Abstract
Phosphoglycerate kinase 2 (PGK2) is a germ cell-specific protein whose mRNA is translationally regulated in the mammalian testis. Using RNA affinity chromatography with the 3′-untranslated region (UTR) of Pgk2 mRNA and adult testis extracts, several associated proteins including a novel isoform of the AU-rich element RNA-binding protein and KH-type splicing regulatory protein (KSRP) were identified. KSRP, a protein of ∼75 kDa, is widely expressed in somatic and germ cells where it is primarily nuclear. In addition to the ∼75-kDa KSRP, a ∼52-kD KSRP, t-KSRP, is present in the cytoplasm of a subpopulation of germ cells. t-KSRP binds directly to a 93-nt sequence (designated the F1 region) of the 3′-UTR of the Pgk2 mRNA and destabilizes Pgk2 mRNA constructs in testis extracts and in transfected cells. We conclude that this testicular variant of the multifunctional nucleic acid–binding protein, KSRP, serves as a decay-promoting factor for Pgk2 mRNA in male germ cells.
INTRODUCTION
Phosphoglycerate kinase (PGK) is a highly conserved and widely expressed glycolytic enzyme. Mammals express two functional PGKs, PGK1, an X-linked ubiquitous protein expressed in somatic cells, oogenic cells and premeiotic and meiotic male germ cells, and a testis-specific isoform, PGK2, expressed in postmeiotic male germ cells and spermatozoa. The latter is believed to have evolved by retroposition to compensate for the loss of PGK1 expression following X-chromosome inactivation during spermatogenesis (1). In mice, Pgk2 mRNA is first detected in early stages of meiotic spermatocytes and dramatically increases in postmeiotic spermatids, while the PGK2 protein is not detected until many days later in late-stage spermatids (2–4). Microarray studies have confirmed the posttranscriptional regulation of Pgk2, demonstrating that Pgk2 mRNAs are initially detected as ribonucleoproteins in meiotic spermatocytes and then move onto polysomes in later stage haploid germ cells (5). This temporal separation of Pgk2 transcription and translation for up to 2 weeks requires both long-term Pgk2 mRNA stabilization and translational activation/degradation, suggesting that trans-acting factors are important in controlling Pgk2 expression.
In the mammalian testis, posttranscriptional regulation of mRNA is essential for the sequential expression of proteins, especially in haploid germ cells because of the termination of transcription as spermatids differentiate (6). The testis contains many RNA-binding proteins, which regulate posttranscriptional events including mRNA processing, transport, localization, stability and translation. The germ cell–specific DNA/RNA-binding Y-box protein, MSY2, and the polypyrimidine tract binding protein 2 (PTBP2) are two proteins that play important roles in germ cell mRNA stabilization (7–9). MSY2 is one of the most abundant RNA-binding proteins in male germ cells, constituting ∼0.7% of total soluble protein. It functions as a global stabilizer/translational suppressor of many germ cell mRNAs. MSY2 specifically recognizes a population of germ cell mRNAs by binding in the nucleus to a consensus promoter sequence present in their genes thereby linking the nuclear events of transcription with the cytoplasmic processes of mRNA storage and stabilization (10). Deletion of MSY2 by gene targeting leads to precocious translation and destabilization/loss of many mRNAs (7,8). PTBP2, a second protein that stabilizes testicular mRNAs, belongs to a multifunctional family of proteins that function as nuclear splicing factors, direct Internal Ribosome Entry Site (IRES)-directed translation initiation and stabilize mRNAs in the cytoplasm (11–17). In vitro assays reveal that PTBP2 increases the stability of Pgk2 mRNA in both testis extracts and in transfected HeLa cells, suggesting that PTBP2 helps maintain the in vivo stability of the Pgk2 mRNA by binding to a regulatory element (9).
To begin to identify proteins that destabilize germ cell mRNAs, RNA affinity chromatography was used to screen testis cytoplasmic extracts with the 3′-untranslated region (UTR) of Pgk2 mRNA. Among several RNA-binding proteins, a novel ∼52-kDa isoform of the multifunctional KH-type splicing regulatory protein (t-KSRP) was identified (9). An ∼75-kDa KSRP that is ubiquitously expressed has been implicated in transcriptional regulation, splicing and mRNA decay in somatic tissues and cultured cells (17–26). Here we demonstrate that t-KSRP binds to an AU-rich sequence in the 3′-UTR of Pgk2 mRNA and destabilizes the Pgk2 mRNA. Moreover, the destabilization of Pgk2 mRNA occurs when t-KSRP and PTBP2 are present together in complexes bound to the 3′-UTR of Pgk2 mRNA.
MATERIALS AND METHODS
Plasmid construction and expression of the ∼52-kDa KSRP
Mass spectrometry sequence analysis of a ∼52-kDa protein purified from an adult testis cytoplasmic extract by RNA affinity chromatography identified 17 peptides mapping to KSRP. The peptides ranged from amino acid 218–684 (9, Table 1). In western blots, an antibody against the C-terminus of the ∼75-kDa KSRP (DB-KS, Figure 4B) recognized both the testicular KSRPs, suggesting that the ∼52-kDa KSRP isoform (t-KSRP) is an N-terminally truncated form of KSRP. Based on the MS-TOF sequences, the western blotting (see below), and the ∼52-kDa electrophoretic mobility of t-KSRP, a recombinant t-KSRP was synthesized starting at methionine 207 and terminating at amino acid 748 (the last amino acid of the ∼75-kDa KSRP). This recombinant N-truncated KSRP was synthesized because it is not known if the smaller KSRP isoform is translated from an internal methionine (at amino acid 207) or is a cleavage product of the ∼75-kDa KSRP. Based on mass spectrometry and estimated protein size, the recombinant protein synthesized is very close in size to a protein containing a peptide starting at amino acid 218 (Table 1). The ∼52-kDa protein, t-KSRP207-748 (NM_010613), was amplified by reverse transcription–polymerase chain reaction (RT–PCR) from adult mouse testis total RNA and subcloned into pET-42a (Novagen, San Diego, CA). Recombinant t-KSRP was expressed in BL21-CodonPlus (DE3)-RP cells (Stratagene, La Jolla, CA), purified with a Glutathione S-transferase (GST) purification kit and, when necessary, digested with thrombin to remove the GST tag. For the transfection studies, the PCR product of t-KSRP (207–748 of full-length KSRP) was digested with HindIII and SalI and cloned into the HindIII/SalI site of pEGFP-C2 (Clontech, Palo Alto, CA) to generate the expression plasmid, pEGFP-KSRP.
Table 1.
MALDI-TOF analysis for 52 kDa KSRP
| Peptide sequence | Location in 75 kDa KSRP (amino acid number) |
|---|---|
| GGPPGQFHDNANGGQNGTV QEIMIPAGK | 218–245 |
| AGLVIGKGGETIK | 246–258 |
| GGETIKQLQER | 253–263 |
| MILIQDGSQNTNVDKPLR | 268–285 |
| ERDQGGFGDR | 306–315 |
| DQGGFGDR | 308–315 |
| VGGGIDVPVPR | 322–332 |
| HSVGVVIGR | 333–341 |
| IQNDAGVR | 349-356 |
| IAHIMGPPDR | 370–379 |
| IINDLLQSLR | 386–395 |
| GRGQGNWGPPGGEMTFSIPTHK | 415–436 |
| AINQQTGAFVEISR | 450–463 |
| GSPQQIDHAK | 480–489 |
| AWEEYYK | 622–628; 648–654 |
| IGQQPQQPGAPPQQDYTK | 630–647 |
| QAQVATGGGPGAPPGSQPDYSA AWAEYYR | 656–684 |
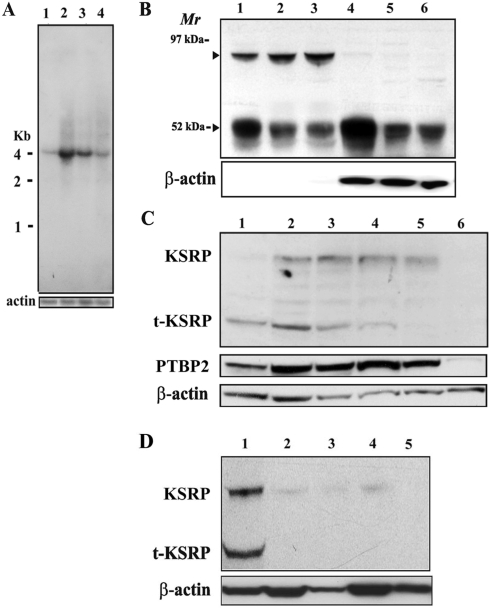
Figure 4.
Two isoforms of KSRP, KSRP and t-KSRP, are expressed from one mRNA in the testis, and t-KSRP is enriched in the cytoplasm of early-stage meiotic cells. (A) Northern blot of KSRP mRNA from brain and prepuberal and adult testes. Total RNAs (15 µg) were electrophoresed in 1% agarose–formaldehyde gels, transferred to Hybond-N + membranes and hybridized with a KSRP 3′-UTR probe. Rehybridization of the blot with a coding region probe for actin served as a loading control. Lane 1, adult brain; lane 2, 17-day-old testis; lane 3, 22-day-old testes; lane 4, adult testes. (B) Western blot of KSRP in nuclear and cytoplasmic fractions of prepuberal and adult testes. Proteins (20 µg) extracted from purified nuclei and cytoplasm were separated by 10% SDS–PAGE and blotted onto polyvinylidene difluoride membranes. Anti-KSRP was used at a dilution of 1:3000. Antiactin served as a loading control. Lanes 1–3 and 4–6 were extracts from nuclei and cytoplasm, respectively. Lanes 1 and 4, 17-day-old testis; lanes 2 and 5, 22-day-old testes; lanes 3 and 6, adult testes. (C) Western blot of KSRP, PTBP2 and actin in extracts of isolated populations of germ cells. Lane 1, preleptotene spermatocytes; lane 2, leptotene/zygotene spermatocytes; lane 3, early pachytene spermatocytes; lane 4, pachytene spermatocytes; lane 5, round spermatids; lane 6, condensing spermatids/residual bodies. The preleptotene spermatocytes, leptotene/zygotene spermatocytes and early pachytene spermatocytes were isolated from the testes of 17-day-old mice, whereas the pachytene spermatocytes, round spermatids and condensing spermatids/residual bodies were isolated from the testes of adult mice. (D) Western blot of t-KSRP in tissue extracts. Lane 1, testis; lane 2, brain; lane 3, liver; lane 4, kidney; lane 5, lung protein extracts (30 µg) analyzed as in (B) with actin serving as a loading control.
Semiquantitative RT–PCR
Total RNA was extracted from the testes of 17-day-old, adult, wild-type mice with Trizol (Invitrogen, Carlsbad, CA), residual genomic DNA was removed with the TurboDNA free kit (Ambion, Austin, TX) and the amount of Pgk2 mRNA was quantitated using qualitative RT–PCR. First-strand complementary DNA was synthesized with random hexamer primers and Superscript III reverse transcriptase following the manufacturer's instructions (Invitrogen). Qualitative PCR reactions were performed with SYBRgreen PCR Master reagents and the 7900HT sequence detector (PE Applied Biosystems, Foster City, CA). The expression level of Pgk2 mRNA was normalized to actin mRNA.
Nuclear run-on assays
Nuclei were isolated, and nuclear run-on reactions were performed as previously described (7). Briefly, a nuclear suspension (100 µl) was mixed with 100 µl of 2 × reaction buffer (10 mM Tris–HCl, pH 8.0, 5 mM MgCl2, 0.3 M KCl, 5 mM Dithiothreitol (DTT), 1 mM Adenosine triphosphate (ATP), 1 mM Cytidine triphosphate (CTP) and 1 mM Guanosine triphosphate (GTP) and 10 µl of [P32]-UTP and incubated at 30°C for 30 min. The reaction was terminated by the addition of 6 µl of 250 mM CaCl2 and 6 µl of RNase-free DNase I (25 µg/ml). After incubation for 10 min at 29°C, the samples were incubated with proteinase K (10 µg/ml) at 42°C for 30 min, and RNA was purified with Trizol (Invitrogen) before hybridization to DNAs bound to nitrocellulose membranes. Hybridizations were carried out for 24 h at 65°C in Quik hybridization solution (Stratagene), and hybridization signals were detected by PhosphorImager (Amersham Biosciences, Piscataway, NJ).
Gel shift assays
Adult testis cytoplasmic extracts (30 µg) were incubated with [32P]-labeled RNA probes as previously described (9) and fractionated in 4% polyacrylamide gels in 0.5 × Tris/Borate/EDTA (TBE) buffer. For supershift assays, antibodies were preincubated with extracts for 5 min before addition of the radiolabeled RNA.
Protein–protein interaction assays
Recombinant PTBP2 (9) containing a His tag at its C-terminus was incubated with His-tag affinity resins for 30 min at 4°C in 200 µl of binding buffer containing 20 mM Tris–HCl, pH 7.6, 100 mM NaCl, 2 mM DTT and 0.5% Tween 20. Adult testis extract was added and incubated for an additional 30 min at 4°C with rotation. The mixture was centrifuged for 1 min at 100g at 4°C, and the pellet was washed three times with binding buffer. The pellets were boiled in sodium dodecyl sulfate (SDS) loading buffer for 3 min, and proteins were resolved in a 10% SDS–polyacrylamide gel. KSRP was detected by western blotting.
For the GST pull-down assay, the ∼52-kDa recombinant KSRP (t-KSRP) containing a GST tag was incubated with recombinant PTBP2 for 30 min at 4°C in 200 µl binding buffer. Glutathione agarose beads were added to the mixture and incubated for 30 min at 4°C. The mixture was centrifuged and washed as described above. The pellets were boiled in SDS loading buffer and electrophoresed in a 10% SDS–polyacrylamide gel. PTBP2 was detected by western blotting.
RNA immunoprecipitation assay
Assays were performed as previously described (9,21). Testis cytoplasmic extracts (10 mg) from sexually mature CD-1 mice were precleared with protein A/G agarose beads (100 µl), preimmune serum (10 µl) and yeast tRNA (100 µg/ml) [prepared in the presence of SUPERase-in (1 U/µl), Ambion] and then incubated with 100 µl anti-KSRP [ab5, raised against the full-length KSRP (27)] and protein G agarose beads at 4°C for 2 h. The mixtures were centrifuged at 400g for 5 min, and the pellets were washed and extracted in Trizol. Immunoprecipitated RNAs were reverse transcribed with random primers with the Superscript III reverse transcription kit (Invitrogen), and PCR assays were performed.
Northern blot analysis
For KSRP northern blots, total testicular RNAs (15 µg) were electrophoresed in 1% agarose–formaldehyde gels and transferred to Hybond-N + membranes (Amersham Biosciences) in alkaline buffer. The probe for KSRP and β-actin was labeled using Ready-to-go DNA labeling beads (Amersham Biosciences), and hybridizations were performed in Quikhyb buffer (Stratagene).
Immunohistochemistry and western blot analyses
Testes from sexually mature CD-1 mice were fixed and processed by the Histological Core Facility of the Children's Hospital of Pennsylvania. Immunohistochemistry was performed as previously described (9). Anti-KSRP and anti-PTBP2 were diluted 1:200 before use. For western blotting, aliquots from tissue lysates or populations of isolated cells (20 µg) were separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and blotted onto polyvinylidene difluoride membranes (Millipore, Bedford, MA). The individual cell types were isolated as previously reported (28). Anti-KSRP [DB-KS (11), raised against the C-terminal of KSRP, or a polyclonal antibody from Abnova (H00008570-A01)], at dilutions of 1:3000, or anti-β-actin (1:10,000 dilution) (Sigma, St Louis, MO) were used as primary antibodies.
Immunodepletion and decay assays
In vitro decay and transfection assays were performed as previously described (9). KSRP was removed from testis cytoplasm extracts by overnight immunoprecipitation at 4°C with anti-KSRP [ab5, raised against the full-length KSRP (27)] that was bound to protein G-Sepharose. Equal amounts of supernatant from extracts, previously incubated with preimmune serum or anti-KSRP was used in the RNA decay assays.
Transfection assays were performed as previously described (9). In brief, reporter vectors, pEGFP-C2-UTR or pEGFP-C2-F3 (9), were transfected into HeLa cells using Liposome 2000 Reagent (Invitrogen) following the manufacturer's protocol. Where noted, reporter vectors were cotransfected into HeLa cells with vectors expressing t-KSRP or PTBP2. A Renilla luciferase phRL-TK vector was used as an internal control for transfection efficiency. To terminate transcription, actinomycin D (10 µM) was added 24 h after transfection. RNAs were purified by Trizol, separated on formaldehyde agarose gels, transferred to Hybond-N + membranes overnight and hybridized with a [P32]dCTP randomly labeled EGFP DNA probe at 68°C for 1 h using the Quikhyb kit (Stratagene).
RESULTS
Pgk2 mRNA accumulates in postmeiotic germ cells
Although Pgk2 mRNAs are transcribed at the onset of meiosis in male germ cells (4), PGK2 protein is first detected many days later in late-stage postmeiotic germ cells (5). Pgk2 mRNA levels increase as the germ cells differentiate (4), and northern blotting and microarrays have revealed a marked increase in Pgk2 mRNA levels in the testes of adult mice compared to prepuberal mice (3,5).
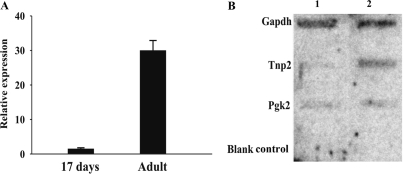
To quantify Pgk2 mRNA levels in prepuberal and adult mice, semiquantitative RT–PCR was used. An increase of Pgk2 mRNA levels of at least 20-fold was detected, consistent with previous reports (Figure 1A) (3,5). To compare Pgk2 mRNA transcription in prepuberal and adult testes, nuclear run-on assays were conducted. RNA was transcribed from isolated testicular nuclei, purified and measured by hybridization to membrane-fixed DNA. DNAs encoding transition protein 2 and glyceraldehyde phosphate dehydrogenase served as control transcripts for postmeiotic and constitutive transcription, respectively (Figure 1B). As reported for run-on transcription assays with nuclei from pachytene spermatocytes and round spermatids (29), similar levels of Pgk2 mRNA transcription were detected in nuclei isolated from the testes of prepuberal (17 day old) and adult mice (Figure 1B). Given that the proportion of spermatocytes in prepuberal testes from 17-day-old mice is similar to that of spermatocytes plus round spermatids in adult testes, these results suggest that the increase in Pgk2 mRNA levels in the adult testis reflects an mRNA buildup resulting from stabilization of Pgk2 mRNA in late-stage meiotic and postmeiotic germ cells (9) rather than an increase in transcription of the Pgk2 gene.
Figure 1.
(A) Quantitation of Pgk2 mRNA levels in prepuberal and adult testes. RT–PCR was used to measure Pgk2 mRNA levels in total RNA prepared from prepuberal (17 days) and adult testes. (B) Nuclear run-on assays were used to determine the transcriptional rate for Pgk2, Tnp2 and Gapdh mRNAs. At the completion of the nuclear run reactions, RNA was purified and hybridized to DNA probes fixed to membranes. Lane 1, nuclei from 17-day-old mice; lane 2, nuclei from adult mice (6- to 8-weeks old).
KSRP is present in a Pgk2 mRNA–protein complex in testis extracts
RNA affinity chromatography with a biotin-labeled RNA representing the 3′-UTR of Pgk2 and adult testis cytoplasmic extracts detected several proteins including an ∼52-kDa isoform of the RNA-binding protein KSRP (9). In somatic cells, a ubiquitously expressed ∼75-kDa KSRP has been implicated in transcriptional regulation, splicing and mRNA decay (17–26). Although the testis contains both KSRP isoforms, the ∼75-kDa KSRP was not detected in repeated chromatography experiments with cytoplasmic extracts consistent with the ∼52-kDa variant being the sole isoform of KSRP in the cytoplasm (see below).
To determine whether KSRP is bound to Pgk2 mRNA in the testis, RNA–protein complexes were immunoprecipitated with anti-KSRP from adult testes extracts. Following purification, RT–PCR was used to assay the precipitated RNAs for Pgk2 mRNA and two control highly abundant germ cell mRNAs that lack known KSRP binding sites. Pgk2 mRNA was immunoprecipitated with anti-KSRP (Figure 2A), while protamine 2 and transition protein 2 mRNAs were not, demonstrating the selective immunoprecipitation of a KSRP–Pgk2 mRNA complex. None of the three mRNAs was precipitated with preimmune serum.
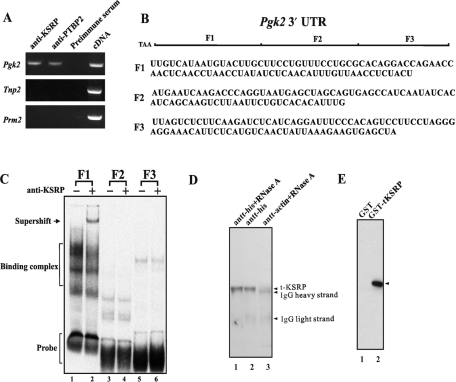
Figure 2.
KSRP binds to Pgk2 mRNA in the testis and in vitro to the F1 region of the Pgk2 mRNA and to PTBP2. (A) Messenger RNA detection by RT–PCR of purified germ cell mRNAs immunoprecipitated from mouse testis extracts with anti-KSRP. (B) A diagram of the three subclones (F1, F2 and F3) of the Pgk2 3′-UTR. (C) KSRP selectively binds to the F1 region to the 3′-UTR of Pgk2 mRNA. Testis extracts (20 µg) were incubated with [32P]-labeled transcripts generated by in vitro transcription from subclones (F1, F2 and F3) of the 3′-UTR of Pgk2 mRNA as previously described (9). RNA–protein complexes were resolved in 4% polyacrylamide gels in 0.5× TBE buffer. Anti-KSRP was added to indicated extracts before the addition of the radiolabeled RNAs. (D) Western blot of endogenous t-KSRP coimmunoprecipitated with r-PTBP2 from testis extracts. His-tagged recombinant PTBP2 (20 µg) was incubated with His-tag affinity resins and testis extract (1 mg). Lane 1, plus RNase A; lane 2, no RNAse; lane 3, antiactin control. Following centrifugation, pellets were solubilized and the proteins resolved in 10% SDS–PAGE. KSRP was detected by western blotting. (E) Recombinant t-KSRP and recombinant PTBP2 interact. GST-tagged r-KSRP was incubated with r-PTBP2. Following the addition of glutathione agarose beads and centrifugation, the pellets were solubilized, electrophoresed in 10% SDS–PAGE and PTBP2 was detected by western blotting. Lane 1, GST; lane 2, t-KSRP tagged with GST.
KSRP binds to the F1 region of the 3′-UTR of Pgk2 mRNA
To determine where KSRP binds in the 3′-UTR of the Pgk2 mRNA, gel shifts were performed with adult testis cytoplasmic extracts and three [P32]-labeled RNA subcloned probes of the entire 3′-UTR [F1 (93 nt), F2 (80 nt) and F3 (86 nt) (Figure 2B) (9)]. An RNA–protein complex was detected with F1 RNA (Figure 2C, lane 2) but not with the F2 or F3 RNA. In gel shifts with subclones of the F1-binding element, KSRP binds to a 34-nt region adjacent to the coding region (data not shown). Moreover, this binding to a specific region of the Pgk2 mRNA was specific because no binding was detected with the F1 subclone or any other part of the 3′-UTR of Pgk1 mRNA (data not shown). PTBP2 also binds directly to the F1 region of the 3′-UTR of the Pgk2 mRNA (9).
t-KSRP and PTBP2 interact
In somatic cells, PTBP and the ∼75-kDa KSRP have been demonstrated to interact as components of nuclear splicing complexes (11,17) and to bind together to the 3′-UTRs of mRNAs they regulate in the cytoplasm (15). To assess whether the ∼52-kDa KSRP interacts with PTBP2 in male germ cells, two approaches were taken. First, His-tagged recombinant PTBP2 was added to and precipitated from testis extracts that had been pretreated with RNase A. Western blotting of the proteins that were precipitated with anti-KSRP detected the ∼52-kDa KSRP (Figure 2D), suggesting a protein–protein interaction between t-KSRP and PTBP2.
To confirm a direct interaction between PTBP2 and t-KSRP (see the Materials and Methods section for the t-KSRP cloning and expression criteria and methodologies), purified recombinant PTBP2 was incubated with purified recombinant GST-tagged t-KSRP and anti-GST. PTBP2 was precipitated in the presence of GST-tagged t-KSRP, but not when GST was substituted for GST-tagged t-KSRP, establishing a direct protein–protein interaction between PTBP2 and t-KSRP (Figure 2E). Thus, t-KSRP can bind both to the F1 region of the 3′-UTR of Pgk2 mRNA and to PTBP2.
A complex of PTBP2 and t-KSRP binds to F1 RNA
Both t-KSRP and PTBP2 selectively bind to a 93-nt region (F1) of the 3′-UTR of Pgk2 mRNA (9, Figure 2C). To determine whether t-KSRP and PTBP2 are bound in the same RNA–protein complex, supershift assays using anti-PTBP2 or anti-KSRP were performed with radiolabeled F1 RNA and testis extracts (Figure 3A) or with recombinant t-KSRP and PTBP2 (Figure 3B). The addition of anti-KSRP to testicular extracts supershifts several F1 RNA–protein complexes from testicular extracts, indicating t-KSRP is a component of multiple complexes with different electrophoretic mobilities (Figure 3A, lane 4). Anti-PTBP2 supershifts multiple complexes, indicating its presence in RNA–protein complexes containing t-KSRP (Figure 3A, lane 3).
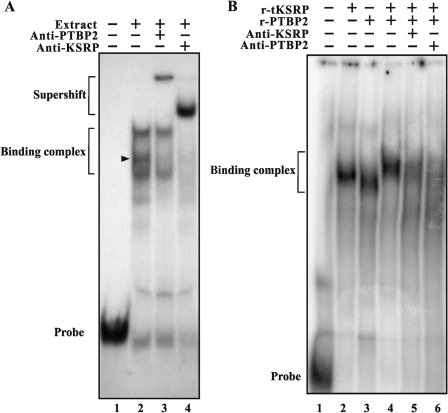
Figure 3.
KSRP and PTBP2 are present in the same complexes with the F1 region of the 3′-UTR of Pgk2 mRNA. RNA gel shifts were performed with testis extracts as in (Figure 2B). (A) Lane 1, transcript F1 without testis extract; lane 2, transcript F1 with extract; lane 3, transcript F1 with extract and anti-PTBP2; lane 4, transcript F1 with extract, anti-PTBP2 and anti-KSRP. The position of one complex shifted by each antibody is shown by an arrow. (B) Recombinant t-KSRP and recombinant PTBP2 (1 µg) interact and form complexes with F1 RNA containing both proteins. Lane 1, transcript F1 without testis extract; lane 2, transcript F1 t-KSRP; lane 3, transcript F1 with PTBP2; lane 4, transcript F1 with PTBP2 and KSRP; lane 5, transcript F1 with PTBP2, t-KSRP and anti-KSRP (Abnova, Taiwan); lane 6, transcript F1 with PTBP2, t-KSRP and anti-PTBP2 (Abnova).
The presence of both t-KSRP and PTBP2 in the same RNA–protein complex(es) is also seen in gel shifts performed with recombinant t-KSRP and recombinant PTBP2 (Figure 3B). Both proteins bind the F1 region of the 3′-UTR of Pgk2 mRNA, yielding complexes with different mobilities (Figure 3B, lanes 2 and 3). A distinct complex of slower electrophoretic mobility is seen when the two proteins are added together with F1 RNA (Figure 3B, lane 4). Addition of anti-KSRP (polyclonal or monoclonal) or anti-PTBP2 dissociates the KSRP–PTBP2–RNA complex (Figure 3B, lanes 5 and 6), demonstrating the presence of t-KSRP and PTBP2 in the same F1 RNA–protein complex.
The ∼52-kDa KSRP is predominantly in the cytoplasm in mouse testis
In somatic cells, the ∼75-kDa KSRP is involved in pre-mRNA splicing in the nucleus and in an mRNA decay pathway in the cytoplasm (17–26). Northern blotting of total RNA from brain and testes of prepuberal and adult mice detects one mRNA of ∼4 kb, in agreement with published reports (18). KSRP mRNA levels are higher in testes from 17-day-old mice than in the testes of 22-day-old and adult mice, suggesting it is more abundant in early-stage meiotic germ cells (Figure 4A, compare lanes 2–4).
Although we only detect one KSRP mRNA of ∼4 kb in both testis and brain (Figure 4A) and an mRNA of this size encodes the ∼75 kDa KSRP in somatic cells (18), western blotting reveals two distinct KSRP protein bands of ∼75 and ∼52 kDa (Figure 4B). Separation of the testis into nuclear and cytoplasmic fractions reveals that the ∼75-kDa KSRP is nuclear (Figure 4B, lanes 1–3) while the majority of t-KSRP is in the cytoplasm (Figure 4B, lanes 4–6). Consistent with KSRP RNA levels (Figure 4A), higher amounts of t-KSRP are seen in the testes of prepuberal mice than in adults (Figure 4B, compare lanes 1 and 4 to lanes 3 and 6). In several RNA affinity chromatography experiments with cytoplasmic testis extracts, t-KSRP was the only KSRP detected (9). The predominance of t-KSRP in the cytoplasm of the testis suggests this ∼52-kDa isoform of KSRP could function as an mRNA-destabilizing protein in the testis (Figure 4B).
t-KSRP is enriched in early stages of meiotic germ cells in mouse testes
Two different approaches, western blotting of proteins from highly purified populations of germ cells and immunohistochemistry, were used to establish the cellular and subcellular locations of KSRP in mouse testes. In western blots of germ cell extracts, t-KSRP was predominant in early meiotic stages (preleptotene, leptotene/zygotene and pachytene spermatocytes) and was not detectable in round spermatids or condensing spermatids/residual bodies, suggesting it functions during meiosis, but not postmeiotically (Figure 4C). In contrast, the nuclear ∼75-kDa KSRP and the Pgk2 mRNA-stabilizing protein, PTBP2, increase during meiosis and are abundant in spermatids.
To better determine the extent of expression of t-KSRP, western blots were prepared with extracts from several somatic tissues. The expression of t-KSRP appears limited because t-KSRP was detected in testis extracts (Figure 4D, lane 1), but not in brain, liver, kidney or lung extracts (Figure 4D, lanes 2–5).
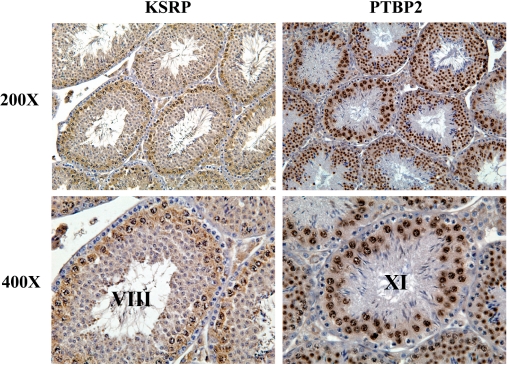
Using affinity-purified antibodies that detect both KSRP isoforms in testes from adult mice by immunohistochemistry, KSRP is primarily present in the nuclei and cytoplasm of early and midstages of meiotic germ cells in stage VIII tubules, consistent with the northern and western blotting data (Figures 4 and 5, left side). In contrast, PTBP2 is more abundant in the nuclei and cytoplasm of later stage meiotic and postmeiotic germ cells in stage XI tubules (Figure 5, right side) (9). Based on the nuclear localization of KSRP and the predominance of t-KSRP in the cytoplasm (Figure 4B), we conclude that t-KSRP and not full-length KSRP is the primary (sole?) KSRP in the cytoplasm of early meiotic stage germ cells (Figure 5).
Figure 5.
Anti-KSRP selectively stains early-stage meiotic germ cells of the adult mouse testis. Immunostaining of testis sections from sexually mature CD-1 mice with (A) affinity-purified anti-KSRP and (B) affinity-purified anti-PTBP2. Anti-KSRP (Abnova, Taiwan) and anti-PTBP2 (from R. Darnell) were used at dilutions of 1:200. Magnification: × 200, × 400. Both antibodies only recognized their respective proteins on western blots.
KSRP destabilizes Pgk2 mRNA in extracts or cells in the presence of PTBP2
Since KSRP serves to destabilize mRNAs in diverse somatic cell types (19–26), we investigated whether t-KSRP could destabilize one of its target mRNAs, the Pgk2 mRNA, using three complementary approaches—in vitro decay assays, transient transfections and protein depletion assays. All confirm a role for t-KSRP in male germ cell mRNA decay.
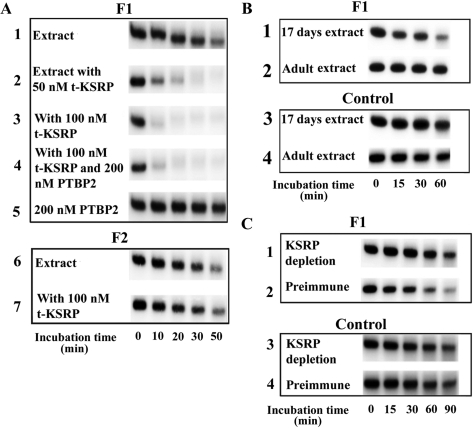
To assess the role of t-KSRP in in vitro decay, 5′-capped [32P]-labeled F1 or F2 transcript was incubated with testis extracts and increasing amounts of recombinant t-KSRP (Figure 6A). Addition of 50 nM of t-KSRP leads to complete F1 RNA degradation within 20 min of incubation (Figure 6A, lane 2), although only a modest RNA reduction was seen in control testis extracts with up to 50 min of incubation (Figure 6A, lane 1). F1 RNA degradation increased when recombinant t-KSRP was increased to 100 nM (Figure 6A, lane 3). In contrast, no increased RNA degradation was seen when 100 nM t-KSRP was added to testis extracts containing the nonbinding F2 RNA (Figure 6A, compare lanes 6 and 7). As previously reported (9), addition of r-PTBP2 completely stabilized F1 RNA in incubations up to 50 min (Figure 6A, lane 5), but addition of up to 200 nM PTBP2 failed to prevent F1 RNA destabilization when exogenous t-KSRP was added (Figure 6A, lane 4). These in vitro decay assays indicate that t-KSRP promotes RNA degradation of a target mRNA and suggest t-KSRP exerts a dominant effect over PTBP2 in testis extracts under the assay conditions tested.
Figure 6.
t-KSRP destabilizes RNAs containing the 3′-UTR of Pgk2 mRNA. (A) t-KSRP destabilizes RNA constructs in testis extracts, and recombinant PTBP2 cannot prevent the destabilization. 5'-capped and 3'-polyadenylated radiolabeled F1 or F2 transcript was incubated at 30°C with testis extracts with or without recombinant t-KSRP (100 or 200 nM) or PTBP2 (200 nM). RNA was purified and analyzed by gel electrophoresis followed by autoradiography. Gels were quantitated by phosphorimaging. The amount of RNA at the beginning of the reaction was set at 100%. All the decay assays were performed at least twice. (B) F1 RNAs are degraded more rapidly in extracts from 17-day-old mice than from adults. Decay assays were performed as in (A) with prepuberal (extracts from testes of 17-day-old mice) or adult extracts except that incubation temperature was set at 25°C. (C) Depletion of KSRP from testis extracts leads to the stabilization of F1 RNA. Decay assays were performed as in (A) with cytoplasmic extracts from which KSRP was depleted.
The increased RNA degradation seen with increasing amounts of recombinant t-KSRP was also seen with endogenous t-KSRP (Figure 6B). Radiolabeled F1 RNA was degraded more rapidly in testis extracts from prepuberal (17-day-old-mice) mice (which contain higher amounts of t-KSRP, Figure 4B) than in testis extracts from adult mice (8–10 weeks, Figure 6B, compare lanes 1 and 2), suggesting the more abundant t-KSRP in 17-day-old mice is consistent with the more rapid RNA degradation. The RNA degradation requires t-KSRP binding to RNA because no degradation was seen when the control nonbinding F2 RNA probe was incubated with prepuberal or adult extracts (Figure 6B, compare lanes 3 and 4). In vitro decay assays with extracts depleted of KSRP by immunoprecipitation provide additional evidence for the destabilizing role for endogenous t-KSRP (Figure 6C). The stability of radiolabeled F1 RNAs in these assays was increased in adult testis extracts that had been depleted of KSRP (Figure 6C, compare lanes 1 and 2). Again this required RNA binding as no differences were seen with the F2 RNA (Figure 6C, compare lanes 3 and 4).
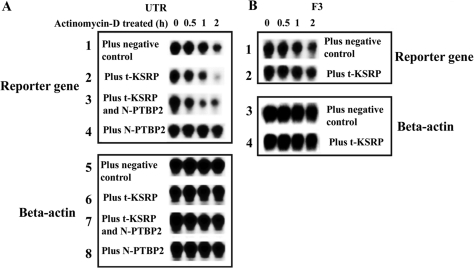
To determine whether t-KSRP can also destabilize mRNAs and be dominant to PTBP2 in living cells, an EGFP construct containing the 3′-UTR of Pgk2 mRNA was cotransfected into HeLa cells with t-KSRP and with or without PTBP2 (Figure 7). For PTBP2 expression, an N-truncated form of PTBP2 was used because this modification targets PTBP2 to the cytoplasm, whereas full-length PTBP2 contains a functional nuclear localization signal directing PTBP2 to nuclei (9). Previous studies have demonstrated that the full-length and truncated forms of PTBP2 are functionally indistinguishable (9). Consistent with the RNA incubations in testis extracts (Figure 6), the transfected RNA is less stable in the transfected cells expressing t-KSRP (Figure 7A, lane 2). Similar to the findings with testis extract incubations, the radiolabeled RNA is stabilized by the expression of PTBP2 (Figure 7A, lane 4), but cotransfection of PTBP2 and t-KSRP (the expression of both was confirmed by western blotting, data not shown) leads to RNA destabilization (Figure 7A, lane 3), The RNA destabilization required RNA binding because there was no destabilization when a control nonbinding F3 RNA was transfected with t-KSRP (Figure 7B, compare lanes 1 and 2). Assays for β-actin mRNA established equal RNA loading per lane and specificity for the RNA destabilization of t-KSRP (see β-actin lanes in Figure 7A and B). We conclude that t-KSRP destabilizes RNAs such as the Pgk2 mRNA in cells and overrules the stabilizing ability of PTBP2 in transfected cells and in testis extracts (Figures 6 and 7).
Figure 7.
t-KSRP destabilizes mRNAs transfected into HeLa cells. (A) HeLa cells were cotransfected with pEGFP-C2-UTR (containing the entire 3′-UTR of Pgk2) and vectors expressing t-KSRP or an N-truncated (cytoplasmic form) of PTBP2. Following actinomycin D addition, cells were collected at intervals, total RNAs were purified and mRNA levels were analyzed by northern blotting. β-Actin mRNA was used as a loading control. (B) The F3 subclone of the 3′-UTR of Pgk2 and β-actin mRNA were used as nonbinding RNA incubation and loading controls, respectively. The F3 RNA was used in place of the F1 RNA because it contained the same polyadenylation signal as the transcript from pEGFP-C2-UTR. Each assay was repeated at least three times with essentially identical results.
DISCUSSION
Here we report the existence of a cytoplasmic isoform of KSRP (t-KSRP) that binds to a class III AU-rich sequence in the 3′-UTR of Pgk2 mRNA and destabilizes the RNA in in vitro and cellular assays. t-KSRP interacts with and is a component of RNA–protein complexes containing PTBP2, another multifunctional RNA-binding protein that serves to stabilize RNAs in the cytoplasm of germ cells. When both PTBP2 and t-KSRP are present, t-KSRP appears to dominate and Pgk2 mRNA is destabilized.
Protein synthesis is dependent on mRNAs whose steady-state levels are regulated by a combination of transcription, stabilization and decay. There is a growing awareness that posttranscriptional events play major regulatory roles in mRNA metabolism and function in eukaryotic cells. Although microarray studies provide great insight into the temporal and cellular transcription of the testicular transcriptome (5), mRNA stabilization is an essential regulatory mechanism in haploid differentiating germ cells (6). Proteins such as the abundant germ cell–specific DNA/RNA-binding protein, MSY2, stabilize many germ cell mRNAs (7), while the PTBP2 selectively stabilizes mRNAs such as the PGK 2 mRNA in the testis (9). Because many male germ cell mRNAs are sequestered into residual bodies, which are subsequently phagocytized by Sertoli cells, major efforts to define mechanisms that degrade male germ cell mRNAs have not been undertaken. In contrast, in somatic cells, mRNA degradation events often dependent on AU-rich sequence elements (AREs) and proteins such as KSRP that bind to them have been well characterized (19,20).
AREs are common cis-acting elements in eukaryotic transcripts that promote rapid mRNA turnover by recruiting KSRP-containing protein complexes to mRNAs (19). In addition to its originally described function(s) as a splicing factor (17,18), KSRP facilitates the degradation of short-lived mRNAs encoding proteins such cytokines, growth factors and protooncogenes through interactions with AREs in their 3′-UTRs (19–26). The tethering of KSRP to HIV-1 mRNA constructs elicits rapid mRNA decay and a dramatic viral reduction by recruiting mRNA decay/machinery factors to the bound KSRP (24). Recently, the chicken protein, ZBP2, a homologue of KSRP, has been demonstrated to bind to sequences that control the subcellular localization of actin mRNAs (30,31). Three classes of AREs have been identified (32) based on the presence of one or more of the AUUUA motifs in their 3′-UTRs (class I), multiple overlapping motifs in their 3′-UTRs (class II) or no obvious AUUUA motifs in their 3′-UTRs (class III).
Although the ∼75-kDa KSRP is often the predominant KSRP in somatic cells, in the testis the ∼52-kDa KSRP, t-KSRP, is the primary isoform of KSRP in the cytoplasm (Figure 4B). Based on the existence of only one size of KSRP mRNA in testis, we believe that t-KSRP is not derived from an independent KSRP mRNA, but is either proteolytically cleaved from the somatic KSRP similar to the apoptotic cleavage of KSRP in liver (33) or t-KSRP is translated from an internal initiation site (methionine 207) in the ∼4-kb KSRP mRNA. Edman protein sequencing of t-KSRP suggests an N-terminus block (data not shown). Sequence analysis of 17 peptides from t-KSRP obtained following RNA affinity chromatography indicates that although t-KSRP lacks part of the first RNA-binding domain, the remaining three KH RNA-binding domains are intact. We find that t-KSRP is functionally active in mRNA binding, consistent with in vitro and in vivo studies demonstrating that the third and fourth KH domains of KSRP are sufficient for promoting mRNA decay in vivo (18,19).
The functions of multifunctional proteins such as KSRP are often defined by their subcellular location(s) (20). Numerous splicing studies with KSRP have been supported by its nuclear localization, and a recent immunofluorescent study concluded KSRP is almost exclusively nuclear in HeLa and neuroblastoma cells (27). Although KSRP shows flexibility in recognition of RNA targets (34), binding of KSRP to AREs in 3′-UTRs of mRNAs is indicative of cytoplasmic roles for KSRP (19–26). To accomplish both splicing and mRNA decay in somatic cells, KSRP would need to shuttle between the nucleus and cytoplasm as has been reported for other RNA-binding proteins such hnRNP K, Translin and Trax (35,36). The N-terminus of KSRP likely contributes to its intracellular movement. The Xenopus homologue of KSRP, which lacks a 58-amino-acid segment near the N-terminus, is present in both the nucleus and the cytoplasm during oogenesis (37), consistent with our observation that in mouse testis the truncated t-KSRP is primarily in the cytoplasm, while the full-length KSRP is solely nuclear (Figure 4). The abundance and cytoplasmic location of t-KSRP and the nuclear location of the ∼75-kDa KSRP support a posttranscriptional mRNA regulatory role for t-KSRP in the mammalian testis.
We have identified a testicular variant of the KSRP, t-KSRP, a protein that we believe facilitates the decay of mRNAs such as Pgk2 mRNA in meiotic germ cells. In contrast to many of the rapidly degraded mRNAs recognized by KSRP in somatic cells, t-KSRP binds to a 93-nt AU-rich class III region in the 3′-UTR of the Pgk2 mRNA, a germ cell mRNA that may be short-lived mRNA during meiosis, but becomes a long-lived stable mRNA in postmeiotic germ cells. This transition appears to be facilitated by another RNA-binding protein in the testis, PTBP2, which functions to maintain Pgk2 mRNA levels (9). Thus, upon reduction of levels of t-KSRP during the transition from spermatocytes to spermatids, the dominant Pgk2 mRNA decay-promoting activity is lost and the transcript-stabilizing activity of PTBP2 prevails, leading to a dramatic increase in the level of Pgk2 transcript in round spermatids in the absence of any increase in transcription rate of the Pgk2 gene.
In vitro assays and transfection studies indicate the PTBP2 stabilizes the Pgk2 mRNA by direct binding to the F1 region of the 3′-UTR of the Pgk2 mRNA, the same part of the 3′-UTR recognized by t-KSRP. Since t-KSRP and PTBP2 interact and both recognize the same RNA-binding region of the Pgk2 mRNA, this raises the question how t-KSRP destabilizes mRNAs in the presence of PTBP2. Although the mechanism resulting in the apparent dominance of the destabilizing effect of t-KSRP over the stabilizing effect of PTBP2 is not known, we do know that the two proteins interact and form F1 RNA–protein complexes that contain both proteins. In the nuclei of WERI-1 retinoblastoma cells, the binding of full-length KSRP to RNA requires the adjacent binding of PTBP2, and the interaction of KSRP, PTBP2 and other factors facilitates the cooperative assembly of an hnRNP complex for splicing (11,17). In the cytoplasm, less is known about the functional interactions between the KSRP and PTBP proteins. As we propose for t-KSRP and PTBP2 in the testis, binding of KSRP and PTBP1 is believed to control the decay and stability of human inducible nitric oxide synthase mRNA, but the mechanism of their molecular interactions is not known (15,21).
The apparent dominance of t-KSRP over PTBP2 may reflect in vivo RNA binding differences among t-KSRP, PTBP2 and accessory proteins/factors. Alternatively, protein availability may be important because during early meiosis when Pgk2 mRNA levels are lowest, t-KSRP is most abundant and PTBP2 levels are low. t-KSRP levels decrease in later meiotic germ cells and round spermatids and is not detectable in condensing spermatids (Figure 4C), consistent with the ∼20-fold increase of Pgk2 mRNA in the testes of adult mice, resulting at least in part from the absence of t-KSRP in later stage postmeiotic germ cells (Figure 1).
Protein variants (isoproteins) are especially common in male germ cells. The testis expresses isoforms of many proteins including lactate dehydrogenase, glyceraldehyde-3-phosphate dehydrogenase, hexokinase, phosphoglycerate mutase, aldolase, pyruvate dehydrogenase E1 alpha, PGK and even KSRP (4). The isoprotein PGK2 is believed to have evolved to replace PGK1 because PGK1 becomes inactivated during meiotic sex-chromosome inactivation. Other isoproteins have been proposed to optimize metabolic activities unique to germ cells. The temporal synthesis patterns of many of these proteins require delayed mRNA utilization, suggesting that t-KSRP and PTBP2 are likely to regulate the cytoplasmic steady-state levels of numerous germ cell mRNAs in addition to Pgk2 mRNA. Sequence analysis of mRNAs precipitated by anti-PTBP2 and anti-KSRP includes Tex27, Ddc8 and Spata6, genes whose mRNAs are translationally regulated similar to Pgk2 mRNA (data not shown) (5).
In summary, Pgk2 mRNA is first detectable in early meiosis in spermatocytes and increases dramatically in spermatids (2–4, Figure 1). t-KSRP is abundant in early meiosis and markedly decreases, becoming undetectable in postmeiotic male germ cells (Figures 4 and 5). The decline of t-KSRP and increase of PTBP2 are concomitant with the increase in Pgk2 mRNA levels. We speculate that in the cytoplasm of early meiotic spermatocytes, t-KSRP promotes the decay of Pgk2 mRNA and as t-KSRP levels decrease, Pgk2 mRNA levels rise from continued transcription and stabilization by protein complexes containing PTBP2 in late-stage meiotic spermatocytes and postmeiotic spermatids. Many additional proteins are likely essential for these processes. In somatic tissues, KSRP stimulates mRNA decay by recruiting a decay-promoting exosomal complex to target mRNAs (19). In testis extracts, t-KSRP interacts with exosomal proteins such as EXOSC7 (Rrp42p) and Rrp40p (data not shown). Thus, a number of trans-acting factors will contribute to the synthesis/stabilization/destabilization of the Pgk2 mRNA during spermatogenesis. Recently, a number of additional germ cell mRNAs have been demonstrated to exhibit temporal RNP/polysomal regulation similar to Pgk2 mRNA (5). The possibility that they represent additional t-KSRP target mRNAs that undergo a coordinated regulation merits investigation.
Funding
National Institutes of Health (grant HD 28832).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr R.B. Darnell (Rockefeller University, New York) for anti-PTBP2 and Dr D.L. Black (University of California, Los Angeles) for anti-KSRP (ab5 and DB-KS).
REFERENCES
- 1.McCarrey JR, Thomas K. Human testis-specific PGK gene lacks introns and possesses characteristics of a processed gene. Nature. 1987;326:501–505. doi: 10.1038/326501a0. [DOI] [PubMed] [Google Scholar]
- 2.Bluthmann H, Cicurel L, Kuntz GW, Haedenkamp G, Illmensee K. Immunohistochemical localization of mouse testis-specific phosphoglycerate kinase (PGK-2) by monoclonal antibodies. EMBO J. 1982;1:479–484. doi: 10.1002/j.1460-2075.1982.tb01194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robinson MO, McCarrey JR, Simon MI. Transcriptional regulatory regions of testis-specific PGK2 defined in transgenic mice. Proc. Natl Acad. Sci. USA. 1989;86:8437–8441. doi: 10.1073/pnas.86.21.8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshioka H, Geyer CB, Hornecker JL, Patel KT, McCarrey JR. In vivo analysis of developmentally and evolutionarily dynamic protein-DNA interactions regulating transcription of the Pgk2 gene during mammalian spermatogenesis. Mol. Cell Biol. 2007;27:7871–7885. doi: 10.1128/MCB.00990-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iguchi N, Tobias JW, Hecht NB. Expression profiling reveals meiotic male germ cell mRNAs that are translationally up- and down-regulated. Proc. Natl Acad. Sci. USA. 2006;103:7712–7717. doi: 10.1073/pnas.0510999103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun RE. Post-transcriptional control of gene expression during spermatogenesis. Semin. Cell Dev. Biol. 1998;9:483–489. doi: 10.1006/scdb.1998.0226. [DOI] [PubMed] [Google Scholar]
- 7.Yang J, Morales CR, Medvedev S, Schultz RM, Hecht NB. In the absence of the mouse DNA/RNA-binding protein MSY2, messenger RNA instability leads to spermatogenic arrest. Biol. Reprod. 2007;76:48–54. doi: 10.1095/biolreprod.106.055095. [DOI] [PubMed] [Google Scholar]
- 8.Yang J, Medvedev S, Yu J, Tang LC, Agno JE, Matzuk MM, Schultz RM, Hecht NB. Absence of the DNA-/RNA-binding protein MSY2 results in male and female infertility. Proc. Natl Acad. Sci. USA. 2005;102:5755–5760. doi: 10.1073/pnas.0408718102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu M, Hecht NB. Polypyrimidine tract binding protein 2 stabilizes phosphoglycerate kinase 2 mRNA in murine male germ cells by binding to its 3'UTR. Biol. Reprod. 2007;76:1025–1033. doi: 10.1095/biolreprod.107.060079. [DOI] [PubMed] [Google Scholar]
- 10.Yang J, Medvedev S, Reddi PP, Schultz RM, Hecht NB. The DNA/RNA-binding protein MSY2 marks specific transcripts for cytoplasmic storage in mouse male germ cells. Proc. Natl Acad. Sci. USA. 2005;102:1513–1518. doi: 10.1073/pnas.0404685102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Markovtsov V, Nikolic JM, Goldman JA, Turck CW, Chou MY, Black DL. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell Biol. 2000;20:7463–7479. doi: 10.1128/mcb.20.20.7463-7479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tillmar L, Carlsson C, Welsh N. Control of insulin mRNA stability in rat pancreatic islets. Regulatory role of a 3'-untranslated region pyrimidine-rich sequence. J. Biol. Chem. 2002;277:1099–1106. doi: 10.1074/jbc.M108340200. [DOI] [PubMed] [Google Scholar]
- 13.Knoch KP, Bergert H, Borgonovo B, Saeger HD, Altkruger A, Verkade P, Solimena M. Polypyrimidine tract-binding protein promotes insulin secretory granule biogenesis. Nat. Cell Biol. 2004;6:207–214. doi: 10.1038/ncb1099. [DOI] [PubMed] [Google Scholar]
- 14.Hamilton BJ, Genin A, Cron RQ, Rigby WF. Delineation of a novel pathway that regulates CD154 (CD40 ligand) expression. Mol. Cell Biol. 2003;23:510–525. doi: 10.1128/MCB.23.2.510-525.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pautz A, Linker K, Hubrich T, Korhonen R, Altenhofer S, Kleinert H. The polypyrimidine tract-binding protein (PTB) is involved in the post-transcriptional regulation of human inducible nitric oxide synthase expression. J. Biol. Chem. 2006;281:32294–32302. doi: 10.1074/jbc.M603915200. [DOI] [PubMed] [Google Scholar]
- 16.Hellen CU, Witherell GW, Schmid M, Shin SH, Pestova TV, Gil A, Wimmer E. Cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl Acad. Sci. USA. 1993;90:7642–7646. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 18.Min H, Turck CW, Nikolic JM, Black DL. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- 19.Gherzi R, Lee KY, Briata P, Wegmuller D, Moroni C, Karin M, Chen CY. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 20.Dean JL, Sully G, Clark AR, Saklatvala J. The involvement of AU-rich element-binding proteins in p38 mitogen-activated protein kinase pathway-mediated mRNA stabilisation. Cell. Signal. 2004;16:1113–1121. doi: 10.1016/j.cellsig.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 21.Linker K, Pautz A, Fechir M, Hubrich T, Greeve J, Kleinert H. Involvement of KSRP in the post-transcriptional regulation of human iNOS expression-complex interplay of KSRP with TTP and HuR. Nucleic Acids Res. 2005;33:4813–4827. doi: 10.1093/nar/gki797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Briata P, Forcales SV, Ponassi M, Corte G, Chen CY, Karin M, Puri PL, Gherzi R. p38-dependent phosphorylation of the mRNA decay-promoting factor KSRP controls the stability of select myogenic transcripts. Mol. Cell. 2005;20:891–903. doi: 10.1016/j.molcel.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 23.Gherzi R, Trabucchi M, Ponassi M, Ruggiero T, Corte G, Moroni C, Chen CY, Khabar KS, Andersen JS, Briata P. The RNA-binding protein KSRP promotes decay of beta-catenin mRNA and is inactivated by PI3K-AKT signaling. PLoS Biol. 2006;5:e5. doi: 10.1371/journal.pbio.0050005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Chou CF, Mulky A, Maitra S, Lin WJ, Gherzi R, Kappes J, Chen CY. Tethering KSRP, a decay-promoting AU-rich element-binding protein, to mRNAs elicits mRNA decay. Mol. Cell Biol. 2006;26:3695–3706. doi: 10.1128/MCB.26.10.3695-3706.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nechama M, Ben-Dov IZ, Briata P, Gherzi R, Naveh-Many T. The mRNA decay promoting factor K-homology splicing regulator protein post- transcriptionally determines parathyroid hormone mRNA levels. FASEB J. 2008;22:3458–3468. doi: 10.1096/fj.08-107250. [DOI] [PubMed] [Google Scholar]
- 26.Winzen R, Thakur BK, Dittrich-Breiholz O, Shah M, Redich N, Dhamija S, Kracht M, Holtmann H. Functional analysis of KSRP interaction with the AU-rich element of interleukin-8 and identification of inflammatory mRNA targets. Mol. Cell Biol. 2007;27:8388–8400. doi: 10.1128/MCB.01493-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall MP, Huang S, Black DL. Differentiation-induced colocalization of the KH-type splicing regulatory protein with polypyrimidine tract binding protein and the c-src pre-mRNA. Mol. Biol. Cell. 2004;15:774–786. doi: 10.1091/mbc.E03-09-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarrey JR, Berg WM, Paragioudakis SJ, Zhang PL, Dilworth DD, Arnold BL, Rossi JJ. Differential transcription of Pgk genes during spermatogenesis in the mouse. Dev. Biol. 1992;154:160–168. doi: 10.1016/0012-1606(92)90056-m. [DOI] [PubMed] [Google Scholar]
- 29.Kumari M, Stroud JC, Anji A, McCarrey JR. Differential appearance of DNase I-hypersensitive sites correlates with differential transcription of Pgk genes during spermatogenesis in the mouse. J. Biol. Chem. 1996;271:14390–14397. doi: 10.1074/jbc.271.24.14390. [DOI] [PubMed] [Google Scholar]
- 30.Gu W, Pan F, Zhang H, Bassell GJ, Singer RH. A predominantly nuclear protein affecting cytoplasmic localization of beta-actin mRNA in fibroblasts and neurons. J. Cell Biol. 2002;156:41–51. doi: 10.1083/jcb.200105133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan F, Hüttelmaier S, Singer RH, Gu W. ZBP2 facilitates binding of ZBP1 to beta-actin mRNA during transcription. Mol. Cell Biol. 2007;27:8340–8351. doi: 10.1128/MCB.00972-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 33.Seok H, Cho J, Cheon M, Park IS. Biochemical characterization of apoptotic cleavage of KH-type splicing regulatory protein (KSRP)/far upstream element-binding protein 2 (FBP2) Protein Pept. Lett. 2002;9:511–519. doi: 10.2174/0929866023408454. [DOI] [PubMed] [Google Scholar]
- 34.García-Mayoral MF, Díaz-Moreno I, Hollingworth D, Ramos A. The sequence selectivity of KSRP explains its flexibility in the recognition of the RNA targets. Nucleic Acids Res. 2008;36:5290–5296. doi: 10.1093/nar/gkn509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siomi H, Matunis MJ, Michael WM, Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho YS, Chennathukuzhi VM, Handel MA, Eppig J, Hecht NB. The relative levels of translin-associated factor X (TRAX) and testis brain RNA-binding protein determine their nucleocytoplasmic distribution in male germ cells. J. Biol. Chem. 2004;279:31514–31523. doi: 10.1074/jbc.M401442200. [DOI] [PubMed] [Google Scholar]
- 37.Kroll TT, Zhao WM, Jiang C, Huber PW. A homolog of FBP2/KSRP binds to localized mRNAs in Xenopus oocytes. Development. 2002;129:5609–5619. doi: 10.1242/dev.00160. [DOI] [PubMed] [Google Scholar]