Abstract
Photoreactive psoralens can form interstrand crosslinks (ICLs) in double-stranded DNA. In eubacteria, the endonuclease UvrABC plays a key role in processing psoralen ICLs. Psoralen-modified triplex-forming oligonucleotides (TFOs) can be used to direct ICLs to specific genomic sites. Previous studies of pyrimidine-rich methoxypsoralen–modified TFOs indicated that the TFO inhibits cleavage by UvrABC. Because different chemistries may alter the processing of TFO-directed ICLs, we investigated the effect of another type of triplex formed by purine-rich TFOs on the processing of 4′-(hydroxymethyl)-4,5′,8-trimethylpsoralen (HMT) ICLs by the UvrABC nuclease. Using an HMT-modified TFO to direct ICLs to a specific site, we found that UvrABC made incisions on the purine-rich strand of the duplex ∼3 bases from the 3′-side and ∼9 bases from the 5′-side of the ICL, within the TFO-binding region. In contrast to previous reports, the UvrABC nuclease cleaved the TFO-directed psoralen ICL with a greater efficiency than that of the psoralen ICL alone. Furthermore, the TFO was dissociated from its duplex binding site by UvrA and UvrB. As mutagenesis by TFO-directed ICLs requires nucleotide excision repair, the efficient processing of these lesions supports the use of triplex technology to direct DNA damage for genome modification.
INTRODUCTION
Psoralens are photoreactive compounds that have been used extensively in the treatment of skin disorders, such as psoriasis (1). These planar, heterocyclic molecules can intercalate into double-stranded DNA, and with absorption of photons at 365 nm, can form adducts with pyrimidine bases. Psoralen molecules contain two photoreactive double bonds, a 3–4 double bond on the pyrone side and a 4′–5′ bond on the furan side that can form DNA interstrand crosslinks (ICLs) preferentially at 5′-TpA-3′ sites in double-stranded DNA (2–4).
In bacteria, the three-component endonuclease, UvrABC, plays an important role in the processing and removal of ICLs. UvrABC has been shown to hydrolyze the 9th phosphodiester bond to the 5′-side and the 3rd phosphodiester bond to the 3′-side of a psoralen ICL (5,6). In this process, dimerized UvrA binds UvrB in a search for damage-induced conformational changes in the double helix. After DNA damage and/or helical distortions are identified, UvrA is released from the DNA and UvrC is recruited to the UvrB-DNA preincision complex. UvrC then cleaves on the 3′-side of the damaged site, followed by a second cleavage on the 5′-side [reviewed in (7,8)].
Triplex-forming oligonucleotides (TFOs) are single-stranded oligonucleotides that can bind specific purine-rich stretches of DNA via Hoogsteen hydrogen bonding through the major groove of the duplex DNA, thereby forming a triplex DNA structure (9,10). Psoralen-modified TFOs can be used to create ICLs at unique sites after irradiation with UVA light making this a useful tool for directing site-specific DNA damage both in vitro and in vivo [reviewed in (11)]. It has been shown that 4′-(hydroxymethyl)-4,5′,8-trimethylpsoralen (HMT) forms primarily 4′–5′-furan-sided monoadducts on the purine-rich strand of the target duplex substrate when targeted with a purine-rich TFO. These monoadducts are readily converted to ICLs with thymidine on the complementary pyrimidine-rich strand (12). In contrast, monoadducts generated on thymidine in the pyrimidine strand are mainly 3,4-pyrone-sided monoadducts which undergo minimal conversion to ICLs (12). In the absence of triplex formation (i.e. with an HMT ICL only) UvrABC preferentially incises the furan-side-adducted strand of an HMT ICL at a 5′-TpA-3′ site (6).
Previous studies of pyrimidine-rich methoxypsoralen (MOP)-modified TFOs have indicated that the presence of the third strand inhibits cleavage by the UvrABC nulcease by as much as 80% compared to the ICL alone (13), presumably due to the third strand blocking the nuclease incision site in the underlying duplex substrate. However, since different binding and crosslinking chemistries may alter the processing of TFO-directed ICLs, here we investigated the effect of purine-rich TFOs on the processing of HMT ICLs by purified UvrA, UvrB and UvrC subunits. Whereas pyrimidine-rich TFOs used previously bind to the purine-rich strand of the target duplex in the same 5′–3′ orientation as the purine-rich strand and require acidic conditions for binding, purine-rich TFOs bind in the major groove at physiological pH in an antiparallel orientation such that the third strand TFO has the opposite 5′–3′ orientation as the purine-rich duplex strand.
Our studies demonstrate efficient cleavage within the TFO binding site of the TFO-directed psoralen ICL by UvrABC compared to the ICL alone, in contrast to published work utilizing a pyrimidine-rich MOP-modified TFO (13). Furthermore, we show that binding of UvrA and UvrB to the TFO-directed ICL results in dissociation of the TFO from its duplex binding site. Our results suggest that the TFO does not hinder cleavage by the UvrABC complex, and therefore supports the use of triplex technology as a powerful means to induce site-specific DNA damage to facilitate genome modification.
MATERIALS AND METHODS
Oligonucleotides
The psoralen-modified oligonucleotide (psoAG30) and psoralen-modified disulfide linked oligonucleotide (pso-SS-AG30) were synthesized and HPLC purified by Midland Certified Reagent Co. (Midland, TX, USA). The 5′-psoralen-modified oligonucleotides were synthesized with the derivative 2-[4′-(hydroxymethyl)-4,5′,8-trimethylpsoralen]-hexyl-1-O-(2-cyanoethyl)-(N,N-diisopropyl)-phosphoramidite. The disulfide-linked oligonucleotide was designed with a disulfide bridge between the psoralen moiety and the oligonucleotide. Primers for PCR preparations of duplex substrate were synthesized by Integrated DNA Technologies (Coralville, IA, USA). Primers were designed to amplify a 120-bp region of the pSupFG1 plasmid encompassing the supF TFO target site, which has been shown to be bound by nucleotide excision repair (NER) factors in the presence of a TFO-directed HMT ICL (14).
Substrate preparation
Target duplex DNA was synthesized by PCR amplification of a 120-bp region of the pSupFG1 plasmid containing the supF TFO target site (14). To create target duplex that was 5′-radiolabeled on either the purine-rich strand or the pyrimidine-rich strand, the appropriate PCR primer was 5′-end-labeled using [γ-32P]ATP and T4 polynucleotide kinase prior to the PCR reaction. The 120-bp PCR product was gel purified on a 15% native polyacrylamide gel. The 5′-end-labeled substrate was incubated 12–16 h in triplex binding buffer (10 mM MgCl2, 10 mM Tris, pH 7.6) with a 50- to 100-fold excess of psoAG30 or pso-SS-AG30. Samples were irradiated with 1.8 J/cm2 of 365 nm UVA light to crosslink the psoralen-modified TFO to the duplex substrate. Substrates crosslinked to pso-SS-AG30 were incubated with 100 mM DTT at 65°C for 3 h to reduce the disulfide bond and remove the TFO. Substrates containing the crosslink only (ICL) or the crosslink with the TFO covalently attached (TFO-ICL) were purified from denaturing 6% polyacrylamide gels, electroeluted and purified over MicroSpin G-25 columns (GE Healthcare, Buckinghamshire, UK). To reanneal crosslinked substrates, samples were heated to 95°C for 5 min and allowed to cool to room temperature (∼5 h) in 10 mM Tris, pH 7.8, 20 mM NaCl. Substrates were repurified over MicroSpin G-25 columns and reincubated in triplex-binding buffer for at least 2 h.
The 3′-end-labeled substrate was made by labeling duplex DNA using deoxyadenosine 5′-triphosphate, 3′[α-32P]-(cordycepin 5′-triphosphate) (Perkin Elmer Life Sciences, Boston, MA, USA) and terminal transferase. The 3′-end-labeled duplex substrate was either gel purified directly on a native polyacrylamide gel or digested with MluI to remove the 3′-end label on the pyrimidine strand prior to gel purification. Both digested and undigested substrates were incubated in triplex-binding buffer with psoAG30 followed by irradiation with UVA as mentioned above. Crosslinked substrate was isolated as described earlier.
To create a substrate labeled on the 3′-end of the TFO, psoAG30 was labeled using deoxyadenosine 5′-triphosphate, 3′[α-32P]-(cordycepin 5′-triphosphate) and terminal transferase. Labeled TFO was incubated in triplex-binding buffer with duplex DNA followed by irradiation with UVA, as above. Crosslinked substrate was isolated as described earlier.
The positive control (F2650) is a 50-bp duplex with a fluorescein-adducted thymine at base 26 in the duplex. It has previously been shown to be an efficient substrate of UvrABC (15,16). F2650 was prepared by labeling the 5′-end of the fluorescein-adducted strand as described earlier. The fluorescein-adducted strand was then purified over a MicroSpin G-25 column and annealed to its complementary strand.
UvrABC protein
Wild-type Bacillus caldotenax UvrA, B. caldotenax UvrB and Thermatoga maritima UvrC subunits were prepared as previously described (17). Proteins were stored at −20°C.
DNase footprinting
Substrates were incubated for 5 min at 22°C with 0.001 U DNase I (Roche Applied Science, Indianapolis, IN, USA) in triplex-binding buffer in a total volume of 10 μl. The reactions were stopped by addition of EDTA (80 mM final concentration) and Herring sperm DNA (40 μg). Samples were heated to 95°C in formamide loading buffer, subjected to polyacrylamide gel electrophoresis (PAGE) on a denaturing 12% gel, and analyzed using the Storm 820 Phosphorimager (Molecular Dynamics, Sunnyvale, CA, USA).
TFO binding assay
Target duplex DNA was 5′-end-labeled with [γ-32P]ATP and incubated with increasing concentrations of TFO (psoAG30) at 37°C for 12–16 h in triplex-binding buffer (10 mM MgCl2, 10 mM Tris, pH 7.6). Samples were either run directly on a 12% TBM (89 mM Tris, 89 mM Boric acid and 10 mM MgCl2) native polyacrylamide gel at 70 V or were incubated in UvrABC buffer (50 mM Tris, pH 7.5, 50 mM KCl, 10 mM MgCl2, 5 mM DTT, 1 mM ATP) at 55°C for 30 min prior to gel loading. Samples were protected from light during the procedure to prevent crosslink formation and were visualized by autoradiography.
UvrABC incision assay
UvrA, UvrB and UvrC proteins were preheated at 65°C for 10 min in UvrABC buffer. Substrate DNA was then incubated with varying concentrations of UvrA as indicated and 100 nM UvrB in UvrABC buffer for 15 min at room temperature. UvrC was added to a final concentration of 50 nM, and the reaction mix was incubated at 55°C for 30 min. Reactions were stopped by addition of EDTA to a final concentration of 20 nM. Samples were heated to 95°C in formamide loading buffer, subjected to PAGE on a denaturing 6% gel and analyzed using the Storm 820 Phosphorimager.
UvrA and UvrB protein binding assay
Target duplex was 5′-end-labeled on the pyrimidine-rich strand. Duplex, ICL and TFO-ICL substrates were prepared as described earlier. Substrate DNA was incubated alone, with 20 nM UvrA, or with 20 nM UvrA and 100 nM UvrB in UvrABC buffer at room temperature for 15 min followed by incubation for 30 min at 55°C. Loading dye was added to each sample to a final concentration of 8% glycerol, and samples were run on a native 4% polyacrylamide gel (89 mM Tris, 89 mM Boric acid, 2 mM EDTA, 1 mM ATP, 10 mM MgCl2) at 100 V (5 V/cm) at room temperature. Alternatively, in order to characterize a quickly migrating band in the TFO-ICL + UvrA+UvrB lane, samples were heated to 95°C for 5 min following incubation with UvrA or UvrA+UvrB and allowed to cool at room temperature for 5 min prior to the addition of loading buffer. Assays were analyzed using the Storm 820 Phosphorimager.
RESULTS
Experimental design
The 120-bp duplex target was designed to contain a 30-bp TFO binding site and a psoralen 5′-AT-3′ crosslinking site (Figure 1). TFO-ICL substrate was created by incubating the 120-bp duplex with the psoralen-modified TFO (psoAG30) followed by irradiation with UVA at 1.8 J/cm2 to crosslink the psoralen to the target duplex. ICL-only substrate was constructed using a psoralen-disulfide-linked TFO (pso-SS-AG30). Following incubation with pso-SS-AG30 and UVA irradiation, the sample was treated with DTT to reduce the disulfide bond to release the TFO. Crosslinked samples were then purified by denaturing PAGE. The fluorescein-adducted duplex F2650, which has previously been shown to be efficiently incised by UvrABC nuclease (15,16), was used as a positive control.
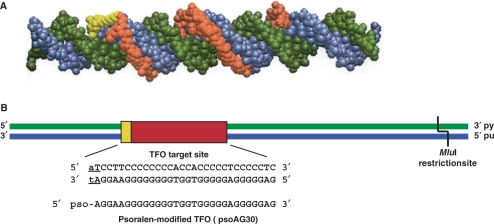
Figure 1.
The target duplex and psoralen-modified TFO. (A) Space-filling model of a psoralen-modified TFO bound to its target duplex. The psoralen moiety is shown in yellow and the TFO is shown in red bound to the purine-rich strand of the target duplex (blue) in the major groove. The pyrimidine-rich strand is shown in green (21). (B) Psoralen-modified TFO and TFO binding site. The TFO binding site is shown in red, the psoralen crosslinking site is shown in yellow and the pyrimidine-rich (py) and purine-rich (pu) strands are depicted in green and blue, respectively. The sequence of the TFO binding site is shown in bold capital letters, the psoralen crosslinking site is underlined and psoAG30, the psoralen-conjugated TFO, is depicted in an antiparallel orientation relative to the purine-rich strand of the target duplex. The MluI restriction site was used to remove the 3′-radiolabel on the pyrimidine strand. Figure 1A reprinted with permission from Vasquez et al., Biochemistry, 35, 10712–10719, Copyright 1996 American Chemical Society.
The triplex structure is present following substrate purification
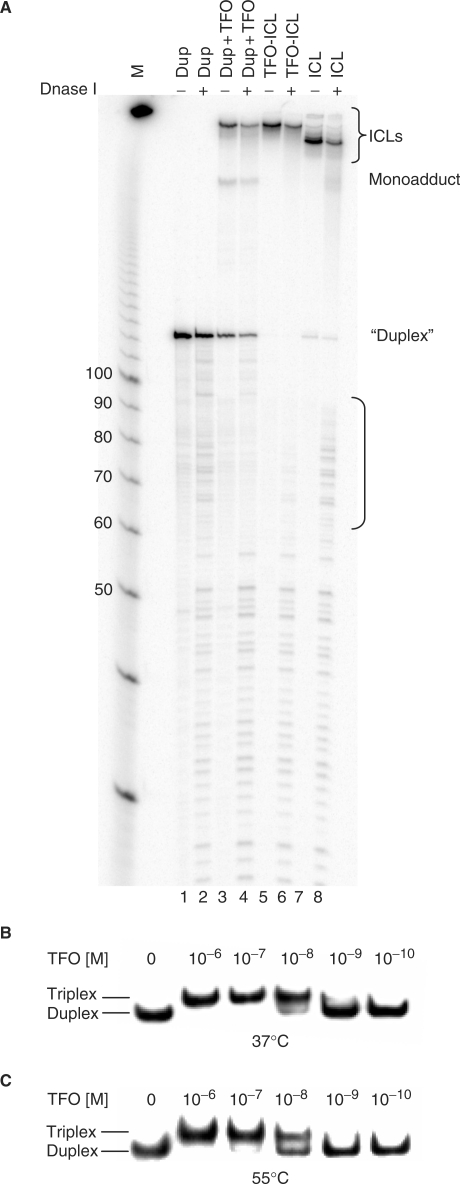
In order to confirm the presence of the triplex structure following gel purification and reannealing of TFO-ICL substrate, DNase footprinting was performed on duplex, ICL only, TFO-ICL and unpurified TFO-ICL substrates (Figure 2A). Digestion of the TFO-ICL substrate was inhibited from approximately 60–90 bases (Figure 2A, lane 6), as expected based on the presence of the bound 30-base TFO, consistent with the protected region observed with the unpurified TFO-ICL substrate (Figure 2A, lane 4).
Figure 2.
Triplex formation occurs under conditions of the UvrABC incision assay. (A) DNase footprint to verify presence of the triplex structure. Substrate labeled on the 5′-end of the purine-rich strand was incubated −/+ DNase I as indicated. Lanes 1 and 2 contain duplex, lanes 3 and 4 contain unpurified TFO-ICL substrate, lanes 5 and 6 contain gel-purified TFO-ICL and lanes 7 and 8 contain gel-purified ICL only. A bracket indicates the protected region. (B) and (C) TFO-binding assay. Target duplex DNA was end-labeled with [γ-32P]ATP and incubated with increasing concentrations of TFO as indicated. Samples were either incubated in standard triplex binding buffer at 37°C and subjected to native PAGE on a 12% TBM gel (B) or were incubated in UvrABC buffer at 55°C for 30 min prior to loading (C). Triplex formation occurs under both conditions (binding affinity of ∼10−8 M), indicating that the reaction conditions did not affect TFO binding.
Triplex formation occurs under the conditions of the UvrABC incision assay
Since the thermophilic UvrABC nuclease functions at 55°C, we wanted to assure that the triplex structure was stable at this temperature. Target duplex DNA was end-labeled and incubated with increasing concentrations of TFO in triplex-binding buffer, or in UvrABC incision assay buffer, as indicated (Figure 2B and C). Samples were either run directly on a 12% TBM native gel or were incubated in UvrABC buffer at 55°C for 30 min prior to gel loading. Under both conditions the TFO demonstrated a binding affinity of ∼10−8 M for its target duplex. To verify that the TFO was not simply reannealing to the duplex in the gel following incubation at 55°C, target duplex was mixed with increasing concentrations of TFO in triplex-binding buffer and run immediately on the gel without incubation. No binding was observed even at high concentrations (data not shown), suggesting that the TFO does not reanneal to its target duplex throughout the course of the gel mobility shift assay. These results suggest that the reaction conditions do not have a substantial effect on TFO binding, a finding consistent with published reports on the thermal stability of triplex DNA structures (18–20).
UvrABC incises TFO-directed psoralen ICLs with a higher efficiency but similar pattern to that of psoralen ICLs alone
Substrate DNA was alternately labeled on the purine-rich strand, pyrimidine-rich strand or the third strand TFO in order to monitor processing by the UvrABC nuclease. Substrate DNA was then incubated with 20 nM UvrA and 100 nM UvrB in UvrABC buffer for 15 min at room temperature. UvrC was added to a final concentration of 50 nM, and the reaction mixture was incubated at 55°C for 30 min. Samples were then subjected to denaturing PAGE to determine the amount and types of incision products generated.
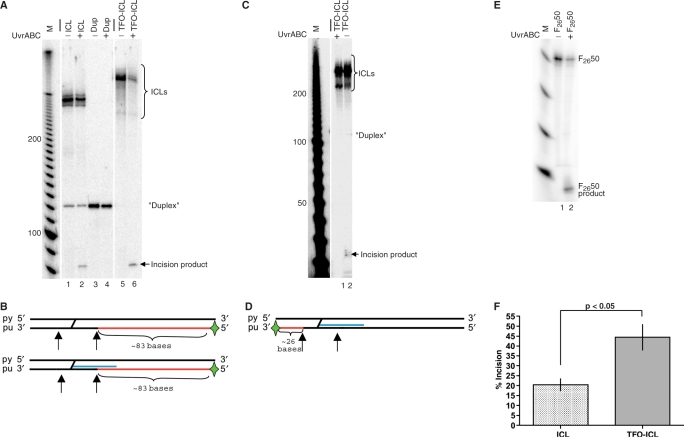
Results from experiments with the substrate 5′-end-labeled on the purine-rich strand are shown in Figure 3A. UvrABC has been shown to incise the purine-rich strand containing the furan adduct more efficiently than the pyrimidine strand (containing the pyrone adduct) in similar studies (6,13). Following incubation with the UvrABC nuclease, both TFO-ICL and ICL-only substrates produced incision products migrating slightly slower than 80 bases (Figure 3A, lanes 2 and 6). This is consistent with cutting of the purine strand ∼9 bases on the 5′-side of the ICL (Figure 3B), which would yield an 83-base product, placing the incision site within the TFO-binding region. No incision product was observed with nondamaged duplex (Figure 3A, lane 4). The fluorescein-adducted substrate F2650 was used as a positive control in the incision assays though may not be shown in all figures for clarity of presentation. A representative assay is shown in Figure 3E. Incision efficiencies observed were similar to those reported previously on ICL-containing substrates in the absence of a third strand TFO (15,16). Interestingly, analysis of the incision activity of the UvrABC nuclease on the TFO-ICL and ICL-only substrates indicated that 5′ incision of the TFO-ICL substrate is more efficient than 5′ incision of the ICL-only substrate (Figure 3F; 44 ± 12.8% and 21 ± 5.2%, respectively, P < 0.05 using Student's t-test; the values obtained for incision activity on the TFO-ICL and ICL substrates were normalized to the incision activity on the F2650 substrate). This is in contrast to published work utilizing a pyrimidine-rich MOP-modified TFO (13). These data suggest that the purine-rich TFO does not inhibit the 5′-incision activity of the UvrABC nuclease under the conditions of our assay.
Figure 3.
UvrABC incision assay with the radiolabel on the purine-rich strand of the target duplex. (A) UvrABC incision assay with 5′-end-labeled purine-rich duplex target strand. Substrate DNA was incubated with purified UvrA (20 nM), UvrB (100 nM) and UvrC (50 nM) as described in the Materials and Methods section and subjected to denaturing PAGE on a 6% gel. Lanes 1 and 2 contain purified psoralen crosslinked duplex substrate treated to remove the TFO (labeled as ICL) −/+ UvrABC, lanes 3 and 4 contain nondamaged duplex DNA (labeled as Dup) −/+ UvrABC and lanes 5 and 6 contain purified TFO-directed psoralen ICL substrate with the TFO covalently attached (labeled as TFO-ICL) −/+ UvrABC. The bar (−) indicates lanes that have been removed for clarity of presentation. (B) Expected incision sites for the UvrABC nuclease on ICL and TFO-ICL substrates. The expected incision product is shown in red, the TFO is depicted in blue and incision sites are marked by arrows. A green star indicates the position of the radiolabel. The pyrimidine-rich and purine-rich strands of the target duplex are labeled py and pu, respectively. The incision product migrating between 80 and 90 bases corresponds with cleavage on the purine-rich strand ∼9 phosphodiester bonds on the 5′-side of the ICL, within the triplex binding site. (C) UvrABC incision assay with the purine-rich strand of the target duplex 3′-end-labeled. Lanes 1 and 2 contain TFO-ICL +/− UvrABC. The incision product migrating between 20 and 30 bases corresponds with cleavage on the purine-rich strand ∼3 phosphodiester bonds on the 3′-side of the ICL. (D) Expected UvrABC incision sites on the TFO-ICL substrate. The figure is marked as in (B). (E) UvrABC incision assay with fluorescein-adducted positive control (F2650) 5′-end-labeled on the fluorescein-adducted strand. Lanes 1 and 2 contain F2650 −/+ UvrABC. (F) Histogram indicating the average incision (as a percent of the total substrate) observed for ICL and TFO-ICL substrates by the UvrABC nuclease. Incision assays were repeated at least three times. Data presented are ±SD, P < 0.05 using Student's t-test.
To gain more information on the nature of the incision sites, the target duplex was 3′-end-labeled on the purine-rich strand of the target duplex. After incubation with the UvrABC nuclease, samples were subjected to denaturing PAGE and an incision product migrating between 20 and 30 bases was observed in the TFO-ICL lane (Figure 3C, lane 1). This product size corresponds with UvrABC incision on the purine-rich strand ∼3 phosphodiester bonds on the 3′-side of the ICL (Figure 3D), which would generate a 26-base product, consistent with previous studies (13).
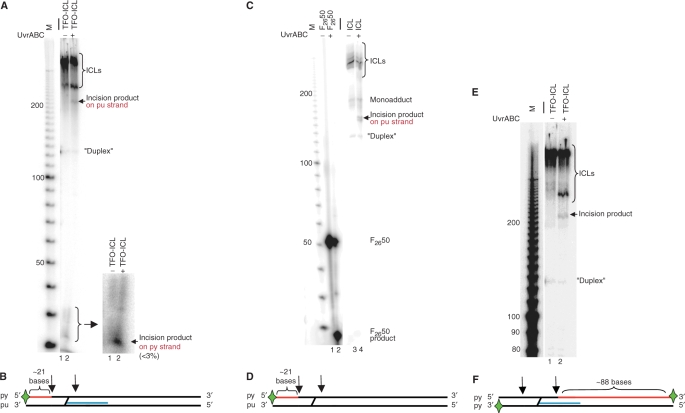
To determine whether the UvrABC nuclease incises the pyrimidine-rich strand of the crosslinked target duplexes, the 5′-end of the pyrimidine-rich strand was labeled and subjected to the UvrABC incision assay. For both TFO-ICL and ICL-only substrates 5′-incision efficiency on the pyrimidine-rich strand was <3% (Figure 4). However, an incision product of ∼220 bases was observed by denaturing PAGE in the sample containing the TFO-ICL substrate and the UvrABC nuclease (Figure 4A, lane 2), likely due to purine-rich strand incision. Similarly, incubation of the ICL-only substrate with UvrABC produced two products migrating between 140 and 150 bases (Figure 4C, lane 4), again suggesting that these products were the result of cutting on the purine-rich strand of the target duplex only (Figure 4B and D). Labeling of the 3′-ends likewise revealed an incision product of ∼220 bp (Figure 4E, lane 2), but no detectable product of ∼88 bases as might be expected with cutting on the 3′ side of the ICL on the pyrimidine-rich strand (Figure 4F). These results point to inefficient incision by UvrABC on the pyrimidine-rich strand compared to the purine-rich strand of the target duplex on both the TFO-ICL and the ICL-only substrates.
Figure 4.
UvrABC incision assay with the radiolabel on the pyrimidine-rich strand of the target duplex. (A) Samples were incubated with 20 nM UvrA, 100 nM UvrB and 50 nM UvrC as described in the Materials and methods section and subjected to denaturing PAGE on a 6% gel. Lanes 1 and 2 contain TFO-ICL −/+ UvrABC. The insert contains a magnified view of the incision product. The incision product migrating just above 20 bases corresponds to cutting on the pyrimidine-rich strand of the target duplex. (B) Expected incision sites for UvrABC on the TFO-ICL substrate. The expected incision product is shown in red, the TFO is depicted in blue and incision sites are marked by arrows. A green star indicates the position of the radiolabel. The pyrimidine-rich and purine-rich strands of the target duplex are labeled py and pu, respectively. (C) UvrABC incision assay with the ICL-only substrate 5′-end-labeled on the pyrimidine-rich strand of the target duplex. Lanes 1 and 2 contain the F2650 positive control, and lanes 3 and 4 contain ICL-only substrate. (D) Expected incision sites for ICL-only substrate. The figure is marked as in (B). (E) UvrABC incision assay with substrate 3′-end-labeled on both strands of the target duplex. Lanes 1 and 2 contain TFO-ICL −/+ UvrABC. (F). Expected incision sites for the UvrABC nuclease on the TFO-ICL substrate. The figure is marked as in (B).
Since the presence of the TFO did not appear to inhibit the incision activity of the UvrABC nuclease on the purine-rich strand of the crosslinked triplex substrate, we 3′-end-labeled the TFO to observe its processing by UvrABC. A product was observed at ∼220 bases, the same size as the product seen when the pyrimidine-rich strand was labeled. However, no products of the size expected from TFO cleavage (<22 bases) were identified as UvrABC incision products (data not shown), indicating that UvrABC does not incise the TFO itself prior to incision of the purine-rich strand of the target duplex. This suggests that the intact TFO remains covalently linked (via the ICL) to the substrate following processing by the UvrABC nuclease and that the TFO may be displaced from the duplex target site during processing.
UvrA and UvrB binding displaces the TFO from the target duplex
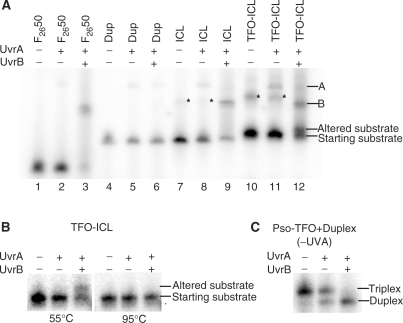
It is known that UvrA2B binds damaged DNA, with UvrA loading UvrB onto the site. UvrA is then released, leaving the UvrB-DNA preincision complex (7). In order to investigate the effects of the TFO on damage recognition and binding by UvrA and UvrB, a gel mobility shift assay was performed with radiolabeled substrates containing an ICL or a TFO-ICL in the presence of purified UvrA and UvrB proteins. We found that binding of UvrA to the ICL and to the TFO-ICL substrates was similar (<7%), and that incubation of the substrates with both UvrA and UvrB resulted in ∼40% binding by UvrB (Figure 5A, lanes 9 and 12), suggesting that the increased incision efficiency of the TFO-ICL substrate was not due to an increase in recognition of or binding by the UvrA2B complex. This is consistent with results reported by Duval-Valentin et al. (13), with a pyrimidine-rich TFO. As expected, incubation of F2650 with both UvrA and UvrB resulted in efficient (∼55%) binding by UvrB (Figure 5A, lane 3). Interestingly, an additional product migrating faster than UvrB-bound TFO-ICL and slightly slower than unbound TFO-ICL was observed on the gel in the sample containing TFO-ICL in the presence of both UvrA and UvrB (Figure 5A, lane 12, band labeled ‘altered substrate’). We speculated that this product may be the result of either binding by partially degraded protein and/or displacement of the TFO by UvrA and/or UvrB. To test this, substrates were subjected to either proteinase K treatment or heat denaturation following incubation with either UvrA or UvrA and UvrB. The product was sensitive to heat denaturation (Figure 5B) but not to proteinase K (data not shown), consistent with structural modifications of the TFO-ICL (such as altered hydrogen bonding of the TFO induced by UvrA and/or UvrB). To substantiate that formation of the UvrB preincision complex was displacing the TFO, a binding reaction was performed with nonirradiated TFO + duplex samples, to form a noncovalent triplex substrate. We found that the TFO was displaced from its binding site on the duplex DNA substrate slightly by UvrA, but dramatically in the presence of UvrA and UvrB (Figure 5C). Taken together, it is likely that the additional product observed is due to displacement of the TFO from its target duplex binding site during formation of the UvrB–DNA complex.
Figure 5.
UvrA or UvrA+UvrB protein binding assay. UvrA or UvrA+UvrB protein binding assays with 5′-end-labeled pyrimidine-rich duplex target strand. (A) Substrate DNA was incubated with purified UvrA (20 nM) or UvrA (20 nM) and UvrB (100 nM) as described in the Materials and Methods section and subjected to 4% native PAGE gel. Lanes 1–3 contain F2650 positive control with no protein, UvrA or UvrA+UvrB, respectively. Lanes 4–6 contain nondamaged duplex DNA with no protein, UvrA or UvrA+UvrB, respectively. Lanes 7–9 contain purified ICL only with no protein, UvrA or UvrA+UvrB, respectively. Lanes 10–12 contain purified TFO-ICL substrate with no protein, UvrA or UvrA+UvrB, respectively. UvrB binding was similar with ICL and TFO-ICL substrates (∼40%, lanes 9 and 12). An ‘altered substrate’ was observed when the TFO-ICL substrate was incubated with UvrA+UvrB (lane 12). Asterisks mark an artifact occurring in lanes containing crosslinked substrate. (B) UvrA or UvrA+UvrB protein-binding assay to test for possible structural modifications of TFO-ICL resulting in an ‘altered substrate’ following incubation with UvrA or UvrA+UvrB. Substrate in the presence or absence of UvrA and/or UvrB was either loaded directly on the gel following incubation at 55°C or heated to 95°C prior to loading. The ‘altered substrate’ disappeared with heat denaturation, consistent with structural modifications of the TFO-ICL. (C) UvrA or UvrA+UvrB protein-binding assay to test for displacement of noncrosslinked TFO. Noncrosslinked triplex substrate (pso-TFO + duplex in the absence of UVA irradiation) was incubated with UvrA or UvrA+UvrB as described and subjected to native PAGE (4% gel). Triplex substrate was denatured to duplex form after incubation with UvrA+UvrB, indicating displacement of the TFO from its target duplex binding site.
UvrA concentration affects the incision efficiencies of TFO-ICL and ICL substrates differently
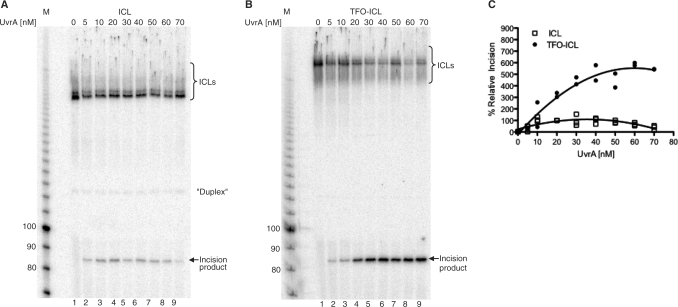
It has been reported that the cleavage efficiencies of both ICLs and TFO-ICLs depend on the concentration of UvrA, with the presence of a pyrimidine-rich third strand inhibiting cleavage at low UvrA concentrations (<70 nM) (13). To investigate the effects of UvrA concentration on the cleavage efficiencies of the purine-rich TFO-ICL substrate compared to the ICL alone, UvrABC incision assays were performed with concentrations of UvrA ranging from 0 nM to 70 nM on substrates labeled on the 5′-end of the purine-rich strand (Figure 6A and B). As observed previously (13), the incision efficiency of the ICL only increased with UvrA concentration to reach a plateau between ∼20 nM and 40 nM. At higher concentrations, the level of incision began to decrease. The incision efficiency of the TFO-ICL substrate was similar to the incision efficiency of the ICL-only substrate at lower concentrations of UvrA (<20 nM). However, with UvrA concentrations of 20 nM and higher, the incision efficiency of the TFO-ICL was substantially higher than the incision efficiency of the ICL only (Figure 6C).
Figure 6.
UvrA concentration-dependent incision efficiency of ICL and TFO-ICL substrates. ICL only (A) and TFO-ICL (B) substrates were labeled on the 5′-end of the purine-rich strand. Substrates were incubated with increasing concentrations of UvrA (0–70 nM as indicated for each lane), 100 nM UvrB and 50 nM UvrC for 30 min at 55°C. (C) Trend lines for ICL and TFO-ICL incision efficiencies with increasing concentrations of UvrA. Relative incision values were determined by calculating the amount of product as a percent of total sample and normalizing to the incision efficiency of ICL-only substrate at 20 nM UvrA.
DISCUSSION
Psoralens are photoreactive compounds that can form ICLs preferentially at 5′-TA-3′ sites in double-stranded DNA. Psoralen-modified TFOs can be used to target psoralen molecules to specific sites in double-stranded DNA (21,22), providing a tool for targeting genes and directing site-specific DNA damage. However, despite the potential utility of psoralen-modified TFOs in the biomedical and biotechnological fields, much remains to be learned about the processing of these lesions by DNA repair proteins. Here, we investigated the effect of a purine-rich TFO on the processing of psoralen ICLs by the UvrABC nuclease. We used substrates containing either a psoralen ICL alone or a TFO-directed ICL in the presence of the purified UvrABC protein complex to compare processing of psoralen ICLs in the presence or absence of the third strand TFO. Our results show nearly identical incision patterns of the ICL with or without the TFO, in concordance with a previous study of TFO-ICLs by Duval-Valentin et al. (13). However, while this previous study demonstrated that a pyrimidine-rich TFO inhibited incision by the UvrABC nuclease, we observed efficient incision of a purine-rich TFO-ICL compared to the ICL alone, likely due to the displacement of the TFO by the UvrB–DNA complex.
In our study, the UvrABC nuclease generated incision products consistent with cleavage on the purine-rich strand of the target duplex approximately nine phosphodiester bonds 5′, and three phosphodiester bonds 3′ to the ICL in the presence or absence of the TFO. Pyrimidine strand cleavage by the UvrABC nuclease was nearly undetectable (<3%) with either substrate. This is in agreement with previous work showing that UvrABC cuts predominantly on the furan-adducted side of an HMT ICL (6), and that furan-sided adducts are produced on the purine-rich strand of the target duplex when HMT is coupled to a purine-rich TFO (12). Of note, the incision made on the 5′-side of the ICL is within the TFO binding site, suggesting that the UvrABC nuclease would either have to incise the TFO first, or displace it prior to cleavage of the purine-rich strand of the underlying target duplex. The incision assays using a TFO-ICL substrate 3′-end-labeled on the TFO showed no evidence of incision of the TFO (data not shown), suggesting that the TFO may be displaced from the duplex during processing by the UvrABC nuclease. The UvrABC complex has been shown to release short oligonucleotides annealed to single-stranded DNA, and the length and amount of oligonucleotide released increased in the presence of bulky DNA adducts (23,24). It is hypothesized that the release of the oligonucleotide is due to strand destabilization activity of the UvrABC nuclease. A similar destabilization of the TFO could allow access to the target incision site on the purine-rich strand of the duplex. Consistent with this, our gel mobility shift assay revealed an additional product migrating faster than UvrB-bound TFO-ICL and slightly slower than unbound TFO-ICL in the TFO-ICL + UvrA+UvrB lane (Figure 5A, lane 12, band labeled ‘altered substate’). The product was sensitive to heat denaturation, suggesting that it resulted from structural modifications of the TFO-ICL (such as altered hydrogen bonding of the TFO). Repetition of the gel mobility shift assay with noncrosslinked TFO + duplex samples (i.e. the TFO was not covalently linked to the substrate) indicated that the TFO was displaced from the duplex DNA substrate by the UvrA2B complex. Taken together, it is likely that the additional product observed is due to displacement of the TFO during the formation of the stable UvrB–DNA complex. Crystal structure studies indicate that formation of the UvrB–DNA complex is marked by insertion of the β-hairpin motif of UvrB between the strands of duplex DNA (25,26). Insertion of the β-hairpin into the DNA may disrupt TFO binding, thereby displacing the TFO and allowing access to the incision site on the target duplex.
At concentrations of UvrA ≥20 nM, the UvrABC nuclease showed more efficient incision activity on the TFO-ICL than the ICL alone. The increase in incision observed with the purine-rich TFO-ICL substrate over that of the ICL alone could be due to better recognition and binding of the UvrA2B protein complex to the purine-rich TFO-ICL compared to the ICL only. Some studies have indicated that the binding affinity of UvrA2B is higher for bulkier substrates, and UvrA2B binding correlates with incision activity (27,28). However, we found that binding to both TFO-ICL and ICL substrates by UvrA and UvrB was similar, demonstrating that the increased incision efficiency of the TFO-ICL substrate was not likely due to an increase in recognition of or binding by the UvrA2B complex. This is consistent with results reported by Duval-Valentin et al. (13) with a pyrimidine-rich TFO and with other reports that binding of UvrA2B complex does not always correlate with incision efficiency (29,30). Since the increased incision efficiency observed in the presence of the triplex does not appear to be the result of increased binding by UvrA2B, it may be that binding and incision by UvrC is facilitated by the presence of the third strand.
The efficient incision of TFO-directed ICLs observed here contrasts with previous work indicating that the presence of a pyrimidine-rich TFO inhibits incision of a psoralen ICL by UvrABC (13). A possible explanation for the discrepancy observed in the two studies could be the nature of the psoralens and TFOs used. While the authors of the previous study used a MOP-modified pyrimidine-rich TFO, we used an HMT-modified purine-rich TFO. Purine-rich TFOs bind in the major groove of the target duplex DNA at physiological pH in an antiparallel orientation to the purine-rich duplex strand, while pyrimidine-rich TFOs require acidic conditions for triplex formation and bind in the same 5′–3′ orientation as the purine-rich duplex strand (31). Additionally, Duval-Valentin et al. (13) reported that the furan side of the psoralen derivative formed adducts primarily with the pyrimidine-rich strand of its target duplex. With the substrate used in this study, the furan side of the psoralen derivative forms adducts primarily on the purine-rich strand of its target duplex. It has been reported that sequence context, psoralen orientation and adduct stereochemistry can affect UvrABC incision patterns and efficiencies (29,30,32). Since our results with a 3′-end-labeled TFO suggest that the TFO is being displaced rather than incised, it is possible that the chemical and physical differences in the TFO-ICL structures under study may affect the ability of UvrABC to displace the TFO after damage recognition. Consistent with this, we demonstrated that higher concentrations of UvrA resulted in increased incision efficiencies of the purine-rich TFO-ICLs compared to the ICLs alone, perhaps due to increased displacement of the TFO.
Another explanation for the discrepancy in the incision efficiencies between the studies could be the result of high temperature effects due to the use of thermophilic UvrABC nuclease in this study. Assays in the current study were performed at 55°C, and the elevated temperature might have affected TFO binding. However, previous thermodynamic studies have shown that triplexes formed with purine-rich TFOs are stable at elevated temperatures (≥65°C), and TFO binding can even stabilize the target duplex at high temperatures (18–20). Consistent with this, the gel mobility shift assay suggested that the reaction conditions do not affect the binding affinity of the purine-rich TFO used in this study.
In conclusion, psoralens are currently used in medicine and research for their ability to form ICLs. TFOs are one way to target such ICLs to specific sites in the genome, providing a means to study ICL repair in vivo. However, if psoralen-modified TFOs are to be used for research and medical purposes, it is important to understand how the presence of the TFO will influence the processing of psoralen ICLs in both bacterial and mammalian systems. Previously, we have shown that TFO-directed ICL lesions are bound by the human NER damage/distortion recognition proteins (XPA-RPA and XPC-RAD23B) with high affinity and specificity (14,33,34), and is it known that NER is required for TFO-induced mutagenesis (35). Similarly, NER is required for excision of polyamine adducts in both Escherichia coli and human cell-free extracts (36); and NER, in conjunction with proteolytic degradation, has been implicated in the repair of DNA–protein crosslinks in bacterial and mammalian systems (37–39). In this study, we demonstrate that the purine-rich TFO enhances incision of the ICL by the UvrABC complex. Further, we show that the presence of the purine-rich TFO does not alter the incision pattern of the ICL and that the TFO is displaced in the presence of UvrA and UvrB. Together, these results indicate that TFO-directed psoralen ICLs are recognized and processed by the NER mechanism in both bacterial and mammalian systems, supporting the use of triplex technology as a powerful tool to induced site-specific DNA damage to facilitate genome modification.
FUNDING
National Institutes of Health/National Cancer Institute (CA097175 and CA093729 to K.M.V.); National Institutes of Environmental Health Sciences Center (ES007784). Funding for open access charge: P01 CA097175.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Ms Sarah Henninger for help in the preparation of the article, and Dr Richard Wood for useful discussions.
REFERENCES
- 1.Stern RS. Psoralen and ultraviolet a light therapy for psoriasis. N Engl J. Med. 2007;357:682–690. doi: 10.1056/NEJMct072317. [DOI] [PubMed] [Google Scholar]
- 2.Song PS, Tapley K.J., Jr Photochemistry and photobiology of psoralens. Photochem. Photobiol. 1979;29:1177–1197. doi: 10.1111/j.1751-1097.1979.tb07838.x. [DOI] [PubMed] [Google Scholar]
- 3.Cimino GD, Gamper HB, Isaacs ST, Hearst JE. Psoralens as photoactive probes of nucleic acid structure and function: organic chemistry, photochemistry, and biochemistry. Annu. Rev. Biochem. 1985;54:1154–1193. doi: 10.1146/annurev.bi.54.070185.005443. [DOI] [PubMed] [Google Scholar]
- 4.Shi Y-B, Hearst JE. Wavelength dependence for the photoreactions of DNA-psoralen monoadducts. 2. Photo-cross-linking of monoadducts. Biochemistry. 1987;26:3792–3798. doi: 10.1021/bi00387a009. [DOI] [PubMed] [Google Scholar]
- 5.Van Houten B, Gamper H, Hearst JE, Sancar A. Construction of DNA substrates modified with psoralen at a unique site and study of the action mechanism of ABC excinuclease on these uniformly modified substrates. J. Biol. Chem. 1986;261:14135–14141. [PubMed] [Google Scholar]
- 6.Van Houten B, Gamper H, Holbrook SR, Hearst JE, Sancar A. Action mechanism of ABC excision nuclease on a DNA substrate containing a psoralen crosslink at a defined position. Proc. Natl Acad. Sci. USA. 1986;83:8077–8081. doi: 10.1073/pnas.83.21.8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Truglio JJ, Croteau DL, Van Houten B, Kisker C. Prokaryotic nucleotide excision repair: the UvrABC system. Chem. Rev. 2006;106:233–252. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 8.Van Houten B, Croteau DL, DellaVecchia MJ, Wang H, Kisker C. ‘Close-fitting sleeves’: DNA damage recognition by the UvrABC nuclease system. Mutat. Res. 2005;577:92–117. doi: 10.1016/j.mrfmmm.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 9.Beal PA, Dervan PB. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science. 1991;251:1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- 10.Cooney M, Czernuszewicz G, Postel EH, Flint SJ, Hogan ME. Site-specific oligonucleotide binding represses transcription of the human c-myc gene in vitro. Science. 1988;241:456–459. doi: 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]
- 11.Jain A, Wang G, Vasquez KM. DNA triple helices: biological consequences and therapeutic potential. Biochimie. 2008;90:1117–1130. doi: 10.1016/j.biochi.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gasparro FP, Havre PA, Olack GA, Gunther EJ, Glazer PM. Site-specific targeting of psoralen photoadducts with a triple helix-forming oligonucleotide: characterization of psoralen monoadduct and crosslink formation. Nucleic Acids Res. 1994;22:2845–2852. doi: 10.1093/nar/22.14.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duval-Valentin G, Takasugi M, Helene C, Sage E. Triple helix-directed psoralen crosslinks are recognized by Uvr(A)BC excinuclease. J. Mol. Biol. 1998;278:815–825. doi: 10.1006/jmbi.1998.1728. [DOI] [PubMed] [Google Scholar]
- 14.Vasquez KM, Christensen J, Li L, Finch RA, Glazer PM. Human XPA and RPA DNA repair proteins participate in specific recognition of triplex-induced helical distortions. Proc. Natl Acad. Sci. USA. 2002;99:5848–5853. doi: 10.1073/pnas.082193799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Truglio JJ, Croteau DL, Skorvaga M, DellaVecchia MJ, Theis K, Mandavilli BS, Van Houten B, Kisker C. Interactions between UvrA and UvrB: the role of UvrB's domain 2 in nucleotide excision repair. EMBO J. 2004;23:2498–2509. doi: 10.1038/sj.emboj.7600263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DellaVecchia MJ, Croteau DL, Skorvaga M, Dezhurov SV, Lavrik OI, Van Houten B. Analyzing the handoff of DNA from UvrA to UvrB utilizing DNA-protein photoaffinity labeling. J. Biol. Chem. 2004;279:45245–45256. doi: 10.1074/jbc.M408659200. [DOI] [PubMed] [Google Scholar]
- 17.Croteau DL, DellaVecchia MJ, Wang H, Bienstock RJ, Melton MA, Van Houten B. The C-terminal zinc finger of UvrA does not bind DNA directly but regulates damage-specific DNA binding. J. Biol. Chem. 2006;281:26370–26381. doi: 10.1074/jbc.M603093200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svinarchuk F, Paoletti J, Malvy C. An unusually stable purine(purine-pyrimidine) short triplex. The third strand stabilizes double-stranded DNA. J. Biol. Chem. 1995;270:14068–14071. doi: 10.1074/jbc.270.23.14068. [DOI] [PubMed] [Google Scholar]
- 19.Svinarchuk F, Monnot M, Merle A, Malvy C, Fermandjian S. The high stability of the triple helices formed between short purine oligonucleotides and SIV/HIV-2 vpx genes is determined by the targeted DNA structure. Nucleic Acids Res. 1995;23:3831–3836. doi: 10.1093/nar/23.19.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pilch DS, Levenson C, Shafer RH. Structure, stability, and thermodynamics of a short intermolecular purine-purine-pyrimidine triple helix. Biochemistry. 1991;30:6081–6088. doi: 10.1021/bi00239a001. [DOI] [PubMed] [Google Scholar]
- 21.Vasquez KM, Wensel TG, Hogan ME, Wilson JH. High-efficiency triple-helix-mediated photo-cross-linking at a targeted site within a selectable mammalian gene. Biochemistry. 1996;35:10712–10719. doi: 10.1021/bi960881f. [DOI] [PubMed] [Google Scholar]
- 22.Wang G, Levy DD, Seidman MM, Glazer PM. Targeted mutagenesis in mammalian cells mediated by intracellular triple helix formation. Mol. Cell Biol. 1995;15:1759–1768. doi: 10.1128/mcb.15.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skorvaga M, Theis K, Mandavilli BS, Kisker C, Van Houten B. The beta-hairpin motif of UvrB is essential for DNA binding, damage processing, and UvrC-mediated incisions. J. Biol. Chem. 2002;277:1553–1559. doi: 10.1074/jbc.M108847200. [DOI] [PubMed] [Google Scholar]
- 24.Gordienko I, Rupp WD. The limited strand-separating activity of the UvrAB protein complex and its role in the recognition of DNA damage. EMBO J. 1997;16:889–895. doi: 10.1093/emboj/16.4.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Theis K, Chen PJ, Skorvaga M, Van Houten B, Kisker C. Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair. EMBO J. 1999;18:6899–6907. doi: 10.1093/emboj/18.24.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Truglio JJ, Karakas E, Rhau B, Wang H, DellaVecchia MJ, Van Houten B, Kisker C. Structural basis for DNA recognition and processing by UvrB. Nat. Struct. Mol. Biol. 2006;13:360–364. doi: 10.1038/nsmb1072. [DOI] [PubMed] [Google Scholar]
- 27.Snowden A, Van Houten B. Initiation of the UvrABC nuclease cleavage reaction. Efficiency of incision is not correlated with UvrA binding affinity. J. Mol. Biol. 1991;220:19–33. doi: 10.1016/0022-2836(91)90378-j. [DOI] [PubMed] [Google Scholar]
- 28.Hoare S, Zou Y, Purohit V, Krishnasamy R, Skorvaga M, Van Houten B, Geacintov NE, Basu AK. Differential incision of bulky carcinogen-DNA adducts by the UvrABC nuclease: comparison of incision rates and the interactions of Uvr subunits with lesions of different structures. Biochemistry. 2000;39:12252–12261. doi: 10.1021/bi0013187. [DOI] [PubMed] [Google Scholar]
- 29.Ramaswamy M, Yeung AT. Sequence-specific interactions of UvrABC endonuclease with psoralen interstrand cross-links. J. Biol. Chem. 1994;269:485–492. [PubMed] [Google Scholar]
- 30.Jiang GH, Skorvaga M, Croteau DL, Van Houten B, States JC. Robust incision of Benoz[a]pyrene-7,8-dihyrodiol-9,10-epoxide-DNA adducts by a recombinant thermoresistant interspecies combination UvrABC endonuclease system. Biochemistry. 2006;45:7834–7843. doi: 10.1021/bi052515e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vasquez KM, Wilson JH. Triplex-directed modification of genes and gene activity. Trends Biochem. Sci. 1998;23:4–9. doi: 10.1016/s0968-0004(97)01158-4. [DOI] [PubMed] [Google Scholar]
- 32.Jones BK, Yeung AT. DNA base composition determines the specificity of UvrABC endonuclease incision of a psoralen cross-link. J. Biol. Chem. 1990;265:3489–3496. [PubMed] [Google Scholar]
- 33.Thoma BS, Wakasugi M, Christensen J, Reddy MC, Vasquez KM. Human XPC-hHR23B interacts with XPA-RPA in the recognition of triplex-directed psoralen DNA interstrand crosslinks. Nucleic Acids Res. 2005;33:2993–3001. doi: 10.1093/nar/gki610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reddy MC, Christensen J, Vasquez KM. Interplay between human high mobility group protein 1 and replication protein A on psoralen-cross-linked DNA. Biochemistry. 2005;44:4188–4195. doi: 10.1021/bi047902n. [DOI] [PubMed] [Google Scholar]
- 35.Wang G, Seidman MM, Glazer PM. Mutagenesis in mammalian cells induced by triple helix formation and transcription-coupled repair. Science. 1996;271:802–805. doi: 10.1126/science.271.5250.802. [DOI] [PubMed] [Google Scholar]
- 36.Nakano T, Katafuchi A, Shimizu R, Terato H, Suzuki T, Tauchi H, Makino K, Skorvaga M, Van Houten B, Ide H. Repair activity of base and nucleotide excision repair enzymes for guanine lesions induced by nitrosative stress. Nucleic Acids Res. 2005;33:2181–2191. doi: 10.1093/nar/gki513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minko IG, Kurtz AJ, Croteau DL, Van Houten B, Harris TM, Lloyd RS. Initiation of repair of DNA-polypeptide cross-links by the UvrABC nuclease. Biochemistry. 2005;44:3000–3009. doi: 10.1021/bi0478805. [DOI] [PubMed] [Google Scholar]
- 38.Reardon JT, Sancar A. Repair of DNA-polypeptide crosslinks by human excision nuclease. Proc. Natl Acad. Sci. USA. 2006;103:4056–4061. doi: 10.1073/pnas.0600538103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakano T, Morishita S, Katafuchi A, Matsubara M, Horikawa Y, Terato H, Salem AM, Izumi S, Pack SP, Makino K, et al. Nucleotide excision repair and homologous recombination systems commit differentially to the repair of DNA-protein crosslinks. Mol. Cell. 2007;28:147–158. doi: 10.1016/j.molcel.2007.07.029. [DOI] [PubMed] [Google Scholar]