Abstract
Rotavirus RNA-dependent RNA polymerase, VP1, catalyzes RNA synthesis within a subviral particle. This activity depends on core shell protein VP2. A conserved sequence at the 3′ end of plus-strand RNA templates is important for polymerase association and genome replication. We have determined the structure of VP1 at 2.9 Å resolution, as apoenzyme and in complex with RNA. The cage-like enzyme is similar to reovirus λ3, with four tunnels leading to or from a central, catalytic cavity. A distinguishing characteristic of VP1 is specific recognition, by conserved features of the template-entry channel, of four bases, UGUG, in the conserved 3′ sequence. Well-defined interactions with these bases position the RNA so that its 3′ end overshoots the initiating register, producing a stable but catalytically inactive complex. We propose that specific 3′ end recognition selects rotavirus RNA for packaging and that VP2 activates the auto-inhibited VP1/RNA complex to coordinate packaging and genome replication.
Introduction
Viral RNA-dependent RNA polymerases (RdRPs) share a common catalytic mechanism and exhibit highly conserved architectures, sequence motifs, and catalytic residues (O'Reilly and Kao, 1998). Despite their similarities, individual viral RdRPs differ in the ways they engage RNA templates, prime RNA synthesis, and coordinate their activities with other processes in the infected cell. RdRPs of segmented double-stranded RNA (dsRNA) viruses of the Reoviridae family catalyze RNA synthesis while contained within subviral particles (Lawton et al., 2000). The structures and functions of Reoviridae RdRPs are expected to reflect this particle-associated RNA synthesis mechanism (Butcher et al., 2001; Tao et al., 2002).
Rotaviruses, members of the Reoviridae family, are nonenveloped particles with three concentric protein layers surrounding 11 segments of dsRNA (Estes and Kapikian, 2007; Prasad et al., 1996). The outermost layer is shed during cell entry, resulting in a transcriptionally active, double-layered particle (DLP). The core of the DLP is built from 60 asymmetrical dimers of shell protein VP2, which forms the DLP inner layer (Lawton et al., 1997; Prasad et al., 1996; Yeager et al., 1990). Copies of VP1, the viral RdRP, and VP3, the RNA capping enzyme, are tethered within the core, near each fivefold axis (Chen et al., 1999; Liu et al., 1992; Pizarro et al., 1991; Valenzuela et al., 1991). Using dsRNA genome segments as templates, the tethered VP1-VP3 complexes synthesize capped, non-polyadenylated, plus-strand (+RNA) molecules that are extruded from the DLP into the cytoplasm of the infected cell (Lawton et al., 1997; Lawton et al., 2000; Prasad et al., 1996). There, the +RNAs function as mRNAs for translation and, after association with VP1 and incorporation into cores (see below), as templates for minus-strand (−RNA) synthesis and formation of genomic dsRNA.
Studies of rotavirus replication intermediates (RIs) suggest that core assembly is initiated by the interaction of VP1 and VP3 with +RNA, forming a pre-core RI that lacks polymerase activity (Gallegos and Patton, 1989; Patton, 1996). VP2 multimers then interact with the pre-core RI to form a core RI, which can initiate −RNA synthesis to produce dsRNA (Gallegos and Patton, 1989; Patton and Gallegos, 1990). The VP2-dependent activity of VP1 coordinates genome replication and RNA packaging (Estes and Kapikian, 2007), preventing the formation of naked dsRNAs in the infected cell (Labbe et al., 1994; Patton, 1996; Zeng et al., 1998).
Experiments with recombinant proteins have characterized the requirements for initiation of rotavirus dsRNA synthesis. Formation of a -RNA initiation complex in vitro is a salt-sensitive process requiring VP1, VP2, GTP, Mg2+, and +RNA template (Chen and Patton, 2000; Tortorici et al., 2003). VP1 interacts with +RNA in the absence of VP2, demonstrating that the role of VP2 extends beyond assisting in recruitment of template RNA to the polymerase (Chen and Patton, 2000; Patton, 1996; Tortorici et al., 2003). A cis-acting +RNA element, the 3′ consensus sequence (3′CS+: 5′-UGUGACC-3′), is critical for template recognition by VP1. The UGUG tetranucleotide of the 3′CS+ is a polymerase recognition signal that mediates high-affinity interactions with VP1 (Chen et al., 2001; Tortorici et al., 2003). The terminal ACC segment of the 3′CS+ is important for initiating minus-strand synthesis, but it is dispensable for specific VP1 binding.
Once dsRNA synthesis is complete, the polymerase switches to a transcriptional mode, with the −RNA as template. During transcription, VP1 initiates +RNA synthesis templated from the 3′ end of −RNA (3′CS-: 5′-(A/U)7CC-3′); thus, the RdRP can clearly function on non-UGUG-containing templates. While high-affinity binding of VP1 to the 3′CS+ UGUG nucleotides is apparently critical for pre-core RI formation and for initiation of dsRNA synthesis, it may not be so critical during initiation of transcription, for which polymerase and template are both sequestered within the confines of the core.
How does VP1 engage the UGUG nucleotides of the 3′CS+ and what is the significance of the resulting interactions during dsRNA synthesis? We have determined the atomic structures of the VP1 apoenzyme and of its complexes with various RNA oligonucleotides. The structures of the VP1/RNA complexes show that VP1 has sequence-specific interactions with the UGUG bases of the 3′CS+ and that these contacts stabilize the template in the catalytic site, one nucleotide past the initiation register. This auto-inhibited VP1/+RNA complex must undergo a structural re-organization following engagement by the core shell protein VP2, leading to activation of the RdRP. The results presented here thus provide a mechanism for coordination of rotavirus RNA packaging and genome replication during infection.
Results
Structure of the VP1 apoenzyme
Recombinant VP1 with a hexahistidine (His) tag fused at either its N- or C-terminus (nVP1 and cVP1, respectively) was purified to >95% homogeneity as a soluble, monomeric, 125-kDa protein. While nVP1 catalyzed only low levels of dsRNA synthesis in vitro, the activity of cVP1 was like that of untagged recombinant VP1 with an incorporation rate of ∼180 nt/min (Supplementary Fig. 1). Because of this robust activity, cVP1 was used for structural analyses. We obtained crystals of cVP1 (hereafter referred to as VP1) using low ionic strength crystallization conditions, similar to those used in assaying VP1 in vitro for polymerase activity. The space group of the apoenzyme crystals was P212121, a= 77.7, b= 112.0, c= 142.2, with one molecule per asymmetric unit. The VP1 structure was determined from data extending to a minimum Bragg spacing of 2.9 Å. Experimental phases were obtained from multiple wavelength anomalous dispersion (MAD) using crystals of selenomethionine (SeMet) substituted VP1 (Table 1). Experimental maps, improved by density modification, allowed unambiguous placement of 90% of the VP1 polypeptide chain. The final, refined atomic model of VP1 contains 1073 out of 1095 amino-acid residues. The missing regions include the N-terminal methionine, two flexible loops (aa 18–22 and 346–358), and the C-terminal His tag.
Table 1. Data Collection Statistics for Native Structure Determination.
| Data Set | Native | SeMet Derivative #1 | SeMet Derivative #2 | SeMet Derivative #3 | |||||
|---|---|---|---|---|---|---|---|---|---|
| Peaka | Inflection | Peak | Inflection | Remote | Peak | Inflection | Remote | ||
| Wavelength (Å) | 1.1808 | 0.9794 | 0.9797 | 0.9794 | 0.9797 | 0.9641 | 0.9794 | 0.9797 | 0.9641 |
| Number of unique reflections | 27999 | 33959 | 25264 | 30517 | 29663 | 24300 | 22928 | 24042 | 22768 |
| Completeness (%)b | 99.9 (99.6) | 84.3 (70.6) | 88.5 (78.3) | 92.8 (71.8) | 89.8 (61.5) | 94.0 (81.0) | 65.1 (53.7) | 73.3 (70.0) | 67.3 (54.0) |
| Resolution (Å) | 30-2.9 | 50-3.2 | 50-3.6 | 50-3.4 | 50-3.4 | ||||
| I/σb | 25.0 (3.5) | 7.9 (2.2) | 6.7 (2.4) | 4.8 (1.2) | 5.1 (1.1) | 5.5 (1.5) | 4.8 (1.3) | 5.1 (1.5) | 4.8 (1.3) |
| Rmergeb,c | .055 (41.0) | 0.083 (.39) | 0.098 (.36) | 0.167 (.64) | 0.161 (.67) | 0.141 (.55) | 0.125 (.51) | 0.135 (.52) | 0.130 (.50) |
| Isomorphous Rcullisd (centric/accentric) | 0.76/0.75 | - | 0.95/0.94 | 0.96/0.91 | 0.93/0.90 | 1.02/0.98 | 0.92/0.91 | 0.92/0.91 | 0.91/0.90 |
| Anomalous Rcullisd (accentric) | - | 0.79 | 0.96 | 0.87 | 0.97 | 0.96 | 0.93 | 0.99 | 0.96 |
| Isomorphous Phasing Powere (centric/accentric) | 0.82/0.88 | - | 0.29/0.31 | - | 0.33/0.33 | 0.24/0.26 | - | 0.21/0.24 | 0.22/0.25 |
| Anomalous Phasing Powere (accentric) | - | 1.15 | 0.5 | 0.73 | 0.41 | 0.44 | 0.58 | 0.32 | 0.36 |
| Figure of merit (centric/accentric) | 0.20/0.28 | ||||||||
|
| |||||||||
| Refinement statistics | 50-2.9 | ||||||||
| R | 23.3% for 24785 reflections | ||||||||
| Rfree | 28.6% for 2136 reflections | ||||||||
| Rms bond length, bond angles | 0.0067 Å, 1.14° | ||||||||
This data set was used as the reference data set in phase refinement using SHARP.
Numbers in parentheses are for the highest-resolution bins.
Rmerge = ΣhΣi |Ihi - 〈Ih〉 |/ΣhΣi Ihi
RCullis = Σh| |Fp(h) - FPH(h)| - |FHcalc(h)| | / Σh|Fp(h) - FPH(h) |, where FP, FPH and FH are the amplitudes of the native, SeMet derivatives, and Se atoms, respectively.
Phasing power = rms(FH/E), where E is the lack of closure error.
VP1 is a compact, globular protein, ∼70 Å in diameter. There are three distinct domains: (i) an N-terminal domain (aa 1–332), (ii) a polymerase domain comprising fingers, palm, and thumb subdomains (aa 333–78), and (iii) a C-terminal “bracelet” domain (aa 779–1089) (Fig. 1). Together, the N- and C-terminal domains sandwich most of the polymerase domain, creating a cage structure with the catalytic region located within a largely hollow center. The structure of VP1 is very similar to that of the reovirus RdRP, λ3 (Tao et al., 2002). Comparison of VP1 and λ3 reveals shared features, as well as features unique to VP1 (Supplementary Fig. 2). As in λ3, four tunnels lead to the VP1 catalytic center (Fig. 1b, c); comparison with the λ3 elongation complexes allows the functions of these tunnels to be assigned (Fig. 1c).
Figure 1. Structure of the VP1 apoenzyme.
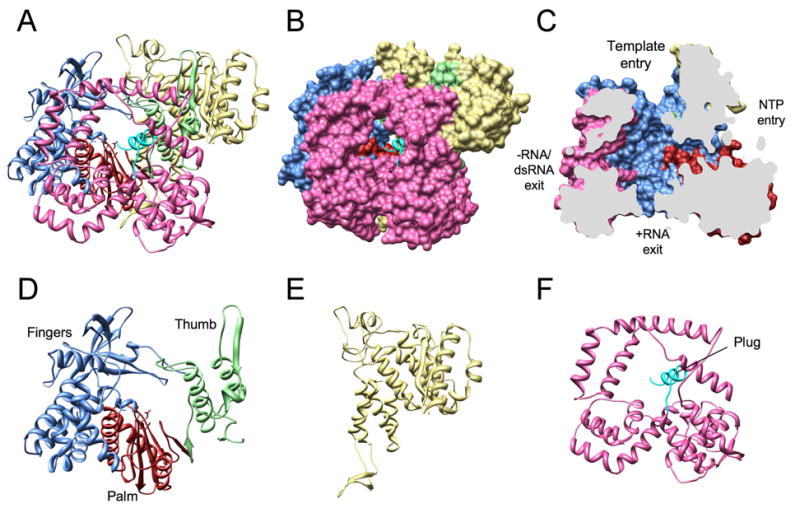
Ribbon diagram (A) and surface rendering (B) of the complete VP1 polypeptide chain. The N-terminal domain is in yellow, the C-terminal (bracelet) domain in pink, and the C-terminal plug in cyan. The conventionally designated subdomains of the polymerase domain are in light blue (fingers), red (palm), and green (thumb). (C) Sagittal cutaway of the image in (B), after rotation to the left by 90°, showing the four tunnels extending into the central cavity. (D-F) Ribbon diagrams of the VP1 domains: polymerase, N-terminal, and C-terminal domains.
The VP1 polymerase domain includes the six canonical motifs A–F shared by RdRPs (Supplementary Fig. 3b) and resembles the polymerase domains of HIV reverse transcriptase(Kohlstaedt et al., 1992), hepatitis C virus NS5B(Ago et al., 1999; Bressanelli et al., 1999; Lesburg et al., 1999), and bacteriophage φ6 P2(Butcher et al., 2001), in addition to reovirus λ3 (Supplementary Fig. 2)(Tao et al., 2002). The palm subdomain (aa 489-523, 596-685) is a four-stranded, antiparallel β sheet supported by three α helices (Fig. 1d). It includes the conserved acidic residues of the active site (Asp520 and Asp525 of motif A; Asp631 and Asp632 of motif C)(Doublie and Ellenberger, 1998; Joyce and Steitz, 1994). At the junction of the palm and thumb subdomains, a small sheet with a β-turn forms a substructure found in other polymerases and known as the “primer grip”(Jacobo-Molina et al., 1993) (Supplementary Fig. 2c, d). There is also, as in λ3 (aa 557–567), a so-called “priming loop”, a flexible structural element that binds the triphosphates of the priming nucleotide during initiation. The priming loop is an exception to the similarities of the palm subdomains of polymerases from many different kinds of viruses. In VP1, the putative priming loop (aa 489–499) bends away from the nucleotide-binding site by about 90° (Supplementary Fig. 2e, f), leaving it in a retracted (‘down’) state incapable of supporting an initiation nucleotide in the priming (P) site.
The VP1 fingers subdomain (aa 333–488, 524–595) lacks a four-stranded β sheet that protrudes from the surface of the λ3 fingers (λ3 aa 400–419, 617–643) (Supplementary Fig. 2d). Moreover, the structural elements of the VP1 fingers that comprise one side of the template entry tunnel (VP1 aa 395–420; λ3 aa 455–491) are somewhat different in their three-dimensional arrangment from those in λ3, consistent with the likely role of these segments in UGUG recognition (see below). The VP1 thumb subdomain (aa 686–778), like that of λ3, includes a β strand followed by three α helices. A loop at the tip of the VP1 thumb (aa 697–700) interacts with the tip of the fingers, enclosing the catalytic site on the palm and creating a constriction in the template entry tunnel (Fig. 1d). The fingers-thumb contact appears to maintain VP1 in a closed conformation even in its resting state. A β-ribbon of the fingers subdomain (aa 445–466) reaches beyond the thumb interface. This structure, representing motif F, includes three arginine residues (Arg452, Arg457, Arg460) that project toward the binding site for the incoming nucleotide (N site) (Supplementary Fig. 2e). In the case of λ3, similar motif-F arginine residues act with conserved aspartic-acid residues of the palm subdomain, and with residues of the extended priming loop to stabilize nucleotides in the P and N sites during initiation(Tao et al., 2002) (Supplementary Fig. 2f).
Surrounding the VP1 polymerase core domain are the N- and C-terminal domains (Fig. 1). The N-terminal domain covers one side of the active site cleft and reinforces the closure induced by the fingers and thumb subdomains. The C-terminal bracelet domain of VP1, absent in other known viral RdRP structures with the exception of λ3, encircles the exit tunnel for the dsRNA product of replication and for the −RNA template of transcription (Fig. 1f). The C-terminal residues of VP1 (aa 1072–1089) form an α-helical plug, which extends 15 Å into the −RNA/dsRNA exit tunnel (Fig. 1b, f). In λ3, the corresponding residues are part of the C-terminal bracelet domain, away from the exit tunnel (Supplementary Fig. 2b). The plug is also present in structures from crystals of nVP1, showing that its observed position in cVP1 is not an artifact of the C-terminal His tag (Fig. 2a).
Figure 2. Polymerase activity of VP1 plug mutants.
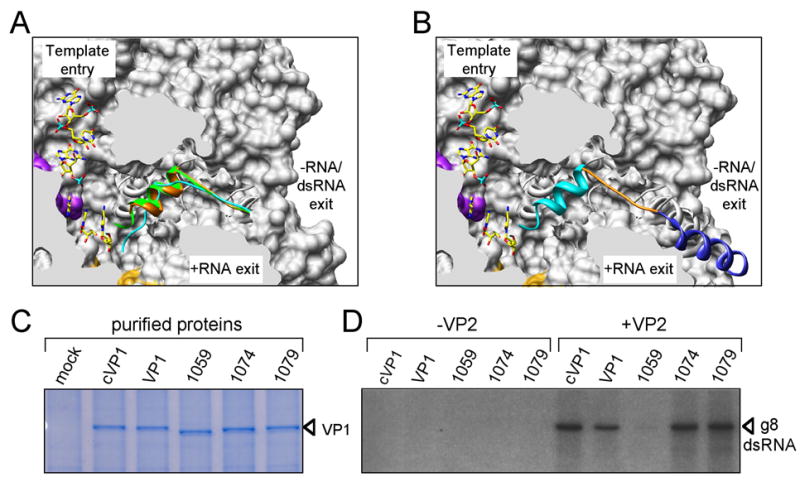
(A) Sagittal cutaway of a surface rendering of VP1, showing the positions of the C-terminal plug in the -RNA/dsRNA exit tunnel. The plugs for the cVP1 apoenzyme (orange), the cVP1/3′CS+ complex (green), and the nVP1 apoenzyme (cyan) are superimposed. A 3′CS+ oligonucleotide is shown in the template entry tunnel, and surfaces of motif F (purple) and the priming loop (gold) are colored for reference. (B) Same image as in (A), but showing only the locations of the cVP1 plug and the connecting α-helix in the C-terminal bracelet (dark blue). The region deleted in each VP1 mutant is indicated by color: 1079 (turquoise), 1074 (orange), and 1059 (dark blue). (C) VP1 proteins were purified using differential centrifugation and analyzed for relative purity by SDS-PAGE and Coomassie blue staining. Wildtype, His-tagged VP1 (cVP1) and untagged (VP1) served as controls for assaying the untagged plug mutants (1059, 1074, and 1079). (D) In vitro dsRNA synthesis. Reaction mixtures contained either wildtype or mutant VP1 and gene 8 +RNA, in the presence or absence of VP2. Radiolabeled dsRNA products were resolved by SDS-PAGE and detected by autoradiography.
Without the intruding C-terminus, the −RNA/dsRNA exit tunnel of VP1 (Fig. 2a) has a diameter of about 20 Å, sufficient to accommodate dsRNA. In contrast, the presence of the plug reduces the diameter to 10–12Å, and the plug must therefore move away for egress of dsRNA. It can clearly do so, as the C-terminal His tag was exposed during purification of the protein. We generated mutant VP1 proteins containing C-terminal deletions of 36, 21, and 16 residues (designated mutants 1059, 1074, and 1079, respectively) and assayed them for polymerase activity in vitro in the presence and absence of VP2 (Fig. 2b, c). Mutants 1074 and 1079 supported dsRNA synthesis at levels indistinguishable from wildtype VP1 and remained dependent on VP2 for activity (Fig. 2d). Thus, activation by VP2 must do more than simply expel the C-terminal plug. The mutated VP1 (1059) containing a deletion of both the plug and an α-helix of the bracelet domain showed diminished but still VP2-dependent activity (Fig. 2d).
VP1 specifically recognizes +RNA templates
We soaked VP1 apoenzyme crystals with an RNA oligonucleotide representing the 3′CS+ (5′-UGUGACC-3′) (Fig. 3, and Supplementary Fig. 4). For describing the structure, we number the nucleotides sequentially from 3′ to 5′ (e.g., C1 to U7). Soaked crystals retained P212121 symmetry and the VP1/3′CS+ structure was determined by molecular replacement using the apoenzyme form as a phasing model (Table 2). Difference maps from crystals soaked with the 3′CS+ oligonucleotide showed strong electron density in the template entry tunnel, and we could position individual nucleotides unambiguously (Fig. 3 and Supplementary Fig. 4). A network of ten hydrogen bonds to the phosphates and ribose rings and eight hydrogen bonds to the bases hold the oligonucleotide in position (Fig. 4a). VP1 residues contributing to this network are predominantly those of the fingers subdomain, but residues of the N-terminal domain also contribute. As in the various RNA oligonucleotide complexes of reovirus λ3, RNA binding produces only subtle conformational shifts in VP1. A loop of the fingers subdomain (aa 397–404) retracts from the central cavity, and Ser398 and Ser401 of the retracted loop form hydrogen bonds with the backbone of the RNA (Fig. 4c). In addition, an antiparallel β hairpin (Pβ1–2: aa 403–417) and an α-helix (Pα6: aa 420–430) connected to the loop has moved away from the active site. Extensive interactions between U7 of the 3′CS+ and Phe416 fix the position of the β-hairpin (Fig. 4c).
Figure 3. Structure of VP1 in complex with RNA oligonucleotides.
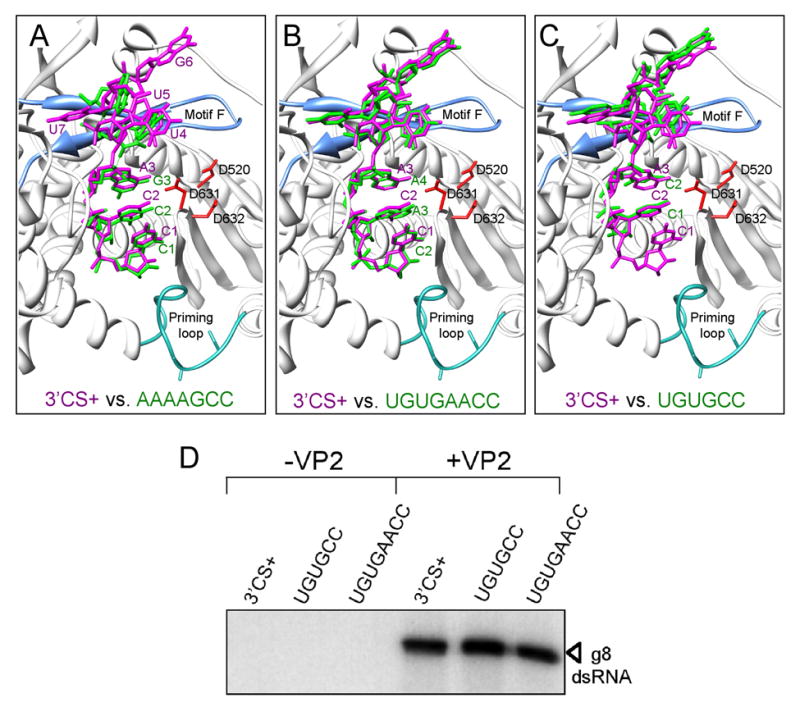
(A-C) The wildtype 3′CS+ oligonucleotide (magenta) in the template entry tunnel with its 3′ C1 residue in a one-nucleotide overshot position. In each panel, one other oligonucleotide is superimposed (green): (A) AAAAGCC, (B) UGUGAACC, or (C) UGUGCC The C1 residue could not be resolved in the VP1/ UGUGAACC complex. The priming loop (turquoise; sticks indicate Ser495 and Ser497), motif F (light blue), and catalytic residues Asp520, Asp631, and Asp632 (red) are shown. (D) In vitro dsRNA synthesis. Template activities of gene 8 +RNAs ending in the conventional 3′CS+ sequence, UGUGCC, or UGUGAACC, were assayed in reaction mixtures containing cVP1, in the presence or absence of VP2. Radiolabeled dsRNA products were resolved by SDS-PAGE and detected by autoradiography.
Table 2. Data Collection Statistics for Structure Determination of RNA Oligonucleotide/VP1 Complexes.
| Experiment | Apoenzyme | 0.5 mM (+)- 3′CS | 0.5 mM (+)- 3′CS + 1mM 3′-deoxy GTP | 0.5 mM (+)- 3′CS A3-deletion | 0.5 mM (+)- 3′CS A3-insertion | 0.5 mM (+)- 5′CS | 0.5 mM (-)- 3′CS |
|---|---|---|---|---|---|---|---|
| RNA Sequence 5′->3′ | UGUGACC | UGUGACC | UGUGCC | UGUGAACC | GGCUUU | AAAAGCC | |
| Soak time (hours) | 24 | 4 | 24 | 24 | 24 | 24 | |
| Space group | P212121 | P212121 | P21 | P212121 | P212121 | P212121 | P212121 |
| a=76.38, | a=76.35, | a=76.0, | a=77.12, | a=76.65, | a=76.45, | a=76.74, | |
| Unit cell dimensions | b=112.47, | b=112.75, | b=143.766, | b=112.66, | b=112.53, | b=112.31, | b=112.15, |
| c=143.05 | c=143.79 | c=112.833 | c=144.84 | c=143.58 | c=143.72 | c=144.31 | |
| α = β = γ = 90 | α = β = γ = 90 | α = γ = 90, β = 90.65 | α = β = γ = 90 | α = β = γ = 90 | α = β = γ = 90 | α = β = γ = 90 | |
| Resolution (Å) | 30-2.9 | 30-2.6 | 30-2.8 | 30-3.2 | 30-3.0 | 30-2.8 | 30-3.0 |
| Completeness (%)* | 99.9 (99.6) | 90.1 (45.1) | 81.9 (62.2) | 88.9 (85.7) | 91.8 (60.3) | 91.4 (47.6) | 92.4 (40.9) |
| I/σ* | 25.0 (3.5) | 18.1 (1.9) | 17.2 (2.7) | 13.7 (3.5) | 16.1 (1.7) | 18.0 (1.5) | 11.9 (1.2) |
| Rmerge (%)* | 5.5 (41.0) | 7.3 (39.7) | 5.1 (33.9) | 9.8 (40.4) | 7.6 (58.2) | 6.5 (53.7) | 10.5 (53.6) |
| Total observed reflections | 109,200 | 124,070 | 138,830 | 59,470 | 106,010 | 125,720 | 121,920 |
| Unique reflections | 28,000 | 35,449 | 49,581 | 18,536 | 23,558 | 28,572 | 23,905 |
| Refinement Statistics | |||||||
| R (%) | 23.32 | 23.7 | 22.5 | 21.53 | 23.5 | 22.8 | 23.4 |
| Rfree (%) | 28.56 | 28.7 | 27.85 | 28.18 | 28.1 | 27.9 | 28.9 |
| Rms bond length | 0.007 | 0.009 | 0.007 | 0.008 | 0.008 | 0.009 | 0.009 |
| Rms bond angles | 1.14 | 1.28 | 1.25 | 1.24 | 1.31 | 1.46 | 1.41 |
The numbers in parentheses refer to the highest-resolution shell.
Figure 4. VP1-template interactions.
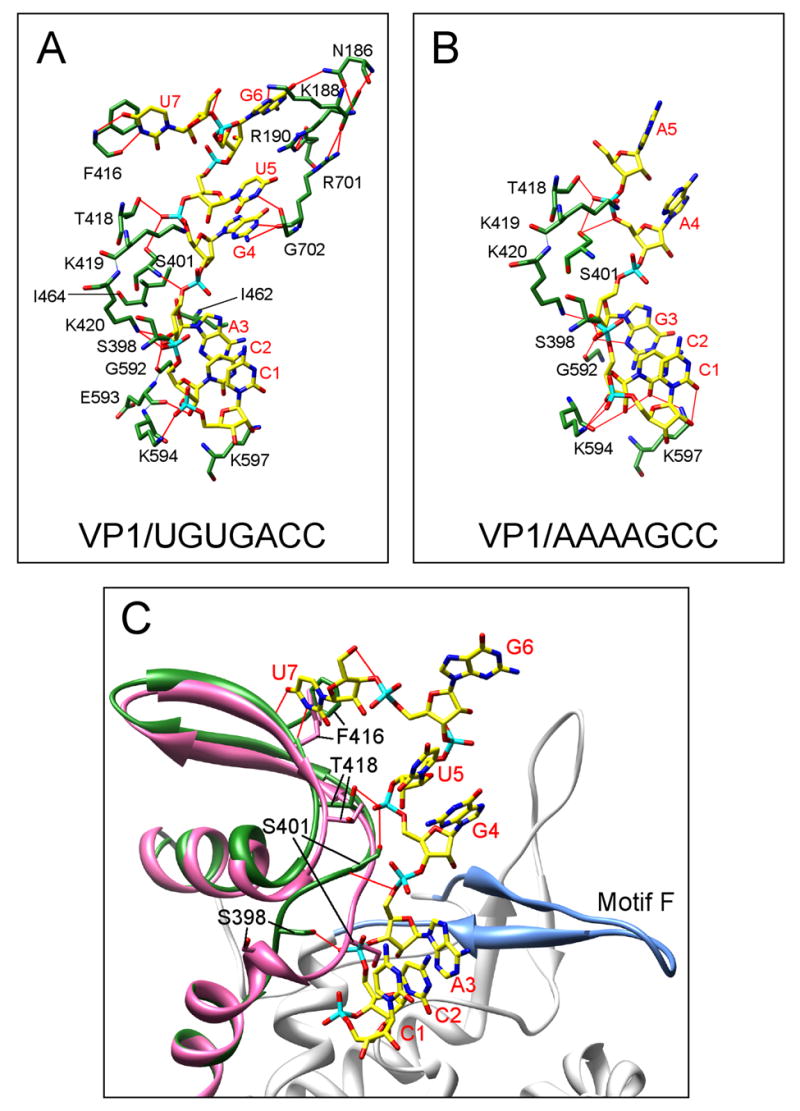
Identification of VP1 residues that engage (A) the 3′CS+ (UGUGACC) or (B) the 3′CS-(AAAAGCC). The relevant amino acids and nucleotides are labeled, and hydrogen bonds are indicated as red lines. Labeled residues are in the fingers subdomain, except for Arg186, Lys188, and Arg190, which are in the N-terminal domain. Both complexes are stabilized, in part, by stacking of the C1 and C2 bases on each other. The VP1/3′CS+ complex is further stabilized by the stacking of G4 and U5 bases on each other, the interaction of the U7 base with Phe416, and the interaction of the G6 base with Arg190 and Arg701. (C) Merged structures of the VP1 apoenzyme (pink) and the VP1/3′CS+ complex (green), showing a shift of a loop-sheet element in the fingers subdomain that accompanies RNA binding. Amino-acid residues and nucleotides are labeled, and hydrogen bonds are indicated as red lines.
Superposition of the VP1/3′CS+ complex and the λ3 initiation complex aligns C2 and A3 of the rotavirus 3′CS+ oligonucleotide with C1 and G2 of the reovirus oligonucleotide, 5′-AUUAGC-3′ (Fig. 5). Thus, C1 of the 3′CS+ overshoots the register for initiation by one nucleotide, presenting C2 and A3 in positions opposite the P and N sites, respectively. Peptide carbonyls of VP1 residues Gly592, Glu593 and Lys594 accept hydrogen bonds from the 2′-OH groups of nucleotides C2 and A3, consistent with the preference of this enzyme for RNA templates (Fig. 4a). The 3′ base, C1, stacks against C2, and there are hydrogen bonds from the ε-amino groups of Lys594 and Lys597 to the C2 ribose and phosphate. The ribose and base of the nucleotide in the templating position (A3) stacks tightly under the side chains of VP1 residues Ile462 and Ile464. As in other polymerase-template complexes, these interactions force the downstream template to bend away from the catalytic pocket and into the template entry channel. Contacts between the fingers and thumb subdomains contribute to the aperture between the template tunnel and the active site.
Figure 5. Comparison of the VP1/3′CS+ and the λ3 initiation complexes.
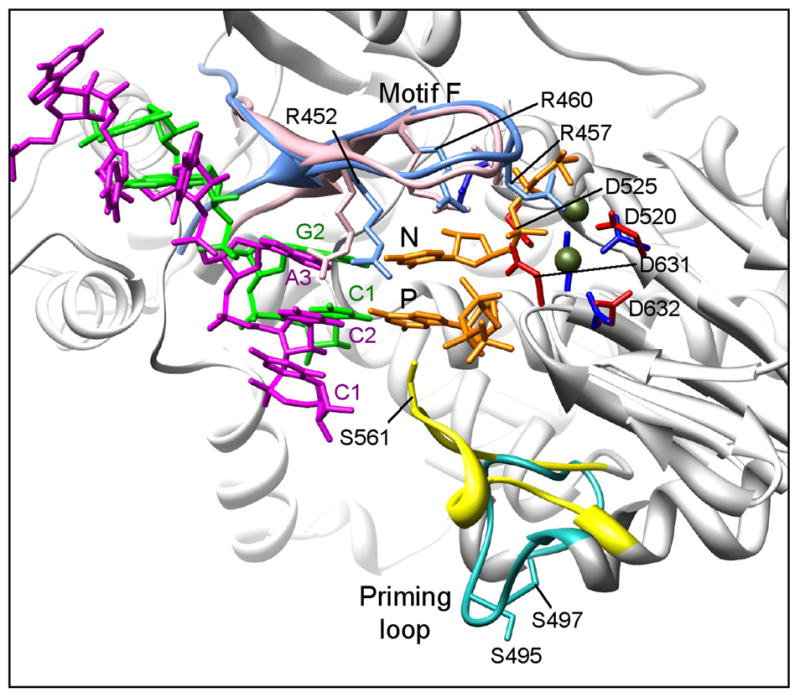
Elements of the rotavirus VP1/3′CS+ complex are superimposed on a ribbon representation of the reovirus λ3 initiation complex (light gray). The priming (P) and incoming (N) nucleotide positions are indicated, and Mg2+ ions are shown in olive. The template RNAs of rotavirus (UGUGACC, magenta) and reovirus (UAGC, green) and the incoming nucleotides (orange) are shown. The priming loop (λ3, yellow; VP1, turquoise) and motif F (λ3, pink; VP1, light blue) are colored, and the location of several key residues, including the catalytic aspartates (λ3, dark blue; VP1, red) and motif F arginines are indicated.
Unexpected features of the VP1/+RNA structure are the defined positions and hydrogen-bonding interactions of the UGUG nucleotides in the template entry channel (Fig. 4a). The hydrogen bonds fix the upstream bases in place and establish preferences for their identity. N1 of G4 and N3 of U5 donate hydrogen bonds to the peptide carbonyl groups of Arg701 and Gly702, respectively; the guanine base of G6 and the guanidinium groups of Arg190 and Arg701 form a three-layer stack, with the sandwiched Arg190 guanidinium group buttressed by the carboxylate of Asp127. Hydrogen bonds to G6 from the ε-amino group of Lys188 (to N7) and from the side-chain amide of N186 (to O6) together specify a guanine. The O4 of U7 accepts a hydrogen bond from the peptide amide group of Phe416, while its N3 donates a hydrogen bond to the peptide carbonyl of the same residue (Fig. 4a). In addition, the pyrimidine ring of U7 forms stacking interactions with the phenyl ring of Phe416. Such well-defined, base-specific interactions distinguish the rotavirus VP1/3′CS+ complex from those of other RdRPs studied to date.
To compare the interaction of VP1 with the 3′ end of its +RNA to that of its −RNA, VP1 apoenzyme crystals were soaked with an oligonucleotide representing the 3′CS− (5′-AAAAGCC-3′) (Fig. 3a and Supplementary Fig. 4). A network of nine hydrogen bonds, all mediated by residues of the fingers subdomain, holds this oligonucleotide in place (Fig. 4b). Seven of these same residues (Ser398, Ser401, Lys419, Lys420, Gly592, Lys594, Lys597) are also involved in anchoring the 3′CS+ into the template entry tunnel. Unlike the UGUG portion of the 3′CS+, the AAAA portion of the 3′CS− is not recognized specifically by the polymerase. The A4 and A5 residues are the only ordered components of the AAAA stretch, and they adopt orientations different from the corresponding bases (G4 and U5) of the 3′CS+ (Fig. 4b and Supplementary Fig. 4). Moreover, the only potentially base-specific interaction links the ε-amino group of Lys597 and O2 of the C2 base. Difference maps show that, like the 3′CS+, the 3′CS− oligonucleotide is in a one-nucleotide overshot register (Fig. 3a and Supplementary Fig. 4). Results from soaking other oligonucleotides (e.g., 5′-GGCUUU-3′) into VP1 crystals show that they bind in-register (Supplementary Fig. 4), excluding the possibility that all RNAs that move into the template entry channel are stabilized in an overshot position.
The 3′CS+ sequences of group A rotaviruses (5′-UGUGACC-3′) and group C rotaviruses (5′-UGUGGCU-3′) share the UGUG motif recognized by SA11 VP1. The VP1 amino-acid residues that interact with the 3′CS in the template channel are conserved not only in VP1s of essentially all group A viruses but also in VP1s of group C, which otherwise have only about 20% sequence identity with those of group A. Genomic segments from group B rotaviruses have a different four-base consensus sequence (5′-ACCC-3′) at the 3′ termini of their positive strands; the corresponding recognition residues of group B VP1s are also less conserved, perhaps corresponding to an alternative specificity. These patterns of conservation are consistent with our conclusion, that UGUG recognition defines a mechanistically significant state of the polymerase-template complex.
UGUG recognition by VP1 defines template overshoot
To examine directly the contribution of UGUG recognition in the template entry channel to the overshot register of the 3′CS+, apoenzyme crystals were soaked with either (i) an oligonucleotide containing an insertion of an adenine between C2 and A3 (5′-UGUGAACC-3′) or (ii) an oligonucleotide with a deletion of A3 (5′-UGUGCC-3′) (Table 2). The A-inserted oligonucleotide reflects the 3′-end of some rotavirus gene 5 +RNAs (strain SA11); this variant 3′CS+ is therefore biologically functional. Density difference maps show that the UGUGA portions of the A-inserted oligonculeotide (5′-UGUGAACC-3′) and the 3′CS+ oligonucleotide are closely aligned in the template entry tunnel, revealing a similar pattern of hydrogen bonding with both templates (Fig. 3b and Supplementary Fig. 4). In this arrangement, the extra A and the C2 residues of the A-inserted oligonucleotide end up occupying the same positions as do the C1 and C2 residues of the 3′CS+ oligonucleotide, thereby leaving the 3′ end of the longer template fragment displaced by two positions beyond the initiation register. Difference maps show that the longer oligonucleotide has an extra bubble of density just beyond its C2 position, possibly accounting for the undefined C1 nucleotide (Supplementary Fig. 4). Despite the doubly overshot position of the 3′ end, +RNAs with a 3′CS+ that includes the inserted A (5′-UGUGAACC-3′) are efficient templates for dsRNA synthesis in vitro (Fig. 3d).
Analysis of VP1 complexes formed with the A3-deleted oligonucleotide (5′-UGUGCC-3′) show that VP1 and the UGUG segment interact just as in the complexes with wild-type (5′-UGUGACC-3′) and A-inserted (5′-UGUGAACC-3′) 3′CS+ (Fig. 3c). Because of the A3 deletion, however, C1 aligns in the initiation register. Weak difference density at the location occupied by C1 of the undeleted template suggests that a small percentage of the A3-deleted oligonucleotide may have aligned in the overshot register (Supplementary Fig. 4), and we propose that the template can shift between the initiation register and the overshot register within the entry tunnel. Recognition of the UGUG bases clearly shifts the equilibrium towards the initiation register in the case of the A3-deleted template.
VP1 recognition of the UGUG bases of +RNAs thus appears to create a stable auto-inhibited polymerase/template complex. We carried out in vitro replication assays using +RNA with an A3-deleted 3′-end (5′-UGUGCC-3′) as template for dsRNA synthesis, to determine whether a correct register is sufficient to generate an active complex. The template activity of A3-deleted +RNA remains VP2 dependent (Fig. 3d). We conclude that the role of VP2 in promoting initiation complex formation extends beyond simply moving the 3′ end of the template into register for initiation.
Attempts to recapitulate an initiation complex by soaking VP1 apoenzyme crystals with the 3′CS+ oligonucleotide, Mg2+, and GTP were not successful. While density maps revealed the presence of the 3′CS+ in the entry tunnel, Mg2+ could not be detected, and only poorly resolved additional density was noted in the catalytic pocket, presumably representing weakly bound GTP. Moreover, the C1 position of the 3′CS+ oligonucleotide remained overshot by one position and the putative priming loop remained in the retracted position. The failure of these soaks to produce functional initiation complexes is consistent with reconstitution assays, which have consistently shown that VP2 is necessary for forming VP1/+RNA complexes that can support dsRNA synthesis. Thus, the response to VP2-directed activation of VP1 is likely to include a shift of the primiing loop from a retracted to an extended conformation, a back translocation of the 3′CS+ into initiation register, and perhaps further concerted changes. The RdRP of the dsRNA bacteriophage φ6 also binds template RNA in an overshot register, determined by a pocket that accepts the 3′ nucleotide (Butcher et al., 2001). In that case, however, addition of the complementary nucleoside triphosphates is sufficient to reposition the template and initiate phosphodiester bond formation.
Cap binding
The surface of reovirus λ3 has a binding site for the N7-methyl-GpppG cap on +RNA; this site also binds GTP (Tao et al., 2002). We soaked the VP1 crystals in GTP and detected a bound nucleotide with its purine ring in roughly the location corresponding to the unmethylated purine site on λ3 (data not shown). An uncapped oligonucleotide with the 5′ sequence of the +RNA, 5′-GGCUUU-3′, binds in the template tunnel (Supplementary Fig. 4d). Thus, the uncapped sequence alone is insufficient to dictate binding proximal to the cap site, and the cap (not present in the soaked oligonucleotide) appears to be the primary element by which VP1 grasps and recognizes the 5′ end of a plus strand.
Discussion
The rotavirus polymerase domain resembles closely that of other viral RdRPs, and its N- and C-terminal domains are very similar to those of reovirus λ3. Moreover, both the conformation of the bound template and the way in which the templating bases are wedged against a segment of the fingers domain are conserved among all polymerases with a right-handed core for which the structure of a catalytic complex is known. Sequence-specific recognition of an RNA template sequence has not previously been documented, however, and the binding of template in an overshot register is also unusual. These similarities and differences allow us to correlate structural features with various aspects of VP1 activity, both in its enzymatic mechanism and in its broader biological function.
A model summarizing events in dsRNA synthesis by VP1 can be developed from comparisons of our crystallographic results with those obtained earlier for λ3 initiation and elongation complexes (Fig. 6). The rotavirus polymerase specifically recognizes its +RNA template by forming hydrogen bonds with the bases of the 3′CS+ UGUG residues. These interactions, combined with additional contacts to the sugar-phosphate backbone, anchor the 3′CS+ such that its C1 residue lies one nucleotide beyond the initiation register. This recognition complex is catalytically inactive. During assembly of progeny cores, VP1/+RNA complexes interact with VP2, inducing conformational changes that lead to initiation. The expected changes include a shift in the priming loop from a retracted to an extended position, allowing the initiating nucleotide to be stabilized in the P site. Nucleotides at the P and N positions may help bring C1 and C2 into alignment for initiation, and conformational changes elsewhere in the molecule must also have a role, as the priming loop does not communicate directly with the molecular surface. Correct alignment and base pairing will lead to formation of the first phosphodiester bond of the −RNA product. Once the initiating dinucleotide has formed, the extended priming loop will impede transfer of the RNA product. Retraction of the priming loop has been seen in λ3 as the polymerase transitions from initiation to elongation mode (Tao et al., 2002). In λ3, the elongating dsRNA product can move through the −RNA/dsRNA exit tunnel without impediment. In VP1, the C-terminal plug must be displaced from this exit tunnel. Forward translocation of the dsRNA product, driven by nucleotide hydroylsis, may be sufficient to open the channel, as the His-tagged plug in our protein preparations, which bind Ni-NTA, appears to emerge spontaneously. The plug does not fill the tunnel, and there is enough space to accommodate a bypassing single-stranded RNA. If it remained in the −RNA/dsRNA exit tunnel during transcription, the plug could have a regulatory role in the switch from replication to transcription.
Figure 6. Proposed model of RNA synthesis by VP1.
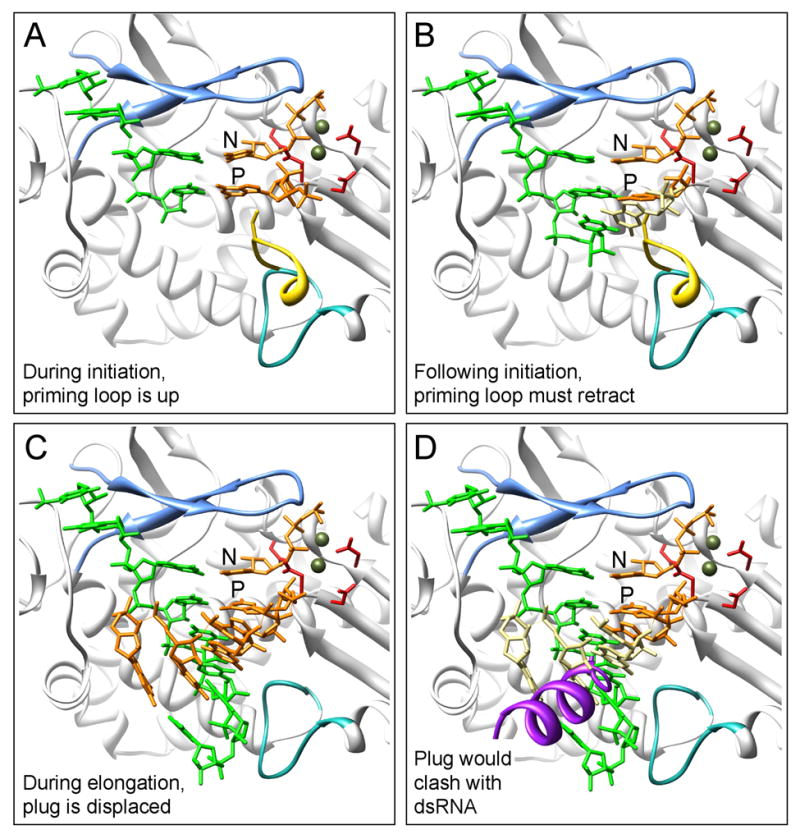
Superimposed on the VP1 structure are elements from the reovirus λ3 (A) initiation complex, PDB-1MWH (B) short elongation complex, PDB-1N38 and (C-D) long elongation complex, PDB-1N35. The reovirus template is shown in green, the incoming nucleotides, in orange, the Mg2+ ions, in olive, and the extended λ3 priming loop, in yellow. The priming (P) and incoming (N) nucleotide positions are indicated in panel (A). Important VP1 structural elements are shown, including: motif F (light blue), the priming loop (turquoise), and the catalytic aspartates (red). (A) During initiation of RNA synthesis, an extended priming loop is proposed to support the priming nucleotide, as seen for λ3. (B) Following initiation, translocation of the dinucleotide requires retraction of the priming loop to prevent a clash between the elongating product (beige) and the extended priming loop. (C) During elongation, a dsRNA product would emerge through the large exit tunnel when the VP1 C-terminal plug is assumed to be displaced. (D) An inserted C-terminal plug (purple) would conflict with passage of the dsRNA product (affected nucleotides in beige), requiring either repositioning of the plug during replication and/or redirection of the +RNA product during transcription.
Following synthesis, the dsRNA products of replication remain in cores, where they act as templates for multiple rounds of transcription. The 3′CS- (AAAAGCC-3′) of the −RNA template, although anchored in the template entry tunnel in our crystalline complex, does not contact VP1 in a sequence-specific manner. Thus, the rotavirus polymerase specifically recognizes only the 3′CS+ of the +RNA template for packaging and dsRNA synthesis. Highly specific screening of the −RNA templates for transcription may not be needed, as they are confined to the interior space of the core. Moreover, retention of the capped 5′ end of the +RNA in the cap-binding site will position the 3′ end of the −RNA near the template entry channel and favor its selective insertion.
All viruses of the many Reoviridae genera have similarly structured cores (see, for example, Lawton et al, 2000), with a tightly packed shell composed of 60 asymmetric dimers of VP2 or a homologous core shell protein (e.g., λ1 in orthoreoviruses and VP3 in orbiviruses). The shell is an icosahedrally symmetric array of 12 core shell protein decamers, each centered on a fivefold axis and each associated with a single (internal) polymerase molecule. The structures described in this paper and the properties of the viral cores suggest a simple mechanism for specific, +RNA packaging in a rotavirus infection. (1) Recognition of the 3′CS+ establishes stable, but inactive, VP1/+RNA complexes. (2) Association of this initial complex with a decamer of VP2 (or VP2 homolog) creates the basic assembly unit of the core. In all Reoviridae family members, one or more copies of an enzyme required for capping the transcript (e.g., VP3 of rotaviruses) must also be recruited. Association of the polymerase with the core shell protein subunits could also precede RNA recognition. (3) Twelve such basic assembly units (e.g., VP1-VP3-RNA-(VP2)10 for rotaviruses) come together to form a functional core. (4) Additional proteins (e.g., VP6, which forms the second “layer” of the rotavirus DLP) then stabilize the core and mediate interaction with outer-shell components (e.g., rotavirus VP4 and VP7). Recombinant polymerases from other Reoviridae family members (e.g., reovirus and bluetongue virus) have modest activity in vitro in the absence of the core shell protein. Whether the purified forms of the reovirus and bluetongue virus polymerases represent the structural and functional equivalents of the VP1 apoenzyme, or possibly a later activated form, remains to be determined.
Experimental Procedures
Expression and purification of proteins
Recombinant baculoviruses expressing untagged VP1 and VP2 and His-tagged VP1 proteins (cVP1 or nVP1) were generated as described elsewhere (Patton et al., 1997; Tortorici et al., 2006). To prepare untagged C-truncated VP1, cDNA encoding residues 1–1059, 1–1074, or 1–1079 were subcloned into pCR-Bac vectors. Recombinant baculoviruses were formed with the Bac-N-Blue baculovirus expression system (Invitrogen). His-tagged proteins were recovered from clarified lysates by ammonium sulfate precipitation and cobalt-affinity (Talon) column chromatography (Tortorici et al., 2006). Untagged VP1 and VP2 were purified as described earlier (Patton et al., 1997).
Expression of SeMet-substituted VP1
We adapted the methionine deprivation method (Bellizzi et al., 1999) to obtain selenomethionine (SeMet) derivatives of VP1. Because high concentrations of the analog was inhibitory to VP1 overexpression, SeMet was introduced in two steps, initially at a concentration of 5 mg/ml of culture, and then, 13 h later, in amounts sufficient to bring the final SeMet concentration to 50 mg/ml. The incorporation of SeMet, as determined by mass spectrometry, was about 85%.
Crystallization and data collection
Crystals were obtained by starting with 1 μl of VP1 at 10 mg/ml in 25 mM Na-HEPES, pH 7.8, 100 mM NaCl mixed with 2 μl crystallization buffer [25 mM Na-MES, pH 6.5, 1.5% (w/v) PEG 3350] and allowing the drop to equilibrate at 12°C by hanging-drop vapor diffusion with a well solution identical in composition to the drop except for the protein. With micro-seeding, thin, plate-like crystals appeared after 1 day and grew to full size (∼15 × 200 × 200 μm3) in about two weeks. Crystals were transferred into cryoprotectant [25% (v/v) ethylene glycol, 8.3 mM Na-HEPES, pH 7.8, 16.7 mM Na-MES, pH 6.5] and flash frozen in liquid nitrogen. X-ray diffraction data were collected at the Advanced Light Source stations 7.2.1 and 7.2.2 and at the Advanced Photon Source beamlines ID-19 and ID-24. Data were reduced and scaled using HKL2000 (Table 1) (Otwinowski and Minor, 1997).
Structure determination
Multi-wavelength anomalous dispersion (MAD) data sets were collected at three wavelengths, corresponding to peak, inflection, and remote points of the Se K-edge profile. Direct methods (Shake-and-Bake: (Weeks and Miller, 1999)) were used to locate Se positions. A run with a 3.2 Å resolution peak-wavelength data set yielded 12 Se positions, and the remaining 21 positions were located in difference maps phased from the initial 12 sites using CNS (Brunger et al., 1998) and validated using Shake-and-Bake. The Se coordinates were refined with SHARP (Bricogne et al., 2003), using data from three SeMet substituted crystals and a native (unsubstituted) crystal (Table 1).
The electron density map calculated with experimental phases was improved by solvent flattening, histogram matching, and phase resolution extension, using the program DM (Cowtan and Main, 1998). Comparison with the structure of reovirus λ3 allowed 90% of the polypeptide chain to be traced unambiguously, using the program O (Jones et al., 1991). Most of the remaining residues were added during iterative rounds of refinement in CNS and rebuilding in O. The final model as analyzed with PROCHECK (Laskowski et al., 1998) has over 87% of the residues in the most favored regions of the Ramachandran plot and only four (0.4%), all in poorly defined loops, in disallowed regions. The final Rwork and Rfree (the latter based on a 7.5% test set) are 23.3% and 28.6%, respectively.
Soaking experiments
Crystals of apoenzyme were transferred to a stabilizing solution [1% (w/v) PEG 3350, 8.3 mM Na-HEPES, pH 7.8, 16.7 mM Na-MES, pH 6.5, 25% (v/v) ethylene glycol] containing 0.5 mM RNA oligonucleotide and incubated for 24 h before freezing (Table 2). The structure of the apoenzyme was used as a phasing model. Initial rigid-body refinement yielded maps with interpretable density for the oligonucleotide, which was built with O; the structures of the various complexes were refined with CNS.
In vitro dsRNA synthesis
Polymerase chain reaction was used to prepare gene 8 cDNAs with a 5′-terminal T7 promoter and a wild type or mutated 3′-terminus (Tortorici et al., 2006). Gene 8 +RNAs were transcribed from the cDNAs using the Ambion T7 MEGAscript system. The reaction mixtures for replication assays (20 μl) included 4 pmols of VP1, 16 pmols of VP2, 8 pmols of gene 8 +RNA, 50 mM Tris-HCl, pH 7.1, 1.5% PEG-8000, 2 mM dithriothreitol, 1.5 U RNasin, 10 mM magnesium acetate, 2 mM MnCl2, 1.25 mM each CTP, UTP, and ATP, 5 mM GTP, and 10 μCi of [α-32P]UTP. Reaction mixtures were incubated for 5 h at 37°C, and 32P-labeled dsRNA products were analyzed by SDS-PAGE and autoradiography.
Preparation of figures
Figures were prepared with Molscript (Kraulis, 1991), Povscript (Fenn et al., 2003), Povray (http://www.povray.org), UCSF Chimera (Pettersen et al., 2004), Raster3D (Merritt and Bacon, 1997), Spock (J.A. Christopher, http://quorum.tamu.edu/spock), and Alscript (Barton, 1993).
Accession codes
Coordinates and structure factors have been deposited with the PDB. Accession numbers are as follows: cVP1 (apo), 2R7Q; nVP1 (apo), 2R7O; cVP1 (UGUGACC), 2R7R; cVP1 (UGUGCC), 2R7S; cVP1 (UGUGAACC), 2R7T; cVP1 (AAAAGCC), 2R7U; cVP1 (GGCUUU), 2R7V; cVP1 (mRNA 5′-cap), 2R7W; cVP1 (UGUGACC + GTP), 2R7X.
Supplementary Material
Acknowledgments
We thank Teresa Broering, Sophia Rits-Volloch, Rachelle Gaudet, Kelly Arnett, Hongyan Yang, Harish Ramanathan, and Tamara Bar-Magen for technical assistance, and Philip Dormitzer, Ethan Settembre, and Piotr Sliz for advice. We also express our appreciation to the staff at ALS (beamlines 7.2.1 and 7.2.2) and APS (beamlines ID-19 and ID-24) for help in data collection. This work was supported by NIH grants CA13202 (to SCH) and AI47904 (to MLN), and by the Intramural Research Program of the NIH, National Institutes of Allergy and Infectious Diseases (JTP, SMM, MAT, and RVC). SCH is an Investigator in the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ago H, Adachi T, Yoshida A, Yamamoto M, Habuka N, Yatsunami K, Miyano M. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure (London, England. 1999;7:1417–1426. doi: 10.1016/s0969-2126(00)80031-3. [DOI] [PubMed] [Google Scholar]
- Barton GJ. ALSCRIPT - a tool to format multiple sequence alignment. Prot Eng. 1993;6:37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
- Bellizzi JJ, Widom J, Kemp CW, Clardy J. Producing selenomethionine-labeled proteins with a baculovirus expression vector system. Structure (London, England. 1999;7:R263–267. doi: 10.1016/s0969-2126(00)80020-9. [DOI] [PubMed] [Google Scholar]
- Bressanelli S, Tomei L, Roussel A, Incitti I, Vitale RL, Mathieu M, De Francesco R, Rey FA. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:13034–13039. doi: 10.1073/pnas.96.23.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr D Biol Crystallogr. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI. A mechanism for initiating RNA-dependent RNA polymerization. Nature. 2001;410:235–240. doi: 10.1038/35065653. [DOI] [PubMed] [Google Scholar]
- Chen D, Barros M, Spencer E, Patton JT. Features of the 3′-consensus sequence of rotavirus mRNAs critical to minus strand synthesis. Virology. 2001;282:221–229. doi: 10.1006/viro.2001.0825. [DOI] [PubMed] [Google Scholar]
- Chen D, Luongo CL, Nibert ML, Patton JT. Rotavirus open cores catalyze 5′-capping and methylation of exogenous RNA: evidence that VP3 is a methyltransferase. Virology. 1999;265:120–130. doi: 10.1006/viro.1999.0029. [DOI] [PubMed] [Google Scholar]
- Chen D, Patton JT. De novo synthesis of minus strand RNA by the rotavirus RNA polymerase in a cell-free system involves a novel mechanism of initiation. RNA (New York, NY. 2000;6:1455–1467. doi: 10.1017/s1355838200001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan K, Main P. Miscellaneous algorithms for density modification. Acta Crystallogr D Biol Crystallogr. 1998;54:487–493. doi: 10.1107/s0907444997011980. [DOI] [PubMed] [Google Scholar]
- Doublie S, Ellenberger T. The mechanism of action of T7 DNA polymerase. Current opinion in structural biology. 1998;8:704–712. doi: 10.1016/s0959-440x(98)80089-4. [DOI] [PubMed] [Google Scholar]
- Estes MK, Kapikian AZ. Rotaviruses. In: Knipe DM, Howley PM, editors. Fields Virology. 5th. Lippincott Williams & Wilkins; 2007. pp. 1917–1974. [Google Scholar]
- Fenn TD, Ringe D, Petsko GA. POVScript+: a program for model and data visualization using persistence of vision ray-tracing. J Appl Crystallogr. 2003;36:944–947. [Google Scholar]
- Gallegos CO, Patton JT. Characterization of rotavirus replication intermediates: a model for the assembly of single-shelled particles. Virology. 1989;172:616–627. doi: 10.1016/0042-6822(89)90204-3. [DOI] [PubMed] [Google Scholar]
- Jacobo-Molina A, Ding J, Nanni RG, Clark AD, Jr, Lu X, Tantillo C, Williams RL, Kamer G, Ferris AL, Clark P, et al. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Joyce CM, Steitz TA. Function and structure relationships in DNA polymerases. Annual review of biochemistry. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science (New York, NY. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- Kraulis PJ. Molscript: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- Labbe M, Baudoux P, Charpilienne A, Poncet D, Cohen J. Identification of the nucleic acid binding domain of the rotavirus VP2 protein. The Journal of general virology. 1994;75(Pt 12):3423–3430. doi: 10.1099/0022-1317-75-12-3423. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Thornton JM. Validation of protein models derived from experiment. Curr Opin Struct Biol. 1998;8:631–639. doi: 10.1016/s0959-440x(98)80156-5. [DOI] [PubMed] [Google Scholar]
- Lawton JA, Estes MK, Prasad BV. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nature structural biology. 1997;4:118–121. doi: 10.1038/nsb0297-118. [DOI] [PubMed] [Google Scholar]
- Lawton JA, Estes MK, Prasad BV. Mechanism of genome transcription in segmented dsRNA viruses. Advances in virus research. 2000;55:185–229. doi: 10.1016/S0065-3527(00)55004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesburg CA, Cable MB, Ferrari E, Hong Z, Mannarino AF, Weber PC. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nature structural biology. 1999;6:937–943. doi: 10.1038/13305. [DOI] [PubMed] [Google Scholar]
- Liu M, Mattion NM, Estes MK. Rotavirus VP3 expressed in insect cells possesses guanylyltransferase activity. Virology. 1992;188:77–84. doi: 10.1016/0042-6822(92)90736-9. [DOI] [PubMed] [Google Scholar]
- Merritt EA, Bacon DJ. Raster 3D: photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- O'Reilly EK, Kao CC. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology. 1998;252:287–303. doi: 10.1006/viro.1998.9463. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Patton JT. Rotavirus VP1 alone specifically binds to the 3′ end of viral mRNA, but the interaction is not sufficient to initiate minus-strand synthesis. Journal of virology. 1996;70:7940–7947. doi: 10.1128/jvi.70.11.7940-7947.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton JT, Gallegos CO. Rotavirus RNA replication: single-stranded RNA extends from the replicase particle. The Journal of general virology. 1990;71(Pt 5):1087–1094. doi: 10.1099/0022-1317-71-5-1087. [DOI] [PubMed] [Google Scholar]
- Patton JT, Jones MT, Kalbach AN, He YW, Xiaobo J. Rotavirus RNA polymerase requires the core shell protein to synthesize the double-stranded RNA genome. Journal of virology. 1997;71:9618–9626. doi: 10.1128/jvi.71.12.9618-9626.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pizarro JL, Sandino AM, Pizarro JM, Fernandez J, Spencer E. Characterization of rotavirus guanylyltransferase activity associated with polypeptide VP3. The Journal of general virology. 1991;72(Pt 2):325–332. doi: 10.1099/0022-1317-72-2-325. [DOI] [PubMed] [Google Scholar]
- Prasad BV, Rothnagel R, Zeng CQ, Jakana J, Lawton JA, Chiu W, Estes MK. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature. 1996;382:471–473. doi: 10.1038/382471a0. [DOI] [PubMed] [Google Scholar]
- Tao Y, Farsetta DL, Nibert ML, Harrison SC. RNA synthesis in a cage--structural studies of reovirus polymerase lambda3. Cell. 2002;111:733–745. doi: 10.1016/s0092-8674(02)01110-8. [DOI] [PubMed] [Google Scholar]
- Tortorici MA, Broering TJ, Nibert ML, Patton JT. Template recognition and formation of initiation complexes by the replicase of a segmented double-stranded RNA virus. The Journal of biological chemistry. 2003;278:32673–32682. doi: 10.1074/jbc.M305358200. [DOI] [PubMed] [Google Scholar]
- Tortorici MA, Shapiro BA, Patton JT. A base-specific recognition signal in the 5′ consensus sequence of rotavirus plus-strand RNAs promotes replication of the double-stranded RNA genome segments. RNA (New York, NY. 2006;12:133–146. doi: 10.1261/rna.2122606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela S, Pizarro J, Sandino AM, Vasquez M, Fernandez J, Hernandez O, Patton J, Spencer E. Photoaffinity labeling of rotavirus VP1 with 8-azido-ATP: identification of the viral RNA polymerase. Journal of virology. 1991;65:3964–3967. doi: 10.1128/jvi.65.7.3964-3967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks CM, Miller R. Optimizing Shake-and-Bake for proteins. Acta Crystallogr D Biol Crystallogr. 1999;55:492–500. doi: 10.1107/s0907444998012633. [DOI] [PubMed] [Google Scholar]
- Yeager M, Dryden KA, Olson NH, Greenberg HB, Baker TS. Three-dimensional structure of rhesus rotavirus by cryoelectron microscopy and image reconstruction. The Journal of cell biology. 1990;110:2133–2144. doi: 10.1083/jcb.110.6.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng CQ, Estes MK, Charpilienne A, Cohen J. The N terminus of rotavirus VP2 is necessary for encapsidation of VP1 and VP3. Journal of virology. 1998;72:201–208. doi: 10.1128/jvi.72.1.201-208.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


