, C).
The remaining 146 ORFs were further analyzed by manual alignment and BLASTP searches to confirm that they are bacterial RTs rather than nonspecific BLAST hits. In total, 121 ORFs were alignable with RT motifs, but three of these were discarded as eukaryotic RTs. In the end, 80 RTs were assigned to 14 additional groups based on reciprocal local BLAST best-hits, shared features outside of the RT domains, and in some cases phylogenetic support (Figures 1 and 2). Thirty-eight RTs remain unclassified because they form less convincing groups or groups of two members.
Figure 2.
A maximum likelihood tree of group II introns and group II-like (G2L) RTs. Group II introns include representatives of the major intron classes [B–F, mitochondria-like (ML), chloroplast-like (CL)] as well as one that is currently unclassified (G2). Group II-like classes (G2L1–G2L5) are highlighted with gray ovals. Nodes that have a bootstrap value ⩾70 are indicated with black circles. The tree file with GenBank gene identification numbers is available in Supplementary Data S2.
Classes of RTs in bacteria
Group II introns
The largest category of RTs in bacteria is the family of group II intron-encoded proteins, with 742 sequences. Of these, approximately 219 are full-length, unique introns (www.fw.ucalgary.ca/group2introns/), and the remaining are either truncated copies, multiple copies within genomes or redundant GenBank entries. Even so, they comprise the largest group of bacterial RTs. Based on phylogenetic analyses of the RT, the introns fall into eight classes (A, B, C, D, E, F, ML and CL) with each class being associated with a distinct RNA secondary structure (45). All of these classes are expected to possess the canonical properties of group II introns, namely ribozyme-based splicing and retromobility.
Group II intron-like RTs
A number of RTs initially appeared to be group II introns, but no associated RNA structures could be found. While it is not possible to exclude the existence of an associated ribozyme, there is no secondary structure motif similar to the highly conserved catalytic domain 5 of known group II introns. Hence, if these RTs are associated with introns, their ribozyme structure would have to be significantly different from known group II introns, or their introns might be encoded elsewhere in the genome.
A phylogenetic tree of the group II-like (G2L) RTs along with a selection of group II intron RTs shows that they fall into five classes (G2L1–G2L5) (Figure 2; Supplementary Data S2). It is important to note that G2L1 is paraphyletic; however, all members share a conserved C-terminal extension and other genomic features that suggest a similar function (below). A notable feature of the G2L classes is the conservation of the maturase (splicing) motif of domain X in three groups (G2L3, G2L4 and G2L5) despite the apparent lack of an intron RNA structure. In addition, G2L5 RTs have a conserved sequence change at the active site of the protein in domain 5 (DD to DN), suggesting that the protein has lost polymerase activity, and possibly has acquired another activity in conjunction with its approximately 500 amino acids N-terminal extension.
While RTs of G2L3-5 have few clues to suggest specific properties, all RTs of G2L1 and G2L2 are associated with CRISPR elements (Clustered Regularly Interspaced Short Palindromic Repeats). CRISPR elements are a recently identified type of genetic cassette and repetitive array found widely across eubacteria and archaea (46,47). The function of CRISPR elements is to provide resistance against phage infections. After a CRISPR-containing bacterium is infected by phage, resistant bacteria emerge which have phage sequence integrated into the CRISPR arrays, and the bacteria are resistant to future infections by that phage (48). Resistance is believed to be mediated by an enzymatic complex composed of several proteins and an RNA generated from the CRISPR array (46). The mechanism for sequence integration into CRISPR arrays remains unknown.
Essentially all CRISPR elements contain a CRISPR-associated gene (cas1), which is a putative integrase and contains the conserved domain COG1518. In G2L1 ORFs, the COG1518 domain is fused to the RT ORF as approximately 400 amino acids C-terminal extension, while for G2L2 RTs this domain is a free-standing ORF located 31- to 501-bp downstream of the RT gene. It is tempting to speculate that G2L1 and G2L2 RTs are involved in the insertion of new phage sequences into the CRISPR arrays using a mechanism analogous to TPRT insertion of group II introns (49). However, only a minority of CRISPR elements have an associated RT gene (50), and so such a mechanism is unlikely to be general. A final note is that the paraphyletic nature of the CRISPR-associated RTs (Figure 2) indicates that RTs were independently acquired by CRISPR elements at least twice. Consistent with this, the RT-associated COG1518 domains are themselves not a monophyletic lineage (50).
Retrons/Retron-like RTs
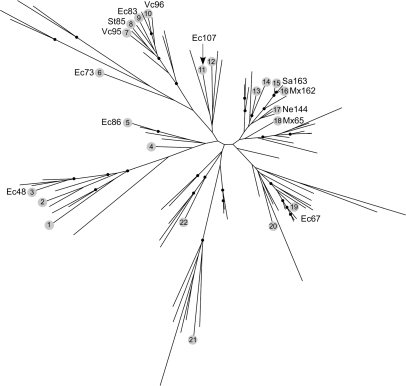
The retron group is the second largest in bacteria with 113 members, of which 97 are unique and 16 are highly similar sequences usually from related strains. Members of this group share the retron-specific regions X and Y (15). Region Y lies within RT domain 7 and has been shown to be functionally important for recognizing the msRNA structure for the priming reaction (32). The role of region X, which lies between domains 2 and 3, is less clear but also seems to be important for the synthesis of msDNA. The retron/retron-like group is highly diverse in sequence, making it difficult to ascertain whether all of its members have the same properties as characterized retrons. We did not attempt to identify potential msRNA/msDNA secondary structures as a means of verification because we considered the likelihood of false positives to be too great. Phylogenetic analysis of the RTs shows that characterized retrons are found throughout much of the tree (Figure 3; Supplementary Data S3). Moreover, RTs identified as retrons in the literature without experimental verification (15) are even more widely distributed. Thus, it seems likely that most RTs in this group are actual retrons. This set of retron RTs is a significant increase from approximately 30 that were previously reported (15).
Figure 3.
A maximum likelihood tree of unique retron and retron-like RTs. Black dots indicate nodes with bootstrap support >70. Gray circled numbers designate sequences that have previously been identified as retrons [see Ref. (15)]. The GenBank gene identification number for each circled number in the figure is as follows: 1 (94310415), 2 (8100799), 3 (1519451), 4 (15925199), 5 (134078), 6 (28882460), 7 (15642382), 8 (39546377), 9 (42501), 10 (22036084), 11 (42775), 12 (19703506), 13 (113475791), 14 (23130511), 15 (115376572), 16 (556277), 17 (17530191), 18 (108757956), 19 (134075), 20 (21233055), 21 (17230453) and 22 (39996465). Those sequences with experimental support are shown with a gray circle and their previously assigned name (15). The tree file with GenBank gene identification numbers is available in Supplementary Data S3.
DGRs
Our set of DGRs includes 36 RTs found in eight phyla of bacteria and two bacteriophages. All of the RT sequences are flanked by potential TR–VR repeats, consistent with known DGR features. These DGRs are expected to possess equivalent diversity-generating properties for their target VR sequence as shown for the BPP-1 phage of B. pertussis (29). As previously noted (18), at least some of the DGRs appear not to be phage-associated, suggesting that the target genes are not restricted to phage tail proteins.
Other RT groups
We identified three groups of Abi RTs whose members are putatively involved in defense against phage infections. The three groups are relatively small, having one, three and eight members, respectively for AbiA, AbiK and Abi-P2 groups. None of the three groups have lengthy domains appended to the RT. Shorter amino acid extensions can be found (less than 200 amino acids) which may encode additional functions, but these sequences are not conserved across bacteria.
Nine classes of ‘unknown group’ RTs were defined (UG1-9) having 3–14 members each. Four of these groups have C-terminal extensions ranging in length from approximately 200–800 amino acids, which may contribute novel biochemical activities (Figure 1). The appended domains are conserved in sequence within the groups with the exception of UG6. The C-terminal domains are not highly similar to proteins of known function, although the UG5 extension gives a very weak match to a carbon–nitrogen hydrolase domain.
Thirty-eight RTs remained unclassified because they fell into less convincing groupings or formed pairs of related RTs. Ten of these RTs are obviously truncated, while we predict that many of the remainder are functional due to the absence of stop codons in their long ORFs. Most unusual among these RTs are two single examples (gi|118579436 and gi|42524098) with long extensions, of which one (gi|42524098) has similarity to a primase at the N-terminus (Figure 1). These two RT examples are likely to possess novel activities or to be involved in novel processes because of the very lengthy sequences appended to the RT domains.
In Kojima and Kanehisa (38), eight novel RT groups were identified in bacteria, based partially on fused and flanking genes. The RT groups identified here are consistent with that study, although we identify an additional nine groups (G2L3, 4, 5, UG2, 4, 7, AbiA, AbiK and Abi-P2), and 38 unclassified ORFs [see Figure 1 legend for correspondence with (38)]. Interestingly, Kojima and Kanehisa (38) observed that RTs of UG3 and UG8 are exclusively found in pairs, in a retroelement they call Group G, and this observation holds true with the slightly expanded number of RTs in our set. Another goal of that study was to examine gene neighborhoods. The most significant observations were four RT lineages reported to be flanked by genes for DNA polymerase A, a primase or a hydrolase. We examined the immediate flanking sequences (±10 000 kb) of the new groups of RTs found in our study, but did not find any conserved flanking genes that would further characterize the retroelements.
Diversity of bacterial RTs
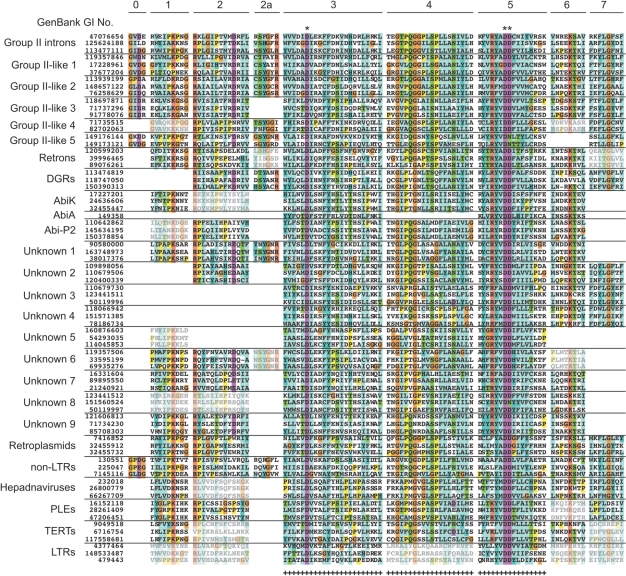
The set of compiled bacterial RTs is remarkably diverse, with only 59 amino acids alignable across the entire set (Figure 4). Table 1 presents data on sequence diversity for each of the RT groups. The most valid measure may be the average pairwise distance of the RTs within each group, using only the 59 characters alignable across all RTs. By that measure, the retron/retron-like group is more diverse than group II introns (i.e. it has a greater average pairwise distance), while DGRs have roughly the same diversity. Seven other classes have equal or greater pairwise distances compared to group II introns, suggesting at least equal diversity despite fewer members in the groups.
Figure 4.
Alignment of RT domains. An alignment of RT domains 0–7 is shown for representative bacterial and eukaryotic RTs. The predicted three catalytic residues at the RT active site are indicated with asterisks. Residues judged to be alignable across all bacterial RTs (and used in distance calculations for Table 1) are indicated with a plus sign at the bottom of the alignment. Absence of sequence in the figure indicates the lack of alignability rather than the absence of amino acids. Sequences considered ambiguously aligned (based on a more comprehensive set of RTs than those depicted in the figure) are indicated with light shading.
Table 1.
RT groups in bacteria
| RT class | Numbera | ORF length (amino acids) |
Average pairwise distance |
|||
|---|---|---|---|---|---|---|
| Range | Average | Alignable charactersb | Across classesc | Within classd | ||
| Group II introns | 217e | 282–769e | 498e | 177 | 0.50f | 0.56f |
| Retrons/retron-like | 96 | 265–658 | 405 | 157 | 0.56 | 0.65 |
| DGRs | 33 | 303–504 | 377 | 179 | 0.49 | 0.58 |
| Group II-like 1 | 6 | 316–731 | 643 | 194 | 0.40 | 0.52 |
| Group II-like 2 | 4 | 431–447 | 439 | 212 | 0.34 | 0.38 |
| Group II-like 3 | 5 | 443–478 | 466 | 157 | 0.44 | 0.52 |
| Group II-like 4 | 2 | 415–418 | 417 | 218 | 0.51 | 0.50 |
| Group II-like 5 | 2 | 420–930 | 675 | 126 | 0.56 | 0.56 |
| Unknown group 1 | 7 | 1184–1317 | 1255 | 190 | 0.44 | 0.53 |
| Unknown group 2 | 14 | 394–574 | 451 | 207 | 0.48 | 0.57 |
| Unknown group 3 | 5 | 406–444 | 425 | 167 | 0.58 | 0.66 |
| Unknown group 4 | 4 | 278–347 | 324 | 151 | 0.56 | 0.61 |
| Unknown group 5 | 7 | 969–1067 | 1018 | 189 | 0.57 | 0.62 |
| Unknown group 6 | 4 | 448–1062 | 909 | 130 | 0.51 | 0.52 |
| Unknown group 7 | 3 | 505–525 | 517 | 218 | 0.50 | 0.61 |
| Unknown group 8 | 5 | 645–690 | 669 | 298 | 0.39 | 0.61 |
| Unknown group 9 | 4 | 524–605 | 562 | 229 | 0.30 | 0.39 |
| Abi-P2 | 6 | 456–589 | 526 | 218 | 0.35 | 0.53 |
| AbiK | 3 | 590–610 | 600 | 213 | 0.25 | 0.41 |
aThe number of unique full-length RTs. Sequences omitted from the table are redundant copies (⩾90% identical to another RT) and sequences missing more than 50 amino acids at either the N- or C-terminus of the RT domain.
bNumber of alignable characters within RT domains 1–7. In cases where groups lacked strong similarity for one or more domains, sizes were estimated based on comparisons with group II introns (Figure 1).
cAverage pairwise distance for each group based on the 59 characters in RT domains 3–5 that could be aligned across all RTs in this study.
dAverage pairwise distance for each group based on alignable characters within the predicted RT domains 1–7 for that group.
eBased on set of introns in Ref. (35).
fBased on a representative set of group II introns.
A limitation of this measurement is its reliance on a small number of characters, all of which are likely to be highly constrained. To increase the number of characters, pairwise distances were determined based on all alignable positions for RT domains 1–7 within each class (130–298 amino acids; column 5 in Table 1). These distances are based on different character sets for each group, but nevertheless the same conclusion is suggested: retrons are more diverse than group II introns, DGRs roughly equal and seven other groups have at least as much diversity as group II introns. A third measure is simply the number of alignable characters within each class, across RT domains 1–7. Consistent with the previous conclusions, retrons have fewer alignable characters than group II introns (157 versus 177), DGRs have about the same number (179 versus 177) and five other groups have fewer alignable characters (126–167 amino acids). Considering these data together, it is clear that RTs in bacteria are remarkably diverse in sequence, and they may be functionally diverse as well.
Interestingly, a comparison with eukaryotic RT sequences suggests greater diversity among bacterial RTs. This inference is drawn from the observation that domains 1, 2 and 7 are unalignable for many bacterial RTs, while obviously present in most eukaryotic RTs (Figure 4). In addition, some bacterial RTs have extreme amino acid substitutions. For example, in the Y/FxDD motif of the catalytic site, the x is nearly always hydrophobic (A, I, V and M), whereas the bacterial group Abi-P2 has a conserved R.
Mobility of the retroelements
It is striking that among this set, group II introns are the only RTs with clear evidence of retromobility. Mobility of the introns is indicated in sequence data by the many redundant and truncated copies, and specifically by multiple intron copies at different locations in a single genome. Furthermore, the exact boundaries of the mobile DNA unit can be determined when identical introns are found in different exons. Among other RTs, the only other possible example of active retromobility is for UG4, where two RTs (gi|151571385 and gi|151571856) are >90% identical and are found within the same genome (Francisella tularensis subsp. novicida GA99-3548). The flanking sequences are identical for ∼100 bp upstream of the RT ORFs, but there is no discernable identity in their 3′-flanking regions. This example is the only instance apart from group II introns in which we find closely related RTs within the same genome. We looked for additional examples of potential mobility by comparing the genomic locations of highly similar RTs found in closely related organisms. In all cases, these RTs were found in identical genomic locations. In addition, we found no lengthy conserved target site duplications that could signify recent mobility. The lack of evidence for independent mobility of RTs (other than group II introns and UG4) suggests either that they are not active for retromobility, or that they transpose too infrequently to detect by these criteria.
On the other hand, the RTs do not appear to be inherited strictly vertically, as suggested from their patchy distributions. For example, the retrons in our collection are found in a large variety of species, but are on the whole infrequent across bacteria. The distribution of retrons within closely related strains has previously been shown to be highly sporadic for Escherichia coli, Klebsiella, Proteus and Salmonella (50,51), while there appears to be a degree of vertical inheritance in myxobacteria (15). The simplest explanation for this sporadic phylogenetic distribution is RT-independent horizontal transfers, which is also consistent with the finding of some retrons within prophages (53). DGRs and other groups of RTs similarly have wide and irregular distributions, suggesting nonvertical inheritance (31). Kojima and Kanehisa (38) made the same observation for UG1, UG3/8, UG5, UG6 and UG9.
Functions of the retroelements
With the exception of G2L5, all groups in this study possess three aspartate residues in motifs that putatively correspond to the active site (Figure 4; Supplementary Figure S1), which is consistent with having RT activity. We consider it likely that all of these protein families fold into a 3D structure similar to other RTs and have the corresponding enzymatic activities. Although there are a number of examples in which RT domains are unalignable (Figures 1 and 4), in these cases there are sufficient amino acids to correspond to the ‘missing’ domains, with the possible exception of domain 2a. Thus, the absence of domains in Figures 1 and 4 signifies that the sequences cannot be aligned unambiguously, but does not necessarily imply that the structural domains are absent from the proteins.
In general, it is likely that an element with RT activity will survive using one of two strategies (or a combination of these). An RT may persist by helping the element propagate as a selfish DNA, or alternatively by providing a benefit to the host organism. Group II introns are clearly selfish DNAs, because they do not have a discernable host-related function and are mobile. In contrast, five other groups of RTs appear to belong to the second category and confer a useful function. Interestingly, all of these functions pertain to either resistance to phage infections [CRISPRs (G2L1, G2L2), AbiA, AbiK and Abi-P2] or promotion of phage infections (DGRs). The lack of evidence for active retrotransposition of other uncharacterized RTs suggests that they have useful functions, perhaps playing roles in phage–host or phage–phage conflicts.
In considering possible roles of the RTs, it is notable that 11 different extensions are fused to the RT domains (Figure 1), suggesting that a variety of biochemical activities are coupled to reverse transcription. These domains include an endonuclease (group II introns), an integrase (G2L1) and a primase (unclassified), while the remainder have no predicted function. One difficulty in predicting function based on sequence alone is illustrated by CRISPR RTs, which are closely related to group II intron RTs (particularly G2L2; Figure 2), yet are implicated in a very different biological process. This observation cautions against assuming shared properties for related RTs, and greatly expands the plausible range of activities and processes that might be carried out by the collection of RTs.
Phylogenetic distribution of RTs
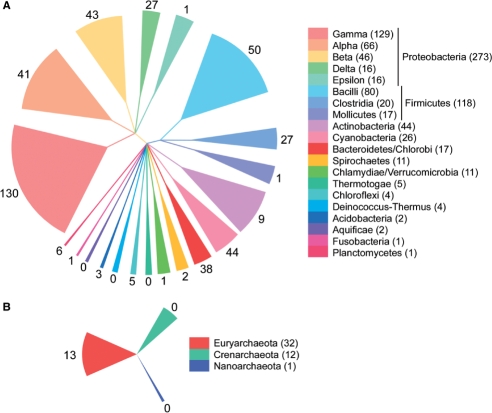
To address the phylogenetic distribution in eubacteria, we considered the RT content of 519 completed genomes. Approximately 25% of the genomes have at least one RT, with 9% containing multiple RTs. These numbers are lower estimates, as they are based on a set of nonredundant RTs, such that very closely related RT copies within a genome (i.e. group II introns and UG4) are counted as a single RT. The RTs are widely distributed across taxa, being found in nearly every eubacterial group (11/14 as defined by NCBI) (Figure 5; Supplementary Table S3). Bacterial groups that lack an RT are also underrepresented in GenBank, as indicated by the number of completed genome sequences (Figure 5). Similarly, the number of RTs in each group is roughly proportional to the number of sequenced genomes, although there are exceptions (compare bacteroidetes/chlorobi group versus mollicutes) (Figure 5). We conclude that RTs do not appear to be excluded from particular bacterial groups or have extreme biases in their distribution.
Figure 5.
Phylogenetic distribution of RTs in eubacteria (A) and archaea (B). The size of each sector represents the number of sequenced genomes in GenBank as of 31 July 2007, with actual numbers shown in parentheses beside the phyla listed to the right. Sector colors specify the bacterial group classification. Numbers beside each sector indicate unique full-length RTs within each bacterial phylum (Supplementary Table S3 and footnotes).
In contrast, RTs are far less frequent in archaea in our data set. Archaeal RTs are mainly restricted to euryarchaeota, and almost all are group II introns, except for a single retron and two full-length unclassified RTs from environmental samples, where the precise organismal classification is unclear (Figure 5; Supplementary Table S2). Interestingly, the RTs found within archaea do not constitute an independent lineage. If archaeal-specific lineages exist, it would seem that they must be too divergent from known RTs to identify by our search strategy, or are sufficiently rare to be missed in our sample size.
Evolutionary implications
The large number of RT types in bacteria raises many new possibilities for the origins of eukaryotic retroelements. It is frequently hypothesized that eukaryotic RTs, and specifically non-LTR elements, are descended from group II introns (54–56). This is based on the similar RT sequences and mobility mechanisms of group II introns and non-LTR retroelements, and also the general increase in complexity of retroelements in eukaryotes. Also supporting this idea is the fact that group II introns are present in organelles, and could be introduced into nuclear chromosomes via the well-known process of endosymbiotic gene transfer (57). Although there are now many more potential bacterial ancestors of non-LTR elements, we still consider group II introns to be the most likely source, because they are the only bacterial RTs that are clearly mobile.
An alternative to a single bacterial ancestor of eukaryotic RTs is multiple introductions of bacterial RTs into eukaryotes. This possibility is made more plausible by the findings in this study. However, due to high sequence divergence, this issue cannot be resolved by phylogenetics alone. A comparison of eukaryotic and prokaryotic sequences in Figure 4 does not reveal to us clear evidence between individual eukaryotic and prokaryotic groups that would suggest a specific bacterial source for a eukaryotic RT. Thus, the available sequence data appear equally consistent with either a single or multiple event introduction of RTs into eukaryotes.
Although reconstruction of global phylogenetic relationships is not possible, the data do suggest that group II introns can readily form derivatives and acquire new functions. This inference is based on the multiple lineages of group II intron-related RTs, of which two have adapted roles in phage defense (G2L1 and G2L2), one has apparently lost RT activity (G2L5), and two have unknown functions. While it is possible that one of these G2L classes was ancestral to the cluster depicted in Figure 2 rather than group II introns, this seems less likely given the increased opportunity that group II introns have to form derivatives, due to their abundance, diversity and mobility. The ability of group II introns to evolve into new forms is consistent with the versatility expected for the putative ancestors of spliceosomal introns.
CONCLUSIONS
Reverse transcriptases in bacteria appear to be fundamentally different from those in eukaryotes. They differ dramatically in prevalence across species and in abundance within genomes. Eukaryotic retroelements are found in nearly every species, and often comprise a large proportion of a genome. In contrast, most bacteria do not contain a RT and when present they are far less abundant within the genome (≤1%). This pattern mirrors that of mobile DNA in bacteria in general. The lower content of bacterial mobile elements is thought to reflect the large population sizes with concomitant increase in efficacy of selection (58). Based on this logic, we predict that a greater proportion of RTs in bacteria will be advantageous to their hosts compared to eukaryotic elements, a hypothesis consistent with data in this study.
Finally, it is clear that with currently accumulated sequence data, we have not yet reached saturation with regard to the number of bacterial RT groups. This conclusion is based on the number of RT types having only one or a few representatives in the data set. It is virtually assured that additional retroelement types are yet to be discovered beyond the 20 groups and 11 architectures set forth here. Based on the unique biochemical reactions carried out by known bacterial retroelements (reverse splicing into DNA, retrohoming, mutagenic homing and msDNA synthesis) and their range of biological processes (proliferation as selfish DNA, tropism switching and abortive phage infection), there is great potential for additional novel properties to be found among the diversity of RTs in bacteria.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institutes of Health Research (MOP-49457). Alberta Ingenuity Foundation (salary support to D.M.S.); Alberta Heritage Foundation for Medical Research (to S.Z.). Funding for open access charge: Canadian Institutes of Health Research (MOP-49457).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Sylvain Moineau (U. Laval) for helpful discussions.
REFERENCES
- 1.Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970;226:1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- 2.Temin HM, Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970;226:1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- 3.Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 4.Eickbush TH. Origin and evolutionary relationships of retroelements. In: Morse SS, editor. The Evolutionary Biology of Viruses. New York: Raven Press; 1994. pp. 121–157. [Google Scholar]
- 5.Haberer G, Young S, Bharti AK, Gundlach H, Raymond C, Fuks G, Butler E, Wing RA, Rounsley S, Birren B, et al. Structure and architecture of the maize genome. Plant Physiol. 2005;139:1612–1624. doi: 10.1104/pp.105.068718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hua-Van A, Le Rouzic A, Maisonhaute C, Capy P. Abundance, distribution and dynamics of retrotransposable elements and transposons: similarities and differences. Cytogenet. Genome Res. 2005;110:426–440. doi: 10.1159/000084975. [DOI] [PubMed] [Google Scholar]
- 7.Boissinot S, Davis J, Entezam A, Petrov D, Furano AV. Fitness cost of LINE-1 (L1) activity in humans. Proc. Natl Acad. Sci. USA. 2006;103:9590–9594. doi: 10.1073/pnas.0603334103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynch M. The Origins of Genome Architecture. Sunderland, MA: Sinauer associates; 2007. pp. 151–192. [Google Scholar]
- 9.Pasyukova EG, Nuzhdin SV, Morozova TV, Mackay TF. Accumulation of transposable elements in the genome of Drosophila melanogaster is associated with a decrease in fitness. J. Hered. 2004;95:284–290. doi: 10.1093/jhered/esh050. [DOI] [PubMed] [Google Scholar]
- 10.Eickbush TH. Telomerase and retrotransposons: which came first? Science. 1997;277:911–912. doi: 10.1126/science.277.5328.911. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura TM, Cech TR. Reversing time: origin of telomerase. Cell. 1998;92:587–590. doi: 10.1016/s0092-8674(00)81123-x. [DOI] [PubMed] [Google Scholar]
- 12.Levis RW, Ganesan R, Houtchens K, Tolar LA, Sheen FM. Transposons in place of telomeric repeats at a Drosophila telomere. Cell. 1993;75:1083–1093. doi: 10.1016/0092-8674(93)90318-k. [DOI] [PubMed] [Google Scholar]
- 13.Lampson BC, Sun J, Hsu MY, Vallejo-Ramirez J, Inouye S, Inouye M. Reverse transcriptase in a clinical strain of Escherichia coli: production of branched RNA-linked msDNA. Science. 1989;243:1033–1038. doi: 10.1126/science.2466332. [DOI] [PubMed] [Google Scholar]
- 14.Lim D, Maas WK. Reverse transcriptase-dependent synthesis of a covalently linked, branched DNA–RNA compound in E. coli B. Cell. 1989;56:891–904. doi: 10.1016/0092-8674(89)90693-4. [DOI] [PubMed] [Google Scholar]
- 15.Lampson BC, Inouye M, Inouye S. Retrons, msDNA, and the bacterial genome. Cytogenet. Genome Res. 2005;110:491–499. doi: 10.1159/000084982. [DOI] [PubMed] [Google Scholar]
- 16.Yamanaka K, Shimamoto T, Inouye S, Inouye M. Retrons. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 784–795. [Google Scholar]
- 17.Lambowitz AM, Zimmerly S. Mobile group II introns. Annu. Rev. Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- 18.Medhekar B, Miller JF. Diversity-generating retroelements. Curr. Opin. Microbiol. 2007;10:388–395. doi: 10.1016/j.mib.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura Y, Kaneko T, Sato S, Ikeuchi M, Katoh H, Sasamoto S, Watanabe A, Iriguchi M, Kawashima K, Kimura T, et al. Complete genome structure of the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1. DNA Res. 2002;9:135–148. doi: 10.1093/dnares/9.4.135. [DOI] [PubMed] [Google Scholar]
- 20.Frankel AD, Young JA. HIV-1: fifteen proteins and an RNA. Annu. Rev. Biochem. 1998;67:1–25. doi: 10.1146/annurev.biochem.67.1.1. [DOI] [PubMed] [Google Scholar]
- 21.Xiong Y, Eickbush TH. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malik HS, Burke WD, Eickbush TH. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- 23.Zimmerly S, Hausner G, Wu X. Phylogenetic relationships among group II intron ORFs. Nucleic Acids Res. 2001;29:1238–1250. doi: 10.1093/nar/29.5.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blocker FJ, Mohr G, Conlan LH, Qi L, Belfort M, Lambowitz AM. Domain structure and three-dimensional model of a group II intron-encoded reverse transcriptase. RNA. 2005;11:14–28. doi: 10.1261/rna.7181105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohr G, Perlman PS, Lambowitz AM. Evolutionary relationships among group II intron-encoded proteins and identification of a conserved domain that may be related to maturase function. Nucleic Acids Res. 1993;21:4991–4997. doi: 10.1093/nar/21.22.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shub DA, Goodrich-Blair H, Eddy SR. Amino acid sequence motif of group I intron endonucleases is conserved in open reading frames of group II introns. Trends Biochem. Sci. 1994;19:402–404. doi: 10.1016/0968-0004(94)90086-8. [DOI] [PubMed] [Google Scholar]
- 27.Zhong J, Lambowitz AM. Group II intron mobility using nascent strands at DNA replication forks to prime reverse transcription. EMBO J. 2003;22:4555–4565. doi: 10.1093/emboj/cdg433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ichiyanagi K, Beauregard A, Lawrence S, Smith D, Cousineau B, Belfort M. Retrotransposition of the Ll.LtrB group II intron proceeds predominantly via reverse splicing into DNA targets. Mol. Microbiol. 2002;46:1259–1272. doi: 10.1046/j.1365-2958.2002.03226.x. [DOI] [PubMed] [Google Scholar]
- 29.Liu M, Deora R, Doulatov SR, Gingery M, Eiserling FA, Preston A, Maskell DJ, Simons RW, Cotter PA, Parkhill J, et al. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science. 2002;295:2091–2094. doi: 10.1126/science.1067467. [DOI] [PubMed] [Google Scholar]
- 30.Chang GS, Hong Y, Ko KD, Bhardwaj G, Holmes EC, Patterson RL, van Rossum DB. Phylogenetic profiles reveal evolutionary relationships within the ‘twilight zone’ of sequence similarity. Proc. Natl Acad. Sci. USA. 2008;105:13474–13479. doi: 10.1073/pnas.0803860105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doulatov S, Hodes A, Dai L, Mandhana N, Liu M, Deora R, Simons RW, Zimmerly S, Miller JF. Tropism switching in Bordetella bacteriophage defines a family of diversity-generating retroelements. Nature. 2004;431:476–481. doi: 10.1038/nature02833. [DOI] [PubMed] [Google Scholar]
- 32.Inouye S, Hsu MY, Xu A, Inouye M. Highly specific recognition of primer RNA structures for 2′-OH priming reaction by bacterial reverse transcriptases. J. Biol. Chem. 1999;274:31236–31244. doi: 10.1074/jbc.274.44.31236. [DOI] [PubMed] [Google Scholar]
- 33.Lampson B, Inouye M, Inouye S. The msDNAs of bacteria. Prog. Nucleic Acid Res. Mol. Biol. 2001;67:65–91. doi: 10.1016/s0079-6603(01)67025-9. [DOI] [PubMed] [Google Scholar]
- 34.Forde A, Fitzgerald GF. Bacteriophage defence systems in lactic acid bacteria. Antonie Van Leeuwenhoek. 1999;76:89–113. [PubMed] [Google Scholar]
- 35.Fortier LC, Bouchard JD, Moineau S. Expression and site-directed mutagenesis of the lactococcal abortive phage infection protein AbiK. J. Bacteriol. 2005;187:3721–3730. doi: 10.1128/JB.187.11.3721-3730.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Durmaz E, Klaenhammer TR. Abortive phage resistance mechanism AbiZ speeds the lysis clock to cause premature lysis of phage-infected Lactococcus lactis. J. Bacteriol. 2007;189:1417–1425. doi: 10.1128/JB.00904-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Odegrip R, Nilsson AS, Haggard-Ljungquist E. Identification of a gene encoding a functional reverse transcriptase within a highly variable locus in the P2-like coliphages. J. Bacteriol. 2006;188:1643–1647. doi: 10.1128/JB.188.4.1643-1647.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kojima KK, Kanehisa M. Systematic survey for novel types of prokaryotic retroelements based on gene neighborhood and protein architecture. Mol. Biol. Evol. 2008;25:1395–1404. doi: 10.1093/molbev/msn081. [DOI] [PubMed] [Google Scholar]
- 39.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swofford DL. PAUP*: Phylogenetic Analysis using Parsimony (*and other methods) 4.0b8. Sunderland, MA: Sinauer Associates; 2003. [Google Scholar]
- 42.Hall T. Bioedit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/nt. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 43.Dimmic MW, Rest JS, Mindell DP, Goldstein RA. rtREV: an amino acid substitution matrix for inference of retrovirus and reverse transcriptase phylogeny. J. Mol. Evol. 2002;55:65–73. doi: 10.1007/s00239-001-2304-y. [DOI] [PubMed] [Google Scholar]
- 44.Stamatakis A, Hoover P, Rougemont JA. Rapid bootstrap algorithm for the RAxML web-servers. Syst. Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 45.Simon DM, Clarke NAC, McNeil BA, Johnson I, Pantuso D, Dai L, Chai D, Zimmerly S. Group II introns in eubacteria and archaea: ORF-less introns and new varieties. RNA. 2008;14:1704–1713. doi: 10.1261/rna.1056108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sorek R, Kunin V, Hugenholtz P. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 48.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 49.Zimmerly S, Guo H, Perlman PS, Lambowitz AM. Group II intron mobility occurs by target DNA-primed reverse transcription. Cell. 1995;82:545–554. doi: 10.1016/0092-8674(95)90027-6. [DOI] [PubMed] [Google Scholar]
- 50.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Herzer PJ, Inouye S, Inouye M, Whittam TS. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 1990;172:6175–6181. doi: 10.1128/jb.172.11.6175-6181.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rice SA, Bieber J, Chun JY, Stacey G, Lampson BC. Diversity of retron elements in a population of rhizobia and other gram-negative bacteria. J. Bacteriol. 1993;175:4250–4254. doi: 10.1128/jb.175.13.4250-4254.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Inouye S, Sunshine MG, Six EW, Inouye M. Retronphage phi R73: an E. coli phage that contains a retroelement and integrates into a tRNA gene. Science. 1991;252:969–971. doi: 10.1126/science.1709758. [DOI] [PubMed] [Google Scholar]
- 54.Boeke JD. The unusual phylogenetic distribution of retrotransposons: a hypothesis. Genome Res. 2003;13:1975–1983. doi: 10.1101/gr.1392003. [DOI] [PubMed] [Google Scholar]
- 55.Eickbush TH. R2 and related site-specific non-long terminal repeat retrotransposons, in Mobile DNA II. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 813–835. [Google Scholar]
- 56.Temin HM. Reverse transcriptases. Retrons in bacteria. Nature. 1989;339:254–255. doi: 10.1038/339254a0. [DOI] [PubMed] [Google Scholar]
- 57.Knoop V, Brennicke A. Promiscuous mitochondrial group II intron sequences in plant nuclear genomes. J. Mol. Evol. 1994;39:144–150. doi: 10.1007/BF00163803. [DOI] [PubMed] [Google Scholar]
- 58.Lynch M. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc. Natl Acad. Sci. USA. 2007;104:8597–8604. doi: 10.1073/pnas.0702207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.