Abstract
Pokeweed antiviral protein (PAP) is a glycosidase of plant origin that has been shown to depurinate some viral RNAs in vitro. We have demonstrated previously that treatment of Brome mosaic virus (BMV) RNAs with PAP inhibited their translation in a cell-free system and decreased their accumulation in barley protoplasts. In the current study, we map the depurination sites on BMV RNA3 and describe the mechanism by which replication of the viral RNA is inhibited by depurination. Specifically, we demonstrate that the viral replicase exhibited reduced affinity for depurinated positive-strand RNA3 compared with intact RNA3, resulting in less negative-strand product. This decrease was due to depurination within the intergenic region of RNA3, between ORF3 and 4, and distant from the 3′ terminal core promoter required for initiation of negative-strand RNA synthesis. Depurination within the intergenic region alone inhibited the binding of the replicase to full-length RNA3, whereas depurination outside the intergenic region permitted the replicase to initiate negative-strand synthesis; however, elongation of the RNA product was stalled at the abasic nucleotide. These results support a role of the intergenic region in controlling negative-strand RNA synthesis and contribute new insight into the effect of depurination by PAP on BMV replication.
INTRODUCTION
Brome mosaic virus (BMV) is a positive-strand RNA plant virus with a genome comprised of three RNAs, designated RNA1, RNA2 and RNA3 (1–3). RNA1 encodes 1a protein with methyltransferase and helicase activity and RNA2 encodes 2a protein bearing motifs of an RNA dependent RNA polymerase (4–6). Both 1a and 2a proteins are essential components of the viral replicase complex, which also contains several host proteins (7,8). RNA3 is dicistronic; the 5′ proximal gene encodes a movement protein that is translated directly from RNA3, whereas the 3′ proximal gene is expressed from a subgenomic RNA4 to produce the coat protein (9–11). Replication of BMV RNAs occurs through a negative-strand RNA intermediate and is performed by the viral replicase complex (2,3).
The cis-acting elements involved in efficient replication of BMV RNAs have been characterized both in vitro, with the isolated replicase, and in vivo, by either transfecting modified RNAs into plant protoplasts or transforming them into yeast cells (12–14). With specific reference to RNA3, these studies have uncovered that the 3′ UTR of this RNA, which forms a tRNA-like structure, bears a stem-loop C that binds the BMV replicase and directs negative-strand synthesis (15,16). In addition, a minimal promoter sequence located at the 3′-end of negative-strand RNA3 interacts with the replicase to direct positive-strand initiation (12,17). The approximately 250-nucleotide intercistronic region of RNA3 contains an enhancer required for replication (18), and a cis-acting sequence essential for replicase assembly in yeast (19,20).
We have shown previously that a glycosidase isolated from the plant Phytolacca americana depurinates BMV RNAs in vitro (21). Known as pokeweed antiviral protein (PAP), this enzyme reduces the titer of several different viruses (22–24). The antiviral activity was initially attributed to host cell toxicity caused by rRNA depurination, as PAP is also a ribosome-inactivating protein (25,26). However, we and others have demonstrated the ability of PAP to directly depurinate viral RNAs (21,27). In addition, pretreatment of only BMV RNA3 with PAP, prior to transfection into barley protoplasts with RNAs 1 and 2, resulted in diminished accumulation of all BMV RNAs. An artificial RNA template containing the minimal binding site for the viral replicase showed that substitution of nucleotides with abasic residues, to mimic depurination, caused the replicase to stall during elongation (28). Here, we investigate the effect of depurination on the activity of the viral replicase using the full-length RNA3 as template. We demonstrate that depurination within the intergenic region of the positive-strand RNA3 inhibited the binding of the replicase complex and decreased the synthesis of RNA both in vitro and in vivo. Therefore, RNA sequence distant from the replicase initiation site for negative-strand synthesis influenced replicase activity. This work provides new understanding of how depurination contributes to the antiviral activity of PAP and also illustrates the utility of this glycosidase to characterize the binding requirements of the viral replicase to its template BMV RNA3.
MATERIALS AND METHODS
Treatment of BMV RNA3 with PAP
Capped, in vitro transcripts of BMV RNA3 (1.5 pmol) were incubated with PAP (34 pmol) in RIP buffer (60 mM KCl, 10 mM Tris–HCl, pH 7.4, 10 mM MgCl2) to a final volume of 50 µl for 30 min at 30°C. Untreated RNA3 was incubated in the same manner in the absence of PAP. Following incubation, RNA was extracted, precipitated in 100% ethanol and resuspended in DEPC-treated water.
Replicase assay
A fraction enriched for the BMV replicase was isolated by gel chromatography from infected barley leaves as described previously (29). Each replicase assay consisted of a 40 µl reaction containing 50 mM sodium glutamate, pH 8.2, 12 mM MgCl2, 8 mM DTT, 1 mM ATP, 1 mM GTP, 1 mM UTP, 0.4% Triton X-100, 4 µCi [α32P]-CTP (10 mCi/ml), 1.5 pmol PAP-treated or untreated BMV RNA3, and 14 µl of RdRp fraction. Following incubation for 90 min at 30°C, the reaction products were extracted and precipitated with 6 volumes 100% ethanol, 0.4 M ammonium acetate, and 10 µg of glycogen. The RNA products were separated in 7 M urea/4.5%, 12% or 20% acrylamide gels, dried and visualized by exposure to X-ray film.
Assessment of RNA3 integrity
PAP-treated and untreated BMV RNA3s (3 pmol) were incubated in replicase buffer only (50 mM sodium glutamate, pH 8.2, 12 mM MgCl2, 8 mM DTT, 1 mM ATP, 1 mM GTP, 1 mM UTP, 0.4% Triton X-100) or in replicase buffer containing RdRp for 30 min at 30°C. Aliquots were removed at various time points (0, 50 and 80 min) and RNA was separated in a 7 M urea/4.5% acrylamide gel and transferred to nylon membrane. To detect BMV RNA3, the membrane was probed with a 200-nucleotide radiolabeled RNA complementary to the 3′ end of BMV RNA3 (pB3HE1; 28). Blots were visualized by exposure to X-ray film.
Mapping of BMV RNA3 depurination sites
Capped, in vitro transcripts of BMV RNA3 were treated with PAP or untreated as described above, and primer extension analysis was used to detect depurination sites. Reverse primers spanning approximately every 200 nucleotides of BMV RNA3 were 5′ end-labeled with T4 polynucleotide kinase for 1 h at 37°C. BMV RNA3 (0.3 pmol) was incubated with 2.5 × 105 cpm of cDNA3 probe and extended with reverse transcriptase for 25 min at 48°C. To identify the sites of depurination, deoxynucleotide sequencing of BMV cDNA3 was conducted with the same reverse primers used for primer extension analysis. The samples were separated in a 7 M urea/6% acrylamide gel and bands were visualized by exposure to X-ray film. Sites of depurination were mapped onto the predicted secondary structure of RNA3 using mfold analysis (30).
Template competition assay
This assay was performed as described for the replicase assay except that 12.5 nM of PAP-treated or untreated BMV RNA3 in vitro transcripts, directing the synthesis of full-length product, were used as template in the RdRp reaction 10 min prior to the addition of 9, 18 or 36 nM of competitor RNA, directing the synthesis of a 200-nucleotide product of the 3′ end of RNA3. The RNA products were extracted, precipitated and resuspended as described for the replicase assay. The products were separated in a 7 M urea/4.5% acrylamide gel, dried and visualized by exposure to X-ray film. The amount of label incorporated into newly synthesized RNAs was quantified with a phosphorimager.
Dot blot assay
The BMV replicase assay was performed as described above using PAP-treated or untreated BMV RNA3 (0.4 pmol) as template and increasing amounts of BMV replicase fraction (0, 5, 10, 15 or 20 µl). Following incubation for 20 min at 30°C, the assay mixture was filtered through a nitrocellulose membrane that was underlaid with a nylon membrane using a dot-blot apparatus. The membranes were probed with an RNA complementary to the 3′ end of BMV RNA3 (pB3HE1; 28). The amount of RNA3 retained on the filter was visualized by exposure to X-ray film.
Filter-binding assay
Capped, internally labeled, BMV RNA3 in vitro transcripts were treated with PAP or untreated as described above. Transcripts were incubated with increasing amounts of BMV replicase fraction (0, 5, 10, 15 or 20 µl) for 2 h before passing the mixture through a nitrocellulose filter. The amount of retained labeled RNA was determined by scintillation counting and corrected by subtracting background counts in the absence of replicase.
Generation of BMV RNA3 constructs
RNA ligase catalyzes a phosphodiester bond between the 5′ acceptor RNA terminating in a 3′ hydroxyl and the 3′ donor RNA terminating in a 5′ monophosphate. Regions of BMV RNA3 were incubated with tobacco acid pyrophosphatase to produce the donor RNAs in a 50 μl reaction containing 5 μl 10 × TAP Buffer (0.5 M sodium acetate, pH 6.0, 10 mM EDTA, 1% β-mercaptoethanol, 0.1% Triton X-100), 1 μl 100 mM ATP, 20 units RNase inhibitor, 10 units TAP and incubated for 1 h at 37°C. Following incubation, RNA was extracted, precipitated in 100% ethanol and resuspended in DEPC-treated water. Regions of BMV RNA3 serving as the donor RNAs were ligated to the regions of RNA3 serving as the acceptor RNAs to produce full-length BMV RNA3. Ligation was performed in a 50 μl reaction: 5 μl 10 × T4 RNA Ligase Buffer (600 mM potassium acetate, 330 mM Tris–acetate, pH 7.8, 100 mM magnesium acetate, 5 mM DTT), 15 units T4 RNA ligase, 20 units RNase inhibitor and incubated for 16 h at 16°C. Following incubation, RNA was extracted, precipitated in 100% ethanol and resuspended in DEPC-treated water. Ligated BMV RNA3 was separated in a 7 M urea/4.5% acrylamide gel and probed by northern blot analysis as described above to visualize full-length RNA3 following ligation. Full-length RNA, pooled from several experiments, was excised and eluted from the gel and RT–PCR was performed followed by sequencing to ensure that the correct alignment of fragments was achieved. Different types of RNA3 constructs (0.5 pmol) were used to perform replicase assays. For the type I construct, the acceptor RNA consisting of nucleotides 1–1913 of BMV RNA3 was incubated with PAP in RIP buffer for 30 min at 30°C. For the type II construct, the donor RNA, which consisted of nucleotides 92–2113 of BMV RNA3, was incubated with PAP, and for the type III construct, the intergenic region consisting of nucleotides 1004–1246 of BMV RNA3 was incubated with PAP. To test the effect of depurination outside the intergenic region yet within the 3′ UTR, 400 nucleotides of the 3′ end of RNA3 (1713–2113) were incubated with PAP prior to ligation to the remaining 5′ region.
Isolation and transfection of barley protoplasts
Protoplasts were isolated from 7-day-old barley leaves by enzymatic digestion as described previously (31). Protoplasts (3 × 105) were transfected with either type I, II or III BMV RNA3 constructs along with BMV RNA 1 and RNA 2 (0.5 µg each RNA) using PEG 1500 as described (28), and incubated at 25°C under constant fluorescent light (165 µmol/m2/s) for 18 h.
Isolation of protoplast nucleic acid and northern blot analysis
Following the 18 h incubation, protoplasts were pelleted at 1000 × g for 20 s and total nucleic acid was isolated by adding 300 µl of Extraction Buffer (2× TE, 1% SDS) and 300 µl phenol:chloroform:isoamyl alcohol. The aqueous layer was re-extracted and nucleic acid was precipitated with 100 µl of 8 M NH4OAC and 1 ml of 95% ethanol and resuspended in DEPC-treated water. Equal amounts of total nucleic acid were separated in a 7 M urea/4.5% acrylamide gel and transferred to nylon membrane. To detect BMV RNAs, the membrane was probed with a 200-nucleotide radiolabeled RNA complementary to the 3′ end of all BMV RNAs and 28S rRNA was also probed as a loading control (28). Blots were visualized by exposure to X-ray film.
RESULTS
Effect of PAP on negative-strand RNA3 synthesis
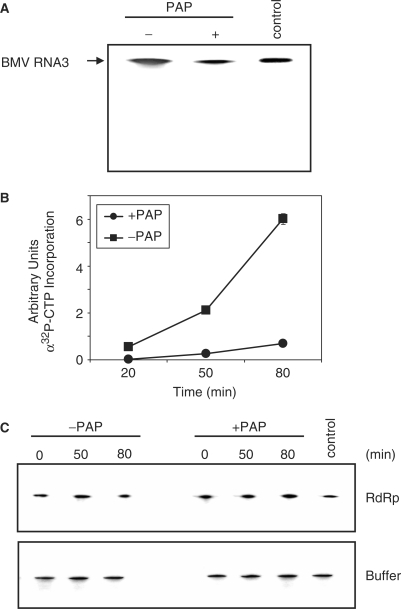
We have shown previously that in vitro PAP-treatment of RNA3 followed by transfection into protoplasts with RNAs 1 and 2 resulted in decreased accumulation of all BMV RNAs (28). To determine if inhibition of replicase activity contributed to this decrease, PAP-treated RNA3 transcript was used as template in an enzyme assay with the BMV replicase complex. Substantially less product was synthesized from PAP-treated template compared with untreated RNA3 (Figure 1A). During this assay, aliquots were removed and the amount of radiolabeled RNA was found to be consistently greater from untreated template, compared with the PAP-treated RNA3 (Figure 1B). To address the possibility that either PAP treatment itself, or subsequent incubation with the replicase complex (which could have contained a co-purified nuclease), may have caused preferential degradation of the template, RNA3, treated or untreated, was incubated over a time course with either replicase complex or buffer. Northern blot analysis indicated no notable difference in the visual integrity of the RNA from treated or untreated samples (Figure 1C).
Figure 1.
Effect of PAP on the synthesis of RNA3 by the viral replicase. (A) PAP-treated and untreated BMV RNA3 in vitro transcripts were used as template for the BMV replicase assay in the presence of radiolabeled CTP. Product RNA3 was separated in a 7 M urea/12% acrylamide gel and visualized by autoradiography. Control is positive-strand, radiolabeled BMV RNA3 in vitro transcript to serve as a size marker. (B) PAP-treated and untreated BMV RNA3 in vitro transcripts were incubated with BMV replicase in the presence of radiolabeled CTP and aliquots were removed at the indicated times. The RNA products were precipitated and the amount synthesized over time was quantified by scintillation counting. Points are means ± SE for three separate experiments. Circles represent PAP-treated template RNA3 and squares represent untreated RNA3. (C) A time-course analysis of the status of PAP-treated BMV RNA3 in vitro transcripts used as template for the replicase assay and primer extension. PAP-treated and untreated BMV RNA3 transcripts were incubated with the replicase (RdRp) or in buffer alone, and aliquots were removed at the indicated times. RNA was analyzed by northern blot for positive-strand BMV RNA3.
Sites of depurination in BMV RNA3
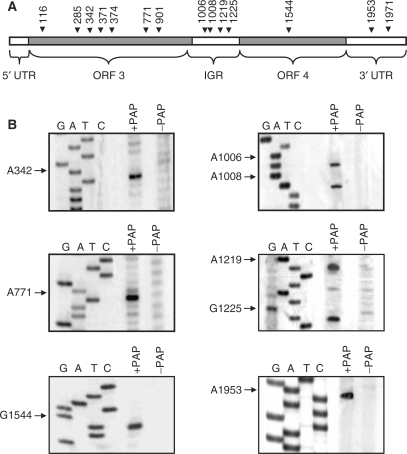
To investigate whether this decrease in RNA synthesis correlated with depurination of the RNA3 template, we mapped the discrete depurination sites along full-length BMV RNA3. The line drawing, illustrating the coding and regulatory regions of this RNA, indicates locations of depurination and the specific nucleotides identified by primer extension analysis (Figure 2A and B). Depurinated nucleotides were found consistently in single-stranded regions of the RNA, as predicted by mfold analysis (30). Interestingly, none of these sites was contained within the core promoter of the 3′ tRNA-like region where synthesis of negative-strand RNA3 initiates. Specifically, the stem loop C of the 3′ tRNA region located within nucleotides 2039–2069 is required for replicase binding and the terminal CCA sequence plus approximately five neighboring nucleotides is recognized by the replicase complex as the site of negative strand initiation (16,32). The decreased amount of RNA3 product synthesized from PAP-treated template suggested that replicase activity was controlled by regions of RNA3 outside these 3′ sequences known to be recognized by the replicase complex.
Figure 2.
In vitro depurination of BMV RNA3 by PAP. (A) Schematic of BMV RNA3 indicating the sites of depurination by their nucleotide number. The 5′ untranslated region (5′ UTR; 1–91 nt), the open reading frame of RNA3 (ORF3; 92–1003 nt), the intergenic region (IGR; 1004–1246 nt), the open reading frame for RNA4 (ORF4; 1247–1813 nt) and the 3′ untranslated region (3′ UTR; 1814–2113 nt) are also indicated. (B) Representative depurination sites determined by primer extension. In vitro transcript of RNA3, either PAP-treated or untreated, was extended with reverse transcriptase using cDNA primers distributed over the length of the viral RNA. Radiolabeled cDNA products were separated in a 7M urea/6% acrylamide gel and visualized by autoradiography. The same primers were used to identify the depurination sites by deoxynucleotide sequencing of BMV DNA3.
The lack of truncation products observed following the replicase assay suggested that rather than initiating on all strands of RNA3 and terminating prematurely when encountering a missing base, the replicase likely only initiated on intact strands of RNA, resulting in full-length product. The observation that some intact product RNA was formed from the PAP-treated template agrees with our earlier findings that less than 100% depurination occurs to RNA treated with PAP at physiological concentration; therefore, some strands of template RNA remained intact following incubation with PAP. The absence of truncated RNA is in contrast to our previous work showing that abasic nucleotides artificially introduced upstream of a minimal core promoter sequence caused the replicase to stall at the missing base (28). We hypothesize that the regulatory regions contained within full-length RNA3 either affected replicase-promoter binding or initiation of RNA synthesis, even though no bases known to be essential for replicase activity in vitro were depurinated.
Effect of PAP on replicase complex binding
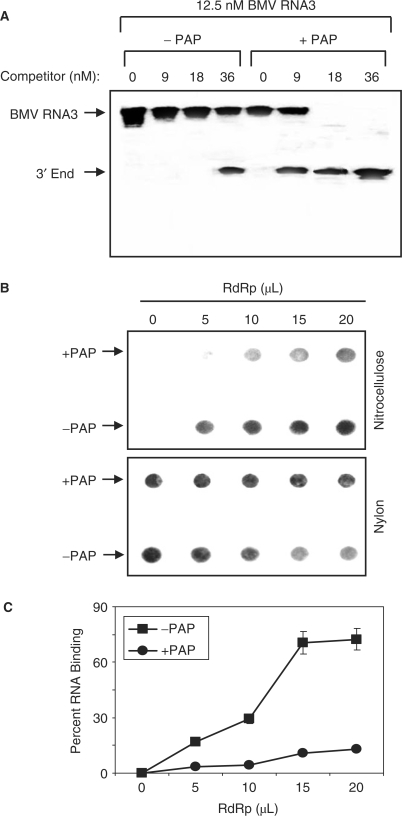
To confirm that the replicase complex exhibited greater activity for untreated RNA compared with the equivalent PAP-treated template, a competition assay was performed in which the enzyme complex was incubated with either template in the presence of increasing concentration of the 3′-terminus of BMV RNA3, which has been shown previously to act as competitor template for the replicase (32). This 200-nucleotide 3′ fragment contains both the promoter and initiator sequences necessary for replicase binding and RNA synthesis. The amount of competitor required to decrease RNA synthesis from the full-length template by 50% (IC50) for PAP-treated RNA3 was 11 nM, compared to 33 nM for untreated RNA3 (Figure 3A). Therefore, the replicase complex exhibited less activity for the PAP-treated BMV RNA3 and was competed away from this template more easily than if untreated RNA3 was provided.
Figure 3.
Replicase interaction with PAP-treated RNA3. (A) PAP-treated and untreated BMV RNA3 in vitro transcripts were used as template RNA with increasing concentration of the 3′ end RNA3 competitor RNA in the replicase assay. Radiolabeled products were separated in a 7 M urea/4.5% acrylamide gel and visualized by autoradiography. (B) PAP-treated and untreated BMV RNA3 in vitro transcripts were incubated with increasing amount of BMV replicase before passage through a nitrocellulose membrane underlaid by a nylon membrane. The retained RNA was visualized by probing the membranes for positive-strand BMV RNA3. (C) Filter-binding assay of PAP-treated and untreated radiolabeled BMV RNA3 transcripts incubated with increasing amount of BMV replicase. The amount of retained RNA was quantified by scintillation counting and corrected by subtracting background counts in the absence of replicase. Points are means ± SE for three separate experiments. Circles represent PAP-treated template RNA3 and squares represent untreated RNA3.
To further test for interaction of the replicase complex with BMV RNA3, either PAP-treated or untreated RNA was incubated with increasing amount of the enzyme complex, passed through nitrocellulose and probed for retained RNA3. A greater amount of untreated RNA3 was trapped by the blot compared with PAP-treated RNA (Figure 3B). As an indicator of the starting amount of RNA in each sample, a nylon membrane was placed under the nitrocellulose and also probed for positive-strand RNA3. This experiment was complemented by a traditional filter-binding assay, which illustrates that the replicase complex bound to untreated RNA3 to a significantly greater extent relative to PAP-treated RNA3 (Figure 3C). These results support the competition assay and indicate that the replicase complex interacted less with full-length PAP-treated RNA3 compared with untreated RNA.
Effect of depurination sites on replicase activity in vitro
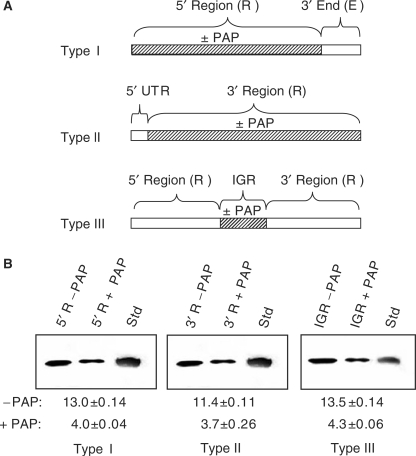
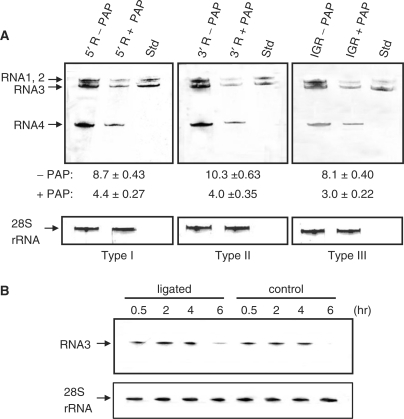
Our observation that prior treatment of RNA3 with PAP reduced the accumulation of RNA product and also reduced the binding of the replicase complex suggested that depurination outside the defined replicase promoter and initiation sites within the 3′ terminus could affect the activity of the enzyme complex. To investigate this possibility, PAP-treated regions of RNA3 were ligated to remaining untreated portions, to regenerate the full-length RNA. For the type I template, the 3′-terminal 200 nucleotides were ligated to the PAP-treated 5′ region of 1913 nucleotides. In addition, two other ligated RNA3 molecules were made, one in which the 5′ UTR (92 nucleotides) was untreated and ligated to the 2021 nucleotides downstream that were first incubated with PAP (type II template), and the third template in which the intergenic region, comprised of nucleotides 1004–1246, was PAP-treated and ligated to the 5′ and 3′ regions (type III template), as illustrated in Figure 4A. Each fragment treated with PAP was assessed by primer extension analysis to confirm the expected depurination sites prior to ligation (data not shown). These ligated RNAs were then used as templates in the replicase assay, to determine whether depurination outside the 5′ and 3′ termini could influence replicase activity. Decreased RNA synthesis was observed with the type I template; therefore, depurination upstream of the characterized replicase binding site at the 3′ end inhibited replicase activity (Figure 4B). Results of the replicase assay using the type II template indicated that depurination downstream of the 5′ UTR also inhibited replicase activity. The decrease in RNA product seen from prior PAP treatment of the 5′ region in type I template and the 3′ region in type II template suggested that depurination within the intergenic region may affect replicase activity. To assess this possibility, type III template was designed to allow PAP treatment only within the intergenic region of RNA3, prior to its ligation to the 5′ and 3′ regions. The observed decrease in amount of RNA3 product indicated that depurination solely within the intergenic region reduced replicase activity (Figure 4C).
Figure 4.
Effect of depurination location on replicase activity in vitro. Regions of RNA3 were treated with PAP and then ligated to the remaining RNA to regenerate full-length RNA3 used as template in the replicase assay. (A) For the type I template, the 5′ region was PAP-treated or untreated and ligated to the remaining 3′ end, which was untreated. For the type II template, the 3′ region was PAP-treated or untreated and ligated to the 5′ UTR, which was untreated. For the type III template, the intergenic region was PAP-treated or untreated and ligated to the remaining 5′ and 3′ regions. (B) Synthesis of RNA3 following the replicase assay using the template RNAs described in (A). The radiolabeled RNA products were separated in a 7 M urea/12% acrylamide gel and visualized by autoradiography. Std represents radiolabeled, positive-strand RNA3 in vitro transcript to serve as a size marker. Values are means of intensities ± SE for three separate experiments quantified with a phosphorimager.
Effect of depurination location on replication in barley protoplasts
Given our current observation that depurination distant from the replicase-binding site on the 3′ end of positive-strand RNA3 could diminish production of RNA in vitro, we investigated whether the same ligated RNAs could alter replication of BMV in vivo. Thus, each type of ligated RNA3 template was transfected into protoplasts with untreated RNAs 1 and 2. Subsequent northern blot analysis of all BMV RNAs showed that type I template, with the portion of RNA3 upstream of the 3′ 200-nucleotide terminus treated with PAP, resulted in diminished replication of all RNAs (Figure 5A). Transfection of the type II template, with the RNA treated downstream of the 5′ UTR with PAP, also inhibited replication of all RNAs. Type III template, with only the intergenic region treated with PAP, also showed decreased accumulation of all RNAs. Therefore, the levels of in vivo replication in protoplasts were consistent with production of RNA in vitro, and support our observation that depurination within the intergenic region of RNA3 inhibits replicase activity. To address the possible instability of the ligated RNAs in cells, a time course analysis was conducted for protoplast samples transfected with either ligated type I template RNA3 or the original RNA3 transcript. No difference in integrity between the two pools of RNAs was observed by northern blot analysis (Figure 5B).
Figure 5.
Effect of depurination location on BMV RNA replication in vivo. (A) Barley protoplasts were transfected with PAP-treated, ligated RNA3 templates described in Figure 4A, plus intact BMV RNA1 and 2 in vitro transcripts and allowed to replicate. Total protoplast RNA was analyzed by northern blot for the presence of positive-strand BMV RNAs. Std is in vitro transcript of positive-strand RNAs 1, 2 and 3 to serve as size markers. Values are means of intensities for RNAs 1, 2 and 3 ± SE for three separate experiments, quantified with a phosphorimager. Blots were also probed for 28S rRNA as a loading control for total RNA. (B) Stability of type I template in barley protoplasts. Type I template (ligated) or non-ligated RNA3 (control) was transfected into barley protoplasts and aliquots of cells were removed at the indicated time points. Total protoplast RNA was probed by northern blot for positive-strand BMV RNA3. The blot was also probed for 28S rRNA as a loading control for total RNA.
Effect of depurination within the intergenic region on replicase binding
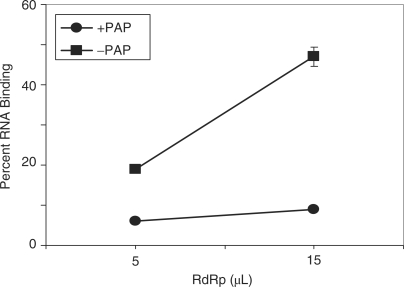
The decreased replicase activity from RNA3 depurinated only within the intergenic region suggested that the replicase does not interact with the RNA template if the intergenic region is depurinated. To test this hypothesis, the replicase complex was incubated with template III. A filter-binding assay showed that the replicase bound significantly less to RNA3 with depurinated intergenic region, compared with untreated, ligated RNA3 (Figure 6).
Figure 6.
Replicase binding to RNA3 depurinated within the intergenic region. The intergenic region of RNA3 was PAP-treated or untreated, prior to ligation to the remaining 5′ and 3′ regions of RNA3. A filter-binding assay was performed with the ligated, radiolabeled BMV RNA3 transcripts and increasing amount of BMV replicase. The amount of retained RNA was quantified by scintillation counting and corrected by subtracting background counts in the absence of replicase. Points are means ± SE for three separate experiments. Circles represent PAP-treated, ligated RNA3 and squares represent untreated, ligated RNA3.
Effect of depurination outside the intergenic region on replicase activity
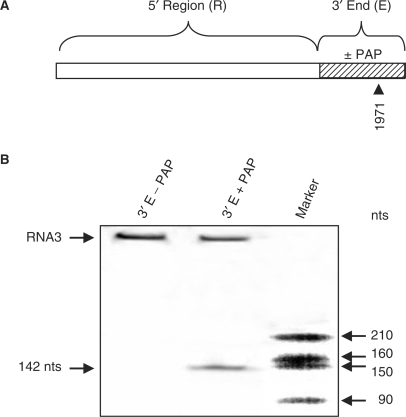
Our previous work showed that abasic deoxynucleotides introduced into a short RNA template containing the minimal replicase promoter allowed the replicase to initiate RNA synthesis, but that the enzyme complex stalled at abasic sites (28). To determine if depurinated nucleotides outside of the intergenic region yet within the 3′ UTR would have the same effect, a portion of the 3′ terminus of RNA3 was incubated with PAP and ligated to the remaining upstream region to regenerate full-length RNA3. This ligated RNA3 was used as template in the replicase assay and truncated RNA3 was observed, corresponding in size to replicase stalling at 1971, the most 3′ depurinated nucleotide (Figure 7). Though some full-length RNA3 was synthesized, indicating less than 100% depurination, the presence of a shorter fragment in PAP-treated RNA3 shows that depurination outside the intergenic region did permit the replicase to bind to the template RNA, but caused replicase stalling when the missing base was encountered. These results agree with our earlier work using a depurinated minimal promoter template and demonstrate that omission of the intergenic region from PAP treatment allowed the replicase to bind to the 3′ end and initiate RNA synthesis.
Figure 7.
Effect of depurination 3′ of the intergenic region on replicase activity in vitro. (A) The 3′ end (terminal 400 nucleotides) of RNA3 was PAP-treated or untreated and ligated to the remaining 5′ region of RNA3. The resulting depurination site following PAP treatment is indicated by the nucleotide number (1971). (B) The RNA3 described in (A) was used as template in the replicase assay and radiolabeled products were separated in a 7 M urea/6% acrylamide gel and visualized by autoradiography. Marker indicates in vitro transcripts of known size.
DISCUSSION
We have shown previously that prior incubation of RNA3 with PAP, followed by transfection with untreated RNA1 and 2 into protoplasts, reduced the level of replication for all BMV RNAs (28). These results implied that depurination of RNA3 inhibited the replicase, either by preventing binding to the template, or inhibiting the synthesis of RNA. The latter possibility was supported by our observation that elongation of negative-strand RNA, from a minimal template sufficient for replicase recognition, was prevented by the presence of artificially introduced abasic deoxynucleotides. The replicase initiated but stalled at the missing base, producing truncated strands of RNA (28). Here, we have worked with full-length RNA3 to gain a more biologically relevant understanding of the effect of depurination on replicase activity. We identified the exact sites of depurination on RNA3 and demonstrated that abasic nucleotides outside the characterized 3′ core promoter for negative-strand synthesis inhibited the replication of RNA, both in vivo and in vitro. Specifically, we have determined that depurination within the intergenic region of RNA3 prevented replicase binding and RNA synthesis in the absence of depurination elsewhere in the full-length RNA3. Therefore, we conclude that the intergenic region is involved in regulating the synthesis of negative-strand RNA3.
The results of our competition assay are consistent with the importance of regions of the RNA outside of the 3′ end promoter for negative-strand synthesis. The fact that the 200-nucleotide 3′-terminus of RNA3 functioned as a competitive RNA indicated that this fragment was sufficient for replicase recognition and RNA synthesis initiation. However, in the presence of full-length RNA3, a 3-fold excess of the 3′ end promoter was required to reduce synthesis from full-length template by 50%, indicating that regions of RNA within the full-length strand enhanced recognition by the replicase. Support for this conclusion comes from previous work showing that deletion of the 18 base poly(A) tract of the intergenic region reduced RNA3 accumulation in vivo (33,34). Additionally, a poly(A) fragment of 16–22 nucleotides competed with a functional core promoter template for RNA synthesis by the replicase, indicating that the replicase interacted with the poly(A) fragment (14). We did not observe depurination within this poly(A) sequence of intact RNA3, however, nucleotides A1219 and G1225 were depurinated and they were 2 and 6 nucleotides 3′ of the poly(A) sequence, respectively. Therefore, depurination near the poly(A) sequence likely affected replicase binding. The optimal sequence for replicase binding to the intergenic region may well exceed the minimal required sequence, as was observed with the 3′ end of RNA3; namely, that stem loop C represents the minimal sequence for replicase binding but that the replicase binds to the 200-nucleotide tRNA-like sequence approximately 3-fold more than to stem loop C alone (32). Moreover, diminished negative-strand synthesis caused by mutation of the initiation sequence within the 200-nucleotide 3′ end of RNA3 was partially rescued by the addition in cis of 5′ sequence of RNA3 (35). The remaining depurinated nucleotides within the intergenic region, namely A1006 and A1008, were located within a 150-base region referred to as the intergenic replication enhancer. Deletion of this region has been shown to decrease RNA3 accumulation in vivo over 100-fold (36). Taken together, these data strengthen the importance of the intergenic region for synthesis of negative-strand RNA3 both in vivo and in vitro. Though distant in linear sequence, the two pairs of depurinated nucleotides come in close proximity when viewed in the secondary structure model for the intergenic region of RNA3 (37). This model bends the intergenic region essentially in half, bringing A1219 and G1225 physically close to A1006 and A1008, with all 4 nucleotides in single-stranded regions, presumably facilitating their depurination by PAP.
Our work shows that depurination of the intergenic region reduced synthesis of full-length RNA3, whereas depurination outside the intergenic region and including the 3′ UTR permitted RNA synthesis but caused the replicase to stall at the abasic site. These results are consistent with the idea that the intergenic region is a regulator of replicase activity; namely, that when present, the intergenic region controls binding and initiation of the replicase at the 3′ end of RNA3. When absent, as is the case with minimal RNA templates containing only the core promoter and initiation site of the 3′ end, negative-strand RNA synthesis occurs regardless, suggesting that the replicase becomes less discriminatory with regards to template choice. The biological role of such a regulator may be to promote initiation from intact templates, that is, replicase binding and initiation at the 3′ end may be facilitated by first or coordinated binding to the intergenic region, which may alter template conformation to favor RNA synthesis. The decreased binding of the replicase complex to RNA3, in which only the intergenic region was depurinated, indicates that synthesis of negative-strand RNA from a full-length template is dependent on interaction of the replicase complex with the intergenic region, in addition to its obvious association with the 3′ end of the RNA.
Further support for the role of replicase binding to the intergenic region comes from previous work showing that a replicase binding domain is required for RNA synthesis from structured templates, whereas short, unstructured RNAs containing only an initiation site without binding domain are sufficient for RNA synthesis by the BMV replicase (38). A lack of RNA product from the structured template without binding domain suggested that inhibition occurred at or before initiation of RNA synthesis. Our data indicating the absence of truncation products and decreased replicase binding when only the intergenic region was depurinated argues that inhibition occurred prior to initiation, at the step of initial replicase interaction with the template RNA. Full-length BMV RNA3 is naturally structured, and therefore, would require both a replicase binding domain and initiation sequence for successful synthesis of RNA. We show that apart from the 3′ end of RNA3, which contains the stem loop C replicase core promoter and initiation sequence, the replicase binding domain of the intergenic region is required for negative-strand synthesis. It has been postulated that the replicase scans an RNA template from the 5′ end in search of an initiation site and that intervening secondary structure, without a replicase binding domain, inhibits this scanning (38). Therefore, we suggest that binding of the replicase to the intergenic region guides the replicase to the 3′ end, bypassing natural secondary RNA structure. Similar scenarios of viral polymerases binding to RNA sequences distant from initiation sites have been described. For example, the bacteriophage Qβ replicase binds to an internal RNA segment more than a kilobase from the 3′ end and long-range basepairing interactions connect the two regions (39). Similarly, influenza virus RNA polymerase activity depends on the presence of 5′ and 3′ genomic viral sequences, suggesting that the polymerase binds both ends of the template RNA (40). Our observations suggest that once binding to the intergenic region is achieved, the replicase accesses the 3′ end of RNA3 to initiate synthesis of negative-strand RNA.
Our data show that transfection of RNA3, depurinated only within the intergenic region, along with untreated RNA1 and 2 into barley protoplasts resulted in decreased accumulation of all RNAs. This decrease was observed in positive-strand RNAs; however, given that the replicase bound less to depurinated RNA3, diminished negative-strand synthesis likely contributed to the decline in total accumulation of RNA3. The intergenic region of RNA3 has also been shown essential for replicase complex assembly in yeast (19); therefore, depurination of the RNA3 intergenic region may also decrease replicase activity in vivo by reducing its assembly, thereby contributing to the observed decline in synthesis of all RNAs. Transfection of RNA1 and 2 with RNA3 bearing mutations in the intergenic region has also been shown to decrease the accumulation of positive-strand RNA1 and 2 several fold (41); therefore damage to the intergenic region of RNA3 may alter RNA1 and 2 levels. In addition to these possible contributing factors, the decline we observed in accumulation of positive-strand RNAs 1 and 2 may be due to decreased synthesis of coat protein, as the level of RNA4 was also diminished, which is synthesized from negative-strand RNA3. Limiting coat protein would result in decreased encapsidation of viral RNAs, leaving them susceptible to nuclease degradation.
Through identification of the nucleotides depurinated by PAP in full-length RNA3, we have contributed new insight into the antiviral activity of this enzyme. Specifically, we have determined that depurination only within the intergenic region of positive-strand RNA3 inhibits the binding of the viral replicase, resulting in decreased initiation of RNA synthesis. Depurination outside the intergenic region does allow the replicase to initiate RNA synthesis; however, elongation is stalled at the missing base. Therefore, PAP inhibits both the replicase interaction with template RNA and its synthesis of new RNA, thereby diminishing viral replication. Further work will focus on how depurination within the intergenic region alters its association with the replicase in vitro and how this depurination may inhibit replicase assembly in vivo.
FUNDING
This work was supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada; a Premier's Research Excellence Award; and infrastructural support was provided by the Canadian Foundation for Innovation (CFI) and the Ontario Innovation Trust (OIT) to K.A.H. Funding for open access charge: Natural Sciences and Engineering Research Council of Canada.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Dr C. C. Kao (Texas A&M University) for providing infectious clones of BMV RNAs and for advice regarding BMV replicase isolation.
REFERENCES
- 1.Goldbach R, LeGall O, Wellink J. Alpha-like viruses in plants. Semin. Virol. 1991;2:19–25. [Google Scholar]
- 2.Ahlquist P. Bromovirus RNA replication and transcription. Curr. Opin. Genet. Dev. 1992;2:71–76. doi: 10.1016/s0959-437x(05)80325-9. [DOI] [PubMed] [Google Scholar]
- 3.Kao CC, Sivakumaran K. Brome mosaic virus, good for an RNA virologist's basic needs. Mol. Plant Pathol. 2000;1:91–97. doi: 10.1046/j.1364-3703.2000.00017.x. [DOI] [PubMed] [Google Scholar]
- 4.Argos P. A sequence motif in many polymerases. Nucleic Acids Res. 1988;16:9909–9916. doi: 10.1093/nar/16.21.9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kong F, Sivakumaran K, Kao CC. The N-terminal half of the brome mosaic virus 1a protein has RNA capping associated activities: specificity for GTP and S-adenosylmethionine. Virology. 1999;259:200–210. doi: 10.1006/viro.1999.9763. [DOI] [PubMed] [Google Scholar]
- 6.Ahola T, den Boon JA, Ahlquist P. Helicase and capping enzyme active site mutations in brome mosaic virus protein 1a cause defects in template recruitment, negative-strand RNA synthesis, and viral RNA capping. J. Virol. 2000;74:8803–8811. doi: 10.1128/jvi.74.19.8803-8811.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kao CC, Quadt R, Hershberger RP, Ahlquist P. Brome mosaic virus RNA replication proteins 1a and 2a from a complex in vitro. J. Virol. 1992;66:6322–6329. doi: 10.1128/jvi.66.11.6322-6329.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quadt R, Kao CC, Browning KS, Hershberger RP, Ahlquist P. Characterization of a host protein associated with brome mosaic virus RNA-dependent RNA polymerase. Proc. Natl Acad. Sci. USA. 1993;90:1498–1502. doi: 10.1073/pnas.90.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller WA, Dreher TW, Hall TC. Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (-)-sense genomic RNA. Nature. 1985;313:68–70. doi: 10.1038/313068a0. [DOI] [PubMed] [Google Scholar]
- 10.Allison R, Thompson C, Ahlquist P. Regeneration of a functional RNA virus genome by recombination between deletion mutants and requirement for cowpea chlorotic mottle virus 3a and coat genes for systemic infection. Proc. Natl Acad. Sci. USA. 1990;87:1820–1824. doi: 10.1073/pnas.87.5.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitz I, Rao ALN. Molecular studies on bromovirus capsid protein. I. Characterization of cell-to- cell movement-defective RNA3 variants of brome mosaic virus. Virology. 1996;226:281–293. doi: 10.1006/viro.1996.0656. [DOI] [PubMed] [Google Scholar]
- 12.Pogue GP, Hall TC. The requirement for a stem-loop structure in brome mosaic virus replication supports a new model for viral positive-strand RNA initiation. J. Virol. 1992;66:674–684. doi: 10.1128/jvi.66.2.674-684.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janda M, Ahlquist P. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell. 1993;72:961–970. doi: 10.1016/0092-8674(93)90584-d. [DOI] [PubMed] [Google Scholar]
- 14.Choi SK, Hema M, Gopinath K, Santos J, Kao C. Replicase-binding sites on plus- and minus-strand brome mosaic virus RNAs and their roles in RNA replication in plant cells. J. Virol. 2004;78:13420–13429. doi: 10.1128/JVI.78.24.13420-13429.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dreher TW, Hall TC. Mutational analysis of the sequence and structural requirements in brome mosaic virus for minus-strand promoter activity. J. Mol. Biol. 1988;201:31–40. doi: 10.1016/0022-2836(88)90436-6. [DOI] [PubMed] [Google Scholar]
- 16.Kim CH, Kao C, Tinoco I. RNA motifs that determine specificity between a viral replicase and its promoter. Nature Struct. Biol. 2000;7:415–423. doi: 10.1038/75202. [DOI] [PubMed] [Google Scholar]
- 17.Sivakumaran K, Kim CH, Tayon, R, Kao CC. RNA sequence and secondary structural determinants in a minimal viral promoter that directs replicase recognition and initiation of genomic plus-strand RNA synthesis. J. Mol. Biol. 1999;294:667–682. doi: 10.1006/jmbi.1999.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sullivan ML, Ahlquist P. Cis-acting signals in bromovirus RNA replication and gene expression: Networking with viral proteins and host factors. Semin. Virol. 1997;73:2622–2632. [Google Scholar]
- 19.Quadt R, Ishikawa M, Janda M, Ahlquist P. Formation of brome mosaic virus RNA-dependent RNA polymerase in yeast requires coexpression of viral proteins and viral RNA. Proc. Natl Acad. Sci. USA. 1995;92:4892–4896. doi: 10.1073/pnas.92.11.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sullivan ML, Ahlquist P. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J. Virol. 1999;73:2622–2632. doi: 10.1128/jvi.73.4.2622-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hudak KA, Wang P, Tumer NE. A novel mechanism for inhibition of translation by pokeweed antiviral protein: depurination of the capped RNA template. RNA. 2000;6:369–380. doi: 10.1017/s1355838200991337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teltow GJ, Irvin JD, Aron GM. Inhibition of herpes simplex virus DNA synthesis by pokeweed antiviral protein. Antimicrob. Agents Chemother. 1983;23:390–396. doi: 10.1128/aac.23.3.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zarling JM, Moran PA, Haffar O, Sias J, Richman DD, Spina CA, Myers DE, Kuebelbeck V, Ledbetter JA, Uckun FM. Inhibition of HIV replication by pokeweed antiviral protein targeted to CD4+ cells by monoclonal antibodies. Nature. 1990;347:92–95. doi: 10.1038/347092a0. [DOI] [PubMed] [Google Scholar]
- 24.Lodge JK, Kaniewski WK, Tumer NE. Broad-spectrum virus resistance in transgenic plants expressing pokeweed antiviral protein. Proc. Natl Acad. Sci. USA. 1993;90:7089–7093. doi: 10.1073/pnas.90.15.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foa-Tomasi L, Campadelli-Fiume G, Barbieri L, Stirpe F. Effect of ribosome-inactivating proteins on virus-infected cells. Inhibition of virus multiplication and of protein synthesis. Arch. Virol. 1982;71:323–332. doi: 10.1007/BF01315062. [DOI] [PubMed] [Google Scholar]
- 26.Endo Y, Tsurugi K, Lambert JM. The site of action of six different ribosome-inactivating proteins from plants on eukaryotic ribosomes: the RNA N-glycosidase activity of the proteins. Biochem. Biophys. Res. Commun. 1988;150:1032–1036. doi: 10.1016/0006-291x(88)90733-4. [DOI] [PubMed] [Google Scholar]
- 27.Rajamohan F, Venkatachalam TK, Irvin JD, Uckun FM. Pokeweed antiviral protein isoforms PAP-I, PAP-II, and PAP-III depurinate RNA of human immunodeficiency virus (HIV)-1. Biochem. Biophys. Res. Commun. 1999;260:453–458. doi: 10.1006/bbrc.1999.0922. [DOI] [PubMed] [Google Scholar]
- 28.Picard D, Kao CC, Hudak KA. Pokeweed antiviral protein inhibits brome mosaic virus replication in plant cells. J. Biol. Chem. 2005;280:20069–20075. doi: 10.1074/jbc.M413452200. [DOI] [PubMed] [Google Scholar]
- 29.Sun JH, Adkins S, Faurote G, Kao CC. Initiation of (-)-strand RNA synthesis catalyzed by the BMV RNA-dependent RNA polymerase: synthesis of oligonucleotides. Virology. 1996;226:1–12. doi: 10.1006/viro.1996.0622. [DOI] [PubMed] [Google Scholar]
- 30.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kroner P, Richards D, Traynor P, Ahlquist P. Defined mutations in a small region of the brome mosaic virus 2a gene cause diverse temperature-sensitive RNA replication phenotypes. J. Virol. 1989;63:5302–5309. doi: 10.1128/jvi.63.12.5302-5309.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chapman MR, Kao CC. A minimal RNA promoter for minus-strand RNA synthesis by the brome mosaic virus polymerase complex. J. Mol. Biol. 1999;286:709–720. doi: 10.1006/jmbi.1998.2503. [DOI] [PubMed] [Google Scholar]
- 33.French R, Ahlquist P. Characterization and engineering of sequences controlling in vivo synthesis of brome mosaic virus subgenomic RNA. J. Virol. 1988;62:2411–2420. doi: 10.1128/jvi.62.7.2411-2420.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smirnyagina E, Hsu YH, Chua N, Ahlquist P. Second-site mutations in the brome mosaic virus RNA3 intercistronic region partially suppress a defect in coat protein mRNA transcription. Virology. 1994;198:427–436. doi: 10.1006/viro.1994.1054. [DOI] [PubMed] [Google Scholar]
- 35.Chapman MR, Rao ALN, Kao CC. Sequences 5′ of the conserved tRNA-like promoter modulate initiation of minus-strand synthesis by the brome mosaic virus RNA-dependent RNA polymerase. Virology. 1998;252:458–467. doi: 10.1006/viro.1998.9473. [DOI] [PubMed] [Google Scholar]
- 36.French R, Ahlquist P. Intercistronic as well as terminal sequences are required for efficient amplification of brome mosaic virus RNA3. J. Virol. 1987;61:1457–1465. doi: 10.1128/jvi.61.5.1457-1465.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baumstark T, Ahlquist P. The brome mosaic virus RNA3 intergenic replication enhancer folds to mimic a tRNA TpsiC-stem loop and is modified in vivo. RNA. 2001;7:1652–1670. [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X, Kim CH, Sivakumaran K, Kao C. Stable RNA structures can repress RNA synthesis in vitro by the brome mosaic virus replicase. RNA. 2003;9:555–565. doi: 10.1261/rna.2190803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klovins J, Berzins V, van Duin J. A long-range interaction in Qbeta RNA that bridges the thousand nucleotides between the M-site and the 3' end is required for replication. RNA. 1998;4:948–957. doi: 10.1017/s1355838298980177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hagen M, Chung TD, Butcher JA, Krystal M. Recombinant influenza virus polymerase: requirement of both 5' and 3' viral ends for endonuclease activity. J. Virol. 1994;68:1509–1515. doi: 10.1128/jvi.68.3.1509-1515.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marsh LE, Huntley CC, Pogue GP, Connell JP, Hall TC. Regulation of (+):(-)-strand asymmetry in replication of brome mosaic virus RNA. Virology. 1991;182:76–83. doi: 10.1016/0042-6822(91)90650-z. [DOI] [PubMed] [Google Scholar]