Abstract
In HIV-1, trans-activation of transcription of the viral genome is regulated by an imperfect hairpin, the trans-activating responsive (TAR) RNA element, located at the 5′ untranslated end of all viral transcripts. TAR acts as a binding site for viral and cellular proteins. In an attempt to identify RNA ligands that would interfere with the virus life-cycle by interacting with TAR, an in vitro selection was previously carried out. RNA hairpins that formed kissing-loop dimers with TAR were selected [Ducongé F. and Toulmé JJ (1999) RNA, 5:1605–1614]. We describe here the crystal structure of TAR bound to a high-affinity RNA aptamer. The two hairpins form a kissing complex and interact through six Watson–Crick base pairs. The complex adopts an overall conformation with an inter-helix angle of 28.1°, thus contrasting with previously reported solution and modelling studies. Structural analysis reveals that inter-backbone hydrogen bonds between ribose 2′ hydroxyl and phosphate oxygens at the stem-loop junctions can be formed. Thermal denaturation and surface plasmon resonance experiments with chemically modified 2′-O-methyl incorporated into both hairpins at key positions, clearly demonstrate the involvement of this intermolecular network of hydrogen bonds in complex stability.
INTRODUCTION
Kissing-loop interactions result from molecular recognition between two nucleic acid hairpins that display partially or fully complementary loops. RNA–RNA loop–loop complexes were identified in both prokaryotic and eukaryotic organisms and were shown to regulate gene expression (1). In Escherichia coli, for instance, the Col E1 plasmid replication is controlled by two plasmid-encoded RNA transcripts, RNAI and RNAII, that form at least three adjacent loop–loop complexes (2,3), with fully complementary loops. In response to oxidative stress, kissing complexes were also identified between the flho mRNA and Oxy S, a small untranslated RNA (4). Loop–loop complexes were also shown to regulate the expression of virulence genes in Staphylococcus aureus (5,6). In retroviruses such as the human immunodeficiency virus type 1 (HIV-1) and the Moloney murine leukemia virus stem-loop structures known as the dimerization initiation site (DIS) trigger retroviral RNA dimerization by forming a transient kissing complex between two DIS elements (7–9). Artificial loop–loop complexes were also identified either by rational design (10) or by in vitro selection of RNA or DNA aptamers against viral targets (11,12) and against the yeast tRNAphe (13).
Kissing complexes have been extensively investigated to understand how nucleic acid bi-molecular complexes engaging from two to up to seven Watson–Crick base pairs can rival longer linear duplexes for stability and specificity (14). In a pioneering work, the temperature-jump method was used to investigate anticodon–anticodon interactions in solution by measuring the thermodynamic and kinetic parameters of tRNAs complexes (15). These results already suggested that an inter-molecular helix could be formed between two anticodons and allowed the definition of solution conditions in which the tRNAs existed mainly as anticodon–anticodon dimers. This was crucial for structural studies. Ten years later, in another solution study that used thermal denaturation monitored by UV-spectroscopy, further insights into RNA hairpin loop–loop complex stability were given with the natural RNAI–RNAII complex in E. coli (16). This work showed that non-canonical structural determinants such as the loop closing residues and specific binding of cations were crucial for complex stability.
In the late 1990's, the solution structure of three RNA–RNA kissing complexes were investigated (10,17–19) but only two were really solved: the loop–loop dimer formed between the TAR–RNA element of HIV-1 and TAR*, a rationally designed RNA hairpin, and the RNAIi/RNAIIi complex with 5′–3′ inverted loops compared with the natural complex RNAI/RNAII. These structures share common features such as stacking of the helices from one stem to the other one through the inter-molecular loop–loop helix. The structures are bent and the three helices are not coaxial.
A limited number of loop–loop dimers have been reported by X-ray crystallography. Apart from the tRNA/tRNA kissing complex formed between two anticodon loops in the tRNAasp crystal structure (20), an intramolecular kissing complex was identified in the 23S RNA of the large ribosomal subunit between residues 412–428 and 2438–2454 (21). Kissing interactions were also described in the crystal structure of the Lysine riboswitch between residues 39–47 and 90–98 (22,23), and in the DIS/DIS dimer of HIV-1 (24). Both 23S/23S and DIS/DIS displayed coaxially aligned helices that contrast, in particular for the DIS/DIS complex, with the bent geometry always observed by NMR spectroscopy (10,18,19). The recently reported solution structure of the HIV-1lai DIS/DIS complex displays (25,26) a very similar overall fold compared to the crystallographic one with three collinear helices (24). This suggests that prior published NMR models with bent helices were likely lacking an appropriate number of constraints.
The RNA hairpin aptamer R06, specific for the trans-activating responsive (TAR) RNA element of HIV-1 was previously identified by in vitro selection (12). This aptamer displays a 5′-GUCCCAGA-3′ consensus sequence and recognizes TAR through loop–loop interactions. The G and A residues closing the aptamer loop were shown to be crucial for complex stability (27). In an attempt to rationalize the molecular basis underlying the stability of this complex, molecular dynamics (MD) were initially used (28). Recently, the liquid-crystal NMR structure of an equivalent complex was solved (29). Both studies revealed structural key elements that contribute to the unusual high stability of the TAR–RNA aptamer kissing complex compared with linear duplexes of same length (14).
In this work, we report the crystal structure of the HIV-1 TAR–RNA element bound to its RNA aptamer at a resolution of 2.9 Å. The structure displays the features of RNA kissing complexes with a continuous helix fold from the TAR stem to the aptamer stem through the inter-molecular loop–loop helix. However, in contrast to previously reported collinear kissing complex crystal structures, the TAR–R06 kissing complex displays an overall kink of 28.1°. Through thermal denaturation experiments and kinetic analysis by surface plasmon resonance, we analyze the hydrogen-bond network seen in the crystal structure and we demonstrate their implication on the stability of the complex. Finally, we are able to compare the overall structure of TAR kissing complexes obtained by X-ray crystallography, NMR and MD.
MATERIALS AND METHODS
RNA synthesis
RNA hairpins, including those with 2′-O-methyl nucleotides and with a biotin at the 3′-end, used for the thermal denaturation and surface plasmon resonance experiments were synthesized on an Expedite 8908 synthesizer. Hairpins containing one BrU residue in the stem (U14 of TAR and U2* of R06) were purchased from Dharmacon and deprotected according to the manufacturer's instructions. The oligonucleotides were purified by electrophoresis on 7 M urea 20% polyacrylamide gels, electro-eluted from the gels and desalted on G25 spin columns. The concentrations were determined on a Nanodrop spectrophotometer using molar extinction coefficients calculated with the Applied Biosystems calculator (http://www.ambion.com/techlib/misc/oligo_calculator.html).
Protein expression and purification
The cDNA encoding bacterial protein ROP was amplified by PCR and cloned into a modified pET-15b Novagen vector using NdeI and BamHI restriction sites. The protein was expressed in Rosetta (DE3) cells at 15°C overnight after induction by 0.25 mM IPTG. Protein purification was performed by a first step of affinity chromatography with His-Select cobalt bound resin followed with an overnight cleavage of the tag with recombinant TEV protease. Protein samples were further purified with Hi-Q sepharose column (Pharmacia, Uppsala, Sweden). The protein was concentrated up to 10 mg/ml, flash-frozen and kept at −80°C in 50 mM Tris–HCl pH 7.5, 100 mM NaCl and 10% glycerol.
NMR titration experiments for the complex formation
RNAs were dialysed for 48 h against a 10 mM sodium phosphate buffer, pH 6.4 at 20°C. Samples were concentrated by lyophilization and resuspended in 90/10 H2O/D2O. Appropriate folding of the hairpins was achieved by heating the samples at 95°C for 2 min and snap-cooling at 4°C. The TAR-aptamer complex was formed by addition of TAR to R06, or the contrary, and monitoring the imino region of 1D spectra at 4°C. The concentration of RNA ranged from 0.7–1 mM (280 µl in Shigemi tubes). Titration of the RNA complex, at 4°C, was achieved by adding increasing amount of protein ROP, prepared in 10 mM sodium phosphate buffer, pH 6.4, containing 100 mM sodium chloride and 1 mM DTT. The imino region of 1D spectra was used to monitor the formation of the protein–nucleic acid complex, as previously described (30).
NMR spectra were recorded at 500 MHz on a Bruker (Wissembourg, France) Avance spectrometer equipped with a z-gradient probe. NMR data were processed using the TopSpin (Bruker) software package. 1H assignments were obtained using standard homonuclear methods. NMR data for exchangeable protons were acquired at 4°C. Solvent suppression for samples in 90/10 H2O/D2O was achieved using the WATERGATE and ‘Jump and Return’ sequences (31,32). Assignment of TAR–R06 kissing complex, based on analysis of NOESY spectra recorded in H2O, has been previously reported (33,34).
X-ray crystallography
The protein–RNA–RNA complex at 0.6 mM was crystallized in 100 mM citric acid pH 4.6, 25–35% MPD. Typical crystals grew over a couple of days at 20°C in sitting drops. Crystals were flash frozen in liquid nitrogen directly from the crystallization drop.
The crystal structure was resolved to 2.9 Å using phases determined from a single anomalous dispersion (SAD) data set on a crystal grown with bromo-2-uridine-substituted RNA. The dataset was reduced using XDS (35). Two bromine sites were located using SHELXD and phases were calculated with SHARP (36). The model was improved by manual docking of bases with Coot (37). Model refinement was achieved with Refmac5 (38). The final model has an R-free value of 22.75% and an R-factor value of 18.67%. Crystallographic statistics are listed in the Table 1.
Table 1.
Crystallographic data phasing and refinement statistics (SAD bromine derivative)
| Beamline | ESRF BM-14 |
|---|---|
| Space group | P213 |
| Cell dimensions | |
| a, b, c (Å) | 85.0, 85.0, 85.0 |
| α, β, γ (°) | 90.0, 90.0, 90.0 |
| X-ray source | BM-14 |
| Wavelength (Å) | 0.91873 |
| Resolution (Å) | 20.0–2.9 |
| Total measurements | 66 483 (10 330)a |
| Unique reflections | 8 723 (1432) |
| Redundancy | 6.43 (6.09) |
| Completeness (%) | 98.9 (96.6) |
| Rsym (%) | 7.8 (38.2) |
| I/σ | 18.66 (5.45) |
| FOM (acentric) | 0.32 |
| Refinement statistics | |
| Base number | 34 |
| R-work (%) | 18.67 (31.7) |
| R-free (%) | 22.75 (32.7) |
| Cruickshank's DPI for coordinate errorb (Å) | 0.371 |
| Rmsd bonds (Å) | 0.009 |
| Rmsd angles (°) | 1.510 |
| B average all atoms (Å2) | 69.17 |
aNumbers in brackets refer to the highest resolution shell (2.9 Å–3.07 Å).
bDPI was calculated according to Cruickshank (49).
Altough formation of TAR–R06–ROP complex was followed by NMR spectroscopy and controlled by electrophoretic mobility shift assay, crystals of either ROP or RNA duplex alone, but not from the ternary complex, were obtained from the same crystallization conditions. No density could be accounted for by the protein in our experimental as well as in our refined electron density maps. A similar observation has also been reported in the case of the crystal structure of RNAI and RNAII (39).
Thermal denaturation experiments
Thermal denaturation of TAR-aptamer dimers was monitored on a Uvikon XL UV/visible spectrophotometer (Bio-Tek Instruments, Winooski, VT, USA) interfaced with a Peltier effect device that controls the temperature of the sample holder within ± 0.1°C. The samples were prepared at 1 µM concentration of each RNA hairpin in a 10 mM sodium phosphate buffer, pH 7.2 at 20°C, containing 20 mM sodium chloride, 140 mM potassium chloride and 0.3 mM magnesium chloride. Each RNA sample was first denaturated by heating at 95°C for 1 min 30 s and snap-cooled on ice for 5 min. Partners were mixed at room temperature in 200 µl quartz cuvettes (Hellma, Müllheim, Germany) and allowed to interact at least for 10 min. Ten drops of paraffin oil were added to minimize sample concentration by evaporation of the aqueous buffer at high temperatures. Denaturation of the samples was achieved by increasing the temperature at a 0.4°C/min rate from 5–95°C and was followed at 260 nm. A cuvette containing only buffer was used as the reference. The melting temperature, Tm, was given as the temperature at the maximum of the first derivative curve of the absorbance versus temperature transitions.
Surface plasmon resonance experiments
Surface plasmon resonance experiments were performed on a BIAcore™ (Uppsala, Sweden) 3000 apparatus (Biacore, GE Healthcare, Uppsala, Sweden). A total of 80–90 resonance units (RU) of biotinylated TAR hairpins, with or without a 2′-O-methyl nucleotide, were immobilized on a streptavidin-coated SA sensorchip (BIAcore™). One flow cell left blank was used as a reference. Binding experiments were performed in the same buffer as that used in the thermal denaturation experiments except that the magnesium chloride concentration was increased from 0.3 mM to 3 mM to increase the stability of the loop–loop complex at room temperature, and 0.005% P20 surfactant was added (running buffer). Aptamer samples were prepared in this buffer and were injected at 20 µl/min across the sensor surface, at 23°C. The experiments were performed by the kinetic titration method (40,41) providing a way to determine the kinetic parameters within one single binding cycle by injecting sequentially increasing amounts of the analyte. The regeneration of the TAR-coated surface was achieved with a 1 min pulse of a 20 mM EDTA solution, as it was shown that magnesium ions were critical for complex stability (27), followed by a 1 min pulse of running buffer to wash the surface.
The association and dissociation rate constants, kon and koff, respectively, were determined from direct curve fitting of the sensorgrams, with BiaEval 4.1 (BIAcore™), assuming a pseudo-first order model corresponding to a single-step reaction of the aptamer binding to the immobilized TAR target, according to Equations 1 and 2, for the association and the dissociation phases, respectively:
| 1 |
| 2 |
where RU is the signal response in resonance units; RUmax, the maximum response level; RUt0, the response at the beginning of the dissociation phase; [Ligand], the molar concentration of the injected aptamer. The apparent binding equilibrium constant, KD, was calculated as koff/kon.
RESULTS AND DISCUSSION
Choice of the hairpin sequences
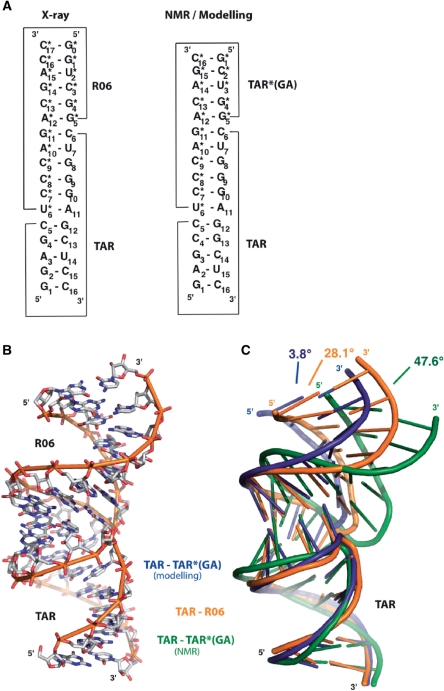
R06 is an RNA aptamer identified by an in vitro selection against the TAR HIV-1 (12). The size of the parent aptamer, a 98-nt long RNA, and of TAR 57-nt long could be optimized by reducing the length of the stems (27). In the first solution studies the upper half part of TAR, 27-nt long, shown to be the minimal domain necessary and sufficient for responsiveness in vivo (42), and an aptamer displaying the 8-nt consensus sequence in the apical loop and an 8-nt base pairs stem were used (14,27,43). The size of the stems could be further reduced to 4 and 5 bp for the aptamer and TAR, respectively, without significant changes in the binding properties (14,34). TAR and R06 used in this work are 16-nt and 18-nt long, respectively, as illustrated in Figure 1A.
Figure 1.
Structure of TAR–R06 aptamer loop–loop complex. (A) Sequence and secondary structure of TAR–R06, TAR–TAR*(GA) from NMR and modelling. Asterisks refer to aptamer residues (28,29). (B) Overall crystal structure of TAR–R06 complex. (C) Superimposition of the TAR–R06 crystal structure (orange) with the TAR–TAR*(GA) liquid-crystal NMR structure (green) and the TAR–TAR*(GA) structure from Molecular Dynamics (blue) was performed with the least-square method using TAR as a reference structure (28,29). Inter-helix angles were measured in MOLecule analysis and MOLecule display (MOLMOL) by generating an axis in each stem over the first four base pairs (50).
In a first attempt to rationalize the higher stability of the TAR–RNA aptamer complex over that formed between TAR and TAR*, the NMR solution structure of TAR–TAR* was used in a previous MD study as a starting structure to simulate the TAR-aptamer kissing complex (28). TAR* is a ligand that was rationally designed with a loop complementary to that of TAR (10) before knowing the crucial role for complex stability played by the GA pair closing the aptamer loop (12,27). TAR* was actually shown to behave in solution as an aptamer-like hairpin when the UA pair next to its loop was replaced by a GA combination (28). Here TAR*(GA) refers to this modified ligand (Figure 1A) used in the MD simulations and liquid-crystal NMR studies (28,29). Despite the slight differences in the stem sequences, all constructs of TAR and the aptamer behaved similarly in the solution for molecular recognition, as far as the loop complementarity and the loop closing residues were maintained (14,27–29,43,44). In particular, all complexes displayed a Tm of 30°C, on average.
Overall structure of TAR–R06 aptamer complex
The TAR–R06 duplex has been crystallized as described in the Material and Methods section and the crystal structure has been resolved by a SAD experiment performed on an RNA duplex substituted by a BrU at positions U14 of TAR and U2* of R06. The structure has been refined to an R-factor of 18.67% and a free R-factor of 22.75% at a resolution of 2.9 Å (Table 1). The observed structure corresponds to a RNA–RNA dimer between TAR and R06 (Figure 1A and B). Both monomers display a similar overall fold with a 1.5 Å overall (all atoms) root-mean-square deviation (RMSD) as calculated with the least-square method. Each monomer folds as a stem-loop structure with a 5 and 6 bp stem for TAR and R06, respectively. Primary recognition between the monomers occurs by Watson–Crick base pairing between the TAR loop and the 6-nt central sequence of the aptamer loop. Each RNA kissing complex stacks against an adjacent molecule through an interaction between the G0*–C17* R06 base pair and the G1–C16 TAR base pair, thus forming an infinite helix. Further packing interactions are observed between the phosphate backbones of orthogonally oriented infinite helices. These packing interactions do not involve residues of the stem-loop. Analysis of the geometrical parameters shows that the complex from X-Ray adopts an A-type conformation, with the exception of the stem-loop junction.
As illustrated in Figure 1C, the X-ray structure (orange) is bent towards the major groove with an inter-helix angle-value of 28.1°, whereas in the liquid-crystal NMR solution structure (green), a strong curvature towards the major groove is observed (47.6°). In contrast, the simulated structure from MD (blue) adopts an overall conformation with an almost coaxial conformation (3.8°). The deviation from ideal helix geometry at stem-loop junctions differs between NMR, X-Ray or MD structures.
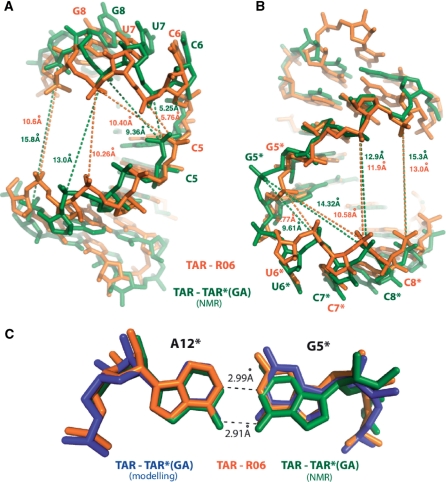
Structure at the stem-loop junctions
The formation of the loop–loop inter-molecular helix induces a deviation of the phosphodiester backbone in the junction region of each molecule. Turns are observed between C5 and C6 bases of TAR, and between G5* and U6* bases of R06 (Figure 2A and B). These structural features have also been described in the structure of kissing complexes with no unpaired residues, solved by NMR (10,34). In TAR–R06 short inter-phosphorus distances are observed between the 5′ phosphate group of C5 and the 5′ phosphate groups of U7 and G8 of TAR (5.76 Å and 10.40 Å respectively), as well as between those of G5* and C7*, C8* of the aptamer (5.77 Å and 10.58 Å respectively) (Figure 2A, B). These distances are different from the corresponding values found in A-type helices where distances between phosphate groups of +2 and +3 residues are 10.4 Å and 12.7 Å, respectively.
Figure 2.
Structure at the stem-loop junctions. (A) Pairwise superimposition of the TAR (X-ray, orange)–TAR (NMR, green) stem-loop junctions is displayed. Distances between C5 phosphate atom and U7 and G8 phosphate are displayed (TAR from the X-ray structure is shown orange, TAR from the NMR is shown in green). (B) Pairwise superimposition of the R06 (orange) and TAR*(GA) is displayed. Distances between G5* and C7* and C8* phosphate atoms are indicated. (C) G5*–A12* kissing loop closing pairs is shown and distances are given in black. TAR–R06, TAR–TAR*(GA) NMR and modelling are displayed as in Figure 1.
Pairwise superimposition of TAR molecules from NMR and X-ray crystallography present an overall rms deviation on all atoms of 1.95 Å (Figure 2A), whereas TAR*(GA) and R06 aptamer have a lower overall rms deviation (1.33 Å) (Figure 2B). The inter-phosphorus distances in the NMR structure compared to the crystal structure between C5(P) and U7(P), G8(P) decreases by 0.48 Å (5.25 Å instead 5.73 Å) and 0.98 Å (9.37 Å instead of 10.35 Å) respectively (Figure 2A and Supplementary Table 1). The major groove of TAR is widened in comparison to the TAR from X-Ray model as exemplified by the larger distances between A2 and G9 phosphate atoms and A3 and G8 phosphates in the NMR structure (15.8 Å and 13.0 Å, respectively) in comparison to G2 and G9 phosphate and A3 and G8 phosphate distances from the X-ray model (10.6 Å and 10.3 Å, respectively).
On the R06 side, the deviation between the X-Ray and the NMR structures is more important. The inter-phosphorus distances between G5* and C7*, C8* are increased by 3.89 Å (9.61 Å instead of 5.77 Å) and 3.74 Å (14.32 Å instead of 10.58 Å), respectively (Figure 2B and Supplementary Table 1). The major groove of TAR*(GA) is severely widened in the NMR structure with width ranges from 12 Å to 13.5 Å compared to 9.1 Å to 11.6 Å for R06. This implies a severe widening of the major groove width of TAR and TAR*(GA) in comparison to their counterparts (TAR and R06), leading to a large difference in the overall bend between the two molecules in the duplex (see also Figure 1C). In comparison, the simulated structure has some distances that are close to those determined in the crystal structure. Other distances agree with the NMR structure or are even larger (Supplementary Table 1).
The GA base pair closing the aptamer loop (residues G5* and A12*) adopts a non-sheared conformation in good agreement with previous studies (28,29,34) (Figure 2C). In this non-canonical base pair, N1 of A12* is hydrogen bonded to N1 of G5* (distance of 2.99 Å) and O6 of G5* is hydrogen bonded to N6 of A12* (distance of 2.91 Å). These distances are mostly similar to those found in the NMR structure (PDB 2RN1), the simulated model (Supplementary Table 1) and the kissing complex formed between TAR and a LNA modified aptamer (PDB 2PN9)(34). The C1′–C1′ distance (12.74 Å) is consistent with the NMR data (12.6 Å) and the simulated structure (13.06 Å) (Supplementary Table 1).
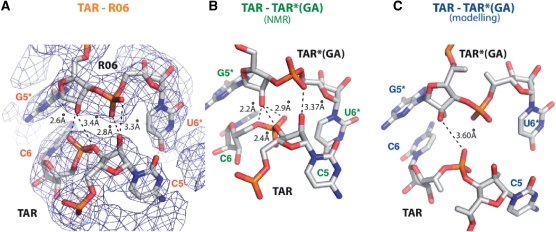
Hydrogen-bonding networks
The presence of additional inter-backbone interactions has been previously proposed and detected in the MD and NMR studies (28,29). In the crystal structure four measured inter-atomic distances are consistent with the establishment of hydrogen bonds and could contribute to non-canonical RNA–RNA interactions (Figure 3A). The G5* (H-O2′) is able to form hydrogen bonds either with C6(O2p) or C5(03′) (distances of 2.57 Å and 3.38 Å, respectively, Supplementary Table 1). The C5(H-O2′) can form hydrogen bonds with U6* (O1p) or U6*(O2p) (distances of 3.33 Å and 2.79 Å, respectively). This hydrogen bonds network differs in the liquid-crystal NMR structure (Figure 3B, Supplementary Table 1) and only one distance is consistent with the formation of a hydrogen bond in the simulated structure (Figure 3C, Supplementary Table 1).
Figure 3.
Hydrogen bonding in TAR–R06 and TAR–TAR*(GA) kissing complexes. (A) TAR C5 and C6 interactions with R06 U6* and G5* are displayed. The Fo–Fc electron density map is contoured at 1 sigma. Hydrogen bonds and distances are displayed. Distances consistent with the formation of hydrogen bonds are shown. Possible hydrogen bonds are indicated by dotted lines. (B and C) Similar to A for TAR C5 and C6 interaction with TAR*(GA) U6* and G5* for the liquid-crystal NMR (NMR) and the Molecular Dynamics (modelling) structures.
In order to assess the contribution on RNA–RNA stability of the inter-backbone H-bonds seen in the crystal structure, 2′-OH groups involved on the aptamer [G5*(O2′)] and TAR [C5(O2′)] sides were chemically modified with 2′-O-methyl moieties. Although adding steric bulk may affect neighbouring interactions, if the 2′-OH are not involved in inter-backbone contacts we expect this substitution not to significantly affect the complex stability. Indeed, it has been shown that the increased stability of a 2′-O-methyl-RNA versus a RNA–RNA dimer duplex is in the range of +0.5°C per modification (45). In addition, the modified ribose adopts a 3′-endo conformation marking this modification as a good RNA mimicry (46).
Thermal denaturation experiments performed on the hairpins alone show that incorporation of a single 2′-O-methyl group either into the aptamer or in the viral RNA has no effect on the intramolecular stability of the hairpins. All aptamer and TAR molecules display a Tm equal, on average, to 71°C and 84°C, respectively (Table 2, RNA alone). In contrast, for bimolecular complexes, replacement of the aptamer G5* 2′–OH by a O-methyl moiety is characterized by a decrease in the melting temperature of the TAR-aptamer complex of 10.8°C with respect to the unmodified complex (Table2 and Supplementary Figure 1), consistent with a recently published work (29). Replacement of the C5 2′-OH of TAR has a milder effect (ΔTm is equal to −6.9°C, Table 2 and Supplementary Figure 1).
Table 2.
Melting temperatures (Tm) for TAR-aptamer complexes
| Hairpins |
Tm (°C) |
||
|---|---|---|---|
| Complexed with TAR | Complexed with TARC5 | RNA alone | |
| R06 | 31.3 ± 0.3 | 24.4 ± 0.4 | 71.5 ± 0.6 |
| R06G5*a | 20.5 ± 0.3 (−10.8°C) | 22.0 ± 0.2 | 69.5 ± 0.6 |
| R06C8* | 33.2 ± 0.6 (+1.9°C) | 25.5 ± 0.2 | 71.3 ± 1.5 |
| R06A12* | 30.3 ± 0.5 (−1.0°C) | 23.9 ± 0.2 | 71.2 ± 1.3 |
| TAR | – | – | 85.3 ± 0.3 |
| TARC5 | – | – | 83.4 ± 0.5 |
aSubscripts refer to 2′-O-methyl residues at the indicated positions. The experiments were performed at 1 µM of each RNA, in a 10 mM sodium phosphate buffer, pH 7.2 at 20°C, containing 140 mM potassium chloride, 20 mM sodium chloride and 0.3 mM magnesium chloride.
The complex formed between TARC5 (subscript refers to 2′-O-methyl residue at the indicated position) and R06G5* (Tm equal to 22.0 ± 0.2°C) behaves mostly as the complex formed between the unmodified TAR and R06G5* (Tm equal to 20.5 ± 0.3°C). Two control experiments were carried out to investigate the specificity of these H-bond interactions. A 2′-O-methyl substitutions at positions where no inter-backbone H-bonds were observed in the crystal structure (C8* 2′-OH and A12* 2′-OH positions of the aptamer) do not affect complex stability in comparison to that observed with the unmodified aptamer whether the complexes were formed with TAR or TARC5 (Table 2 and Supplementary Figure 1).
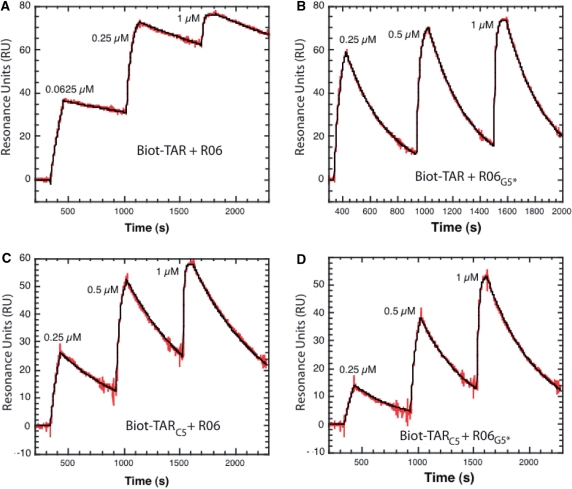
To support further the implication of the 2′-OH groups protons of TAR C5 and of the G5* of the aptamer for generating the inter-molecular network of H-bonds, surface plasmon resonance experiments were performed. Biotinylated TAR and TARC5 were immobilized onto streptavidin-coated sensorchips and binding of the unmodified and G5* modified aptamers were analysed by injecting in a continuous flow across the surface increasing amounts of aptamers as illustrated in Figure 4. The sensorgrams were fitted to a single-step reaction as described in Material and Methods section. The kinetic parameters determined from such an analysis are listed in Table 3. The results demonstrate unambiguously that the 2′-OH proton of the G5* residue of the aptamer closing pair is crucial to avoid rapid dissociation of the complex (Table 3 and Figure 4). The off-rate, koff, increases from 0.29 ± 0.11 s−1 to 3.30 ± 0.01 s−1 when the proton of the G5 2′-OH of R06 is replaced by a methyl group whereas the on-rate, kon, remains unchanged. This leads to a 10-fold increase in the dissociation equilibrium constant (Kd = 44.9 ± 7.4 nM) compared to that of the unmodified RNA–RNA complex (Kd = 3.1 ± 0.1 nM).
Figure 4.
Kinetic analysis of TAR-aptamer complex formation by surface plasmon resonance. A total of 80–90 RU of biotinylated TAR and TARC5 were immobilized onto streptavidin-coated SA sensorchips (BIAcore™). Hairpin aptamers, prepared in the running buffer, were injected across the sensor surface at a 20 µl/min flow rate, at 23°C. The sensorgrams were fitted as described in Materials and Methods section assuming a pseudo-first kinetic model of the aptamer binding to the immobilized TAR hairpins. The red curves represent the recorded data and the black one the fit of the sensorgrams to a kinetic titration dataset of three analyte injections. (A) Injection of the unmodified R06 aptamer (0.0625 µM, 0.25 µM and 1 µM) across the TAR-coated flowcell. (B) Injection of R06G5* (0.25 µM, 0.5 µM and 1 µM) across the TAR-coated flowcell. (C) Injection of the unmodified aptamer (0.25 µM, 0.5 µM and 1 µM) across the TARC5-coated flowcell. (D) Injection of R06G5* (0.25 µM, 0.5 µM and 1 µM) across the TARC5-coated flowcell.
Table 3.
Equilibrium and rate constants for TAR-aptamer complexes
| Complexes | kon (104 M−1 s−1) | koff (10−3 s−1) | Kd (nM) |
|---|---|---|---|
| TAR–R06 | 9.2 ± 0.6 | 0.29 ± 0.11 | 3.1 ± 0.1 |
| TAR–R06G5*a | 7.9 ± 1.5 | 3.30 ± 0.01 | 44.9 ± 7.4 |
| TARC5–R06 | 11.0 ± 0.1 | 1.48 ± 0.02 | 13.3 ± 0.7 |
| TARC5–R06G5* | 5.8 ± 0.1 | 2.19 ± 0.01 | 38.5 ± 1.7 |
aSubscripts refer to 2′-O-methyl residues at the indicated positions. TAR and TARC5 hairpins were immobilized onto streptavidin-coated sensorchips. The kinetic experiments were performed in the buffer used for the thermal denaturation experiments except that magnesium ions concentration was increased from 0.3 mM to 3 mM and 0.005% P20 surfactant was added. The thermodynamic parameters were determined from direct curve fitting of the sensorgrams as described in Materials and Methods section.
The kinetic experiments performed with TARC5 agree well with additional inter-molecular H-bond being established between TAR and the aptamer. However, removal of this C5 H2′-OH promoted H-bond is less destabilizing (Kd = 13.3 ± 0.7 nM) than that involving G5* H2′-OH of the aptamer (Kd = 44.9 ± 7.4 nM) (Table 3). TARC5–R06G5* complex behaves as TAR–R06G5* and displays an equilibrium dissociation constant close to that of the parent RNA–RNA complex. The results obtained by SPR confirm that the effects of a 2′-O-methyl substitution on complex stability are not similar if TAR or the aptamer are chemically modified. Adding steric bulk with the 2′-O-methyl group makes difficult to deconvolute unambiguously the contribution of each hydrogen bonds. The substitution may also affect the neighbouring inter-molecular H-bonds promoted by each of the hydroxyl groups. Interestingly, a stacking interaction is observed between the G5* and C6 bases (Figure 3A). In addition to disturbing the inter-molecular H-bonds network, replacement of the H′2 proton of G5* by a methyl group may affect this stacking interaction and explain why a major effect on complex stability is observed when the substitution is made on the aptamer side.
In summary, the X-ray and liquid-crystal structures differ mainly at the stem loop junctions where a larger major groove is observed for the NMR structure. This induces different angles in the orientation of each molecule within the duplex. The overall bend between the stem helices is about 20° lower in the crystal structure than in the liquid-crystal NMR model. The conformation deduced from MD simulations, even though generated from a starting structure first validated in vitro, is coaxially aligned after a 3 ns trajectory. Although it is difficult to estimate the crystal packing contribution to the overall bend of the molecule, no specific constraints are observed at the stem-loop junctions that would involve neighbouring molecules. Rather, the overall packing favours the formation of an infinite helix by head to tail packing of following molecules. The NMR orienting media (Pf1 filamentous phases) could interfere with the relative orientations of the two helices and could favour a specific relative orientation of the two helices if the kissing complex is not strictly rigid. Both techniques would then reveal two different conformations that may exist in solution. Variations of buffers could also partially explain this overall difference. It is of basic knowledge that divalent cations and specifically magnesium ions are critical in RNA conformation. In TAR–R06, magnesium ions have a stabilizing effect (27). Strong evidence of direct binding of this cation to the DIS of HIV-1 has been reported (47) and was proposed for the RNAI–RNAII complex from the ColE1 plasmid (18) and the TAR–TAR* complex (10). Magnesium ions would bind at the center of two phosphate clusters flanking the major groove of the loop–loop helix. Here, we cannot make conclusions about the contribution of Mg2+ ions due to the limited resolution of the data. Last, although TAR–R06 and TAR–TAR*(GA) behave similarly in solution as quantified by UV-spectroscopy suggesting that stability is controlled by the loop–loop complementarity and the GA closing pair of the aptamer, we cannot totally exclude that the slight differences in the sequence of the stems is responsible for the specific bending of each structure.
Beyond the techniques and the discrepancies that may result from artefacts, the question arises whether the overall conformation of kissing-loop dimers is correlated with the size of the loops and the presence or not of unpaired residues. The backbone has to cross the major groove to connect the loop–loop helix with the stem helices. This implies severe structural constraints. Unpaired residues next to the loop–loop helix help to release these constraints. In fact, the DIS/DIS of HIV-1 and the 23S/23S dimers, that display unpaired residues in the loops, are coaxially aligned (21,24). Kissing-loop dimers with fully complementary loops are then expected to display a larger curvature, the amplitude of which will depend on the length of the loop–loop helix. This is reminiscent of the bending observed between the two stems of pseudoknots (48). Between these extremes, the TAR-aptamer complex might be an intermediate case where a non-canonical loop closing-pair with a large interglycosidic distance contributes to relax the structure at the stem-loop junctions in addition to providing increased stacking interaction. Finally, our data clearly show that inter-molecular hydrogen bonds between ribose 2′-OH and phosphate oxygens contribute to the stability of the complex together with the inter-molecular stacking interaction due to the presence of the G5* residue. Such combination of non-canonical RNA–RNA interactions which have been also reported in the Moloney murine leukemia virus DIS complex (8), play a key role finely tuning molecular recognition for high specificity and stability between partners that can primarily interact through <7 bp.
COORDINATES
The atomic coordinates and structure factors have been deposited in the Protein Data Bank (accession code 2jlt).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Institut National de la Santé et de la Recherche Médicale and the Institut Européen de Chimie et Biologie. Funding for open access charge: INSERM.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS.
NMR experiments were recorded at the Metabolome Pole of Functional Genomics Platform at Bordeaux. We thank Mickaël Maucourt, Catherine Deborde and Dominique Rolin for providing access to the 500 MHz spectrometer located at INRA (UMR Biologie du Fruit), Villenave d'Ornon (FRANCE). We would like to acknowledge support from staff members of the ESRF BM-14 beamline and Brice Kauffmann. We also thank Thierry Dakhli for ROP purification and Nathalie Pierre for oligonucleotides synthesis. We are also grateful to J.J. Toulmé and C. Mackereth for critical reading of the manuscript and E. Ennifar for stimulating discussions. The authors would like to acknowledge the reviewers' contributions to the article.
REFERENCES
- 1.Brunel C, Marquet R, Romby P, Ehresmann C. RNA loop-loop interactions as dynamic functional motifs. Biochimie. 2002;84:925–944. doi: 10.1016/s0300-9084(02)01401-3. [DOI] [PubMed] [Google Scholar]
- 2.Eguchi Y, Tomizawa J. Complex formed by complementary RNA stem-loops and its stabilization by a protein: function of CoIE1 Rom protein. Cell. 1990;60:199–209. doi: 10.1016/0092-8674(90)90736-x. [DOI] [PubMed] [Google Scholar]
- 3.Eguchi Y, Tomizawa J. Complexes formed by complementary RNA stem-loops. Their formations, structures and interaction with ColE1 Rom protein. J. Mol. Biol. 1991;220:831–842. doi: 10.1016/0022-2836(91)90356-b. [DOI] [PubMed] [Google Scholar]
- 4.Argaman L, Altuvia S. fhlA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J. Mol. Biol. 2000;300:1101–1112. doi: 10.1006/jmbi.2000.3942. [DOI] [PubMed] [Google Scholar]
- 5.Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007;21:1353–1366. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geisinger E, Adhikari RP, Jin R, Ross HF, Novick RP. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol. Microbiol. 2006;61:1038–1048. doi: 10.1111/j.1365-2958.2006.05292.x. [DOI] [PubMed] [Google Scholar]
- 7.Paillart JC, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc. Natl Acad. Sci. USA. 1996;93:5572–5577. doi: 10.1073/pnas.93.11.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim CH, Tinoco I., Jr A retroviral RNA kissing complex containing only two G.C base pairs. Proc. Natl Acad. Sci. USA. 2000;97:9396–9401. doi: 10.1073/pnas.170283697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li PT, Bustamante C, Tinoco I., Jr Unusual mechanical stability of a minimal RNA kissing complex. Proc. Natl Acad. Sci. USA. 2006;103:15847–15852. doi: 10.1073/pnas.0607202103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang KY, Tinoco I., Jr The structure of an RNA “kissing” hairpin complex of the HIV TAR hairpin loop and its complement. J. Mol. Biol. 1997;269:52–66. doi: 10.1006/jmbi.1997.1021. [DOI] [PubMed] [Google Scholar]
- 11.Boiziau C, Dausse E, Yurchenko L, Toulme JJ. DNA aptamers selected against the HIV-1 trans-activation-responsive RNA element form RNA-DNA kissing complexes. J. Biol. Chem. 1999;274:12730–12737. doi: 10.1074/jbc.274.18.12730. [DOI] [PubMed] [Google Scholar]
- 12.Ducongé F, Toulmé JJ. In vitro selection identifies key determinants for loop-loop interactions: RNA aptamers selective for the TAR RNA element of HIV-1. RNA. 1999;5:1605–1614. doi: 10.1017/s1355838299991318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scarabino D, Crisari A, Lorenzini S, Williams K, Tocchini-Valentini GP. tRNA prefers to kiss. Embo J. 1999;18:4571–4578. doi: 10.1093/emboj/18.16.4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darfeuille F, Reigadas S, Hansen JB, Orum H, Di Primo C, Toulmé JJ. Aptamers targeted to an RNA hairpin show improved specificity compared to that of complementary oligonucleotides. Biochemistry. 2006;45:12076–12082. doi: 10.1021/bi0606344. [DOI] [PubMed] [Google Scholar]
- 15.Romby P, Moras D, Bergdoll M, Dumas P, Vlassov VV, Westhof E, Ebel JP, Giégé R. Yeast tRNAAsp tertiary structure in solution and areas of interaction of the tRNA with aspartyl-tRNA synthetase. A comparative study of the yeast phenylalanine system by phosphate alkylation experiments with ethylnitrosourea. J. Mol. Biol. 1985;184:455–471. doi: 10.1016/0022-2836(85)90294-3. [DOI] [PubMed] [Google Scholar]
- 16.Gregorian RS, Jr, Crothers DM. Determinants of RNA hairpin loop-loop complex stability. J. Mol. Biol. 1995;248:968–984. doi: 10.1006/jmbi.1995.0275. [DOI] [PubMed] [Google Scholar]
- 17.Dardel F, Marquet R, Ehresmann C, Ehresmann B, Blanquet S. Solution studies of the dimerization initiation site of HIV-1 genomic RNA. Nucleic Acids Res. 1998;26:3567–3571. doi: 10.1093/nar/26.15.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee AJ, Crothers DM. The solution structure of an RNA loop-loop complex: the ColE1 inverted loop sequence. Structure. 1998;6:993–1005. doi: 10.1016/s0969-2126(98)00101-4. [DOI] [PubMed] [Google Scholar]
- 19.Mujeeb A, Clever JL, Billeci TM, James TL, Parslow TG. Structure of the dimer initiation complex of HIV-1 genomic RNA. Nat. Struct. Biol. 1998;5:432–436. doi: 10.1038/nsb0698-432. [DOI] [PubMed] [Google Scholar]
- 20.Westhof E, Dumas P, Moras D. Restrained refinement of two crystalline forms of yeast aspartic acid and phenylalanine transfer RNA crystals. Acta Crystallogr. A. 1988;44 (Pt 2):112–123. [PubMed] [Google Scholar]
- 21.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 22.Serganov A, Huang L, Patel DJ. Structural insights into amino acid and gene control by a lysine riboswitch. Nature. 2008;455:1263–1267. doi: 10.1038/nature07326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garst AD, Heroux A, Rambo RP, Batey RT. Crystal structure of the lysine riboswitch regulatory mRNA element. J. Biol. Chem. 2008;283:22347–22351. doi: 10.1074/jbc.C800120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ennifar E, Walter P, Ehresmann B, Ehresmann C, Dumas P. Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nat. Struct. Biol. 2001;8:1064–1068. doi: 10.1038/nsb727. [DOI] [PubMed] [Google Scholar]
- 25.Baba S, Takahashi K, Noguchi S, Takaku H, Koyanagi Y, Yamamoto N, Kawai G. Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers. J. Biochem. 2005;138:583–592. doi: 10.1093/jb/mvi158. [DOI] [PubMed] [Google Scholar]
- 26.Kieken F, Paquet F, Brulé F, Paoletti J, Lancelot G. A new NMR solution structure of the SL1 HIV-1Lai loop-loop dimer. Nucleic Acids Res. 2006;34:343–352. doi: 10.1093/nar/gkj427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ducongé F, Di Primo C, Toulmé JJ. Is a closing “GA pair” a rule for stable loop-loop RNA complexes. J. Biol. Chem. 2000;275:21287–21294. doi: 10.1074/jbc.M002694200. [DOI] [PubMed] [Google Scholar]
- 28.Beaurain F, Di Primo C, Toulmé JJ, Laguerre M. Molecular dynamics reveals the stabilizing role of loop closing residues in kissing interactions: comparison between TAR–TAR* and TAR-aptamer. Nucleic Acids Res. 2003;31:4275–4284. doi: 10.1093/nar/gkg467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Melckebeke H, Devany M, Di Primo C, Beaurain F, Toulmé JJ, Bryce DL, Boisbouvier J. Liquid-crystal NMR structure of HIV TAR RNA bound to its SELEX RNA aptamer reveals the origins of the high stability of the complex. Proc. Natl Acad. Sci. USA. 2008;105:9210–9215. doi: 10.1073/pnas.0712121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Comolli LR, Pelton JG, Tinoco I., Jr Mapping of a protein-RNA kissing hairpin interface: Rom and Tar-Tar*. Nucleic Acids Res. 1998;26:4688–4695. doi: 10.1093/nar/26.20.4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 32.Sklenar V, Peterson RD, Rejante MR, Feigon J. Two- and three-dimensional HCN experiments for correlating base and sugar resonances in 15N,13C-labeled RNA oligonucleotides. J. Biomol. NMR. 1993;3:721–727. doi: 10.1007/BF00198375. [DOI] [PubMed] [Google Scholar]
- 33.Lebars I, Husson C, Yoshizawa S, Douthwaite S, Fourmy D. Recognition elements in rRNA for the tylosin resistance methyltransferase RlmA(II) J. Mol. Biol. 2007;372:525–534. doi: 10.1016/j.jmb.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 34.Lebars I, Richard T, Di Primo C, Toulmé JJ. NMR structure of a kissing complex formed between the TAR RNA element of HIV-1 and a LNA-modified aptamer. Nucleic Acids Res. 2007;35:6103–6114. doi: 10.1093/nar/gkm655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 1993;26:795–800. [Google Scholar]
- 36.de La Fortelle E, Bricogne G. SHARP: a maximum-likelihood heavy-atom parameter refinement program for the MIR and MAD Methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 39.Klosterman PS, Shah SA, Steitz TA. Crystal structures of two plasmid copy control related RNA duplexes: An 18 base pair duplex at 1.20 Å resolution and a 19 base pair duplex at 1.55 Å resolution. Biochemistry. 1999;38:14784–14792. doi: 10.1021/bi9912793. [DOI] [PubMed] [Google Scholar]
- 40.Karlsson R, Katsamba PS, Nordin H, Pol E, Myszka DG. Interpreting kinetic rate constants from optical biosensor data recorded on a decaying surface. Anal. Biochem. 2006;261:203–210. doi: 10.1006/abio.1998.2744. [DOI] [PubMed] [Google Scholar]
- 41.Di Primo C. Real time analysis of the RNAI-RNAII-Rop complex by surface plasmon resonance: from a decaying surface to a standard kinetic analysis. J. Mol. Recogit. 2008;21:37–45. doi: 10.1002/jmr.860. [DOI] [PubMed] [Google Scholar]
- 42.Jakobovits A, Smith DH, Jakobovits EB, Capon DJ. A discrete element 3′ of human immunodeficiency virus 1 (HIV-1) and HIV-2 mRNA initiation sites mediates transcriptional activation by an HIV trans activator. Mol. Cell. Biol. 1988;8:2555–2561. doi: 10.1128/mcb.8.6.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darfeuille F, Arzumanov A, Gryaznov S, Gait MJ, Di Primo C, Toulmé JJ. Loop-loop interaction of HIV-1 TAR RNA with N3′–>P5′ deoxyphosphoramidate aptamers inhibits in vitro Tat-mediated transcription. Proc. Natl Acad. Sci. USA. 2002;99:9709–9714. doi: 10.1073/pnas.122247199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darfeuille F, Arzumanov A, Gait MJ, Di Primo C, Toulmé JJ. 2′-O-methyl-RNA hairpins generate loop-loop complexes and selectively inhibit HIV-1 Tat-mediated transcription. Biochemistry. 2002;41:12186–12192. doi: 10.1021/bi025974d. [DOI] [PubMed] [Google Scholar]
- 45.Monia BP, Lesnik EA, Gonzalez C, Lima WF, McGee D, Guinosso CJ, Kawasaki AM, Cook PD, Freier SM. Evaluation of 2′-modified oligonucleotides containing 2′-deoxy gaps as antisense inhibitors of gene expression. J. Biol. Chem. 1993;268:14514–14522. [PubMed] [Google Scholar]
- 46.Lubini P, Zurcher W, Egli M. Stabilizing effects of the RNA 2′-substituent: crystal structure of an oligodeoxynucleotide duplex containing 2′-O-methylated adenosines. Chem. Biol. 1994;1:39–45. doi: 10.1016/1074-5521(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 47.Jossinet F, Paillart JC, Westhof E, Hermann T, Skripkin E, Lodmell JS, Ehresmann C, Ehresmann B, Marquet R. Dimerization of HIV-1 genomic RNA of subtypes A and B: RNA loop structure and magnesium binding. RNA. 1999;5:1222–1234. doi: 10.1017/s1355838299990982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giedroc DP, Theimer CA, Nixon PL. Structure, stability and function of RNA pseudoknots involved in stimulating ribosomal frameshifting. J. Mol. Biol. 2000;298:167–185. doi: 10.1006/jmbi.2000.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cruickshank DW. Remarks about protein structure precision. Acta Crystallogr. D Biol. Crystallogr. 1999;55:583–601. doi: 10.1107/s0907444998012645. [DOI] [PubMed] [Google Scholar]
- 50.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14 doi: 10.1016/0263-7855(96)00009-4. 29–32, 51–55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.