Abstract
The dynamic mechanisms by which RNAs acquire biologically functional structures are of increasing importance to the rapidly expanding fields of RNA therapeutics and biotechnology. Large energy barriers separating misfolded and functional states arising from alternate base pairing are a well-appreciated characteristic of RNA. In contrast, it is typically assumed that functionally folded RNA occupies a single native basin of attraction that is free of deeply dividing energy barriers (ergodic hypothesis). This assumption is widely used as an implicit basis to interpret experimental ensemble-averaged data. Here, we develop an experimental approach to isolate persistent sub-populations of a small RNA enzyme and show by single molecule fluorescence resonance energy transfer (smFRET), biochemical probing and high-resolution mass spectrometry that commitment to one of several catalytically active folds occurs unexpectedly high on the RNA folding energy landscape, resulting in partially irreversible folding. Our experiments reveal the retention of molecular heterogeneity following the complete loss of all native secondary and tertiary structure. Our results demonstrate a surprising longevity of molecular heterogeneity and advance our current understanding beyond that of non-functional misfolds of RNA kinetically trapped on a rugged folding-free energy landscape.
INTRODUCTION
Large fractions of all eukaryotic genomes are transcribed into biologically functional, non-protein coding RNAs (1), profoundly impacting our understanding of cellular function and the application of biotechnology (2). Folding of RNA is generally characterized by a rugged-free energy landscape, challenging our understanding of how a given RNA sequence reaches its functional tertiary structure (3–8). Recent single molecule fluorescence resonance energy transfer (smFRET) approaches have provided unprecedented insight into the RNA folding problem by uncovering highly heterogeneous folding kinetics in a number of biologically relevant RNAs (3,7–18). Arguably the most striking example of heterogeneity comes from experiments using the hairpin ribozyme (3,10–12,17,19), a small self-cleaving RNA.
Small self-cleaving RNAs involved in the rolling-circle replication of certain plant, fungus and animal virus satellites are favored model systems to study RNA folding-function relationships as their catalytic function can serve as a practical reporter of a biologically relevant fold (20,21). Various forms of the hairpin ribozyme derived from the tobacco ringspot virus satellite RNA have provided a wealth of structural and biochemical information on how folding impacts the biological activity of RNA (3,10–12,17,19,22–28). The hairpin ribozyme catalyzes site-specific and reversible backbone self-cleavage, following the dynamic formation of a docked structure in which the internal loops of two adjacent domains form well-understood tertiary contacts (Figure 1a) (25,26,28). Ill-understood, however, is the underlying nature of the consistently observed (3,10–12,17,19) multiple sub-populations with dramatically different folding kinetics that result in profoundly biphasic and incomplete catalytic activity (Figure 1b) (3,12). Given our increasing appreciation of RNA's role in biology and biotechnology and the ubiquity of heterogeneities in RNA folding and function, it is critical to ask whether the observed heterogeneities are intrinsic or not.
Figure 1.
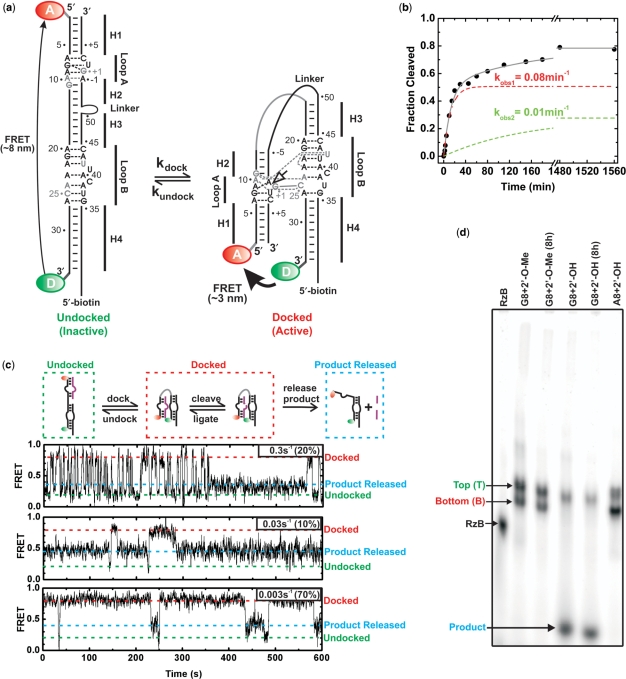
Heterogeneity in folding and function of the hairpin ribozyme. (a) Secondary structure of the 2WJ hairpin ribozyme, composed of the RzA and RzB strands. The docked and undocked conformations of the ribozyme are shown with canonical and non-canonical base pairs indicated by solid and dashed lines, respectively. Nucleotides forming interdomain hydrogen bonds are shown in gray, the cleavage site is indicated by an open arrow. The ribozyme was labeled either with a domain terminal donor(D)–acceptor(A) fluorophore pair and a 5′-biotin, and/or with a 5′- or 3′-end 32P label. (b) Single-turnover cleavage time course of the 3′-32P-labeled ribozyme with a domain terminal donor–acceptor fluorophore pair and a 5′-biotin, as monitored by D-PAGE, was used to determine the indicated cleavage rate constants for the two phases of the reaction (12 mM MgCl2, pH 7.5, 25°C). The contribution of each phase is shown as a dashed line. (c) Heterogeneous undocking kinetics of the catalytically active, trans-cleaving (no Linker), t-2WJ hairpin ribozyme under multiple-turnover conditions. The docked, undocked and product released states are indicated by red, green and blue dashed lines, respectively. Three single molecule time trajectories demonstrating catalytic proficiency of distinct sub-populations (undocking rate constants and the fraction of molecules undocking with this rate constant are given in the upper right corner of each trajectory; see also Supplementary Figure S3). (d) EMSA separation of the 3′-32P-labeled ribozyme using a non-cleavable (2′-O-methyl) or cleavable (2′-OH) synthetic RzB strand, and inactive (transcribed with A8) or active (synthetic with fluorophores and G8) RzA strand, with and without an 8 h preincubation in native buffer. Two bands, termed T and B, are observed for ribozyme containing non-cleavable RzB (45% B) or inactive RzA (66% B). Cleavable ribozyme shows cleavage and subsequent product release for ∼60% of the material during electrophoresis (with <10% B band remaining). Preincubation of the ribozyme results in additional cleavage (>70%), consistent with the final cleavage extent in solution.
Here, we use smFRET, gel electrophoresis, footprinting and mass spectrometry analyses that, taken together, demonstrate that the hairpin ribozyme's observed heterogeneity reflects a deeply furrowed folding landscape that is intrinsic to the RNA. We find that commitment to one of several functional folds occurs unexpectedly high on the RNA folding-free energy landscape, resulting in partially irreversible folding. Ensemble techniques cannot easily differentiate between rapidly interchanging RNA conformers and those that are separated by deeply dividing energy barriers. The intrinsic nature of such deeply dividing energy barriers within a population of functional RNAs for even a relatively small tertiary RNA fold thus has important ramifications for the interpretation of ensemble-averaged RNA structure and function data. Finally, our observations suggest that it may not be necessary, or even possible, to completely avoid folding heterogeneity in the biological evolution and human design of functional RNAs.
MATERIALS AND METHODS
RNA preparation
Synthetic RNA oligonucleotides were purchased from the Howard Hughes Medical Institute Biopolymer Keck Foundation Biotechnology Resource Laboratory (Yale University, New Haven, CT, USA), deprotected and purified by 20% (w/v) denaturing polyacrylamide gel electrophoresis (D-PAGE), in 8 M urea, 2 mM EDTA and 89 mM Tris–Borate and C8-reverse-phase HPLC chromatography, as described previously (20,29).
Transcribed RNA was generated in vitro from partially double-stranded DNA containing a T7 promoter as detailed in Supplementary Data. The RNA sequences are strand RzA (5′-AAA UAG AGA AGC GAA CCA GAG AAA CAC ACG CCA AA-3′), where the transcribed RzA contains one additional G on the 5′-end and the underlined G (G8) was mutated to an A (A8) to inactivate self-cleavage during electrophoretic mobility shift assay (EMSA) of fully transcribed ribozyme. In strand RzB (5′-AU AUA UUU GGC GUG GUA CAU UAC CUG GUA CCC CCU CGC AGU CCU AUU U-3′) a 2′-O-methyl modification is present at A-1 (underlined) for the non-cleavable construct used in most of our assays. The sequence in bold is absent from the synthetic construct prepared for mass spectrometry.
Cleavage assays
Single-turnover ensemble cleavage assays were carried out in 50 mM Tris–HCl (pH 7.5), 12 mM MgCl2, over a 26 h time course at 25°C. Ribozyme (final concentration 100 nM strand RzA with a domain terminal donor–acceptor (Cy3–Cy5) fluorophore pair and <10 nM 3′-32P radiolabeled strand RzB with a 5′-biotin) was pre-annealed by heating to 70°C for 2 min and slow cooling over 5 min to room temperature. After pre-incubation for 15 min at 25°C, 1/10th volume of 10× standard buffer was added to initiate the reaction. Two microliter reaction aliquots were taken at appropriate time intervals and quenched with 13-μl stop solution (80% formamide, 0.025% xylene–cyanol, 0.025% bromophenol blue and 10 mM EDTA). The 3′-cleavage product was separated from uncleaved RzB by 20% (w/v) D-PAGE, in 8 M urea, 2 mM EDTA and 89 mM Tris–Borate. The extent of cleavage was quantified, normalized to the sum of the substrate and product bands, and averaged over three separate cleavage assays using a PhosphorImager Storm 840 with ImageQuant software (Molecular Dynamics). Product formation was fit to the double-exponential first-order rate equation y(t) = y0 + A1(1−e–kobs1*t) + A2(1−e–kobs2*t), where A1 + A2 is the final extent of cleavage and kobs1 and kobs2 are the rate constants.
EMSA
The two strands of the hairpin ribozyme were heat annealed in native buffer containing 50 mM Tris–Acetate (pH 7.5) and either 12 mM Mg-Acetate or 50 mM Na-Acetate, and 10% (v/v) glycerol. Annealed samples were loaded onto a 10% polyacrylamide (19:1 acrylamide:bisacrylamide ratio) gel with 50 mM Tris–Acetate (pH 7.5) and 12 mM Mg-Acetate. For the comparison of cleavable and non-cleavable material the ribozyme was annealed in 50 mM Na-Acetate, and 1 mM Na-EDTA to prevent cleavage prior to electrophoresis. Non-denaturing gels were run at 20 V/cm at 4°C for 14–16 h. Additional details including the quantification of fluorophore labeled RNA bands using a FluorImager are described in Supplementary Data.
D-PAGE refolding
Strands RzA and RzB were annealed at a concentration of 5.7 µM in annealing buffer containing 50 mM Tris–HCl (pH 7.5), 50 mM Na-Acetate and 5 mM Na-EDTA, with trace amounts of 5′-32P radiolabeled strands RzA and RzB. The annealing solution was heated to 70°C for 2 min before cooling to room temperature over 15 min. Annealed samples were then subjected to EMSA and imaged through autoradiography. The samples were eluted by diffusion into a volume approximately three times larger than the excised band containing 50 mM Na-EDTA (pH 8.0). The sample was then precipitated by adding 1/10th volume of 3 M Na-acetate (pH 5.2) and another 2.5 volume of 100% cold ethanol followed by centrifugation. The dried samples were resuspended in buffer containing 50 mM Tris–Acetate (pH 7.5), 50 mM Na-Acetate, 1 mM Na-EDTA, 10% (v/v) glycerol and subjected to 20% (w/v) D-PAGE, in 8 M urea, 2 mM EDTA and 89 mM Tris–Borate for 3 h at ∼55°C and imaged by autoradiography. The four resulting samples were eluted by diffusion into a volume of water approximately three times larger than the excised band. The sample was then precipitated by adding 1/10th vol of 3 M Na-acetate (pH 5.2) and another 2.5 volume of 100% cold ethanol, followed by centrifugation. The samples were resuspended and annealed at 70°C in 50 mM Tris–HCl (pH 7.5), 50 mM Na-Acetate and 1 mM Na-EDTA. These samples were again subjected to EMSA, then imaged and quantified using a PhosphorImager Storm 840 with ImageQuant software.
tr-FRET
Separated material was eluted from the EMSA gel through diffusion into a standard buffer containing 12 mM MgCl2 50 mM Tris–HCl (pH 7.5) and then further exchanged into the same standard buffer using Nap-10™ SephadexTM columns. A frequency-doubled Nd:YVO4 laser (Spectra-Physics Millenia Xs-P, operated at 9.0 W) pumped a frequency-doubled, mode-locked Ti:sapphire laser (Spectra-Physics Tsunami, operated at 1 W) that excited fluorescein at 490 nm with 2 ps width pulses, picked down to 4 MHz. Isotropic emission was detected at 520 nm (10 nm band-pass interference filter) in 4096 sampling channels, with a time increment of 12 ps/channel, up to >40 000 peak counts, as previously described (30). Fluorescence decays were collected in the presence and absence of the acceptor fluorophore. The effect of the acceptor on the decay of fluorescein emission in the doubly labeled complex was then used to determine a distance distribution between the two fluorophores as previously described (30). A Förster distance of 55 Å and an orientation factor κ2 = 2/3 were used to determine distances.
sm-FRET
For single molecule measurements of the unseparated ribozyme the RzA and RzB strands were annealed at a concentration of 500 nM. The annealing buffer containing 50 mM Tris–HCl (pH 7.5) 100 mM NaCl, and 1% 2-mercaptoethanol. The annealing solution was heated to 70°C for 2 min before cooling to room temperature over 15 min. Separated material was eluted from the EMSA gel into a buffer containing 50 mM Tris–HCl (pH 7.5), and 12 mM MgCl2. Ribozyme samples were diluted to a concentration of ∼25 pM and bound to a streptavidin-coated quartz slide utilizing the biotin–streptavidin interaction as previously described (3,10,12). Fluorescence signals from donor and acceptor fluorophores were measured with either 1-s time resolution for non-cleavable EMSA separated ribozyme or 100 ms for cleavable multi-turnover ribozyme, using a total internal reflection fluorescence microscope as described (3,10,12). Single molecule measurements for non-cleavable ribozyme were carried out in 50 mM Tris–HCl (pH 7.5), 12 mM MgCl2, at 22°C, whereas multiple-turnover measurements of the cleavable ribozyme were carried out in 12, 100 or 500 mM MgCl2 with an excess concentration of 1 nM substrate (the 500 mM MgCl2 measurements are shown in Figure 1c). An oxygen scavenger system (10% glucose, 2% β-mercaptoethanol, 750 µg/ml glucose oxidase and 90 mg/ml catalase) was used to slow down fluorophore photobleaching. Dwell times for the (un)docked states and their relative fractions were analyzed as detailed in Supplementary Data.
Footprinting
Tb3+-mediated footprinting (31) was used to partially degrade ribozyme samples separated by EMSA. 32P-labeled ribozyme was separated by the EMSA and located using autoradiography. The material was cut out of the gel and eluted with the addition of buffer containing 50 mM Tris–HCl (pH 7.5), 10 mM Na-EDTA, 50 mM NaCl in a volume approximately three times larger than the excised band. The buffer was then exchanged for 50 mM Tris–HCl (pH 7.5), 50 mM NaCl, using a Nap-10™ Sephadex™ column. Samples were concentrated using Centricon centrifugal filters at 4°C. This preparation does not result in the redistribution of either T or B species as judged by EMSA. The radiolabeled RNA (∼150 000 c.p.m.) was next incubated with RNase or Tb3+ to achieve partial degradation. Tb3+ footprinting was carried out by adding 0.1, 1 and 5 mM Tb3+ to the RNA samples and incubating at 25°C for 1 h. The reaction was quenched with 50 mM EDTA in 20% (v/v) formamide, and immediately analyzed by 20% (w/v) D-PAGE, in 8 M urea, 2 mM EDTA and 89 mM Tris–Borate, alongside T1 and alkaline ladders. The results from the reactions with 5 mM Tb3+ were used for Π analysis (31). For RNase V1 footprinting, the ribozyme was prepared as in Tb3+ footprinting but incubated instead with RNase V1 at room temperature for 15 min. Mg2+ was added to the V1 reaction along with the RNase to a final concentration of 100 µM. A range of RNase concentrations were used to determine optimal concentration for partial degradation. Degradation patterns were analyzed as described in Supplementary Data.
Mass spectrometry
For mass spectrometry a construct in which helix four is truncated was annealed at a concentration of ∼100 µM and separated by EMSA. Separated RNA was located using UV shadowing and eluted from the gel into a buffer containing 10 mM Na-EDTA, 50 mM Tris–HCl (pH 7.5). The RNA was then concentrated using Centricon Plus-20™ and further separated for 3 h using 20% (w/v) D-PAGE, in 8 M urea, 2 mM EDTA and 89 mM Tris–Borate. The samples were again imaged through UV-shadowing and eluted into 50 mM Tris–HCl (pH 7.5), 1 mM Na-EDTA. The RNA was desalted three times and concentrated >100-fold using Centricon Plus-20™ devices according to the manufacturer's instructions, followed by further desalting using Centricon YM-3™ filters according to the manufacturer's instructions. If significant salt adduction was still observed in mass spectra, additional desalting was performed by ethanol precipitation [once or twice; protocol modified from (32)]. For this purpose the RNA was dissolved in water and 1/3 volume of 10 M ammonium acetate (pH 8) and another 2.5 volume of 100% cold ethanol (stored at –80°C until right before use) was added. The mixture was vortexed for a few seconds and stored at –80°C for 3 h followed by centrifugation at 12 500 r.p.m. for 15 min. The supernatant was decanted and 400 µl cold 70% (v/v) ethanol was added to the precipitate, followed by another incubation at –80°C for 2 h. Centrifugation was once again performed at 12 500 r.p.m. for 15 min, the supernatant was decanted and the precipitate was dried down and re-suspended in 50 mM ammonium acetate with 25% (v/v) methanol. Desalting of the ribozyme does not result in redistribution of the B species as determined by EMSA. Reduction of salt adduction is essential since the presence of multiple salt adducts complicates spectra and spreads the total signal over multiple peaks, thereby significantly reducing signal-to-noise ratios.
Samples were electrosprayed at 50 µl/h through an external Apollo II ion source equipped with dual ion funnels in negative ion mode and analyzed by an actively shielded 7 T quadrupole-Fourier transform ion cyclotron resonance mass spectrometer (Bruker Daltonics, Billerica, MA, USA) as described previously (33,34). All mass spectra were acquired with XMASS (version 6.1, Bruker Daltonics) in broadband mode from m/z 500 to m/z 2500 with 512 k data points and summed over 32 or 64 scans. Data processing with one zero fill but no apodization was performed with the MIDAS analysis software (35) prior to fast Fourier transformation. Due to the high molecular weight of the RNAs, average rather than monoisotopic mass was used for spectral interpretation. Mass-to-charge (m/z) values for each observed charge state were calculated from a weighted average of all observed isotopic peaks. Masses were calculated from a weighted average of all charge states. Spectra were externally calibrated using a calibration standard (G2421A; Agilent Technologies, Palo Alto, CA, USA) and a two-term frequency-to-mass calibration equation (36).
RESULTS
Both synthetic and transcribed hairpin ribozymes harbor multiple, catalytically active molecular species
To examine the persistent folding heterogeneity of the hairpin ribozyme, we explored an ensemble EMSA to isolate molecular sub-populations based on their average hydrodynamic radii. During EMSA a catalytically inactivated two-way junction (2WJ) construct (Figure 1a) migrates in two bands of distinct interdomain docking equilibria (Figure 1d). This heterogeneous mobility is observed over a wide range of annealing protocols (Figure 2), in the presence or absence of a domain terminal fluorophore pair, and independent of whether catalysis is blocked by a synthetic 2′-O-methyl modification of the cleavage site A-1 or a ribozyme G8A mutation in fully in vitro transcribed material (Figures 1d, 2, 3a and 4c). The corresponding cleavable 2WJ ribozyme cleaves to 77% completion with biphasic rate constants of 0.08 and 0.01 min−1 (Figure 1b), which have been linked to more and less stably docking sub-populations, respectively (3,12). During EMSA the cleavable 2WJ ribozyme cleaves to >60% completion (Figure 1d), thereby demonstrating native folding during EMSA and necessitating inactivating modifications for the preservation, isolation and further characterization of the two bands. Additionally, we examined smFRET trajectories of a trans-cleaving t-2WJ construct where the substrate strand is separated from the remainder of the RzB strand, such that the products generated upon backbone cleavage rapidly dissociate, giving rise to a distinct FRET signal of the substrate-free ribozyme after catalysis (Figure 1c). We find that smFRET trajectories of these t-2WJ ribozymes with distinct undocking behaviors repeatedly show transitions to the self-cleaved, intermediate FRET state (Figure 1c). We therefore infer that: (i) multiple, catalytically active molecular species exist within both synthetic and transcribed hairpin ribozyme preparations (source independence), and (ii) our catalytically inactivating modifications allow us to separate molecular sub-populations for further study that reproduce the heterogeneous behavior of the catalytically active ribozyme.
Figure 2.
Multiple annealing conditions resulting in similar EMSA distributions. In the presence of monovalents alone (50 mM NaCl) or in the presence of divalents (12 mM MgCl2) 32P-labeled ribozyme (fully synthetic 2′-O-methyl construct used for mass spectrometry analysis) was subjected to the following annealing protocols: (1) 25°C for 20 min; (2) 70°C, 2 min, 25°C, 20 min; (3) 90°C, 2 min, 25°C 20 min; (4) 90°C, 2 min, 70°C, 2 min, 65°C, 2 min, 25°C, 20 min; (5) 90°C, 2 min, 25°C, 10 min repeated three times, then 70°C, 2 min, 25°C, 10 min repeated two times; (6) 90°C, 2 min, 55°C, 30 min, 25°C, 20 min. The distribution between T and B species was then analyzed by EMSA.
Figure 3.
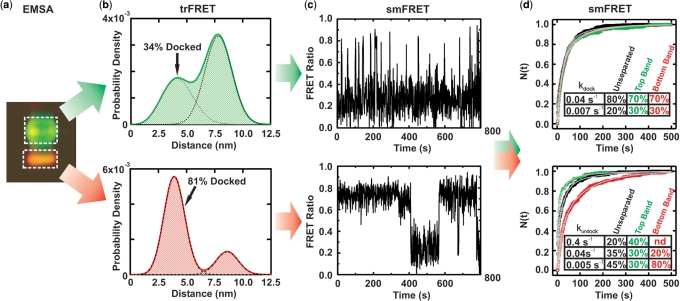
FRET characterization of EMSA separated ribozyme. (a) EMSA-based separation of fluorophore-labeled ribozyme showing differences in gel mobility and FRET (pseudo-color, green donor, red acceptor). RNA from the boxed areas was isolated as T and B species for further analysis. (b) Probability distribution of inter-fluorophore distance in the T and B species as derived by trFRET analysis, with fraction of docked material indicated. (c) Exemplary time trajectories characteristic of the sub-populations that are most enriched in the isolated T and B species. (d) The T and B species exhibit significantly different undocking kinetics in single molecule dwell-time analyses. The normalized fraction of dwell times (N) shorter than time (t) in the undocked (top) and docked states (bottom) are plotted for the isolated T (green) and B (red) species as well as the unseparated ribozyme (black). Solid gray lines indicate the multi-exponential global fits used to determine the (un)docking rate constants; the time-window corrected fraction of molecules (un)docking with the given rate constant is indicated in the table insets.
Figure 4.
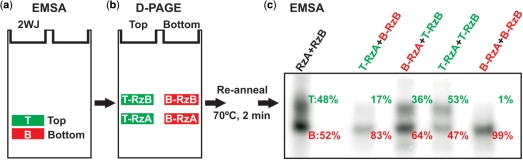
Extraordinary stability of molecular sub-populations of the hairpin ribozyme. Following EMSA separation (a) of fully in vitro transcribed ribozyme into two bands, the T and B species were further separated by D-PAGE (b) into their composite RzA and RzB strands. The four samples T-RzA, T-RzB, B-RzA and B-RzB were then annealed in all four possible combinations to refold the ribozyme, and analyzed by EMSA (c). An image of the second EMSA gel is shown with the fraction of T and B species for each lane indicated to the right of the respective band.
EMSA allows for isolation of molecular species that can be further characterized by ensemble and single molecule FRET
To further examine the underlying molecular sub-populations, we eluted the top (T) and bottom (B) bands of donor–acceptor (fluorescein–tetramethylrhodamine) labeled hairpin ribozyme separately from the EMSA gel (Figure 3a). The isolated T and B species were subjected to time-resolved FRET (trFRET) analysis in native buffer to probe their ensemble-averaged donor–acceptor distance distributions. This analysis shows that both species access the docked conformation, but to different extents (Figure 3b): The more slowly migrating T species is dominated by the extended undocked fold, whereas the faster migrating B species is governed by the more compact docked fold (Figure 3b). These findings reveal a surprising longevity of the observed folding heterogeneity, throughout the isolation process.
Next, we employed smFRET using doubly (Cy3 and Cy5) labeled, EMSA purified ribozyme species, which demonstrates that the T species is enriched in the more rapidly undocking ribozyme sub-populations, while the B species is enriched in the more slowly undocking sub-populations (Figure 3c). Quantification of the relative ribozyme fractions with distinct (un)docking rate constants in the T and B species confirms this qualitative assessment (Figure 3d). Notably, smFRET resolves several molecular sub-populations in both EMSA species, highlighting the enhanced resolution of the single molecule approach; complementarily, ensemble EMSA allows us to physically isolate and individually characterize two kinetically distinct, enriched ribozyme species.
The observed molecular heterogeneity remains associated with the separated composite RNA strands of each EMSA species
To further characterize the thermodynamic stability of the hairpin ribozyme's molecular sub-populations, we isolated T and B species from in vitro transcribed ribozyme by EMSA and then physically separated the two composite strands (RzA and RzB, Figure 1a) by denaturing polyacrylamide gel electrophoresis (D-PAGE, which melts all four helices of the ribozyme), followed by ethanol precipitation and re-suspension in native buffer (Figure 4a and b). The RzA and RzB strands from the T and B species were then recombined in all four possible permutations, heat annealed and analyzed by EMSA (Figure 4c). We find that this refolding protocol results in the expected redistribution of the annealed T-RzA/T-RzB ribozyme. Strikingly, no significant redistribution of the annealed B-RzA/B-RzB ribozyme occurs, which continues to form the dominantly docked B species. Additionally, the mixed (heterologous) T-RzA/B-RzB and B-RzA/T-RzB ribozymes redistribute only partially. These observations suggest that, after folding into the stably docked B species, both the RzA and RzB strands retain a tendency to form the more active B species throughout an isolation procedure that subjects them to harshly denaturing conditions (including >2 h in 8 M urea at ∼55°C during D-PAGE). In the presence of an excess of either fresh RzA or RzB, redistribution of both the isolated T and B species can be achieved, via either heating (>50°C for 2 min) (Supplementary Data and Supplementary Figure 1) or DNA-mediated strand removal using synthetic donor–acceptor labeled ribozyme (Supplementary Data and Supplementary Figure 2). Interconversion in the presence of excess RzA or RzB is likely trivial in nature as it allows one of the two strands in the ribozyme to be replaced, however it does provide further evidence that both strands contribute to the persistent heterogeneity. Conversion of T to B species via heating alone can be achieved in the absence of excess RzA or RzB, but we do not observe conversion of B to T, consistent with our D-PAGE refolding assay (data not shown). Additionally, we observe retention of folding heterogeneity following incubation at 37°C for 4.5 h in a DNA-mediated interconversion assay (Supplementary Data and Supplementary Figure 2), addition of a large molar excess of EDTA followed by exchange into Mg2+-free buffer in preparing material for footprinting, and after extensive desalting required for mass spectrometric analysis, as judged by EMSA. Furthermore, we observe that the sub-populations of the catalytically active trans-cleaving t-2WJ ribozyme retain distinct undocking kinetics following product dissociation, which disrupts all inter-strand contacts in domain A (Figure 1c). Such a pronounced robustness of the observed molecular heterogeneity [‘molecular memory’ (3)] is consistent with retention of the molecular sub-populations over a wide range of ionic conditions (Supplementary Data and Supplementary Figure 3) (10) and upon introduction of various modifications into the ribozyme (12). It is also in agreement with observations of persistent molecular heterogeneity in both less (15) and more complex tertiary folded RNAs (7,8,13).
Footprinting reveals subtle structural differences between the EMSA species
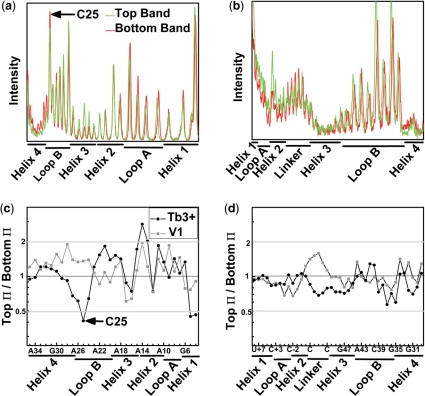
The stability of RNA helices, and the propensity to form alternate base pairing patterns, has previously been invoked as the source of the rugged folding pathways characteristic of RNA (6–8). To probe for even subtle secondary structure differences between the T and B species at single nucleotide resolution, we employed Tb3+ (24) and RNase V1 footprinting (37) to monitor single- and double-stranded RNA segments, respectively. To avoid trivial differences in footprinting pattern due to their distinct docking equilibria, we compared the T and B species side-by-side at low ionic strength, i.e. in the absence of docking (10), where heterogeneity is conserved. The Tb3+ and RNase V1 footprinting patterns are both strikingly similar for the T and B species, suggesting identical secondary structures (Figure 5). This finding is consistent with the paradoxical observation of extensive native structure within a deeply misfolded and partially active conformer of the Tetrahymena group I intron ribozyme, which was proposed to represent a topological isomer of the native state (7,8). For the hairpin ribozyme, however, the presence of topological isomers is inconsistent with our refolding assays, which demonstrate that the persistence of folding heterogeneity is not a consequence of strand entanglement. The most significant, albeit subtle footprinting difference between the hairpin ribozyme's T and B species we find at nucleotide C25 in loop B, which is involved in the critical G+1:C25 docking contact (10,12) (Figure 1a). The B species is slightly more sensitive to Tb3+-mediated scission at C25 than the T species (Figure 4), suggesting a possible structural basis for the different folding behaviors of the T and B species: a local, persistent perturbation in the loop B structure (23).
Figure 5.
Footprinting reveals identical secondary structures in the EMSA separated ribozyme species. Profiles of the Tb3+-mediated cleavage patterns of 32P-radiolabeled strand RzA (a) and strand RzB (b). The high degree of similarity between the T (top) and B (bottom) species indicates a shared secondary structure and suggests that the two species do not arise from gross misfolding of the ribozyme. Ratio of Π values, which represent averaged and normalized fractions of cleavage relative to background, are given for strands RzA (c) and RzB (d). Values <0.5 (gray line) indicate 2-fold greater sensitivity to Tb3+ (circles) or RNase V1 (squares) induced cleavage in the B species. Values >2 (gray line) indicate an at least 2-fold greater sensitivity in the T species.
High-resolution mass spectrometry provides no evidence for chemical modification of the RNA
The difficulty to convert the more stably docking B species into the T species raises the possibility that the observed molecular heterogeneity originates from a chemical modification, such as retention of a protection group from chemical synthesis that favors the most stably docked sub-population(s). The fact that we observe T and B species in both chemically synthesized (Figures 1c, d, 2 and 3a) and in vitro transcribed (Figures 1d and 4) ribozyme preparations, provides evidence against some of these artifactual origins of molecular heterogeneity. To directly probe for even subtle chemical heterogeneity, we subjected the isolated RzA and RzB strands from both the T and B species to high-resolution electrospray ionization (ESI) hybrid quadrupole-Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometry (34). For all four RNA samples, we observe the masses predicted from their complete RNA sequence, with <1 Da error, providing strong evidence against the notion that a stable adduct or chemical modification causes the observed persistent molecular heterogeneity (Figure 6). No additional peaks are observed in the B species, and low abundance peaks observed in the T species are of insufficient abundance and mass to account for the observed heterogeneity (see later). We cannot rule out, however, that the RNA is modified in a mass-neutral fashion, or that an RNA adduct forms that is stable under all solution conditions but dissociates in the gas phase of our mass spectrometry analysis.
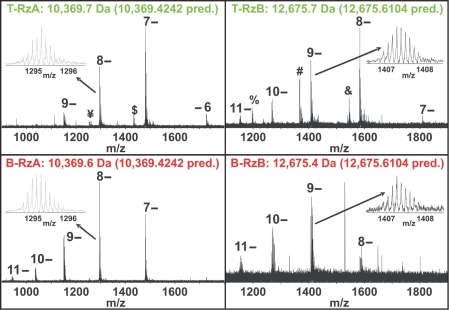
Figure 6.
FT-ICR mass spectrometry analysis of EMSA separated ribozyme. Negative mode ESI FT-ICR mass spectra show peaks generated from intact molecular ions of both RzA and RzB at different charge states as indicated, as well as additional peaks with relatively low abundance. The insets show zoomed in spectra of the 8-charge state for the RzA strand and the 9- charge state for the RzB strand. The experimental average masses are given above the spectra and the predicted masses are indicated in parentheses. Minor peaks ¥ and $ represent the 8- and 7-charge states, respectively, of a fragment resulting from loss of a single adenosine monophosphate from the 5′-end of T-RzA; minor peak & represents the 8-charge state of a fragment resulting from loss of a single uridine from the 3′-end of T-RzB; minor peaks % and # represent the 8- and 7-charge states, respectively, of the 5′-fragment resulting from backbone cleavage between C-2 and A-1.
Low abundance peaks in the T-RzA and T-RzB samples of Figure 6, marked ‘$’, ‘¥’ and ‘&’, represent material that may have been present during EMSA, although backbone cleavage could also occur during the ionization process of mass spectrometry, or more generally at some point following elution from the final denaturing gel. In the T-RzA sample, the low abundance peaks marked ‘$’ and ‘¥’ represent material that is missing adenosine monophosphate on the 5′-end of the RzA strand. Even if this chemical heterogeneity was present during EMSA, the 5′-nucleotide of RzA is at the end of helix 1 and thus remote from the docking loops A and B and the helical junction (Figure 1). In fact, in EMSA and single-molecule experiments using fluorophore-labeled ribozyme the donor fluorophore is present at this position, therefore in these experiments heterogeneous FRET-detected EMSA mobility can only be observed for ribozyme containing the full length RzA. In addition, the ‘$’ and ‘¥’ peaks constitute only ∼5% of the total signal, so are highly unlikely to induce the observed molecular heterogeneity.
In the T-RzB sample, the low abundance peak marked ‘&’ represents uridine loss from the 3′-end of RzB, also at the end of helix 1. The abundance of this peak also constitutes only ∼5% of the total signal. If we assume similar ionization efficiencies between the full length and the 1-nt (one-nucleotide) shorter species ‘$’, ‘¥’ and ‘&’, which is expected due to their very similar size and composition, then the maximum fraction of shorter ribozymes within the T species (either in the RzA or RzB strand) that our mass spectrometry is consistent with is 10%. If chemical modifications were the cause for the appearance of the T species, our mass spectrometry analysis would leave 90% of the chemical modifications of the T species unaccounted for. While it is possible that there are additional modifications of sufficiently low abundance that have a large cumulative effect but escape detection by mass spectrometry, we do not favor this interpretation based on the body of contradicting evidence described in the discussion section and throughout the results section.
The two minor peaks labeled ‘%’ and ‘#’ (Figure 6) represent the 8- and 7-charge states, respectively, of a significantly smaller RNA fragment than the main peak (smaller by 3 097 Da, i.e. 10 nt). It is the 5′-fragment of backbone cleavage between C-2 and A-1 that must have occurred at some point after EMSA and elution from the final denaturing gel, which otherwise would easily have resolved such a large mass difference. It therefore cannot be responsible for the EMSA mobility differences. The size of the peak appears to represent a larger fraction of the signal than the other minor peaks (although still less than 1/3 of the total signal), however, the significantly smaller mass of this species likely leads to a higher ionization efficiency and thus over-representation in the overall signal.
Therefore, all additional peaks in the T-RzA and T-RzB samples are of low abundance and are the result of RNA that is only slightly degraded (−1 nt) at its very end (peaks ‘$’, ‘¥’ and ‘&’) or is a degradation product of such small mass (peaks ‘#’, ‘%’) that it must have been generated after EMSA and subsequent D-PAGE. We conclude that our mass spectrometry data are not consistent with the notion that chemical heterogeneity is responsible for the hairpin ribozyme's molecular heterogeneity. We also note that no signs of covalent modification were observed in a previous tandem mass spectrometry analysis of enzymatically digested hairpin ribozyme (38), and that molecular heterogeneity is observed independently of the buffers used for gel electrophoresis and biochemical assays (data not shown). Interestingly, the higher average charge state distributions observed in our ESI mass spectrometry for B-RzA and B-RzB relative to their T species counterparts is consistent with more stably folded conformations of the B species strands (39), although they could also be due to variations in concentration and desalting efficiency.
DISCUSSION
We show here that molecular heterogeneity is intrinsic to at least the hairpin ribozyme and common to both major routes for the in vitro preparation of RNA, chemical synthesis and in vitro transcription. We demonstrate that multiple folding sub-populations of a functional RNA are unexpectedly resistant to repartitioning following unfolding (denaturation). Furthermore, our mass spectrometry data indicate that the heterogeneity is encoded in the RNA fold and is not a result of chemical modification.
Additional, circumstantial evidence against chemical sources of molecular heterogeneity
Our EMSA analysis indicates that, if present, chemical modifications that destabilize the docked state to form the T species would need to occur either during or prior to chemical or enzymatic synthesis. This idea is rationalized as follows: If destabilizing chemical damage arose subsequent to synthesis, then we would expect to observe an accumulating conversion of B to T species following isolation of the T and B species. However, exposure of the B species to conditions identical to our handling prior to EMSA (including D-PAGE, and ethanol precipitation) does not result in (additive) conversion to T species, so that post-synthesis chemical damage cannot explain the observed heterogeneity. Furthermore, our re-folding assays are inconsistent with the presence of any form of destabilizing chemical damage that favors formation of the T species as the source of the EMSA heterogeneity. Our D-PAGE-based refolding assays demonstrate that strands B-RzA and B-RzB are both fully competent to reform the B species (Figure 4), so if chemical damage led to formation of the T species, then B-RzA and B-RzB have to be presumed undamaged (or at least not damaged in a way that destabilizes docking). We further infer that we would be able to interpret the fractions of T species observed in the heterologous mixtures of T-RzA+B-RzB and B-RzA+T-RzB as the fractions of damaged T-RzA and T-RzB strands, respectively. However, the sum of the fractions of top band observed for T-RzA+B-RzB and B-RzA+T-RzB is only 53% (17%+36%), far less than the 100% necessary to fully explain the T species in terms of chemical damage. From this argument follows that, if we assumed chemical damage to be responsible for the observed molecular heterogeneity, then our refolding assays would suggest that these chemical modifications be present in the B species and therefore fortuitously stabilize the docked (catalytically active) state. This possibility that chemical damage favors the predominantly docked and catalytically most active B species seems unlikely since nearly all chemical modifications of the hairpin ribozyme tested in previous studies significantly destabilize the docked state (12,22).
Potential role of S-turns in highly stable, functional RNA structures
Taking all of our observations together, the energy barrier to interconversion of the hairpin ribozyme's conformational isomers appears to approach or even exceed the barrier to backbone scission. The ergodic hypothesis implies that each functional molecule, given enough time and energy, faithfully reproduces the full range of conformational and functional behaviors observed in a snapshot of the ensemble (40,41). Our observations underscore that the notion of ergodic behavior, which is generally used as a basis to interpret ensemble-averaged data, cannot be safely assumed even for RNAs as simple as the 2WJ hairpin ribozyme. While there may still exist untested conditions under which the conformations do exchange, it appears unlikely that such conditions would be compatible with RNA chemical stability since all attempts at interconversion under conditions that are extreme and yet compatible with chemical stability provide no significant signs of conversion of the B to the T species, unless one of the composite RNA strands is exchanged for fresh material. (Please note that conversion of the T to the B species does appear to occur, Figure 4). Our experiments therefore suggest that a given functional hairpin ribozyme molecule in solution at standard pressure will undergo spontaneous degradation before it can sample all major populations present in a snapshot of the ensemble.
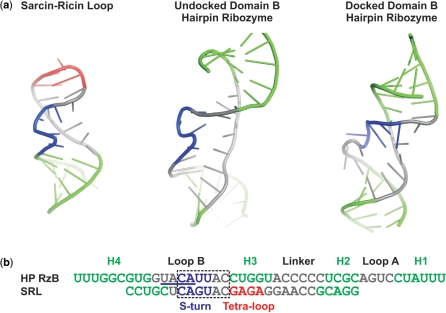
A plausible explanation for our observations is the retention of alternative, non-native, intra-strand structures that arise along the unfolding pathway of the T and B species and are resistant to unfolding under harshly denaturing conditions. While alternative explanations, including mass neutral chemical modifications such as the isomerization of the backbone to a 2′,5′-phosphodiester, cannot be ruled out entirely, we do not currently favor this interpretation due to the observed presence of T and B species in both synthetic and in vitro transcribed ribozymes. A strikingly similar phenomenon was observed in studies of the Sarcin–Ricin loop (SRL) where multiple populations in native gels cannot be interconverted, following even multiple cycles of heating to 90°C (42). The highly persistent heterogeneity in the SRL was unexpected for this small (23 nt) stem loop structure, and as with the hairpin ribozyme the heterogeneity is independent of how the RNA is synthesized. Interestingly, the SRL and loop B of the hairpin ribozyme share sequence homology and an S-turn structural motif, which changes shape in the hairpin ribozyme upon interdomain docking (Figure 7). The S-turn thus arises as a possible source of the deeply dividing energy barriers in both RNAs.
Figure 7.
Structural and sequence homology between the sarcin–ricin loop (SRL) and loop B of the hairpin ribozyme. (a) Cartoon representations of the crystal structure of the SRL (PDB ID 1Q9A), the undocked solution structure of the isolated domain B (PDB ID 1B36) and the crystal structure of domain B after docking with domain A (PDB ID 2OUE). Structural elements are color-coded as follows; helices are green, the GAGA tetra-loop is red, and the S-turn in the SRL and the corresponding nucleotides 39–42 in loop B are blue; in the undocked domain B structure these nucleotides compose the S-turn. The S-turn observed in the docked structure is formed by an alternative sequence (nucleotides 37–40). (b) Alignment of the sequence of the RzB strand used in this study with the SRL sequence from previous EMSA assays (42). The S-turns of the undocked domain B and SRL are aligned and color-coded as in (a) for comparison, whereas the S-turn sequence in the docked domain B is underlined.
One function, multiple folds
The capacity of RNA to adopt multiple non-exchanging functional conformations suggests that, in addition to the paradigm of one-fold-one-function (6), superior fitness of a specific RNA sequence may be achieved by its capability to adopt multiple folds that converge on a single function. Recent cross-linking experiments support the presence of multiple functional folds of the hairpin ribozyme (27). Cross-linking in the presence of cobalt–hexamine indicates the presence of U+2-G36 base stacking that is compatible with catalytic activity, but inconsistent with high-resolution X-ray crystal structures solved in the presence of cobalt–hexamine (28). The crystal structures and cross-linking data appear to reveal at least two significantly different conformations compatible with catalytic activity, consistent with our single-molecule FRET analysis of the catalytically active ribozyme (Figure 1c). Conformational differences around G36 could account for our footprinting data at C25, which is positioned in reasonable proximity to G36 across loop B. Our EMSA refolding experiments now show that the multiple functional conformations of the hairpin ribozyme are separated by unexpectedly high energy barriers.
Our experiments, in conjunction with the widespread observation of multiphasic and incomplete reaction kinetics of functional RNAs in ensemble-averaged experiments, as well as a growing body of evidence for heterogeneous single-molecule behavior, suggest that deeply furrowed free energy landscapes may be an inescapable feature of RNA in biology and biotechnology.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (GM62357 to N.G.W.); National Institutes of Health Molecular Biophysics Training Grant fellowship (to M.A.D.); NSF Career award (CHE-05-47699 to K.H.). Funding for open access publication charge: National Institutes of Health (GM62357).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Gregory Bokinsky and Hazen Babcock for their assistance in originally setting up our single molecule microscope, Daniel Herschlag, Xiaowei Zhuang, Steven Chu, Andrew Feig and Hashim al-Hashimi for helpful discussions, and an anonymous reviewer for making us aware of reference 42.
REFERENCES
- 1.Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- 2.Couzin J. MicroRNAs make big impression in disease after disease. Science. 2008;319:1782–1784. doi: 10.1126/science.319.5871.1782. [DOI] [PubMed] [Google Scholar]
- 3.Zhuang X, Kim H, Pereira MJ, Babcock HP, Walter NG, Chu S. Correlating structural dynamics and function in single ribozyme molecules. Science. 2002;296:1473–1476. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
- 4.Sosnick TR, Pan T. RNA folding: models and perspectives. Curr. Opin. Struct. Biol. 2003;13:309–316. doi: 10.1016/s0959-440x(03)00066-6. [DOI] [PubMed] [Google Scholar]
- 5.Pljevaljcic G, Klostermeier D, Millar DP. The tertiary structure of the hairpin ribozyme is formed through a slow conformational search. Biochemistry. 2005;44:4870–4876. doi: 10.1021/bi047772i. [DOI] [PubMed] [Google Scholar]
- 6.Schultes EA, Spasic A, Mohanty U, Bartel DP. Compact and ordered collapse of randomly generated RNA sequences. Nat. Struct. Mol. Biol. 2005;12:1130–1136. doi: 10.1038/nsmb1014. [DOI] [PubMed] [Google Scholar]
- 7.Russell R, Das R, Suh H, Travers KJ, Laederach A, Engelhardt MA, Herschlag D. The paradoxical behavior of a highly structured misfolded intermediate in RNA folding. J. Mol. Biol. 2006;363:531–544. doi: 10.1016/j.jmb.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 8.Bhaskaran H, Russell R. Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone. Nature. 2007;449:1014–1018. doi: 10.1038/nature06235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhuang X, Bartley LE, Babcock HP, Russell R, Ha T, Herschlag D, Chu S. A single-molecule study of RNA catalysis and folding. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 10.Bokinsky G, Rueda D, Misra VK, Rhodes MM, Gordus A, Babcock HP, Walter NG, Zhuang X. Single-molecule transition-state analysis of RNA folding. Proc. Natl Acad. Sci. USA. 2003;100:9302–9307. doi: 10.1073/pnas.1133280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okumus B, Wilson TJ, Lilley DM, Ha T. Vesicle encapsulation studies reveal that single molecule ribozyme heterogeneities are intrinsic. Biophys. J. 2004;87:2798–2806. doi: 10.1529/biophysj.104.045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rueda D, Bokinsky G, Rhodes MM, Rust MJ, Zhuang X, Walter NG. Single-molecule enzymology of RNA: essential functional groups impact catalysis from a distance. Proc. Natl Acad. Sci. USA. 2004;101:10066–10071. doi: 10.1073/pnas.0403575101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie Z, Srividya N, Sosnick TR, Pan T, Scherer NF. Single-molecule studies highlight conformational heterogeneity in the early folding steps of a large ribozyme. Proc. Natl Acad. Sci. USA. 2004;101:534–539. doi: 10.1073/pnas.2636333100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brender JR, Dertouzos J, Ballou DP, Massey V, Palfey BA, Entsch B, Steel DG, Gafni A. Conformational dynamics of the isoalloxazine in substrate-free p-hydroxybenzoate hydroxylase: single-molecule studies. J. Am. Chem. Soc. 2005;127:18171–18178. doi: 10.1021/ja055171o. [DOI] [PubMed] [Google Scholar]
- 15.Hodak JH, Downey CD, Fiore JL, Pardi A, Nesbitt DJ. Docking kinetics and equilibrium of a GAAA tetraloop-receptor motif probed by single-molecule FRET. Proc. Natl Acad. Sci. USA. 2005;102:10505–10510. doi: 10.1073/pnas.0408645102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Downey CD, Fiore JL, Stoddard CD, Hodak JH, Nesbitt DJ, Pardi A. Metal ion dependence, thermodynamics, and kinetics for intramolecular docking of a GAAA tetraloop and receptor connected by a flexible linker. Biochemistry. 2006;45:3664–3673. doi: 10.1021/bi0520941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu S, Bokinsky G, Walter NG, Zhuang X. Dissecting the multistep reaction pathway of an RNA enzyme by single-molecule kinetic "fingerprinting". Proc. Natl Acad. Sci. USA. 2007;104:12634–12639. doi: 10.1073/pnas.0610597104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pereira MJB, Nikolova EN, Hiley SL, Collins RA, Walter NG. Single molecule FRET microscopy of a complex ribozyme reveals dynamic hierarchical folding toward catalysis. J. Mol. Biol. 2008;382:496–509. doi: 10.1016/j.jmb.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ditzler MA, Aleman EA, Rueda D, Walter NG. Focus on function: single molecule RNA enzymology. Biopolymers. 2007;87:302–316. doi: 10.1002/bip.20819. [DOI] [PubMed] [Google Scholar]
- 20.Walter NG. Structural dynamics of catalytic RNA highlighted by fluorescence resonance energy transfer. Methods. 2001;25:19–30. doi: 10.1006/meth.2001.1212. [DOI] [PubMed] [Google Scholar]
- 21.Bokinsky G, Zhuang X. Single-molecule RNA folding. Acc. Chem. Res. 2005;38:566–573. doi: 10.1021/ar040142o. [DOI] [PubMed] [Google Scholar]
- 22.Walter NG, Hampel KJ, Brown KM, Burke JM. Tertiary structure formation in the hairpin ribozyme monitored by fluorescence resonance energy transfer. EMBO J. 1998;17:2378–2391. doi: 10.1093/emboj/17.8.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butcher SE, Allain FH, Feigon J. Solution structure of the loop B domain from the hairpin ribozyme. Nat. Struct. Biol. 1999;6:212–216. doi: 10.1038/6651. [DOI] [PubMed] [Google Scholar]
- 24.Walter NG, Yang N, Burke JM. Probing non-selective cation binding in the hairpin ribozyme with Tb(III) J. Mol. Biol. 2000;298:539–555. doi: 10.1006/jmbi.2000.3691. [DOI] [PubMed] [Google Scholar]
- 25.Rupert PB, Ferre-D’Amare AR. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 26.Rupert PB, Massey AP, Sigurdsson ST, Ferre-D’Amare AR. Transition state stabilization by a catalytic RNA. Science. 2002;298:1421–1424. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- 27.Lambert D, Heckman JE, Burke JM. Cation-specific structural accommodation within a catalytic RNA. Biochemistry. 2006;45:829–838. doi: 10.1021/bi0513709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salter J, Krucinska J, Alam S, Grum-Tokars V, Wedekind JE. Water in the active site of an all-RNA hairpin ribozyme and effects of Gua8 base variants on the geometry of phosphoryl transfer. Biochemistry. 2006;45:686–700. doi: 10.1021/bi051887k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walter NG. Curr. Protocols Nucleic Acid Chem. 2002. Probing RNA structural dynamics and function by fluorescence resonance energy transfer (FRET). Chapter 11.10, 11.10.1–11.10.23. [DOI] [PubMed] [Google Scholar]
- 30.Pereira MJ, Harris DA, Rueda D, Walter NG. Reaction pathway of the trans-acting hepatitis delta virus ribozyme: a conformational change accompanies catalysis. Biochemistry. 2002;41:730–740. doi: 10.1021/bi011963t. [DOI] [PubMed] [Google Scholar]
- 31.Harris DA, Tinsley RA, Walter NG. Terbium-mediated footprinting probes a catalytic conformational switch in the antigenomic hepatitis delta virus ribozyme. J. Mol. Biol. 2004;341:389–403. doi: 10.1016/j.jmb.2004.05.074. [DOI] [PubMed] [Google Scholar]
- 32.Limbach PA, Crain PF, McCloskey JA. Characterization of oligonucleotides and nucleic acids by mass spectrometry. Curr. Opin. Biotechnol. 1995;6:96–102. doi: 10.1016/0958-1669(95)80015-8. [DOI] [PubMed] [Google Scholar]
- 33.Yang J, Mo J, Adamson JT, Hakansson K. Characterization of oligodeoxynucleotides by electron detachment dissociation fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 2005;77:1876–1882. doi: 10.1021/ac048415g. [DOI] [PubMed] [Google Scholar]
- 34.Mo J, Hakansson K. Characterization of nucleic acid higher order structure by high-resolution tandem mass spectrometry. Anal. Bioanal. Chem. 2006;386:675–681. doi: 10.1007/s00216-006-0614-z. [DOI] [PubMed] [Google Scholar]
- 35.Senko MW, Canterbury JD, Guan S, Marshall AG. A high-performance modular data system for Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun. Mass. Spectrom. 1996;10:1839–1844. doi: 10.1002/(SICI)1097-0231(199611)10:14<1839::AID-RCM718>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 36.Ledford EB, Jr, Rempel DL, Gross ML. Space charge effects in Fourier transform mass spectrometry. Mass calibration. Anal. Chem. 1984;56:2744–2748. doi: 10.1021/ac00278a027. [DOI] [PubMed] [Google Scholar]
- 37.Huntzinger E, Boisset S, Saveanu C, Benito Y, Geissmann T, Namane A, Lina G, Etienne J, Ehresmann B, Ehresmann C, et al. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005;24:824–835. doi: 10.1038/sj.emboj.7600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas JM, Perrin DM. Active site labeling of G8 in the hairpin ribozyme: implications for structure and mechanism. J. Am. Chem. Soc. 2006;128:16540–16545. doi: 10.1021/ja063942y. [DOI] [PubMed] [Google Scholar]
- 39.Guo X, Bruist MF, Davis DL, Bentzley CM. Secondary structural characterization of oligonucleotide strands using electrospray ionization mass spectrometry. Nucleic Acids Res. 2005;33:3659–3666. doi: 10.1093/nar/gki671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Birkhoff GD, Koopman BO. Recent contributions to the Ergodic Theory. Proc. Natl Acad. Sci. USA. 1932;18:279–282. doi: 10.1073/pnas.18.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neumann JV. Physical applications of the Ergodic Hypothesis. Proc. Natl Acad. Sci. USA. 1932;18:263–266. doi: 10.1073/pnas.18.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Korennykh AV, Plantinga MJ, Correll CC, Piccirilli JA. Linkage between substrate recognition and catalysis during cleavage of sarcin/ricin loop RNA by restrictocin. Biochemistry. 2007;46:12744–12756. doi: 10.1021/bi700931y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.