Abstract
The serine recombinase Tn3 resolvase catalyses recombination between two 114 bp res sites, each of which contains binding sites for three resolvase dimers. We have analysed the in vitro properties of resolvase variants with ‘activating’ mutations, which can catalyse recombination at binding site I of res when the rest of res is absent. Site I × site I recombination promoted by these variants can be as fast as res × res recombination promoted by wild-type resolvase. Activated variants have reduced topological selectivity and no longer require the 2–3′ interface between subunits that is essential for wild-type resolvase-mediated recombination. They also promote formation of a stable synapse comprising a resolvase tetramer and two copies of site I. Cleavage of the DNA strands by the activated mutants is slow relative to the rate of synapsis. Stable resolvase tetramers were not detected in the absence of DNA or bound to a single site I. Our results lead us to conclude that the synapse is assembled by sequential binding of resolvase monomers to site I followed by interaction of two site I-dimer complexes. We discuss the implications of our results for the mechanisms of synapsis and regulation in recombination by wild-type resolvase.
INTRODUCTION
Site-specific recombinases promote controlled rearrangements of DNA sequences, by binding, cutting and rejoining the DNA strands at sites that they recognize. The capacity of site-specific recombinases to bring about insertion or excision of defined DNA segments has led to their widespread application in experimental genetics and biotechnology, with further potential uses in gene therapy (1,2). Members of one large family of these enzymes, the serine recombinases, have biological functions that include bacteriophage integration and excision, switching of bacterial gene expression by DNA inversion and transposon cointegrate resolution (3).
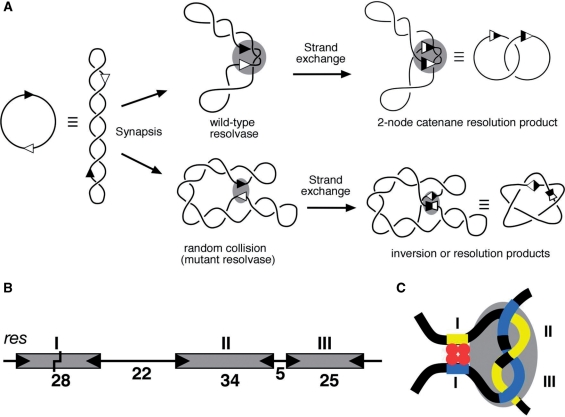
Much of our understanding of the mechanisms of serine recombinases has come from studies on the closely related cointegrate resolvases from the bacterial transposons Tn3 and γδ (4). Tn3 (or γδ) resolvase catalyses recombination between two 114 bp res sites (Figure 1A and B). Each res site contains binding sites for three resolvase dimers. The DNA strands are broken and rejoined at specific bonds near the centre of the 28 bp binding site I; catalysis of these reactions is by the subunits bound to site I. The ‘accessory’ binding sites II (34 bp) and III (25 bp) are also required for recombination. Resolvase dimers bound to sites II and III do not participate in the catalysis of strand cleavage and rejoining, but have an essential role in assembly of the synaptic complex (‘synapse’) of two res sites that is a prerequisite for strand exchange (Figure 1A and C). The synapse has regulatory functions which include restriction of recombination to pairs of res sites that are in the same orientation in a supercoiled DNA molecule, specification of a single round of recombination and specification of the 2-noded catenane recombinant product (5).
Figure 1.
Synapsis and recombination by Tn3 resolvase. (A) Cartoon illustrating synapsis and strand exchange of a supercoiled substrate by wild-type and activated resolvase mutants. The two sites (res or site I) are indicated by arrowheads. Synapsis of two res sites in head-to-tail orientation by wild-type resolvase traps three topological nodes (shaded oval), and strand exchange results in a 2-noded catenane resolution product. Activated mutant resolvases can synapse sites by random collision, giving rise to products of variable topologies; in the example shown, a 5-noded knot inversion product. (B) The Tn3 recombination site res. The boxes represent binding sites for dimers of resolvase, with binding motifs represented by the wedges at the ends of each box. The lengths of the DNA segments are indicated (base pairs). The point within site I at which resolvase breaks and rejoins the DNA is marked by a staggered line. (C) Cartoon showing the path of the DNA in the res × res synapse, and the relationship of the catalytic and regulatory parts. The four resolvase subunits forming the tetramer in the site I synapse component of this complex are shown in red, and the grey shaded oval represents the eight subunits presumed to be bound to the accessory sites II and III.
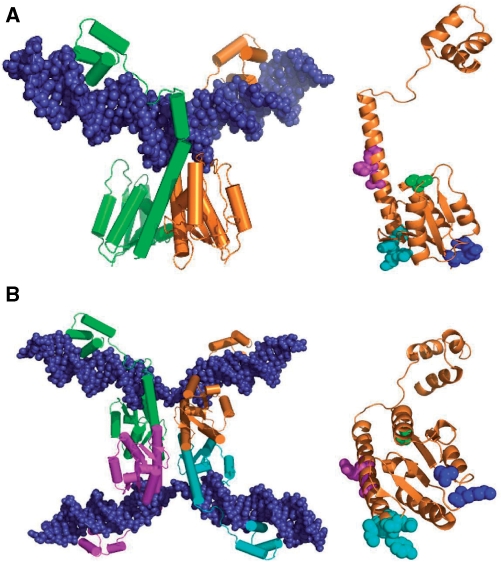
Tn3 resolvase is a 185-amino acid protein consisting of two domains. The C-terminal domains (∼45 amino acids) of resolvase dimers make sequence-specific interactions with the DNA at each end of binding sites I, II and III in res. The N-terminal domain contains the active site for catalysis of strand exchange, and all known interactions between subunits involve residues of this domain (Figure 2). The structure of the N-terminal domain of γδ resolvase, solved by X-ray crystallography (6–9), revealed two important types of interaction between subunits. The 1–2 or dimer interface mediates dimerization of resolvase in solution and when bound to DNA. The 2–3′ interface connects dimers; it has essential functions for wild-type resolvase in assembly of the synapse of two res sites and in activation of recombination (5,7,10). The 1–2 dimer is present in a structure of γδ resolvase bound to site I DNA (11; Figure 2A), but the 2–3′ interaction has not yet been observed in X-ray structures of complexes containing DNA.
Figure 2.
Resolvase structures and activating mutations. (A) Crystal structure of a wild-type γδ resolvase dimer bound to site I [1GDT; (11)]. On the right, residues relevant to the experiments presented here are shown in spacefill on the orange resolvase subunit (in the same orientation); S10 in green; R2 and E56 in blue; G101, E102, M103 and K105 in cyan; A117, R121 and E124 in magenta. The hydroxyl group of S10 is the catalytic nucleophile in recombination. The sidechains of R2 and E56 contribute to the 2–3′ interface (see Introduction section). The other highlighted residues are the sites of activating mutations of Tn3 resolvase. The γδ resolvase residues E102 and K105 correspond to D102 and Q105, respectively, in Tn3 resolvase. (B) Crystal structure of a synaptic tetramer of γδ resolvase with two cleaved site Is [1ZR4; (16)], and on the right the orange resolvase subunit in the same orientation, showing residues in spacefill as in (A), except that the following residues are mutant; A2, K56, S101, Y102, I103, Q124. The images were created with PyMol.
Resolvase variants with ‘activating’ mutations promote recombination at site I in the absence of the accessory binding sites of res (12,13). Certain combinations of activating mutations are more effective than any single mutations, and one double mutant (D102Y E124Q) was shown to have site I × site I recombination activity in vitro (12). The activating mutations affect residues at or close to surfaces involved in subunit interactions, in particular the dimer interface (13). One group of these residues is near the N-terminus of the long E-helix which lies on the dimer interface, and comprises G101, D102, M103 and Q105. M103 is the first residue of the E-helix (Figure 2A). Variants with combinations of activating mutations at these positions have high site I × site I activity in Escherichia coli (13), and have been used in recent analysis of the structures of recombination intermediates. Sarkis et al. (14) demonstrated the formation of a ‘crossover site synapse’ (herein referred to as a ‘site I synapse’) in which two copies of site I are held together by a tetramer of γδ resolvase with several activating mutations. The low-resolution structure of a site I synapse containing similar Tn3 resolvase mutants was solved by small-angle scattering combined with other biophysical techniques (15), and structures of site I synapses with cleaved DNA strands have been solved by X-ray crystallography (16,17; Figure 2B). In these structures, the site I DNA is on the outside of a tetrameric core of the resolvase catalytic domains. How the activating mutations promote the formation of the site I synapse remains unclear (see Discussion section). The conformation of the resolvase subunits in the crystallographic site I synapse is substantially different from that seen in earlier structures; the dimer interface has been radically altered, resulting in a flat hydrophobic interface between pairs of subunits in the tetramer. Strand exchange by resolvase is proposed to involve a relative rotation of the components of the cleaved site I synapse about this flat interface [16; reviewed by Grindley et al. (4)].
Deregulation of recombination by ‘activating’ mutations has been observed in other members of the serine recombinase family, including Sin resolvase (18,19), the DNA invertases Cin, Gin and Hin (20–22) and the bacteriophage φC31 integrase (23). These activated recombinase variants have potential uses as tools for genetic manipulation (2). Chimaeric recombinases comprising activated serine recombinase catalytic domains attached to zinc finger DNA recognition domains can promote recombination at sites recognized by the zinc finger domains, and might allow natural genomic sequences to be chosen as targets for insertions or deletions (24,25).
Despite the substantial interest in highly activated resolvase mutants, their functional properties in vitro have not yet been reported in any detail. Here, we describe the in vitro properties of a number of these mutants, focusing on a set of variants with mutations near the N-terminus of the E-helix, which have particularly striking-activating effects. We show that site I × site I cleavage and recombination by these mutants can be fast and efficient. The mutants have lost the strong topological selectivity of wild-type resolvase to varying extents, and make products via a random collision synapsis pathway. Increased site I × site I activity is correlated with increased stability of a site I synapse, formation of which is much faster than the subsequent DNA cleavage and strand exchange steps of the recombination reaction. We show that one very active resolvase variant is predominantly monomeric in solution, whereas it forms a stable tetramer within the site I synapse. Our results are consistent with synapsis by docking of two site I-bound dimers. The site I synapse component of the wild-type res × res synaptic complex might be formed by a similar pathway, promoted by contacts between resolvase subunits at site I and the accessory sequences.
MATERIALS AND METHODS
Plasmids
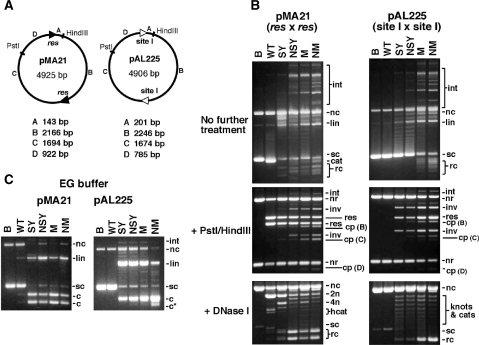
The pBR322 derivatives pMA21 and pAL225 contain two copies of Tn3 res and site I, respectively [Figure 3A; (26)]. Supercoiled plasmid DNA for in vitro assays was purified from transformed E. coli strain DS941 (27), using an alkaline lysis method followed by caesium chloride–ethidium bromide density gradient ultracentrifugation. DNA concentrations were estimated by measuring absorbance at 260 nm. Expression plasmids for resolvase mutants were derived from pSA1101 (12) by replacement of the wild-type resolvase reading frame with mutant versions obtained from previously described plasmids (12,13).
Figure 3.
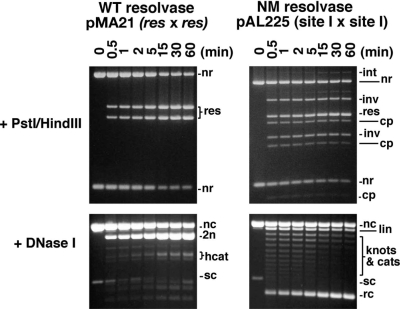
In vitro recombination by wild-type resolvase and activated mutants. (A) Plasmid substrates pMA21 (res × res) and pAL225 (site I × site I). DNA segments between the recombination sites and restriction sites are denoted with letters A–D, and their sizes are given below each diagram. (B) Reactions of pMA21 and pAL225 with Tn3 resolvase and mutants. Following incubation with resolvase for 1 h, each sample was divided into three equal aliquots and separated by agarose gel electrophoresis without further treatment (upper panels), or digested with PstI and HindIII (centre panels), or nicked with DNase I (lower panels; 0.7% gel). DNase I-nicked intermolecular recombination products, with low gel mobility, are not shown. Lanes marked B are controls (incubated without resolvase). (C) Cleavage of pMA21 and pAL225 by Tn3 resolvase and mutants in EG buffer, which contains 40% ethylene glycol. The gels in parts B and C are annotated as follows: sc, supercoiled plasmid substrate; nc, nicked circular substrate; lin, linearized substrate plasmid; cat, supercoiled catenane resolution product; rc, free circular resolution product; int, intermolecular recombinant products; nr, non-recombinant restriction fragment; res, restriction fragment from resolution product; inv, restriction fragment from inversion product; cp, restriction fragment from cleavage product; 2n, 4n, 2-noded or 4-noded nicked catenane resolution product; hcat, 2-node catenane product nicked in only one DNA circle; c, product of cleavage at both recombination sites; c*, product of cleavage at a non-site I position.
Purification of resolvases
Resolvase overexpression was induced by adding isopropylthio-β-galactoside to late log phase cultures of E. coli strain BL21(DE3)pLysS (28) transformed with pSA1101 or derivatives with mutant reading frames. The double mutant G101S D102Y (‘SY’) resolvase used here has a C-terminal hexahistidine tag, but all others do not. Purification was essentially as described in Arnold et al. (12) except that, for the mutant proteins, Tris–HCl was replaced by 50 mM sodium phosphate (pH 7.5) in the buffers used for the ion exchange steps. Resolvase purity was assessed by SDS–PAGE, and concentrations were estimated by comparison to a reference sample of resolvase which had been determined by amino acid analysis.
In vitro recombination and cleavage reactions
For a typical reaction, 2 µl of resolvase, diluted in 20 mM Tris–HCl (pH 7.5), 1 mM DTT, 0.1 mM EDTA, 1 M NaCl, 50% v/v glycerol was added to 20 µl of ‘reaction buffer’ containing 0.4 µg of plasmid DNA. Reaction buffer contains 10 mM MgCl2, 0.1 mM EDTA and 50 mM sodium phosphate (pH 6.0 or 7.0), or 50 mM Tris–HCl (pH 8.0 or 8.2), or 50 mM glycine–NaOH (pH 9.0 or 10.0). ‘Standard’ reactions were at pH 8.2. The final concentration of resolvase was 200 nM, and reactions were at 37°C for 1 h, unless stated otherwise. Assays of resolvase-mediated DNA cleavage were similar except that the plasmid DNA was in ‘EG buffer’ [50 mM Tris–HCl (pH 8.2), 0.1 mM EDTA, 40% v/v ethylene glycol]. Reactions were terminated by heating at 70°C for 5 min, or by adding loading buffer (for cleavage assays). The products were analysed by agarose gel electrophoresis (after digestion with restriction enzymes or DNase I if required). Loading buffer [50% v/v glycerol, 100 mM Tris–HCl (pH 7.5), 0.5% w/v sodium dodecyl sulphate (SDS), 1 mg/ml protease K, 0.5 mg/ml bromophenol blue] was added (25% of the sample volume) prior to loading on the gel. Gels were 1.2% (or 0.7% for nicked samples) agarose, in TAE buffer [40 mM Tris–acetate (pH 8.2), 20 mM sodium acetate, 1 mM EDTA], and were run horizontally in TAE buffer at ∼3 V/cm. The gels were stained with ethidium bromide (0.6 μg/ml) for 40 min, then photographed with a Canon EOSD30 digital camera, a 480 nm bandpass filter (Peca Products, Deloit, Wisconsin, USA) and 254 nm UV transillumination. DNase I nicking reactions were initiated by adding 1/10 volume of a solution containing 20 μg/ml DNase I, 3 mg/ml ethidium bromide, 50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 50% v/v glycerol to the reaction sample. Following incubation at 37°C for 1 h, the reaction was stopped by adding loading buffer (see above; 25% of sample volume). The sample was extracted with 1:1 phenol–chloroform, then chloroform, to remove ethidium prior to loading the samples on a 0.7% agarose gel.
Binding/synapsis assays
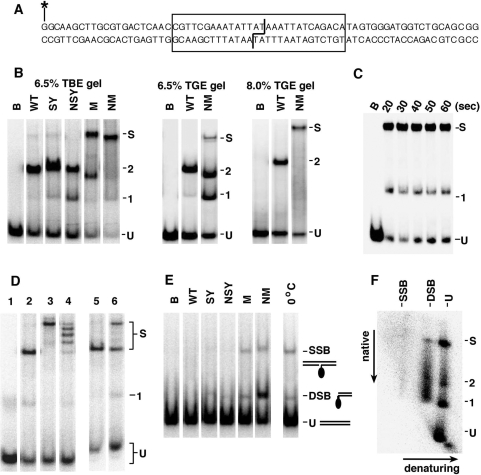
The procedure was modified from Arnold et al. (12) and Sarkis et al. (14). The top strand of the site I oligonucleotide (sequence shown in Figure 6A) was 32P-labelled at the 5′-end with T4 kinase, and annealed with an equimolar amount of unlabelled bottom strand. Diluted resolvase (2.2 µl) was added to 20 µl samples containing 52.5 nM site I DNA, 20 mM Tris–HCl (pH 7.5), 10 µg/ml poly(dI/dC) and 4% w/v Ficoll. The final resolvase concentration was typically 400 nM. Following incubation at 22°C for 10 min, the samples were cooled in ice for 10 min, then loaded onto a 6.5% polyacrylamide gel (30:0.8, acrylamide: bisacrylamide). The buffer in the gel and tanks was either tris–borate (TBE) (100 mM Tris base, 100 mM boric acid, 1 mM EDTA; pH ∼8.3) or tris–glycine (TGE) (50 mM Tris base, 10 mM glycine, 0.1 mM EDTA; pH ∼9.4). Gels were pre-run for 30 min at 200 V and 4°C. Samples were then loaded and the gels were run for a further 3 h at 200 V under the same conditions, without buffer recirculation. Gels were dried, and bands were visualized by phosphorimaging or autoradiography. To observe products containing resolvase–DNA covalent linkages, binding assay mixtures were treated with 0.1% SDS for 30 min, then loaded on gels as described above, except that the buffer was TBE plus 0.1% SDS. For the 2D gels (as in Figure 6F), a single sample was first separated on a gel with TBE buffer as described above. The gel was soaked in TBE plus 0.5% SDS for 1 h at room temperature, then in TBE plus 0.1% SDS for 1 h, then run in the perpendicular direction in TBE plus 0.1% SDS buffer.
Figure 6.
Binding and synapsis of site I by resolvase variants. (A) The site I-containing double-stranded oligonucleotide used in the binding and synapsis experiments. The site I sequence is boxed. The staggered line indicates the bonds cleaved by resolvase. The top strand was 5′-end labelled with 32P (asterisk). (B) Binding and synapsis by wild-type (WT) resolvase and activated mutants, assayed by non-denaturing polyacrylamide gel electrophoresis. Lanes marked B are controls (incubated without resolvase). Bands corresponding to unbound site I DNA, complexes containing 1 or 2 resolvase subunits and site I synapse are indicated by U, 1, 2 and S, respectively [band assignment based on (14,34)]. The SY resolvase used here has a C-terminal hexahistidine tag whereas all the other resolvases do not, which might account for the slightly lower mobilities of the SY complexes. Left panel; 6.5% polyacrylamide gel with TBE buffer. Centre panel; 6.5% gel with TGE buffer. Right panel; 8% gel with TGE buffer. (C) Time course of site I binding/synapsis by NM resolvase. Resolvase was added to samples at 0°C, and aliquots were withdrawn from the mixture at the stated times, then loaded on a running 6.5% TBE gel. (D) Assembly of the site I synapse with different sized resolvases. NM–GFP is NM resolvase with a 23 kDa GFP domain fused to the C-terminus. Binding/synapsis mixtures were separated on a 6.5% TBE gel. Lane 1, no resolvase. Lane 2, +NM resolvase. Lane 3, +NM–GFP resolvase. Lane 4, +a 1:1 mixture of NM and NM–GFP. Lane 5, NM resolvase (400 nM) was added to site I, at 22°C. After 5 min, NM–GFP (400 nM) was added, and the sample was loaded on the gel after a further 10 min. Lane 6, NM and NM–GFP resolvases were added separately to site I, and the two samples were mixed after 5 min at 22°C. The mixture was loaded on the gel after a further 10 min. (E) Cleavage of site I by activated resolvase mutants. Reactions were set up as in (B), and after 30 min at 37°C the resolvase was denatured with 0.1% SDS. Electrophoresis was on a 6.5% polyacrylamide gel in TBE buffer containing 0.1% SDS. The positions of bands containing unbound DNA (U), and DNA with a SSB DSB are indicated. A resolvase subunit is covalently attached to the SSB and DSB species. In the lane on the right, NM resolvase was added to site I at 0°C and 0.1% SDS was added after 5 min. (F) 2D analysis of site I synapse. A sample containing NM resolvase and site I was separated on a 6.5% polyacrylamide–TBE gel, then soaked in SDS-containing buffer to denature resolvase and run in a second dimension in the presence of SDS. Annotation of the gel is as in parts B and E.
Gel filtration chromatography
The multimeric states of wild-type and mutant resolvases in solution were examined by gel filtration chromatography using a Superose 12 10/300 GL column (Amersham Biosciences, Amersham, UK), run at room temperature on an AKTA Purifier HPLC system (Amersham Biosciences). The elution buffer contained 50 mM sodium phosphate (pH 7.5), 1 M NaCl, 5% v/v glycerol, 0.5 mM DTT and 0.1 mM EDTA. Samples (100 μl; resolvase concentration ∼65 μM) were loaded on the column, and elution (0.5 ml/min) was monitored by measurement of absorbance at 215 nm and 280 nm. Molecular weight was estimated by comparison of elution volume at the peak maximum to a standard curve (based on horse cytochrome c, 12.4 kDa; bovine carbonic anhydrase, 29 kDa; bovine serum albumin, 66 kDa; yeast alcohol dehydrogenase, 150 kDa; sweet potato β-amylase, 200 kDa). These proteins, and blue dextran (2000 kDa) for void volume determination, were obtained from Sigma.
RESULTS
Properties of a range of activated mutant resolvases
To begin our in vitro analysis, several of the resolvase variants characterized in vivo and found to be activated by Burke et al. (13) were overexpressed and purified. The purified proteins were tested for recombination activity on plasmid substrates containing two copies of site I or two full res sites. These results are summarized in Table 1, along with the recombination activity of the variants in E. coli [taken from (13)].
Table 1.
Catalytic properties of Tn3 resolvase variants in E. coli and in vitro
|
In vivo recombination |
In vitro recombination |
|||||
|---|---|---|---|---|---|---|
| Mutant | Mutations | res × res | l × l | res × res | l × l | Site I synapsis |
| WT | Wild-type | ++ | − | ++ | − | − |
| N | R2A E56K | − | − | − | − | − |
| S | G101S | ++ | − | ND | ND | ND |
| Y | D102Y | ++ | − | ++ | − | − |
| Q | E124Q | ++ | − | ++ | − | − |
| SY | G101S D102Y | ++ | ++ | ++ | + | − |
| YQ | D102Y E124Q | ++ | + | ++ | + | − |
| NYQ | R2A E56K D102Y E124Q | ++ | ++ | + | + | − |
| NSY | R2A E56K G101S D102Y | ++ | ++ | ++ | + | − |
| NSYQ | R2A E56K G101S D102Y E124Q | ++ | ++ | ++ | ++ | + |
| M | G101S D102Y M103I Q105L | ++ | + | ++ | ++ | + |
| MQ | G101S D102Y M103I Q105L E124Q | ++ | − | ++ | ++ | + |
| NM | R2A E56K G101S D102Y M103I Q105L | ++ | ++ | ++ | ++ | + |
| NMQ | NM + E124Q | ++ | + | ++ | ++ | + |
| C | A117V R121K E124Q | − | − | + | − | − |
| YC | D102Y A117V R121K E124Q | ++ | − | + | + | − |
| MC | M + C | − | − | + | + | ND |
Recombination activity in E. coli was estimated by an assay for resolution as detected by colony colour (13). The results [taken from (13)] are shown as ++ (pale colonies, full resolution), + (pink colonies, partial resolution) or − (red colonies, low or zero resolution). In vitro assays were performed under standard conditions, using the substrates pMA21 (res × res) and pAL225 (site I × site I), as described in the Materials and methods section. The symbols give a rough comparison of extent of recombination, high levels (such as wild-type resolvase on pMA21, or NM resolvase on pAL225) shown as ++, lower levels as + and no observable recombination as −. ND indicates ‘not done’. Synapsis assays used non-denaturing TBE gels as described in Materials and methods section; + indicates the observation of a distinct site I synapse band.
There was a good correspondence between the recombination activities of the variants in vitro and in vivo. No single mutant promotes efficient recombination of a site I × site I substrate, but several multiple mutants do so [as was first shown for D102Y E124Q by Arnold et al. (12)]. Whereas some combinations of activating mutations elicit high site I × site I recombination activity (for example, G101S D102Y, and the group G101S D102Y M103I Q105L), other combinations do not; for example, the A117V R121K E124Q triple mutant is almost inactive and binds poorly to site I. A double mutation at the 2–3′ interface (R2A E56K) abolished all in vitro recombination by wild-type resolvase [as expected; (7)]. However, the 2–3′ mutations were neutral or even stimulated site I × site I recombination in the context of variants which already have this activity (such as D102Y E124Q), as had previously been observed in vivo by Burke et al. (13). The 2–3′ interface is thus not required for the catalytic functions of resolvase at site I. Variants with high site I × site I recombination activity formed a synaptic complex of two site I fragments in a bandshift assay (Table 1; further details below).
In vitro recombination by selected activated mutants
Two activated resolvase variants with multiple mutations near the N-terminus of the E-helix, and their counterparts with additional mutations of residues on the 2–3′ interface, were selected for more detailed analysis. These were the double mutant of adjacent residues G101S D102Y (‘SY’), and the quadruple mutant G101S D102Y M103I Q105L (‘M’), which has higher site I × site I activity than SY in vitro (Table 1). The 2–3′-defective variants of SY and M (‘NSY’ and ‘NM’) have the mutations R2A E56K. NM resolvase was previously shown to form a stable site I synapse (15), and its catalytic domain was used to construct chimaeric recombinases with zinc finger DNA-binding domains (24). The site I synapse crystal structures of Li et al. (16) use a γδ resolvase variant with equivalents of five of the six mutations in NM resolvase (R2A, E56K, G101S, E102Y and M103I), as well as another activating mutation E124Q. The natural γδ resolvase residue K105 corresponds to Q105 in Tn3 resolvase (mutated to L in M and NM).
Wild-type, SY, NSY, M and NM resolvases were tested on plasmid substrates containing either two res sites in direct repeat (pMA21) or two copies of site I (pAL225) (Figure 3A). Following incubation for 1 h at 37°C, reaction samples were analysed by further treatment with restriction enzymes or DNase I, then agarose gel electrophoresis (Figure 3B). All the mutants recombined pAL225 in vitro, whereas wild-type resolvase did not. The mutants retain high sequence specificity; no products indicating reaction at non-site I sequences were observed under these conditions. The 2–3′ interface-defective NM resolvase gave more recombinant products than the 2–3′ proficient M resolvase, whereas the extent of recombination by SY and NSY was similar.
The results in Figure 3 show that recombination activity on pAL225 is correlated with several other effects. First, the activated mutants show partial or complete loss of the wild-type specificity for ‘resolution’ of pMA21, as seen by the presence of products of inversion and intermolecular reactions (Figure 3B). A second, related effect is the loss of the wild-type specificity for a 2-noded catenane product. Various other product topologies (free recombinant circles, knots, catenanes and intermolecular products) were seen following DNase I nicking to remove DNA supercoiling. ‘Ladders’ of topologically complex products were observed on the gels after reaction of pAL225 with SY or NSY resolvase, as expected for recombination following a ‘random collision’ synapsis pathway (12,29). However, M and NM resolvases formed substantial amounts of simple, free circle products in this assay after 1 h of reaction (Figure 3B). We show below (see Figure 4) that this alternative product pattern is the result of multiple rounds of reaction, not a different synapsis pathway. NM resolvase and other highly activated mutants no longer require supercoiling for activity, and recombine linear or oligonucleotide substrates [data not shown; see also (12)].
Figure 4.
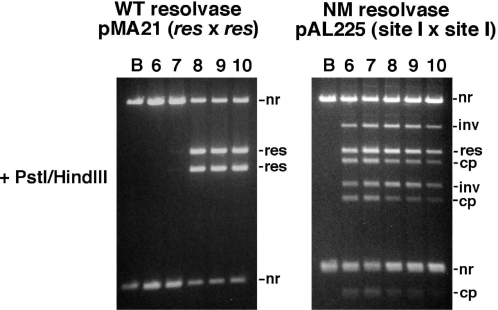
Time courses of recombination by wild-type (WT) and activated mutant (NM) resolvase. Recombination of pMA21 (res × res) by WT resolvase and of pAL225 (site I × site I) by NM resolvase was assayed under standard conditions. Aliquots were stopped at the indicated times. Products were analysed by electrophoresis after PstI + HindIII digestion (1.2% gel; upper panels), or nicking with DNase I (0.7% gel; lower panels). Annotation is as in Figure 3.
Another property of the activated mutants which is apparent in Figure 3 is their increased tendency to form species with double-strand breaks (DSBs) at site I. It was shown previously that the DNA cleavage and rejoining activities of resolvase can be partially decoupled in reaction buffers containing high concentrations of glycerol or ethylene glycol (30,31). We tested for resolvase-mediated cleavage of pMA21 and pAL225 in a buffer containing 40% ethylene glycol (Figure 3C). Wild-type resolvase gave traces of DSB products [as expected; (32)], but only with pMA21 (res × res), whereas the activated mutants gave substantial amounts of DSB products from both substrates.
NM resolvase is the most activated variant of those studied here. In time course assays (Figure 4, and data not shown), NM resolvase-mediated site I × site I (pAL225) recombination was at least as fast as wild-type resolvase-mediated res × res (pMA21) recombination. Wild-type resolvase made exclusively a 2-noded catenane resolution product which accumulated to a high level at later time points, as expected. In contrast, the NM resolvase reaction products at early time points were mainly topologically complex, but recombinant free circles became more abundant over the time course. This change is reflected in the restriction digest; at early time points, the ‘resolution’ and ‘inversion’ recombinant bands were observed in an approximate 1:1 ratio, whereas later time points show an increased proportion of ‘resolution’ products. The final (60 min) time point in Figure 4 corresponds to the data shown in Figure 3B. We conclude that the changes in product topology and restriction digest pattern over the time course are due to multiple rounds of strand exchange (see Discussion section). A substantial amount of ‘non-recombinant’ DNA is observed even at later time points in the pAL225/NM resolvase reaction, which is also consistent with multiple rounds of strand exchange that convert recombinants back to the non-recombinant configuration.
Another surprising feature of the activated resolvase variants is their altered pH sensitivity. SY, NSY, M and NM resolvases are all active on pMA21 (res × res) or pAL225 (site I × site I) over a wide pH range, from <6.0 to >10.0, whereas wild-type resolvase (33) and the single mutant D102Y are almost inactive on pMA21 at pH 7.0 and below (Figure 5 and data not shown). The reasons for this altered pH sensitivity are still unclear. The mutations might induce conformational changes of active site residues, which reduce the sensitivity of catalysis to low pH, or the effects might be due to loss of dependence on pH-sensitive regulatory interactions between the catalytic tetramer and subunits at the accessory sites.
Figure 5.
pH dependence of recombination by wild-type (WT) and activated mutant (NM) resolvase. Recombination of pMA21 (res × res) by WT resolvase and pAL225 (site I × site I) by NM resolvase was assayed in buffers with different pH, as indicated (lanes marked B are controls, not treated with resolvase). Reaction products were analysed by agarose gel electrophoresis after digestion with PstI and HindIII. Annotation is as in Figure 3.
Binding and synapsis by variant resolvases
To assess the ability of the resolvase variants to mediate formation of a synapse of two site I's, we used a gel electrophoresis assay similar to that described in Sarkis et al. (14). The activated M and NM mutant resolvases made a synaptic complex that was observed as a slow-running band [labelled S in Figure 6B; see also (15)]. The site I synapse was not observed following binding by wild-type resolvase or the SY and NSY variants; we presume that any synapse formed by these proteins (or the other synapsis-negative variants reported in Table 1) is too unstable to survive gel electrophoresis. Complexes corresponding to binding of one or two resolvase subunits to a single site I fragment were also observed in all cases, though these bands (especially the 2-subunit complex) were very faint in the NM resolvase lane. Curiously, the mobilities of all types of complex varied slightly depending on which resolvase was used.
NM resolvase-mediated synapsis is fast. Even at 0°C, the maximum amount of site I synapse was formed within 20 s of addition of resolvase (as fast as we could load the samples on the gel) (Figure 6C).
The pattern of complexes observed was strongly affected by the conditions of gel electrophoresis. Much lower amounts of site I synapse were observed when the samples were separated using TGE running buffer [previously used to observe complexes of Tn3 resolvase with res or parts of res; (12,26,34)] instead of TBE buffer. The intensities of the bands corresponding to site I bound by one or two resolvase subunits correspondingly increased, as illustrated for NM resolvase (Figure 6B). We conclude that the site I synapse decomposes on entry into the gel with TGE buffer, to give the observed one-site complexes. The predominance of the NM resolvase site I synapse band was restored when the TGE gel was 8% rather than 6.5% polyacrylamide (Figure 6B).
The stoichiometry of the NM resolvase site I synapse (that is, two copies of site I held together by four subunits of resolvase) was confirmed by experiments analogous to those used by Sarkis et al. (14) to study synapsis by γδ resolvase mutants (Figure 6D). A larger version of NM resolvase (NM–GFP) was created by fusing a green fluorescent protein (GFP) domain to the C-terminus. Site I treated with mixtures of NM and NM–GFP gave five synapse bands, which we assign as containing 0, 1, 2, 3 or 4 NM–GFP subunits. Analogous experiments using mixtures of different sized site I oligonucleotides showed that there were two DNA fragments in the complex [(15), and data not shown]. When NM and NM–GFP resolvases were mixed immediately before addition to the site I DNA, the synapses contained the statistically predicted distribution of the two types of monomers (lane 4 in Figure 6D), and the distribution did not change if the resolvase mixture was kept at −20°C overnight prior to addition to the DNA (data not shown). This result indicates either that there are no stable unbound resolvase multimers in solution, or that the subunits comprising any such species can exchange rapidly; otherwise synapses containing the unmixed NM or NM–GFP multimers would be anomalously abundant. However, once the site I synapse is formed, the resolvase tetramer in it is quite stable. When pre-formed NM resolvase synapse was challenged with excess NM–GFP, no synapses containing mixtures of subunit types were seen (Figure 6D, lane 5). Likewise, when pre-formed NM and NM–GFP site I synapses were mixed, the complexes did not disproportionate (Figure 6D, lane 6). TGE electrophoresis buffer conditions (see above; Figure 6B) destabilize the site I synapse, giving complexes of a single site I with one or two resolvase subunits, but no species that might be a single site I bound to a resolvase tetramer was observed.
These results differ from Sarkis et al. (14), whose data indicated the presence of stable activated mutant γδ resolvase tetramers in solution, which then bound to the DNA. It was previously shown that wild-type Tn3 resolvase is in monomer–dimer equilibrium in solution at low concentrations (<1 μM) (35). We compared wild-type Tn3 resolvase and NM resolvase by gel filtration chromatography, at higher concentrations (∼65 μM), and at the same salt concentration used in the resolvase storage/dilution buffer. Both proteins eluted as single peaks, with apparent molecular weights of 50 kDa (wild-type resolvase) or 26 kDa (NM resolvase) (data not shown). We conclude that wild-type Tn3 resolvase (subunit molecular weight 20.5 kDa) is predominantly dimeric in solution in this experiment, and NM resolvase is predominantly monomeric. A possible reason for destabilization of the dimers of NM resolvase and other activated mutants is noted in the Discussion section.
Resolvase-mediated site I cleavage was observed as single-strand break (SSB) or DSB products with a covalently attached resolvase subunit, following treatment of samples with the protein denaturant SDS (Figure 6E). The resolvase variants that produced the most site I synapse (Figure 6B) gave the highest levels of cleavage products, suggesting that cleavage occurs within the synapse. However, the rate of cleavage by NM resolvase is much lower than the rate of synapsis. Whereas synapsis reached a maximum within 20 s at 0°C (Figure 6C), the site I DNA was mostly uncleaved even after 30 min at 37°C (Figure 6E). To analyse the state of the site I DNA in the NM resolvase synapse as observed by polyacrylamide gel electrophoresis, we performed a 2D experiment. Following electrophoresis in TBE buffer, the gel was soaked in TBE containing SDS, then run in the perpendicular direction (Figure 6F). Separation in the second dimension showed that most of the site I DNA in the synapse band was uncleaved, with small amounts of single-strand and double-strand cleavage products. A larger amount of double-strand cleavage product was observed as a smear running faster than synapse in the first dimension, suggesting decomposition of cleaved site I synapses during electrophoresis.
DISCUSSION
Recombination by activated resolvase variants in vitro; role of the accessory sites of res
‘Activated’ Tn3 resolvase multiple mutants can promote rapid in vitro recombination between two copies of the 28 bp dimer-binding site I or ‘crossover site’ of the 114 bp Tn3 recombination site res. The rate of site I × site I recombination can be as high as that of res × res recombination promoted by wild-type resolvase. Wild-type resolvase is active only on a supercoiled res × res substrate and forms exclusively a 2-noded catenane resolution product, whereas the mutants recombine res × res or site I × site I substrates to give resolution, inversion and intermolecular recombination products of variable topologies, as well as DNA cleavage products. Mutant-catalysed site I × site I cleavage and recombination in a plasmid substrate are faster than cleavage of site I oligonucleotides, suggesting that the reaction is stimulated by negative supercoiling. The topologies of the site I × site I products are consistent with a ‘random collision’ mechanism of synapsis (29), as has been inferred from the products formed by activated mutants of other serine recombinases (18,36,37). The apparent selective formation of free circle site I × site I resolution products at later time points in NM resolvase reactions (Figure 4) can be accounted for by multiple rounds of recombination; under these circumstances products with simpler topology, and therefore lower free energy, will eventually accumulate.
The activating mutations present in the resolvase variants studied here were identified in E. coli screens for res × site I or site I × site I recombination activity; that is, reactions where one or both partners lack the res ‘accessory’ binding sites II and III [Figure 1B; (12)]. The properties of the mutants might therefore provide insights into the functions of the accessory binding sites in the wild-type system. The accessory sites have an established role in bringing about topological selectivity by formation of an intertwined structure (29,38,39), but how do they then stimulate recombination at site I? One hypothesis would be that accessory site synapsis is necessary simply to enhance the overall rate of formation or stability of the complete two-res synapse. However, the activated mutant NM resolvase recombines a site I × site I substrate, with no accessory sites, faster than wild-type resolvase recombines a res × res substrate (Figure 4), suggesting that this is not a sufficient explanation. A more appealing hypothesis is that resolvase subunits at the synapsed, intertwined accessory sites somehow promote the site I-bound resolvase subunits from a catalytically inactive configuration to an active one.
The 2–3′ interface is essential for in vitro catalytic activity of wild-type resolvase (see Introduction section). It has been shown to be involved in contacts between subunits at site I and the accessory sites (5,10), and site I-accessory site 2–3′ interactions have been built into structure-based models for the res × res synapse (10,14,19). However, the 2–3′ interface is dispensable for site I × site I recombination by activated resolvase mutants. A site I synapse made by wild-type resolvase was not observed in our gel-based assays; either it is not formed, or it is too unstable to survive electrophoresis. Site I synapse formation leading to catalysis by wild-type resolvase might require 2–3′ contacts from subunits at the accessory sites, whereas activating mutations allow site I synapse formation without 2–3′ contacts (Figure 6B), bypassing this regulatory mechanism.
Formation and stability of the site I synapse
Formation of a synapse of two site I oligonucleotides by NM resolvase is fast (Figure 6C), but subsequent DNA cleavage is much slower (Figure 6E). The synapse bands seen on our gels contain largely uncleaved site I DNA (Figure 6F). The site I synapse is held together by a tetramer of resolvase (Figure 6D; 14), which is stable whilst it is in the synapse but is not present as a long-lived species in solution or bound to a single site I (Figure 6B, Figure 6D and gel filtration results). Based on these results and previous evidence (5,15,34), our favoured model for assembly of the site I synapse is that resolvase monomers bind cooperatively to site I to form dimers, and two site I-dimer complexes then interact with each other. This pathway might be advantageous in the context of the biology of the wild-type resolution system because it can fully exploit the potential for cooperativity in synapse formation, though mutants other than NM, or other related resolvases, might well behave differently (e.g., see 14,17). A similar pathway for synapsis was deduced by Sanders and Johnson (40) from an elegant study of synaptic complexes and strand exchange intermediates formed by an activated H107Y mutant of Hin, a DNA invertase related to resolvase. Hin H107Y forms a synapse comprising two hix recombination sites and four Hin subunits which can be observed by gel electrophoresis. The hix DNA in the synapse has been cleaved by Hin H107Y to give DSBs, and this cleavage is required for the stability of the complex, in contrast to the Tn3 resolvase site I synapse (Figure 6F). Further studies on the Hin H107Y synapse showed that the Hin subunits translocate with the DNA half-sites during strand exchange, supporting a subunit rotation model for the mechanism of catalysis (41).
How do activating mutations exert their effects?
The activating mutations in the resolvase variants studied here were identified as conferring a ‘gain of function’ phenotype; that is, ability to recombine substrates lacking one or both sets of res accessory sites. However, mutations in general are most likely to result in loss of function, so it might be better to regard the effect of activating mutations as loss of a regulatory function of wild-type resolvase. That is, wild-type resolvase has evolved so that its catalytic activity is strictly dependent on formation of a synapse involving two complete res sites, and the mutants have lost this dependence. Our in vitro experiments show that the mutations have various additional effects on resolvase activity (see above).
The crystal structures of wild-type γδ resolvase and activated γδ resolvase mutants (both unbound and in complexes with site I) should provide clues to the nature of the regulation, and why it is defective in the activated mutants. The structures of the site I synaptic intermediates [1ZR2, 1ZR4, 2GM4; (16,17)] and of an activated variant catalytic domain without DNA [2GM5; (17)] contain a tetrameric arrangement of the catalytic domains which is not found in any of the structures of wild-type resolvase [2RSL, 1GDR, 1GDT; (6,8,9,11)] (Figure 2B). However, the structures do not provide an obvious explanation of how the mutations stabilize the tetramer, or why it is not formed by wild-type resolvase. Li et al. (16) make two hypotheses. First, the observed bond angles for the wild-type resolvase residue G101 are allowed only for glycine, so substitution of this residue by serine in the activated mutants might destabilize the dimer by forcing a change in the conformation of this part of the protein, the ‘hinge’ connecting the globular N-terminus of the catalytic domain to the E-helix. Second, some of the mutated residues (e.g. Y102) make new hydrophobic contacts in the tetrameric structures.
To relate the properties of the activated mutants to the mechanism of the wild-type resolution system, it is important to establish whether the changes in resolvase conformation between the crystal structures of the (wild-type) dimer bound to site I and the (activated mutant) tetramer in a cleaved site I synapse occur before or after initial synapsis. An ‘early’ synaptic interface between two resolvase dimers in approximately wild-type conformation has been proposed (13,14,16), involving residues near the N-terminus of the E-helix including G101, D102 and M103, which are mutated in the activated variants studied here. However, this hypothetical interface has not been observed by crystallography. The rapidly formed, uncleaved site I synapses observed in this study (and in previous studies using catalytically defective resolvase mutants or chemically modified DNA; 14,15) are distinct from all the current crystallographic structures which contain cleaved site I DNA, and therefore might contain the hypothetical early synaptic interface, stabilized by the activating mutations. Alternatively, the activating mutations might promote pre-synapsis conversion of the dimer into a conformation resembling that seen in the crystallographic tetramer. In this latter scenario, there would be no equivalent of the hypothetical early synaptic interface. Further work will be needed to distinguish between these two models, and to determine whether either of them is applicable to the natural res × res recombination system.
FUNDING
U.K. Commonwealth Academic Staff Scholarship Award (to F.J.O); a Carnegie Trust Ph.D Studentship (to S.V.C.T.W); Wellcome Trust (Grant reference numbers 039542, 072552). Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are very grateful to Phoebe Rice for helpful comments on the manuscript.
REFERENCES
- 1.Wirth D, Gama-Norton L, Riemer P, Sandhu U, Schucht R, Hauser H. Road to precision: recombinase-based targeting technologies for genome engineering. Curr. Opin. Biotechnol. 2007;18:411–419. doi: 10.1016/j.copbio.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 2.Akopian A, Stark WM. Site-specific recombinases as instruments for genomic surgery. Adv. Genet. 2005;55:1–23. doi: 10.1016/S0065-2660(05)55001-6. [DOI] [PubMed] [Google Scholar]
- 3.Rowland SJ, Stark WM. Site-specific recombination by the serine recombinases. In: Mullany P, editor. The Dynamic Bacterial Genome. New York, NY, USA: Cambridge University Press; 2005. pp. 121–150. [Google Scholar]
- 4.Grindley NDF, Whiteson KL, Rice PA. Mechanism of site-specific recombination. Ann. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 5.Grindley NDF. The movement of Tn3-like elements: transposition and cointegrate resolution. In: Craig N, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA II, Chap. 14. Washington, DC: ASM Press; 2002. pp. 272–302. [Google Scholar]
- 6.Sanderson MR, Freemont PS, Rice PA, Goldman A, Hatfull GF, Grindley NDF, Steitz TA. The crystal structure of the catalytic domain of the site-specific recombination enzyme γδ resolvase at 2.7 Å resolution. Cell. 1990;63:1323–1329. doi: 10.1016/0092-8674(90)90427-g. [DOI] [PubMed] [Google Scholar]
- 7.Hughes RE, Hatfull GF, Rice PA, Steitz TA, Grindley NDF. Cooperativity mutants of the γδ resolvase identify an essential interdimer interaction. Cell. 1990;63:1331–1338. doi: 10.1016/0092-8674(90)90428-h. [DOI] [PubMed] [Google Scholar]
- 8.Rice PA, Steitz TA. Model for a DNA-mediated synaptic complex suggested by crystal packing of γδ resolvase subunits. EMBO J. 1994;13:1514–1524. doi: 10.2210/pdb1gdr/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rice PA, Steitz TA. Refinement of γδ resolvase reveals a strikingly flexible molecule. Structure. 1994;2:371–384. doi: 10.1016/s0969-2126(00)00039-3. [DOI] [PubMed] [Google Scholar]
- 10.Murley LL, Grindley NDF. Architecture of the γδ resolvase synaptosome: oriented heterodimers identify interactions essential for synapsis and recombination. Cell. 1998;95:553–562. doi: 10.1016/s0092-8674(00)81622-0. [DOI] [PubMed] [Google Scholar]
- 11.Yang W, Steitz TA. Crystal structure of the site-specific recombinase γδ resolvase complexed with a 34 bp cleavage site. Cell. 1995;82:193–208. doi: 10.1016/0092-8674(95)90307-0. [DOI] [PubMed] [Google Scholar]
- 12.Arnold PH, Blake DG, Grindley NDF, Boocock MR, Stark WM. Mutants of Tn3 resolvase which do not require accessory binding sites for recombination activity. EMBO J. 1999;18:1407–1414. doi: 10.1093/emboj/18.5.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burke ME, Arnold PH, He J, Wenwieser S.VCT, Rowland SJ, Boocock MR, Stark WM. Activating mutations of Tn3 resolvase marking interfaces important in recombination catalysis and its regulation. Mol. Microbiol. 2004;51:937–948. doi: 10.1046/j.1365-2958.2003.03831.x. [DOI] [PubMed] [Google Scholar]
- 14.Sarkis GJ, Murley LL, Leschziner AE, Boocock MR, Stark WM, Grindley NDF. A model for the γδ resolvase synaptic complex. Mol. Cell. 2001;8:623–631. doi: 10.1016/s1097-2765(01)00334-3. [DOI] [PubMed] [Google Scholar]
- 15.Nöllmann M, He J, Byron O, Stark WM. Solution structure of the Tn3 resolvase-crossover site synaptic complex. Mol. Cell. 2004;16:127–137. doi: 10.1016/j.molcel.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 16.Li W, Kamtekar S, Xiong Y, Sarkis GJ, Grindley NDF, Steitz TA. Structure of a synaptic γδ resolvase tetramer covalently linked to two cleaved DNAs. Science. 2005;309:1210–125. doi: 10.1126/science.1112064. [DOI] [PubMed] [Google Scholar]
- 17.Kamtekar S, Ho RS, Cocco MJ, Li W, Wenwieser S.VCT, Boocock MR, Grindley NDF, Steitz TA. Implications of structures of synaptic tetramers of γδ resolvase for the mechanism of recombination. Proc. Natl Acad. Sci. USA. 2006;103:10642–10647. doi: 10.1073/pnas.0604062103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowland SJ, Boocock MR, Stark WM. Regulation of Sin recombinase by accessory proteins. Mol. Microbiol. 2005;56:371–382. doi: 10.1111/j.1365-2958.2004.04550.x. [DOI] [PubMed] [Google Scholar]
- 19.Mouw KW, Rowland SJ, Gajjar MM, Boocock MR, Stark WM, Rice PA. Architecture of a serine recombinase-DNA regulatory complex. Mol. Cell. 2008;30:145–155. doi: 10.1016/j.molcel.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haffter P, Bickle TA. Enhancer-independent mutants of the Cin recombinase have a relaxed topological specificity. EMBO J. 1988;7:3991–3996. doi: 10.1002/j.1460-2075.1988.tb03287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klippel A, Cloppenborg K, Kahmann R. Isolation and characterisation of unusual gin mutants. EMBO J. 1988;7:3983–3989. doi: 10.1002/j.1460-2075.1988.tb03286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merickel SK, Haykinson MJ, Johnson RC. Communication between Hin recombinase and Fis regulatory subunits during coordinate activation of Hin-catalyzed site-specific DNA inversion. Genes Dev. 1998;12:2803–2816. doi: 10.1101/gad.12.17.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rowley PA, Smith MC, Younger E, Smith MCM. A motif in the C-terminal domain of φC31 integrase controls the directionality of recombination. Nucleic Acids Res. 2008;36:3879–3891. doi: 10.1093/nar/gkn269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akopian A, He J, Boocock MR, Stark WM. Chimeric site-specific recombinases with designed DNA sequence recognition. Proc. Natl Acad. Sci. USA. 2003;100:8688–8691. doi: 10.1073/pnas.1533177100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordley RM, Smith JD, Gräslund T, Barbas C.F., III Evolution of programmable zinc finger-recombinases with activity in human cells. J. Mol. Biol. 2007;367:802–813. doi: 10.1016/j.jmb.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 26.Bednarz AL, Boocock MR, Sherratt DJ. Determinants of correct res site alignment in site-specific recombination by Tn3 resolvase. Genes Dev. 1990;4:2366–2375. doi: 10.1101/gad.4.12b.2366. [DOI] [PubMed] [Google Scholar]
- 27.Summers DK, Sherratt DJ. Resolution of ColE1 dimers requires a DNA sequence implicated in the three-dimensional organization of the cer site. EMBO J. 1988;7:851–858. doi: 10.1002/j.1460-2075.1988.tb02884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 29.Stark WM, Boocock MR. Topological selectivity in site-specific recombination. In: Sherratt DJ, editor. Mobile Genetic Elements (Frontiers in Mol. Biology series). Oxford, UK: Oxford University Press; 1995. pp. 101–129. [Google Scholar]
- 30.Reed RR, Grindley NDF. Transposon-mediated site-specific recombination in vitro: DNA cleavage and protein-DNA linkage at the recombination site. Cell. 1981;25:721–728. doi: 10.1016/0092-8674(81)90179-3. [DOI] [PubMed] [Google Scholar]
- 31.Boocock MR, Zhu X, Grindley NDF. Catalytic residues of γδ resolvase act in cis. EMBO J. 1995;14:5129–5140. doi: 10.1002/j.1460-2075.1995.tb00195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McIlwraith MJ, Boocock MR, Stark WM. Tn3 resolvase catalyses multiple recombination events without intermediate rejoining of DNA ends. J. Mol. Biol. 1997;266:108–121. doi: 10.1006/jmbi.1996.0765. [DOI] [PubMed] [Google Scholar]
- 33.Stark WM, Grindley NDF, Hatfull GF, Boocock MR. Resolvase-catalysed reactions between res sites differing in the central dinucleotide of subsite I. EMBO J. 1991;10:3541–3548. doi: 10.1002/j.1460-2075.1991.tb04918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blake DG, Boocock MR, Sherratt DJ, Stark WM. Cooperative binding of Tn3 resolvase monomers to a functionally asymmetric binding site. Curr. Biol. 1995;5:1036–1046. doi: 10.1016/s0960-9822(95)00208-9. [DOI] [PubMed] [Google Scholar]
- 35.Nöllmann M, Byron O, Stark WM. Behavior of Tn3 resolvase in solution and its interaction with res. Biophys. J. 2005;89:1920–1931. doi: 10.1529/biophysj.104.058164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klippel A, Kanaar R, Kahmann R, Cozzarelli NR. Analysis of strand exchange and DNA binding of enhancer-independent Gin recombinase mutants. EMBO J. 1993;12:1047–1057. doi: 10.1002/j.1460-2075.1993.tb05746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crisona NJ, Kanaar R, Gonzalez TN, Zechiedrich EL, Klippel A, Cozzarelli NR. Processive recombination by wild-type Gin and an enhancer-independent mutant. Insight into the mechanisms of recombination site selectivity and strand exchange. J. Mol. Biol. 1994;243:437–457. doi: 10.1006/jmbi.1994.1671. [DOI] [PubMed] [Google Scholar]
- 38.Watson MA, Boocock MR, Stark WM. Characterisation of the synaptic intermediate in site-specific recombination by Tn3 resolvase. J. Mol. Biol. 1996;257:317–329. doi: 10.1006/jmbi.1996.0165. [DOI] [PubMed] [Google Scholar]
- 39.Kilbride E, Boocock MR, Stark WM. Topological selectivity of a hybrid site-specific recombination system with elements from Tn3 res/resolvase and bacteriophage P1 loxP/Cre. J. Mol. Biol. 1999;289:1219–1230. doi: 10.1006/jmbi.1999.2864. [DOI] [PubMed] [Google Scholar]
- 40.Sanders ER, Johnson RC. Stepwise dissection of the Hin-catalyzed recombination reaction from synapsis to resolution. J. Mol. Biol. 2004;340:753–766. doi: 10.1016/j.jmb.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 41.Dhar G, Sanders ER, Johnson RC. Architecture of the Hin synaptic complex: the recombinase subunits translocate with the DNA strands. Cell. 2004;119:33–45. doi: 10.1016/j.cell.2004.09.010. [DOI] [PubMed] [Google Scholar]