Abstract
Plasmids are ubiquitous mobile elements that serve as a pool of many host beneficial traits such as antibiotic resistance in bacterial communities. To understand the importance of plasmids in horizontal gene transfer, we need to gain insight into the ‘evolutionary history’ of these plasmids, i.e. the range of hosts in which they have evolved. Since extensive data support the proposal that foreign DNA acquires the host's nucleotide composition during long-term residence, comparison of nucleotide composition of plasmids and chromosomes could shed light on a plasmid's evolutionary history. The average absolute dinucleotide relative abundance difference, termed δ-distance, has been commonly used to measure differences in dinucleotide composition, or ‘genomic signature’, between bacterial chromosomes and plasmids. Here, we introduce the Mahalanobis distance, which takes into account the variance–covariance structure of the chromosome signatures. We demonstrate that the Mahalanobis distance is better than the δ-distance at measuring genomic signature differences between plasmids and chromosomes of potential hosts. We illustrate the usefulness of this metric for proposing candidate long-term hosts for plasmids, focusing on the virulence plasmids pXO1 from Bacillus anthracis, and pO157 from Escherichia coli O157:H7, as well as the broad host range multi-drug resistance plasmid pB10 from an unknown host.
INTRODUCTION
Plasmids are commonly found in bacteria, and often confer various phenotypes to their host such as resistance to antibiotics and heavy metals, production of toxins and other virulence factors, biotransformation of hydrocarbons, symbiotic nitrogen fixation, etc. (1). The overuse of antibiotics in the treatment of infectious diseases of humans and animals and for non-therapeutic purposes in agriculture has contributed to the emergence and spread of antibiotic-resistant pathogens, and this is partly due to horizontal gene transfer via conjugative plasmids (2,3). Many plasmids can transfer horizontally by conjugation, and replicate and be maintained in either a limited or much wider range of hosts (narrow versus broad host range) (4). Transmissible plasmids can be categorized into two types, according to their mobilization ability: (i) ‘self-transmissible’ or ‘conjugative’ plasmids that encode their own transfer machinery, and (ii) ‘mobilizable’ plasmids that can only transfer if provided with the transfer machinery by a co-resident self-transmissible plasmid (4). In spite of the critical role of plasmids in the spread of drug resistance and virulence factors, we have limited insight into their ecology and evolution. In particular we know little about which hosts serve as reservoirs of drug resistance, virulence and other plasmids.
A first assessment of the bacterial hosts that are potential long-term carriers of specific plasmids can be based on plasmid-chromosome sequence comparison. By August 2008, there were 1490 completely sequenced plasmid genomes in the NCBI database (http://www.ncbi.nlm.nih.gov). Many of these plasmid sequences were obtained during whole bacterial genome sequencing projects. Among the 1490 plasmids, 1355 (90.9%) are derived from Bacteria, 57 (3.8%) from Archaea and 39 (2.6%) from Eukaryota. Thirty-nine plasmids or 2.6% of the sequences currently archived in GenBank come from uncultured bacteria, and therefore their hosts are unknown. This fraction is expected to increase because of the current rapid rise in metagenomic and other cultivation-independent studies that generate plasmid sequences, including those of the human microbiome (5–8). For example, a plasmid genome sequence analysis project currently performed by us will more than triple this number in a very short time. Thus, methods that can suggest candidate hosts of plasmids based on DNA sequence data alone would help us assess their evolutionary history.
Extensive data support the proposal that foreign DNA will acquire the host's nucleotide composition during long-term residence, which is often referred to as ‘genome amelioration’ (9,10). Amelioration may result from restrictions in DNA conformation and mutational biases of replication and repair machinery in the host. The same pressures that homogenize the nucleotide composition throughout the chromosome will also drive a plasmid sequence's nucleotide composition towards that of the host (11). It follows that similar nucleotide compositions between a plasmid and a host's chromosome may indicate long-term evolution of the plasmid in that host, whereas a dissimilar nucleotide composition suggests independent evolutionary histories.
Native genes have been distinguished from recently acquired foreign genes using compositional features such as G+C content and frequencies of short oligonucleotides (di-, tri- and tetra-nucleotides) (12–16). One such feature is the dinucleotide relative abundance, which is defined as the ratio of the observed to expected dinucleotide frequency (11,17–28). The profile of 16 dinucleotide relative abundance values is relatively constant throughout the genome, except for regions that were recently acquired via horizontal transfer, such as genomic islands. Additionally, closely related species have more similar profiles than distantly related species. This profile of the dinucleotide composition therefore has been termed a ‘genomic signature’ (17,29).
The average absolute dinucleotide relative abundance difference, termed δ-distance, has been commonly used to measure the genomic signature difference between DNA sequences. Previous applications of the δ-distance to plasmids and their host chromosomes have led to contradictory conclusions. Campbell et al. (11) provided qualitative evidence that plasmids have similar genomic signatures to their known hosts using δ-distances averaged over 50 kb segments of the host chromosome. Van Passel et al. (25) used a more quantitative method taking into account the variability in composition around the chromosome, and concluded that plasmids were not more similar to their hosts than genomic aberrations such as horizontally transferred DNA and rRNA gene clusters. The results from van Passel's study indicate that the δ-distance can be improved upon by including information about the variability among dinucleotide relative abundance values along the bacterial chromosome. This motivated us to consider the Mahalanobis distance (30–32), which is well known in multivariate statistical analysis (e.g. discriminant analysis), but has not been considered so far for measuring genomic signature differences. The Mahalanobis distance corrects for the variability in the data (here, dinucleotide abundance changes along the chromosome), as well as for the covariance among the variables. It does this by giving less weight to correlated variables, proportional to their degree of correlation. Indeed, because the dinucleotide relative abundance is calculated on both strands of the DNA, the frequencies of the reverse complements of each dinucleotide are highly correlated. The Mahalanobis distance adjusts for this correlation, whereas the δ-distance does not.
Understanding the host in which plasmids evolved is important because (i) for an increasing number of drug resistance and other plasmids the host is not known since they were obtained by cultivation-independent methods, and (ii) even for plasmids that were found in specific strains, these hosts may not always be the long-term hosts. The goal of this study was to use the Mahalanobis distance to measure the genomic signature difference between bacterial plasmids and chromosomes, and to use it as a tool to propose candidate long-term hosts of plasmids. We first compared the performance of the Mahalanobis distance with the commonly used δ-distance in detecting the host in which a particular plasmid was found (designated as ‘known host’). We then focused on the virulence plasmids pXO1 from Bacillus anthracis, and pO157 from Escherichia coli O157:H7 to illustrate that this method can be used to generate hypotheses about the potential long-term reservoirs of virulence plasmids. Finally, we proposed candidate long-term hosts for the broad host range multi-drug resistance plasmid pB10. Neither the recent nor long-term hosts of this plasmid are known because it was captured from a waste water treatment plant by a cultivation-independent method. The Mahalanobis distance performed better than the δ-distance in identifying the known plasmid hosts among 230 bacterial strains, and in proposing candidate long-term hosts that are plausible given our empirical knowledge of plasmid host range. The approach thus generates testable hypotheses about the bacteria that may act as reservoirs for plasmids of interest.
MATERIALS AND METHODS
Software
Genome analyses were conducted using the G-language Genome Analysis Environment version 1.8.3 (33,34), available at http://www.g-language.org/. Statistical tests and graphics were implemented in the R version 2.6.0 (35), available at http://www.r-project.org/.
Genome sequences
Completely sequenced genomes in GenBank format (36) of bacterial plasmids and their corresponding host chromosomes were downloaded from the NCBI ftp site (ftp://ftp.ncbi.nih.gov/genomes/Bacteria). In cases where the host has multiple chromosomes, only the largest chromosome was used for the analysis because the definition of second chromosomes versus megaplasmids is still unclear (20). The final data set included 504 plasmids and 230 chromosomes. Complete listings of the plasmid genomes used in this study are shown in Supplementary Table S1.
Analyses
Representation of dinucleotide composition of a DNA sequence
Dinucleotide composition of a DNA sequence was represented by a vector, which consists of 16 dinucleotide relative abundance values (29). The dinucleotide relative abundance value (xij) is defined as the observed dinucleotide frequency divided by the expected dinucleotide frequency, which is the product of the component mononucleotide frequencies:
where fi and fj denote the frequency of the mononucleotide i and j, respectively (i, j ∈ A, C, G, T) and fij the frequency of the dinucleotide ij. These values combine counts from both strands of the sequence.
Measurement of dinucleotide composition difference between DNA sequences
A plasmid genome sequence was compared with non-overlapping 5-kb segments spanning a bacterial chromosomal sequence. The difference in dinucleotide composition, or ‘genomic signature’, between the plasmid and chromosome sequences was quantified by the following two distance measures. A high value of these distances represents a large difference in genomic signature between the DNA sequences.
First, the average absolute dinucleotide relative abundance difference (δ-distance) between a plasmid and a set of 5-kb chromosomal segments was calculated as:
where xij is the relative abundance value of the dinucleotide ij for the plasmid, yij is the mean of relative abundance values of dinucleotide ij calculated from the chromosomal segments, and the sum extends over all 16 dinucleotides.
Second, the Mahalanobis distance (D2) between a plasmid and a set of 5-kb chromosomal segments was calculated as:
where X is a vector of dinucleotide relative abundance values for a plasmid, Y is a mean vector of dinucleotide relative abundance values calculated from the chromosomal segments, S is the variance–covariance matrix of the dinucleotide relative abundances calculated from the chromosomal segments (S−1 is the inverse matrix of S), and the superscript T is the transposition operator. In the S matrix, the variance values indicate the degree of variability in relative abundance of each dinucleotide among the chromosome segments, and the covariance values reflect the correlations among relative abundances of the dinucleotides. The higher the correlation the greater the covariance so that multiplying the difference matrix (X – Y) by the inverse of S reduces the influence of highly correlated dinucleotides. Because the dinucleotides are counted on both strands, there is a high degree of correlation between the reverse complements of each dinucleotide (i.e. CC/GG, TT/AA, TG/CA, AG/CT, AC/GT and GA/TC). The Mahalanobis distance therefore adjusts for double counting dinucleotides and takes into account the variability along the chromosome due to the presence of genomic islands, etc.
To provide a P-value for the distance values (Mahalanobis or δ-distance), an empirical distribution of the distances for each chromosome segment was derived for each chromosome. This empirical distribution was constructed using each 5-kb chromosome segment as xij or X in the equations for the δ- or Mahalanobis distance, respectively. The advantage of using the P-values is that it places all values between 0 and 1, whereas Mahalanobis distance and δ-distance have no upper bound. P-values close to 1 indicate small distances and similar dinucleotide compositions between a plasmid and chromosome, whereas P-values close to 0 indicate large distances and dissimilar dinucleotide compositions between a plasmid and chromosome.
Performance comparison of Mahalanobis distance and δ-distance
The Mahalanobis distance and δ-distance values between each of 504 plasmids and 230 chromosomes were determined. These values were then used to rank different chromosomes with respect to similarity to a given plasmid: a chromosome ranking the highest (1) was most similar in dinucleotide composition and that ranking lowest (230) was most dissimilar. Since each plasmid was originally sequenced as part of a whole-genome sequencing project for a particular bacterial strain, each plasmid had one ‘known’ host. It was previously shown that the dinucleotide composition similarities between plasmids and the chromosomes of their known host tend to rank high (11). The performances of the Mahalanobis and δ-distances were evaluated for their ability to rank the known host chromosome highest among 230 bacterial chromosomes.
RESULTS
Performance of Mahalanobis and δ-distances in identifying plasmid hosts based on genomic signature similarity
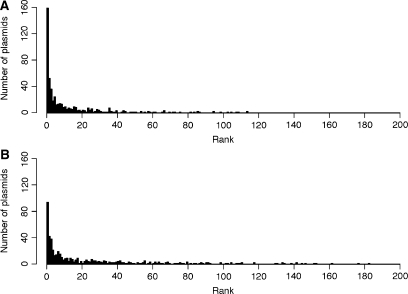
Compared to the δ-distance method, the Mahalanobis distance is an improved measure of the similarity in dinucleotide composition (here briefly called ‘genomic signature’) between plasmids and chromosomes because it takes into account variability of the data along the chromosome and correlations among dinucleotide relative abundance values on both DNA strands. Our first goal was to compare the ability of the Mahalanobis and δ-distance methods to identify among 230 bacteria the ‘known host’ of 504 plasmids, that is, the host in which the plasmid was found. Even though some plasmids may not have evolved in their known host, it was previously shown that dinucleotide composition similarities between plasmids and host chromosomes usually tend to rank high (11). Performance was measured by ranking the distances in genomic signature between each plasmid and all 230 chromosomes. Rank 1 represents the plasmid–host pair with the most similar signature (‘highest ranking’), and 230 corresponds to the most dissimilar pair. The distribution of ranks for all 504 plasmids and their known hosts is presented in Figure 1 as histograms, based on the Mahalanobis distance (Figure 1A) and δ-distance (Figure 1B) (see Supplementary Table S1 for details). Both distributions were heavily skewed toward high ranks, supporting previous findings that the similarities in signatures between each plasmid and its known host are among the highest (11). Of the 504 pairs of plasmids and their known hosts, 159 (32%) ranked number 1 based on the Mahalanobis distance, while only 94 (19%) ranked 1 based on the δ-distance. Thus for one in three plasmids, Mahalanobis distance identified the known plasmid host as the host with the most similar dinucleotide abundance among 230 strains. In 63% of cases, the Mahalanobis distance had a higher rank than the δ-distance, in 17% of cases, the δ-distance had the higher rank, and in 20% of cases did the two measures have the same rank. The median value of the ranks based on the Mahalanobis distances (four) was higher than that based on the δ-distance (eight). A Wilcoxon signed rank test, which compared the ranks based on the two methods, was highly significant (P < 2.2 × 10−16). Thus, in general, the genomic signature similarities between plasmids and their known hosts tended to rank higher when using the Mahalanobis distance than when using the δ-distance.
Figure 1.
Histograms showing the distribution of ranks of genomic signature similarities between 504 plasmids and their known hosts based on Mahalanobis distance (A) and δ-distance (B).
We tested the robustness of our result that Mahalanobis distance performs better than the conventional δ-distance by varying chromosomal segment size, word size (e.g. tri- and tetra-nucleotides) (12,29,37) and composition of the host data set. Campbell et al. (11) and Wong et al. (20) used 50-kb chromosomal segments, while van Passel et al. (25) used plasmid-size chromosomal segments; that is, the segment size was not fixed but depended on the plasmid size. In the present study, we used a fixed segment size. We found that our results remained similar when partitioning chromosomal sequences into different sizes of segments; e.g. 2, 5, 10 and 20 kb in length (data not shown). Among these, 5 kb was selected because the median rank of the genomic signature similarities between plasmids and their known hosts was maximized. The results were also consistent with those obtained when using relative abundances of different word sizes (data not shown). We also demonstrated that our results were robust to the composition of the host data set by testing different subsets of bacteria, e.g. Proteobacteria, Firmicutes, Gram-positive bacteria, Gram-negative bacteria and also when only one representative was randomly selected in the case of species for which multiple strains have been sequenced (data not shown). Thus, Mahalanobis distance performed better than δ-distance in identifying plasmid hosts based on genomic signature similarity, regardless of the datasets used.
Degree of similarity in genomic signature between a plasmid and its host
The ranks of the Mahalanobis distances indicate the similarity in genomic signature between a plasmid and its host relative to all other bacteria in the data set, but do not provide a measure of the degree of similarity within the genomes, and are very much biased by the available genome sequences. Additionally, the Mahalanobis distance between a specific plasmid and a host chromosome has no upper bound and is therefore hard to interpret. Therefore we expressed the degree of similarity in genomic signature between a plasmid and a particular host chromosome with a more intuitively meaningful value. The Mahalanobis distance between a plasmid and the collective set of chromosomal segments of each host was converted to a P-value associated with the empirical distribution of the Mahalanobis distances between the individual and the complete set of chromosomal segments of that host. Likewise, the δ-distance between a plasmid and a set of chromosomal segments was also converted to the P-value associated with an empirical distribution of the δ-distances between individual chromosomal segments and the mean for all chromosomal segments. P-values close to zero reflect large distances and dissimilar genomic signatures between plasmid and chromosome, whereas values close to one reflect small distances and similar genomic signatures. Plasmids whose hosts ranked first did not necessarily have P-values close to 1. For example, using the Mahalanobis distance, the known host of pXFPD1.3, Xylella fastidiosa Temecula1, ranked first among the 230 bacteria used in this study but had a P < 0.05 (Supplementary Table S1). This suggests that even though this host ranked first there must be other hosts, not included in this study, that have genomic signatures much more similar to that of plasmid pXFPD1.3, and therefore are more likely the long-term host of this plasmid. When all plasmids were compared to all 230 bacterial chromosomes, each plasmid showed P < 0.05 with 143 or more bacteria (at least 62%), and the median number of hosts with P < 0.05 was 198 (86%). This indicates that the vast majority of the bacteria considered here can be rejected as potential long-term hosts of these plasmids.
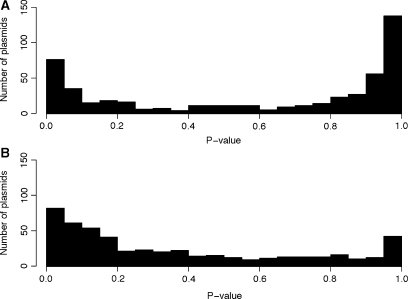
The distribution of P-values for all 504 plasmids and their known hosts is presented in Figure 2 as histograms, based on the Mahalanobis distance (Figure 2A) and δ-distance (Figure 2B) (see Supplementary Table S1 for details). In 78% of cases, the Mahalanobis distance had a higher P-value than the δ-distance, and in 20% of cases, the δ-distance had the higher P-value. The median value of the P-values based on the Mahalanobis distances (0.77) was higher than that based on the δ-distance (0.23). A Wilcoxon signed rank test comparing the P-values based on the Mahalanobis distance and the δ-distance was significant (P < 2.2 × 10−16). Of the 504 plasmids tested here, 138 (27%) had P > 0.95 based on the Mahalanobis distance, while only 42 (8%) had P > 0.95 based on the δ-distance. This indicates that Mahalanobis distance more frequently than δ-distance identifies the known plasmid host as the one with the most similar genomic signature. Of the 504 plasmids tested here, 76 (15%) had P < 0.05 based on the Mahalanobis distance, while 82 (16%) had P < 0.05 based on the δ-distance. Thus, fewer than one in six plasmids showed a significantly different genomic signature from that of their host chromosome, and apparently have not (yet) ameliorated their genomes to that of the host they were found in.
Figure 2.
Histogram showing the distribution of P-values derived from Mahalanobis distances (A) and δ-distances (B) between 504 plasmids and their known hosts.
Correlation of plasmid-host genomic signature difference with plasmid size
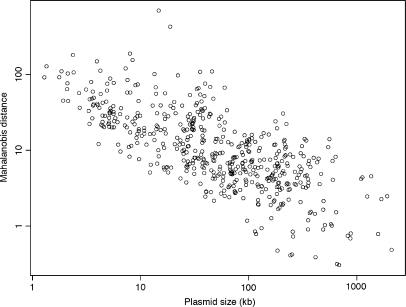
We reassessed the correlation of the genomic signature differences between plasmids and their host chromosomes with plasmid size, which was analyzed by van Passel et al. (25). Among the 504 plasmids tested here, the genome size ranged from 1286 bp (Xylella fastidiosa 9a5c plasmid pXF1.3) to 2 094 509 bp (Ralstonia solanacearum GMI1000 plasmid pGMI1000MP). As shown in Figure 3, the Mahalanobis distance between a plasmid and its known host was negatively correlated with the plasmid genome size (Spearman's rank correlation coefficient, r = −0.72; P < 2 × 10−16). Thus, the larger the plasmid, the more similar was its genomic signature to that of the host chromosome. This observation is unlikely to be caused by the greater sensitivity of small sequences to small changes in dinucleotide relative abundance values, because all plasmid genomes tested here were larger than 1 kb.
Figure 3.
Plot of Mahalanobis distances between 504 plasmids and their known hosts, against plasmid sizes.
Our result is in contrast with van Passel et al. (25), who concluded that there is a positive relationship between plasmid size and difference between plasmid and host in genomic signature. It is hard to define exactly why our results are contradictory because there are a sufficient number of differences between our study and the former study [the data set, now not excluding plasmids >100 kb as was done in previous study, chromosomal segment size (5 kb versus plasmid-size), and distance metric]. We examined the effect of plasmid size, chromosomal segment size and distance metric. First, in our dataset 35% of the plasmids were >100 kb, but the negative correlation between Mahalanobis distance and plasmid size was still observed when applied only to plasmids <100 kb. Second, the negative correlation was also observed when using δ-distance instead of Mahalanobis distance. Third, when we used plasmid-size chromosomal segments and our data set, there was no longer a significant correlation between plasmid size and either the Mahalanobis distance or the P-value associated with the distance (data not shown). Thus, we conclude that when taking into account variability along the chromosome by using the Mahalanobis distance method and fixed chromosomal segments sizes (5–20 kb), larger plasmids tend to be more similar in genomic signature to their host than smaller plasmids.
Proposing candidate long-term hosts of virulence plasmids
To illustrate our analysis, we compared the two methods in detail by focusing on plasmids derived from Bacillus anthracis ‘Ames Ancestor’ as examples. B. anthracis—the causative agent of anthrax—is a member of the B. cereus sensu lato group (B. cereus, B. anthracis and B. thuringiensis). The Ames Ancestor strain contains two plasmids, pXO1 and pXO2, which code for toxin production and encapsulation, respectively (38,39), and completely define the pathogenic potential of B. anthracis. Table 1 lists the 10 highest ranking bacterial strains based upon their Mahalanobis distance and δ-distance from plasmid pXO1. Seven bacterial strains of the B. cereus sensu lato group showed the smallest Mahalanobis distance from plasmid pXO1, and all top-10 strains are members of the Firmicutes (Table 1). In contrast, the top-10 smallest δ-distances from pXO1 were found among strains of the Cyanobacteria, Proteobacteria, Bacteroidetes, and Firmicutes. Given the known narrow host range of this plasmid (S. Khan, personal communication) (40), the first three phyla identified by δ-distance are unlikely to be true hosts (Table 1). Thus, among the top-10 ranking strains the Mahalanobis distance only identified strains that are very plausible hosts of the plasmids, while the δ-distance identified several bacteria in which the plasmids have not been shown to replicate. The known host of pXO1, B. anthracis Ames Ancestor, ranked 5 based on the Mahalanobis distance and only 12 based on the δ-distance (Table 1). The P-value for plasmid pXO1 and its known host B. anthracis Ames Ancestor was higher (0.99) when using the Mahalanobis distance than when using the δ-distance (0.38). Thus, the known host of plasmid pXO1 ranked higher and was estimated to be more similar in genomic signature to pXO1 when using the Mahalanobis distance than when using the δ-distance.
Table 1.
Ten highest ranking bacterial strains based on Mahalanobis distance and δ-distance for plasmid pXO1 from B. anthracis str. Ames Ancestor
| Bacterial strain | Phylum | D2 | P(D2) | δ | P(δ) |
|---|---|---|---|---|---|
| Sorted by Mahalanobis distance | |||||
| Bacillus cereus ATCC 14579 | Firmicutes | 2.17 | 0.994 | 47.5 | 0.517 |
| Bacillus thuringiensisstr. Al Hakam | Firmicutes | 2.83 | 0.987 | 50.6 | 0.415 |
| Bacillus cereus E33L | Firmicutes | 2.86 | 0.986 | 50.0 | 0.415 |
| Bacillus thuringiensis serovar konkukian str. 97-27 | Firmicutes | 2.91 | 0.989 | 51.0 | 0.423 |
| Bacillus anthracis str. Ames Ancestora | Firmicutes | 3.26 | 0.988 | 52.2 | 0.379 |
| Bacillus cereus ATCC 10987 | Firmicutes | 3.42 | 0.977 | 50.9 | 0.406 |
| Bacillus cereus subsp. cytotoxis NVH 391-98 | Firmicutes | 4.44 | 0.960 | 59.2 | 0.226 |
| Lactobacillus salivarius UCC118 | Firmicutes | 5.43 | 0.926 | 50.8 | 0.378 |
| Staphylococcus epidermidis RP62A | Firmicutes | 5.72 | 0.887 | 55.5 | 0.306 |
| Staphylococcus epidermidis ATCC 12228 | Firmicutes | 7.03 | 0.858 | 57.3 | 0.216 |
| Sorted by δ-distance | |||||
| Nostoc sp. PCC 7120 | Cyanobacteria | 53.01 | 0.018 | 41.7 | 0.440 |
| Anabaena variabilis ATCC 29413 | Cyanobacteria | 62.56 | 0.012 | 42.7 | 0.429 |
| Buchnera aphidicola str. APS (Acyrthosiphon pisum) | Proteobacteria | 19.64 | 0.211 | 43.1 | 0.523 |
| Bacillus cereus ATCC 14579 | Firmicutes | 2.17 | 0.994 | 47.5 | 0.517 |
| Bacillus cereus E33L | Firmicutes | 2.86 | 0.986 | 50.0 | 0.415 |
| Bacillus thuringiensis str. Al Hakam | Firmicutes | 2.83 | 0.987 | 50.6 | 0.415 |
| Lactobacillus salivarius UCC118 | Firmicutes | 5.43 | 0.926 | 50.8 | 0.378 |
| Bacillus cereus ATCC 10987 | Firmicutes | 3.42 | 0.977 | 50.9 | 0.406 |
| Bacillus thuringiensis serovar konkukian str. 97-27 | Firmicutes | 2.91 | 0.989 | 51.0 | 0.423 |
| Bacteroides fragilis NCTC 9343 | Bacteroidetes | 9.91 | 0.711 | 51.2 | 0.297 |
D2, Mahalanobis distance; P(D2), P-value based on Mahalanobis distance; δ, δ-distance; P(δ), P-value based on δ-distance.
aKnown host.
The P-values are not completely negatively correlated with the distances because they are based on empirical distributions that differ between bacterial chromosomes.
To further illustrate the utility of the Mahalanobis distance measure, we focused on enterohemorrhagic Escherichia coli O157:H7, which is the predominant causative agent of hemorrhagic colitis, a bloody diarrhea. The virulence plasmid pO157 is an F-like plasmid found in most O157:H7 strains, and is composed of a number of potential virulence genes (41). Table 2 lists the 10 highest ranking bacterial strains based upon their Mahalanobis distance and δ-distance for plasmid pO157 of the E. coli strain O157:H7 EDL933. The top-10 strains based on the Mahalanobis distance were all members of the Proteobacteria, more specifically of the family Enterobacteriacae, which are the typical hosts of narrow-host-range plasmids with F-like replication and maintenance features such as pO157 (42). In contrast, the top-10 strains based on the δ-distance included a Staphylococcus saprophyticus strain belonging to the Firmicutes, a very unlikely host of this plasmid. The known host of pO157 ranked 10 based on the Mahalanobis distance and only 42 based on the δ-distance. The P-value for plasmid pO157 and its known host was much higher (0.93) when using the Mahalanobis distance than when using the δ-distance (0.24). Interestingly, plasmid pO157 showed a smaller Mahalanobis distance (and higher P-values, ranging between 0.95 and 0.97) with seven Yersinia pestis and two Y. pseudotuberculosis strains than with the E. coli host strain. This suggests a potential long-term relationship between pO157-like plasmids and Yersinia, and requires further study.
Table 2.
Ten highest ranking bacterial strains based on Mahalanobis distance and δ-distance for plasmid pO157 from E. coli O157:H7 EDL933
| Bacterial strain | Phylum | D2 | P(D2) | δ | P(δ) | |
|---|---|---|---|---|---|---|
| Sorted by Mahalanobis distance | ||||||
| Yersinia pestis Antiqua | Proteobacteria | 3.79 | 0.972 | 32.8 | 0.724 | |
| Yersinia pestis KIM | Proteobacteria | 4.31 | 0.962 | 33.2 | 0.704 | |
| Yersinia pestis Angola | Proteobacteria | 4.33 | 0.962 | 32.8 | 0.700 | |
| Yersinia pestis CO92 | Proteobacteria | 4.41 | 0.966 | 33.3 | 0.696 | |
| Yersinia pestis biovar Microtus str. 91001 | Proteobacteria | 4.42 | 0.968 | 33.7 | 0.687 | |
| Yersinia pestis Nepal516 | Proteobacteria | 4.44 | 0.967 | 33.2 | 0.713 | |
| Yersinia pestis Pestoides F | Proteobacteria | 4.49 | 0.963 | 33.6 | 0.673 | |
| Yersinia pseudotuberculosis IP 31758 | Proteobacteria | 4.62 | 0.948 | 34.3 | 0.667 | |
| Yersinia pseudotuberculosis IP 32953 | Proteobacteria | 4.73 | 0.955 | 35.3 | 0.631 | |
| Escherichia coli O157:H7 EDL933a | Proteobacteria | 5.51 | 0.926 | 59.3 | 0.238 | |
| Sorted by δ-distance | ||||||
| Yersinia pestis Antiqua | Proteobacteria | 3.79 | 0.972 | 32.8 | 0.724 | |
| Yersinia pestis Angola | Proteobacteria | 4.33 | 0.962 | 32.8 | 0.700 | |
| Yersinia pestis Nepal516 | Proteobacteria | 4.44 | 0.967 | 33.2 | 0.713 | |
| Yersinia pestis KIM | Proteobacteria | 4.31 | 0.962 | 33.2 | 0.704 | |
| Yersinia pestis CO92 | Proteobacteria | 4.41 | 0.966 | 33.3 | 0.696 | |
| Yersinia pestis Pestoides F | Proteobacteria | 4.49 | 0.963 | 33.6 | 0.673 | |
| Yersinia pestis biovar Microtus str. 91001 | Proteobacteria | 4.42 | 0.968 | 33.7 | 0.687 | |
| Yersinia pseudotuberculosis IP 31758 | Proteobacteria | 4.62 | 0.948 | 34.3 | 0.667 | |
| Yersinia pseudotuberculosis IP 32953 | Proteobacteria | 4.73 | 0.955 | 35.3 | 0.631 | |
| Staphylococcus saprophyticus subsp. saprophyticus ATCC 15305 | Firmicutes | 338.76 | 0.000 | 40.6 | 0.525 |
D2, Mahalanobis distance; P(D2), P-value based on Mahalanobis distance; δ, δ-distance; P(δ), P-value based on δ-distance.
aKnown host.
See Table 1 legend for explanation of P-values
Proposing candidate long-term hosts of multi-drug resistance plasmids
Since at least 50% of the plasmid–host pairs ranked first, second, third or fourth based upon the Mahalanobis distance, we can use this method to propose putative hosts for plasmids from unknown hosts. Therefore, we compared the Mahalanobis and δ-distance measures for a broad host range plasmid for which the host is not known, but whose host range has been investigated experimentally. Several broad host range plasmids of the IncP-1 group have been captured from bacterial communities using ‘exogenous plasmid isolation’ methods (43,44), which do not retrieve the plasmid host. Empirical work has shown that IncP-1 plasmids such as pB10 (45) can transfer and replicate in most species of Gram-negative bacteria, mostly only α-, β- and γ-Proteobacteria, and typically not in bacteria outside of these groups (46,47). We compared pB10's genomic signature with 663 complete bacterial chromosome sequences available from the NCBI ftp site (ftp://ftp.ncbi.nih.gov/genomes/Bacteria) in August 2008. Table 3 lists the 10 highest ranking bacterial strains based upon their Mahalanobis distance and δ-distance from plasmid pB10. The top-10 strains based on the Mahalanobis distance are members of the β- (first 9) and γ-Proteobacteria with P-values ranging between 0.80 and 0.98. The top-10 strains based on the δ-distance included strains belonging to the Actinobacteria and the Chlorobi group. The results from the Mahalanobis distances are consistent with the known host range of IncP-1 plasmids, while those from the δ-distance are not (46,47). Moreover, Ralstonia eutropha JMP134 (currently reclassified as Cupriavidus necator) is the known host of the IncP-1 plasmid pJP4, which is very closely related to plasmid pB10 (45). This strain ranked 14 in our comparison with pB10 using the Mahalanobis distance, and only 38 when the δ-distance was used (data not shown). Nine of the top-10 strains had a higher G+C content than plasmid pB10 (64.2%) when the ranking was based on the Mahalanobis distance, compared to only four based on the δ-distance. Taking into account the finding that plasmids tend to have a lower G + C content than their hosts (25,48), the Mahalanobis distance again performed better than the δ-distance in identifying potential hosts of plasmid pB10. In conclusion, the Mahanalobis distance more correctly assessed the potential range of long-term hosts of this drug resistance plasmid than the δ-distance.
Table 3.
Ten highest ranking bacterial strains based on Mahalanobis distance and mean δ-distance for broad host range plasmid pB10 from an unknown host (%GC = 64.2)
| Bacterial strain | Phylum | D2 | P(D2) | δ | P(δ) | %GC |
|---|---|---|---|---|---|---|
| Sorted by Mahalanobis distance | ||||||
| Chromobacterium violaceum ATCC 12472 | Proteobacteria | 3.07 | 0.984 | 44.1 | 0.604 | 64.8 |
| Ralstonia eutropha H16 | Proteobacteria | 6.05 | 0.923 | 69.6 | 0.133 | 66.5 |
| Ralstonia solanacearum GMI1000 | Proteobacteria | 6.10 | 0.886 | 73.9 | 0.152 | 67.0 |
| Azoarcus sp. EbN1 | Proteobacteria | 6.32 | 0.922 | 93.6 | 0.061 | 65.1 |
| Bordetella petrii DSM 12804 | Proteobacteria | 6.44 | 0.902 | 66.0 | 0.226 | 65.5 |
| Azoarcus sp. BH72 | Proteobacteria | 7.31 | 0.856 | 47.3 | 0.481 | 67.9 |
| Dechloromonas aromatica RCB | Proteobacteria | 7.45 | 0.863 | 33.9 | 0.687 | 59.2 |
| Acidovorax avenae subsp. citrulli AAC00-1 | Proteobacteria | 7.46 | 0.844 | 64.4 | 0.243 | 68.5 |
| Bordetella bronchiseptica RB50 | Proteobacteria | 7.84 | 0.802 | 76.8 | 0.098 | 68.1 |
| Xanthomonas campestris pv. vesicatoria str. 85-10 | Proteobacteria | 8.09 | 0.823 | 93.9 | 0.062 | 64.7 |
| Sorted by δ-distance | ||||||
| Dechloromonas aromatica RCB | Proteobacteria | 7.45 | 0.863 | 33.9 | 0.687 | 59.2 |
| Renibacterium salmoninarum ATCC 33209 | Actinobacteria | 116.15 | 0.000 | 36.4 | 0.471 | 56.3 |
| Pseudomonas fluorescens PfO-1 | Proteobacteria | 13.02 | 0.445 | 39.7 | 0.547 | 60.5 |
| Chromobacterium violaceum ATCC 12472 | Proteobacteria | 3.07 | 0.984 | 44.1 | 0.604 | 64.8 |
| Chlorobaculum parvum NCIB 8327 | Chlorobi | 44.06 | 0.037 | 44.1 | 0.379 | 55.8 |
| Pseudomonas aeruginosa PAO1 | Proteobacteria | 10.74 | 0.657 | 44.1 | 0.482 | 66.6 |
| Pseudomonas aeruginosa UCBPP-PA14 | Proteobacteria | 8.28 | 0.790 | 44.4 | 0.486 | 66.3 |
| Pseudomonas syringae pv. syringae B728a | Proteobacteria | 30.23 | 0.065 | 46.5 | 0.369 | 59.2 |
| Pseudomonas aeruginosa PA7 | Proteobacteria | 9.05 | 0.758 | 46.9 | 0.432 | 66.4 |
| Hahella chejuensis KCTC 2396 | Proteobacteria | 31.08 | 0.077 | 47.0 | 0.353 | 53.9 |
D2, Mahalanobis distance; P(D2), P-value based on Mahalanobis distance; δ, δ-distance; P(δ), P-value based on δ-distance; %GC, genome G + C content defined as 100 × (G + C)/(A + T + G + C).
See Table 1 legend for explanation of P-values.
DISCUSSION
Differences in dinucleotide composition (genomic signature) between bacterial chromosomes and plasmids have previously been measured using the δ-distance (11,20,25,49). Here, we introduce the Mahalanobis distance for this purpose. The Mahalanobis distance has an advantage over the δ-distance because it takes into account both variances and covariances among the 16 dinucleotide relative abundance values along the bacterial chromosome. We conclude that the Mahalanobis distance performs better than the δ-distance because it more often identified the known hosts of the plasmids as the host with greatest genomic signature similarity, and in contrast to δ-distance, its top-10 ranking strains were always plausible hosts based on empirical host range knowledge.
We have converted the Mahalanobis distances to P-values so that we may provide more intuitively meaningful descriptions of the relationship between a plasmid and its known host. It is hard to know whether a distance of 20 reflects a large or a small difference in genomic signature, but it is easy to interpret that a P-value close to 1 indicates highly similar signatures, and values close to 0 indicate highly dissimilar signatures. We hypothesize that plasmid–host pairs with low Mahalanobis distances and very high P-values represent plasmids that have acquired the hosts’ signature through genome amelioration, and thus have been long-term residents of that host. More than a quarter of the plasmids tested here fit into this category, with P > 0.95 (Figure 2). They may have been exchanged rarely between hosts or only between closely related hosts with similar genomic signatures. Moreover, ∼85% of all plasmid–host pairs showed P > 0.05, indicating that the known host cannot be rejected as putative long-term host.
Coincident with the observation that many plasmids share their host's dinucleotide composition is the fact that large plasmids seem to be more similar in genomic signature to their hosts than small plasmids (Figure 3). Even though our results are in contradiction with those of van Passel et al. (25) (Results section), they can be explained based on known plasmid biology. Large plasmids are often not self-transferable, and even difficult to mobilize with a helper plasmid by conjugation, or they have a narrow host range (4,50). They are also less likely than smaller plasmids to persist as intact molecules outside a cell or to be taken up by transformation. It seems likely therefore that large plasmids spend most of their evolutionary time in a single bacterial host. For example, Sinorhizobium meliloti megaplasmid pSymB (1 683 333 bp) is not self-transmissible, and has not been successfully cured from S. meliloti (51). It has been suggested that pSymB has acquired genes essential to the host's viability and that its genomic signature became similar to that of the main chromosome due to long-term residence in that host (20). Similarly, large Rhizobium sym plasmids were shown to be strongly associated with specific chromosomal types, suggesting limited horizontal transfer (52,53).
Only 15% of plasmids tested here showed P < 0.05 (Figure 2) due to a high Mahalanobis distance with their known host, suggesting significantly different genomic signatures. If genome amelioration is a general process, our results imply that 15% of our plasmids were recently acquired by the hosts they were found in. Based on the analysis of plasmid size versus plasmid-host signature similarity (Figure 3), these dissimilar plasmids are relatively small (smaller than 63 kb, Supplementary Table S1). Since smaller plasmids have a higher probability of being mobilized or transformed, or of actively transferring by conjugation than very large plasmids, the strain they were found in may be only one of several recent hosts. The genomic signature analysis can thus generate hypotheses about the evolutionary history of plasmids, which can then be empirically tested. As more genome sequences of plasmids and their hosts become available, it should be possible in the near future to test the relationship between plasmid-host signature similarity and plasmid characteristics such as size, transferability and host range.
Another example of how the genomic signature analysis can be used to generate hypotheses about the evolutionary history of plasmids is shown in Table 1. The P-values for plasmid pXO1 and members of B. cereus sensu lato group (B. cereus, B. anthracis and B. thuringiensis) were greater than 0.95, suggesting that not only the known host (B. anthracis) but also the other species (B. cereus and B. thuringiensis) could be the hosts of plasmid pXO1. This finding is strongly supported by the following two facts. First, the different species comprising the B. cereus sensu lato group are largely defined by differences in plasmids, while the chromosomes have been shown to be similar in both gene content and gene order (54,55). Second, conjugation studies have shown that plasmid pXO1 can be transferred from B. anthracis to B. cereus (56). A second example, as shown in Table 2, is plasmid pO157, which was more similar in genomic signature to Y. pestis—the causative agent of plague—than its known host E. coli O157:H7. Moreover pO157 is very similar in genomic signature to plasmid pCD1 of Y. pestis (data not shown). These results led us to hypothesize that plasmid pO157 was acquired by E. coli O157:H7 from Y. pestis. This is supported by evidence for the transfer of genes (57) and plasmids (58) between E. coli and Y. pestis.
The genomic signature analysis can also be used to propose candidate long-term hosts of plasmids taken directly from clinical or environmental samples using exogenous plasmid isolation methods (44) or metagenomic approaches (8). For example, the top-9 ranking bacteria identified as potential hosts of plasmid pB10 because of their very similar genomic signatures, are members of the β-Proteobacteria. The suggestion that β-Proteobacteria are the most likely hosts of pB10 is completely in agreement with the fact that other IncP-1 plasmids very similar to pB10 are often found in β-Proteobacteria (4). Thus, the genomic signature analysis using the Mahalanobis distance provides a list of potential hosts of plasmids, which can then be used to experimentally test the host range of that plasmid. It may also result in finding better cloning vectors for specific species.
An interesting observation is the low similarity in genomic signature between Buchnera aphidicola plasmids pTrp1 and pBBp1 and their B. aphidicola host chromosomes (Supplementary Table S1). Since B. aphidicola is an obligate intracellular symbiont of aphids, with which it has co-evolved, and since its genome shows no or little indication of horizontal gene acquisition (15), it is not expected to exchange much genetic material with distantly related bacteria. Yet, these two small plasmids must have been recently acquired by these two Buchnera strains from other Buchnera strains with very different genomic signatures as previously suggested by van Passel et al. (25,26). The only alternative explanation is the absence of genome amelioration over evolutionary time, which would be exceptional considering that a third B. aphidicola plasmid, pLeu, shows clear indications of similar genomic signature with its host chromosome. This case is a good example of how results of the Mahalanobis distance analysis of dinucleotide relative abundance can form the basis of further research to better understand the evolutionary history of plasmids.
The approach we present here still has some limitations. First, an important caveat to this approach is the mosaic nature of plasmids, and the often large fraction of foreign accessory genes in plasmid genomes. A better approach might be to include only orthologous core genes (essential plasmid backbone genes involved in replication, maintenance and transfer) in this analysis. Determining this precise set of genes, however, can be difficult if phylogenetically closely related plasmids are not available (59–61). Even though we did not exclude accessory plasmid genes in our analysis, the similarity in genomic signature with that of the known plasmid hosts was still very high for many plasmids. Second, we cannot rule out the possibility that plasmids recently acquired by the host could have a similar genomic signature with that host by chance. To exclude these false positives, results should be double-checked by other criteria; for example, taking into account the finding that plasmids tend to have a lower G+C content than their hosts (25,48). We must also realize that several strains, even between species, may have very similar genomic signatures, and that findings are always biased by which host genome sequences are available in the databases.
In conclusion, we showed that the Mahalanobis distance performs better than the conventional δ-distance in measuring genomic signature differences between plasmids and chromosomes of potential hosts. In the future, the genomic signature analysis using the Mahalanobis distance can be applied to (i) inferring potential hosts of mobile genetic elements, including plasmids, phages and transposons obtained through cultivation-independent methods such as metagenomics and plasmid capture (5,8,62), and (ii) detecting anomalous genomic regions, e.g. genomic islands including pathogenicity islands that were recently acquired by horizontal transfer (18,22). The combined use of the genomic signature analysis and complementary analyses (e.g. analyses of G+C content, synonymous codon usage, amino acid usage and experimental evidence of host range) will improve our understanding of the potential long-term hosts and host range of plasmids, and how this is shaped by plasmid–host interactions.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
FUNDING
National Science Foundation (EF-0627988); National Institutes of Health (P20RR016454, P20RR16448). Funding for open access charges: National Science Foundation (EF-0627988).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of IBEST (Initiative for Bioinformatics and Evolutionary Studies) at the University of Idaho, Azusa Kuroki for useful discussions and Kazuharu Arakawa for his technical advice on the G-language Genome Analysis Environment. We are also grateful to the editor and three anonymous reviewers because their comments greatly improved our article.
REFERENCES
- 1.Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 2005;3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 2.Croft AC, D’Antoni AV, Terzulli SL. Update on the antibacterial resistance crisis. Med. Sci. Monit. 2007;13:RA103–RA118. [PubMed] [Google Scholar]
- 3.Martinez JL. Antibiotics and antibiotic resistance genes in natural environments. Science. 2008;321:365–367. doi: 10.1126/science.1159483. [DOI] [PubMed] [Google Scholar]
- 4.Sota M, Top EM. In: Plasmids: Current Research and Future Trends. Bayreuth G, editor. Norfolk, UK: Caister Academic Press; 2008. p. 111.p. 181. [Google Scholar]
- 5.Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 2004;68:669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones BV, Marchesi JR. Transposon-aided capture (TRACA) of plasmids resident in the human gut mobile metagenome. Nat. Methods. 2007;4:55–61. doi: 10.1038/nmeth964. [DOI] [PubMed] [Google Scholar]
- 7.Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, Takami H, Morita H, Sharma VK, Srivastava TP, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ono A, Miyazaki R, Sota M, Ohtsubo Y, Nagata Y, Tsuda M. Isolation and characterization of naphthalene-catabolic genes and plasmids from oil-contaminated soil by using two cultivation-independent approaches. Appl. Microbiol. Biotechnol. 2007;74:501–510. doi: 10.1007/s00253-006-0671-4. [DOI] [PubMed] [Google Scholar]
- 9.Lawrence JG, Ochman H. Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- 10.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 11.Campbell A, Mrazek J, Karlin S. Genome signature comparisons among prokaryote, plasmid, and mitochondrial DNA. Proc. Natl Acad. Sci. USA. 1999;96:9184–9189. doi: 10.1073/pnas.96.16.9184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dufraigne C, Fertil B, Lespinats S, Giron A, Deschavanne P. Detection and characterization of horizontal transfers in prokaryotes using genomic signature. Nucleic Acids Res. 2005;33:e6. doi: 10.1093/nar/gni004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Vallve S, Guzman E, Montero MA, Romeu A. HGT-DB: a database of putative horizontally transferred genes in prokaryotic complete genomes. Nucleic Acids Res. 2003;31:187–189. doi: 10.1093/nar/gkg004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Vallve S, Romeu A, Palau J. Horizontal gene transfer in bacterial and archaeal complete genomes. Genome Res. 2000;10:1719–1725. doi: 10.1101/gr.130000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura Y, Itoh T, Matsuda H, Gojobori T. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat. Genet. 2004;36:760–766. doi: 10.1038/ng1381. [DOI] [PubMed] [Google Scholar]
- 16.Rosas-Magallanes V, Deschavanne P, Quintana-Murci L, Brosch R, Gicquel B, Neyrolles O. Horizontal transfer of a virulence operon to the ancestor of Mycobacterium tuberculosis. Mol. Biol. Evol. 2006;23:1129–1135. doi: 10.1093/molbev/msj120. [DOI] [PubMed] [Google Scholar]
- 17.Karlin S, Burge C. Dinucleotide relative abundance extremes: a genomic signature. Trends Genet. 1995;11:283–290. doi: 10.1016/s0168-9525(00)89076-9. [DOI] [PubMed] [Google Scholar]
- 18.Karlin S. Detecting anomalous gene clusters and pathogenicity islands in diverse bacterial genomes. Trends Microbiol. 2001;9:335–343. doi: 10.1016/s0966-842x(01)02079-0. [DOI] [PubMed] [Google Scholar]
- 19.Jernigan RW, Baran RH. Pervasive properties of the genomic signature. BMC Genomics. 2002;3:23. doi: 10.1186/1471-2164-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong K, Finan TM, Golding GB. Dinucleotide compositional analysis of Sinorhizobium meliloti using the genome signature: distinguishing chromosomes and plasmids. Funct. Integr. Genomics. 2002;2:274–281. doi: 10.1007/s10142-002-0068-0. [DOI] [PubMed] [Google Scholar]
- 21.Coenye T, Vandamme P. Use of the genomic signature in bacterial classification and identification. Syst. Appl. Microbiol. 2004;27:175–185. doi: 10.1078/072320204322881790. [DOI] [PubMed] [Google Scholar]
- 22.van Passel MW, Bart A, Thygesen HH, Luyf AC, van Kampen AH, van der Ende A. An acquisition account of genomic islands based on genome signature comparisons. BMC Genomics. 2005;6:163. doi: 10.1186/1471-2164-6-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Hill K, Singh S, Kari L. The spectrum of genomic signatures: from dinucleotides to chaos game representation. Gene. 2005;346:173–185. doi: 10.1016/j.gene.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 24.Quirke AM, Reen FJ, Claesson MJ, Boyd EF. Genomic island identification in Vibrio vulnificus reveals significant genome plasticity in this human pathogen. Bioinformatics. 2006;22:905–910. doi: 10.1093/bioinformatics/btl015. [DOI] [PubMed] [Google Scholar]
- 25.van Passel MW, Bart A, Luyf AC, van Kampen AH, van der Ende A. Compositional discordance between prokaryotic plasmids and host chromosomes. BMC Genomics. 2006;7:26. doi: 10.1186/1471-2164-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Passel MW, Kuramae EE, Luyf AC, Bart A, Boekhout T. The reach of the genome signature in prokaryotes. BMC Evol. Biol. 2006;6:84. doi: 10.1186/1471-2148-6-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mrazek J, Karlin S. Distinctive features of large complex virus genomes and proteomes. Proc. Natl Acad. Sci. USA. 2007;104:5127–5132. doi: 10.1073/pnas.0700429104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Srividhya KV, Alaguraj V, Poornima G, Kumar D, Singh GP, Raghavenderan L, Katta AV, Mehta P, Krishnaswamy S. Identification of prophages in bacterial genomes by dinucleotide relative abundance difference. PLoS ONE. 2007;2:e1193. doi: 10.1371/journal.pone.0001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karlin S, Campbell AM, Mrazek J. Comparative DNA analysis across diverse genomes. Annu. Rev. Genet. 1998;32:185–225. doi: 10.1146/annurev.genet.32.1.185. [DOI] [PubMed] [Google Scholar]
- 30.Crawford JR, Allan KM. The Mahalanobis Distance index of WAIS-R subtest scatter: psychometric properties in a healthy UK sample. Br. J. Clin. Psychol. 1994;33:65–69. doi: 10.1111/j.2044-8260.1994.tb01094.x. [DOI] [PubMed] [Google Scholar]
- 31.Lachenbruch PA. Discriminant diagnostics. Biometrics. 1997;53:1284–1292. [PubMed] [Google Scholar]
- 32.Wu TJ, Burke JP, Davison DB. A measure of DNA sequence dissimilarity based on Mahalanobis distance between frequencies of words. Biometrics. 1997;53:1431–1439. [PubMed] [Google Scholar]
- 33.Arakawa K, Mori K, Ikeda K, Matsuzaki T, Kobayashi Y, Tomita M. G-language genome analysis environment: a workbench for nucleotide sequence data mining. Bioinformatics. 2003;19:305–306. doi: 10.1093/bioinformatics/19.2.305. [DOI] [PubMed] [Google Scholar]
- 34.Arakawa K, Tomita M. G-language System as a platform for large-scale analysis of high-throughput omics data. J. Pesticide Sci. 2006;31:282–288. [Google Scholar]
- 35.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2007. [Google Scholar]
- 36.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pride DT, Wassenaar TM, Ghose C, Blaser MJ. Evidence of host-virus co-evolution in tetranucleotide usage patterns of bacteriophages and eukaryotic viruses. BMC Genomics. 2006;7:8. doi: 10.1186/1471-2164-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okinaka R, Cloud K, Hampton O, Hoffmaster A, Hill K, Keim P, Koehler T, Lamke G, Kumano S, Manter D, et al. Sequence, assembly and analysis of pX01 and pX02. J. Appl. Microbiol. 1999;87:261–262. doi: 10.1046/j.1365-2672.1999.00883.x. [DOI] [PubMed] [Google Scholar]
- 39.Okinaka RT, Cloud K, Hampton O, Hoffmaster AR, Hill KK, Keim P, Koehler TM, Lamke G, Kumano S, Mahillon J, et al. Sequence and organization of pXO1, the large Bacillus anthracis plasmid harboring the anthrax toxin genes. J. Bacteriol. 1999;181:6509–6515. doi: 10.1128/jb.181.20.6509-6515.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tinsley E, Khan SA. A novel FtsZ-like protein is involved in replication of the anthrax toxin-encoding pXO1 plasmid in Bacillus anthracis. J. Bacteriol. 2006;188:2829–2835. doi: 10.1128/JB.188.8.2829-2835.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burland V, Shao Y, Perna NT, Plunkett G, Sofia HJ, Blattner FR. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 1998;26:4196–4204. doi: 10.1093/nar/26.18.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guiney DG. Host range of conjugation and replication functions of the Escherichia coli sex plasmid Flac. Comparison with the broad host-range plasmid RK2. J. Mol. Biol. 1982;162:699–703. doi: 10.1016/0022-2836(82)90397-7. [DOI] [PubMed] [Google Scholar]
- 43.Schlueter A, Szczepanowski R, Puehler A, Top EM. Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. FEMS Microbiol. Rev. 2007;31:449–477. doi: 10.1111/j.1574-6976.2007.00074.x. [DOI] [PubMed] [Google Scholar]
- 44.Top E, De Smet I, Verstraete W, Dijkmans R, Mergeay M. Exogenous isolation of mobilizing plasmids from polluted soils and sludges. Appl. Environ. Microbiol. 1994;60:831–839. doi: 10.1128/aem.60.3.831-839.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schlueter A, Heuer H, Szczepanowski R, Forney LJ, Thomas CM, Puehler A, Top EM. The 64 508 bp IncP-1β antibiotic multiresistance plasmid pB10 isolated from a waste-water treatment plant provides evidence for recombination between members of different branches of the IncP-1β group. Microbiology. 2003;149:3139–3153. doi: 10.1099/mic.0.26570-0. [DOI] [PubMed] [Google Scholar]
- 46.De Gelder L, Vandecasteele FP, Brown CJ, Forney LJ, Top EM. Plasmid donor affects host range of promiscuous IncP-1β plasmid pB10 in an activated-sludge microbial community. Appl. Environ. Microbiol. 2005;71:5309–5317. doi: 10.1128/AEM.71.9.5309-5317.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas CM, Smith CA. Incompatibility group P plasmids: genetics, evolution, and use in genetic manipulation. Annu. Rev. Microbiol. 1987;41:77–101. doi: 10.1146/annurev.mi.41.100187.000453. [DOI] [PubMed] [Google Scholar]
- 48.Rocha EP, Danchin A. Base composition bias might result from competition for metabolic resources. Trends Genet. 2002;18:291–294. doi: 10.1016/S0168-9525(02)02690-2. [DOI] [PubMed] [Google Scholar]
- 49.van Passel MW, van der Ende A, Bart A. Plasmid diversity in Neisseriae. Infect. Immun. 2006;74:4892–4899. doi: 10.1128/IAI.02087-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szpirer C, Top E, Couturier M, Mergeay M. Retrotransfer or gene capture: a feature of conjugative plasmids, with ecological and evolutionary significance. Microbiology. 1999;145:3321–3329. doi: 10.1099/00221287-145-12-3321. [DOI] [PubMed] [Google Scholar]
- 51.Oresnik IJ, Liu SL, Yost CK, Hynes MF. Megaplasmid pRme2011a of Sinorhizobium meliloti is not required for viability. J. Bacteriol. 2000;182:3582–3586. doi: 10.1128/jb.182.12.3582-3586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turner SL, Bailey MJ, Lilley AK, Thomas CM. Ecological and molecular maintenance strategies of mobile genetic elements. FEMS Microbiol. Ecol. 2002;42:177–185. doi: 10.1111/j.1574-6941.2002.tb01007.x. [DOI] [PubMed] [Google Scholar]
- 53.Wernegreen JJ, Harding EE, Riley MA. Rhizobium gone native: unexpected plasmid stability of indigenous Rhizobium leguminosarum. Proc. Natl Acad. Sci. USA. 1997;94:5483–5488. doi: 10.1073/pnas.94.10.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoffmaster AR, Ravel J, Rasko DA, Chapman GD, Chute MD, Marston CK, De BK, Sacchi CT, Fitzgerald C, Mayer LW, et al. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc. Natl Acad. Sci. USA. 2004;101:8449–8454. doi: 10.1073/pnas.0402414101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rasko DA, Altherr MR, Han CS, Ravel J. Genomics of the Bacillus cereus group of organisms. FEMS Microbiol. Rev. 2005;29:303–329. doi: 10.1016/j.femsre.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 56.Green BD, Battisti L, Thorne CB. Involvement of Tn4430 in transfer of Bacillus anthracis plasmids mediated by Bacillus thuringiensis plasmid pXO12. J. Bacteriol. 1989;171:104–113. doi: 10.1128/jb.171.1.104-113.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamamoto T, Taneike I. The sequences of enterohemorrhagic Escherichia coli and Yersinia pestis that are homologous to the enteroaggregative E. coli heat-stable enterotoxin gene: cross-species transfer in evolution. FEBS Lett. 2000;472:22–26. doi: 10.1016/s0014-5793(00)01414-9. [DOI] [PubMed] [Google Scholar]
- 58.Hinnebusch BJ, Rosso ML, Schwan TG, Carniel E. High-frequency conjugative transfer of antibiotic resistance genes to Yersinia pestis in the flea midgut. Mol. Microbiol. 2002;46:349–354. doi: 10.1046/j.1365-2958.2002.03159.x. [DOI] [PubMed] [Google Scholar]
- 59.Charlebois RL, Doolittle WF. Computing prokaryotic gene ubiquity: rescuing the core from extinction. Genome Res. 2004;14:2469–2477. doi: 10.1101/gr.3024704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uchiyama I. MBGD: microbial genome database for comparative analysis. Nucleic Acids Res. 2003;31:58–62. doi: 10.1093/nar/gkg109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Uchiyama I. MBGD: a platform for microbial comparative genomics based on the automated construction of orthologous groups. Nucleic Acids Res. 2007;35:D343–D346. doi: 10.1093/nar/gkl978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Edwards RA, Rohwer F. Viral metagenomics. Nat. Rev. Microbiol. 2005;3:504–510. doi: 10.1038/nrmicro1163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.