Abstract
In natural systems, host species are often co-infected by multiple pathogen species, and recent work has suggested that many pathogens can infect a wide range of host species. An important question therefore is what determines the host range of a pathogen and the community of pathogens found within a given host species. Using primates as a model, we show that infectious diseases are more often shared between species that are closely related and inhabit the same geographical region. We find that host relatedness is the best overall predictor of whether two host species share the same pathogens. A higher frequency of pathogen host shifts between close relatives or inheritance of pathogens from a common ancestor may explain this result. For viruses, geographical overlap among neighbouring primate hosts is more important in determining host range. We suggest this is because rapid evolution within viral lineages allows host jumps across larger evolutionary distances. We also show that the phylogenetic pattern of pathogen sharing with humans is the same as that between wild primates. For humans, this means we share a higher proportion of pathogens with the great apes, including chimpanzees and gorillas, because these species are our closest relatives.
Keywords: host specificity, transmission mode, phylogeny, geographical overlap, parasites, pathogen taxonomy
1. Introduction
Our understanding of the ecology and evolution of infectious diseases has increased dramatically over the past 20 years (Anderson & May 1991; Hudson et al. 2001; Nunn & Altizer 2006). However, much of this work has focused on single host–single pathogen interactions; an unlikely model of natural populations. Within wild primates, it is estimated that over 60% of pathogens can infect multiple host species (Pedersen et al. 2005), and in humans over 75% of pathogens classified as emerging are shared with animal reservoirs (Taylor et al. 2001). A critical question is what determines the breadth of host species that a pathogen can infect (host range). First, differences in pathogen biology may influence the range of susceptible host species (Cleaveland et al. 2001; Taylor et al. 2001; Pedersen et al. 2005). In wild primates, viruses tend to have broad host ranges, with nearly 50% capable of infecting hosts from multiple mammalian orders (Pedersen et al. 2005), consistent with patterns found in other mammals (Cleaveland et al. 2001; Taylor et al. 2001). By contrast, half of the recorded helminth pathogens are specific to a single primate species (Pedersen et al. 2005). Second, differences in host biology are also likely to be important, but have been less studied (Roy 2001). Host species differ in the size and composition of their pathogen communities (Nunn et al. 2003; Nunn & Altizer 2006; Lindenfors et al. 2007), yet little is known about the host characteristics that determine the distribution of pathogens. Here, we explore patterns of pathogen sharing by contrasting the pathogen communities of primate hosts.
There are two broad explanations for pathogen sharing among host species (Page 2003). First, host species may inherit their pathogen communities from a common ancestor (shared by descent). Second, pathogens may expand their host range by infecting novel host species, which we term ‘host shifts’ following Antonovics et al. (2002). For both cases, we might predict that closely related host species harbour similar pathogen communities owing to their common evolutionary heritage. In the former case, pathogens are inherited directly, in a manner analogous to the inheritance of genetically determined biological traits. In the latter case, related hosts may share pathogens because they have similar immunological responses and life-history traits (Pfennig 2000; Ricklefs & Fallon 2002; Perlman & Jaenike 2003), facilitating pathogen range expansion among more closely related hosts. However, if host shifts are common, even distantly related hosts may share pathogens, in which case geographical proximity between hosts might be important, for example, by influencing contact rates and, thereby, opportunities for pathogen transfer (Antonovics et al. 2002).
Many of the most virulent emerging infectious diseases (EIDs) in humans and wildlife are thought to be largely a product of host shifts, for example SARS and HIV in humans (Daszak et al. 2000), Ebola in African apes (Walsh et al. 2003), canine distemper virus in wild dogs and lions (Roelke-Parker et al. 1996) and Batrachochytrium dendrobatidis fungus in global amphibian populations (Daszak et al. 1999). Disease emergence following a host shift can result in rapid spread and high virulence because naive hosts may lack appropriate immune responses (Osterhaus 2001; Altizer et al. 2003). A better understanding of the determinants of pathogen community similarity will be a critical first step in predicting the likelihood of future host shifts, and thereby provide the potential to reduce mortality, morbidity and high economic costs frequently associated with disease emergence.
To date, paucity of data on parasite occurrence and host species attributes has prevented large-scale comparative studies. Here, we use newly compiled datasets on the distribution, phylogeny and pathogen communities of wild primates to explore, for the first time, the importance of evolutionary relatedness and geographical overlap between host species in determining pathogen community similarity in a multi-host–multi-pathogen system. Finally, we contrast patterns of pathogen sharing between wild primates with a dataset of pathogen sharing between wild primates and humans to explore whether similar evolutionary and ecological factors might structure the pathogen community of humans. Our results may therefore provide insight for understanding the diversity of human pathogens and the potential for host shifts between humans and wildlife.
2. Material and methods
(a) Primate host and pathogen data
Evolutionary relationships among host species and divergence times were derived from Bininda-Emonds et al. (2007). Primate species distributions were obtained from Grenyer et al. (2006). Pathogen species occurrences were obtained from the Global Mammal Parasite Database (Nunn & Altizer 2005; www.mammalparasites.org), comprising 2462 records representing 415 pathogen species across 117 of the 232 wild primate species recognized in the phylogenetic tree of Bininda-Emonds et al. (2007). Here, we focus on associations within viruses, protozoans and helminths because these groups account for almost 80% of the recorded pathogens from wild primates (Nunn & Altizer 2005; Pedersen et al. 2005). The pathogen species in the database represents a small but unknown fraction of the total pathogen community that infect wild primates. The number of pathogens recorded per primate host is positively correlated with sampling effort, even for well-studied species (Altizer & Pedersen 2008). Taylor et al. (2001) suggested that human pathogens may number over 1400, an order of magnitude larger than current records for wild primates. It is possible that the unique ecological status and distribution of humans provides a poor model for generalizing to the pathogen communities of primates; nonetheless, our number of recorded primate pathogens is likely to be a substantial underestimate of total pathogen species richness. In the following, we discuss the potential for biases due to incomplete sampling.
We partitioned transmission mode into two categories: ‘direct’ and ‘indirect’ (derived from Pedersen et al. 2005). Direct transmission includes any pathogen that can be transmitted by physical or sexual contact between an infected and a susceptible individual. Indirect transmission includes pathogens transmitted only via vectors, intermediate hosts or through the environment. In addition to the database of naturally occurring wild primate parasites, we also recorded pathogen co-occurrence in wild primates and humans by comparing the primate records with a separate database of human pathogens (Taylor et al. 2001), comprising 1415 species pathogenic to humans, of which 112 species also infect wild primates.
(b) Statistical analysis
First, we used partial Mantel tests to determine the relationship between geographical overlap and host relatedness with pathogen sharing. Second, we performed sensitivity analyses using a standard generalized linear model (GLM) approach (Crawley 2002) across all unique host pair combinations (6786 pairwise comparisons).
We treated pathogen sharing as an index of community similarity, measuring the difference in pathogen species composition between two (or more) assemblages (i.e. host species), frequently referred to as beta diversity. We employ the following derivation of the Jaccard index (βj) because it is one of the most frequently adopted in the literature and its statistical properties are well understood (Koleff et al. 2003):
Jaccard (1912), where a is the number of pathogen species found on two host species, X and Y; b is the number of pathogen species on host X that are not found on host Y; and c is the number of pathogen species on host Y that are not found on host X. Our measure, βj, is independent of the relative sizes of the pathogen species pools of X and Y (Koleff et al. 2003); this is important as sample sizes of pathogens frequently differ between primate species due to variation in sampling intensity. We quantified host geographical range overlap in two ways: first, as a binary variable, scoring each host pair as either sympatric or allopatric (sympatry); second, we used the relative proportion of overlap among sympatric host pairs only (proportional overlap), calculated as
where X and Y represent the distributions of the respective host species in each pairwise comparison (Barraclough et al. 1999).
(c) Mantel tests
Partial correlation coefficients of host relatedness and geographical overlap with pathogen sharing were estimated by partial Mantel tests in PASSaGE (Beta Version 2.0.6.17; Rosenberg 1998–2008) and significance was assessed by permutation. Model coefficients are reported in table 1 and table S1 in the electronic supplementary material.
Table 1.
Summary of the results from the partial Mantel tests of shared pathogens (βj) against host divergence time and sympatry for each of the pathogen subsets.
| model | pathogen | R | explanatory variable | z | p |
|---|---|---|---|---|---|
| 1 | total | −0.27 | divergence time | −6686 | 0.001 |
| 0.24 | sympatry | 74.51 | 0.001 | ||
| 2 | protozoan | −0.31 | divergence time | −11 765 | 0.001 |
| 0.22 | sympatry | 104.52 | 0.001 | ||
| 3 | helminth | −0.10 | divergence time | −1428 | 0.001 |
| 0.16 | sympatry | 29.70 | 0.001 | ||
| 4 | virus | −0.08 | divergence time | −2789 | 0.12 |
| 0.14 | sympatry | 62.05 | 0.001 | ||
| 5 | direct | −0.15 | divergence time | −3125 | 0.001 |
| 0.11 | sympatry | 28.53 | 0.001 | ||
| 6 | indirect | −0.24 | divergence time | −7002 | 0.001 |
| 0.25 | sympatry | 94 | 0.001 |
(d) Generalized linear models
For each subset of pathogens and all pathogens combined, we regressed βj against host relatedness and one of the two measures of geographical overlap. It was not possible to include pathogen type into the model because the response variable, proportion of shared pathogens, varied with each pathogen subset. All GLMs were constructed in the statistical package R (R: a programming environment for data analysis and graphics, v. 2.4.0; http://www.r-project.org/), with βj as the response variable, assuming binomial errors with a logit link function. Pseudo-r2 for the models were estimated as the proportional increase in residual deviance when removing the term of interest from the model. Because each host species was included in multiple pairwise comparisons in the latter analysis, model assumptions regarding independence of data points may be violated. Model coefficients should therefore be interpreted cautiously.
As originally formulated, Jaccard's index of community similarity (Jaccard 1912) assumes that all species have been sampled exhaustively; however, this is almost certainly not the case for the pathogen communities included within our analysis (Altizer & Pedersen 2008). We therefore performed model sensitivity by repeating the set of GLMs including the total number of pathogens recorded for each host (i.e. a+b and a+c, where c<b) as covariates to account for variation in pathogen sample sizes. We did not attempt to directly correct for differences in pathogen sampling intensity, for example, by using scientific citation counts (Nunn et al. 2003), because we were not interested in absolute pathogen numbers, but rather the frequency of shared pathogens.
In total, we ran two GLMs for all pathogens combined (total) and each pathogen subset (protozoa, helminth, virus; direct and indirect transmissions). Model outputs for the GLMs of divergence time and sympatry are summarized in the electronic supplementary material, table S2. Table S3 in the electronic supplementary material includes sample sizes as additional explanatory variables in the models.
(e) Pathogen sharing with humans
To evaluate whether humans demonstrate phylogenetic patterns of pathogen sharing similar to wild primates, we constructed an additional GLM of the relationship between pathogen sharing and relatedness between humans and wild primates from the subset of pathogens shared between them (data from Taylor et al. 2001). We then incorporated pairwise comparisons between wild primates and used Akaike's information criteria (AIC) to compare the fit of the regression model with and without allowing a separate slope and intercept for comparisons between wild primates and humans, and for comparisons of pathogen sharing exclusively among wild primates. We considered humans as sympatric with all wild primates; therefore, for equivalence, we included only sympatric wild primate pairs in this analysis. It is possible that directed screening for human diseases within hominids (the great apes) could influence our results; unfortunately, we did not have sufficient information to evaluate potential study biases within our dataset.
3. Results
The mean pathogen community size was 6.9 pathogens per host, with 28% of host pairs sharing at least one pathogen. The maximum number of shared pathogens (17) was between gorilla (Gorilla gorilla) and chimpanzee (Pan troglodytes), which represented 41 and 40% of their parasite communities, respectively (βj=0.25). Eight host pairs, for example, within the genus Macaca (macaques) and Callicebus (titi monkeys), had identical pathogen communities (βj=1.0), but in each case pathogen community size was a single pathogen species. The mean proportion of shared pathogens across all 6786 pairwise comparisons was approximately 2%. The analysis of primate distribution maps revealed 14% of host pairs to be sympatric in part of their geographical range, although most (84%) primates were sympatric with at least one other primate species included in the analysis.
(a) Host relatedness, geographical overlap and pathogen community similarity
For all pathogens, we found both geographical overlap and phylogenetic distance to be significant predictors of pathogen sharing between primate host species (model 1, table 1). The results from the GLMs were broadly consistent with the Mantel tests (table S2 in the electronic supplementary material). However, we re-emphasize that model coefficients from the GLMs should be interpreted cautiously owing to non-independence of the pairwise comparisons, and we focus discussion on results from the Mantel tests.
Divergence time was consistently the stronger explanatory variable, with more recently diverged hosts sharing a greater proportion of their total pathogen communities. Host species that overlapped in their geographical distributions also tended to share a greater proportion of pathogens; however, including the magnitude of overlap (proportional overlap) did not provide a better fit to the model (table S1 in the electronic supplementary material). The inclusion of pathogen sample sizes into the GLMs explained an additional 9–16% of the variation in pathogen sharing independent from that explained by geography and phylogeny, which remained the dominant explanatory variables for all models (models 1–6; table S3 in the electronic supplementary material).
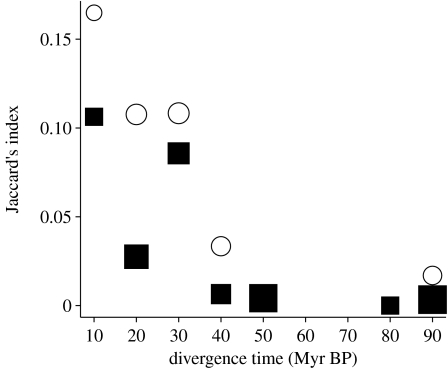
Geographical overlap between host species may covary with host relatedness, confounding simple comparisons between pathogen sharing and either predictor variable alone. However, the link between overlap and phylogeny is complex (Webb et al. 2002). It is possible that more closely related species overlap less in their geographical ranges, either due to recent speciation in allopatry or competitive displacement among close relatives (Webb et al. 2002; Davies et al. 2007). We might also predict very distantly related species to overlap less in their distributions, owing to divergence in habitat preferences or occurrence of geographical barriers preventing secondary contact. The precise relationship between overlap and phylogeny will therefore depend upon evolutionary rates, host dispersal ability and geographical contingency. Nonetheless, by controlling first for evolutionary relatedness, and then for geographical overlap, using partial Mantel tests, we show both to be significant independent predictors of pathogen sharing. Furthermore, the slope of the relationship between divergence time and pathogen sharing was similar for both sympatric and allopatric comparisons in the GLMs, but sympatric hosts shared a higher proportion of pathogens (figure 1).
Figure 1.
Relationship between divergence time and proportion of shared pathogens. Squares (allopatric host pairs) and circles (sympatric host pairs) represent mean pathogen community similarity (βj; Jaccard 1912) between host pairs in 10 Myr bins; the size of the symbol is proportional to the number of pairwise contrasts within each time interval. This figure is illustrative only and follows the approach of Gilbert & Webb (2007); model fitting was performed using the individual pairwise contrasts (§2).
(b) Pathogen taxonomy and transmission strategy
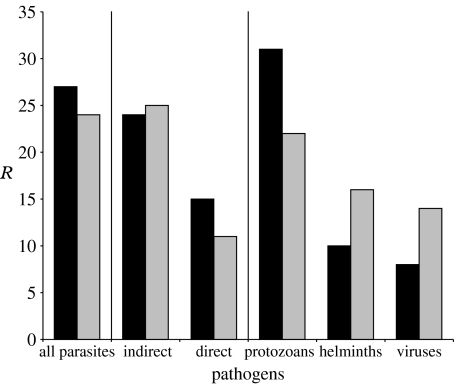
Previous work has shown significant variation in host specificity among pathogen taxonomic groups and among transmission modes (Cleaveland et al. 2001; Taylor et al. 2001; Pedersen et al. 2005). We found that the key predictors of pathogen community similarity differed with pathogen taxonomy (figure 2). Divergence time was the better predictor of protozoan community similarity, although geography was also significant (model 2; table 1; tables S1–S3 in the electronic supplementary material). By contrast, geography was the better predictor of viral communities (model 4; table 1; tables S1–S3 in the electronic supplementary material), whereas the key predictor of helminth community similarity varied between models (model 3; table 1; tables S1–S3 in the electronic supplementary material). We did not find a strong difference between the predictors of pathogen sharing with transmission strategies: divergence time was marginally the better predictor of pathogens with direct transmission strategies, while the relative importance of divergence time and geography differed between models for pathogens with indirect transmission strategies (models 5 and 6; table 1; table S1 in the electronic supplementary material). The results from the GLMs were generally consistent with the Mantel tests (models 5 and 6; tables S2 and S3 in the electronic supplementary material).
Figure 2.
Variation in explanatory power with pathogen taxonomy and transmission mode. Bar heights represent correlation coefficients (R) from the partial Mantel tests. Divergence time is negatively correlated with pathogen sharing. Black bars, divergence time; grey bars, sympatry.
(c) Pathogen sharing between wild primates and humans
We found that humans also share more pathogens with closely related primate species (pseudo-r2=0.29, p<0.001). Humans are therefore almost four (3.8) times more likely to share pathogens with chimpanzees (P. troglodytes), which last shared a common ancestor with humans 8.6 Myr BP, than with colobus monkeys (Colobus), which diverged over 34.4 Myr BP. Furthermore, the relationship between phylogenetic distance and pathogen sharing with humans did not significantly differ from that between wild primates (ΔAIC=2.9). We did not consider the relationship between geographical overlap and pathogen sharing in this analysis owing to the highly dynamic and cosmopolitan nature of human distributions.
4. Discussion
Using a broad comparative approach, we identify two key ecological and evolutionary determinants of pathogen community similarity. First, more recently diverged hosts (relatives) are more likely to share similar pathogen communities than more distant relatives. Second, sympatric hosts (neighbours) are also more likely to share a greater proportion of pathogen species, but the magnitude of range overlap is not important; hosts co-occurring over a large proportion of their range were no more likely to share pathogens than hosts with limited geographical overlap. Our analysis is the first to demonstrate the role of both phylogeny and geography in pathogen sharing in mammals and across diverse pathogen taxa.
We used both partial Mantel tests (which compute the first-order partial correlation on three or more distance matrices; Smouse et al. 1986; see also Legendre 2000) and GLMs to explore pattern in our data. Because host species were included multiple times in our pairwise comparisons, the GLMs may violate assumptions of independence of data. However, the GLM approach allowed us to evaluate the influence of both evolutionary and geographical distances simultaneously, and perform additional sensitivity analyses. We therefore referred to the results from both approaches, but reported only significance values for the Mantel tests in §3. Critically, the results from the Mantel tests and the GLMs were highly congruent. Sensitivity analysis demonstrated that our results were not an artefact of phylogenetic non-independence or differences in pathogen sampling intensity. We view the strength of our results to be remarkable, considering the multiple interacting factors that are probably important in determining host susceptibility (Anderson & May 1991; Nunn & Altizer 2006). Furthermore, topological uncertainty in the phylogenetic tree of primates, miscalibration of divergence times and shifts in geographical distributions will tend to reduce signal strength, making our analyses conservative.
There are two possible explanations for the link between host phylogeny and pathogen sharing: first, a higher frequency of host shifts between closely related and hence biologically similar species; second, coinheritance of pathogens from a common ancestor (shared by descent). Host shifts pose the greatest threat for disease emergence. However, distinguishing host shifts from coinheritance is difficult, typically requiring detailed phylogenetic information on both hosts and pathogens (e.g. Page 2003). Furthermore, even close matching between host and pathogen phylogenies might simply be a reflection of more frequent host shifts between adjacent branches of the host phylogenetic tree, rather than pathogen–host coevolution (Roy 2001; Charleston & Roberston 2002).
We suggest that the significant relationship between geographical overlap and pathogen sharing is best explained by host shifts. Potential for contact between a pathogen and a novel host is likely to be greatest where infected and non-infected species occur in sympatry, increasing the opportunity for host shifts (Fenton & Pedersen 2005). By including both overlap and divergence times within our model, we demonstrate that our results were not an artefact of range overlap covarying with host relatedness. A correlation between range overlap and pathogen community similarity may seem intuitive; however, to the best of our knowledge, it has not previously been demonstrated in a natural system. Furthermore, we note that our study might help explain previous observations of a positive correlation between within-host pathogen species richness and host sympatry (Nunn et al. 2004). If pathogens are more likely to be shared between geographically overlapping host species, then the more hosts that overlap in space, the greater the opportunity for pathogen transfer, and hence greater within-host pathogen species richness.
The consequences of pathogen host shifts can vary. The infection of the new host may be transient, sometimes referred to as ‘spillover’ (Daszak et al. 2000), in which case the pathogen will eventually be lost from the host without repeated reintroductions via cross-species transmission. Alternatively, the pathogen may be maintained independently in both species resulting in a host range expansion and potentially leading to the emergence of the disease in the new host species (Fenton & Pedersen 2005). While our data cannot distinguish between recent spillover events and sustained transmission within a novel host species, we would predict the two to be strongly correlated. We suggest that the likelihood of pathogen establishment in a novel host increases with multiple spillover events in much the same way as the invasion success of a species beyond its native range increases with multiple introductions (Kolar & Lodge 2001; Lockwood et al. 2005). The recent emergence of HIV in humans provides an important case in point. Over 36 strains of SIV have been identified from non-human primates (Peeters 2004), and researchers have documented at least seven separate spillover events into human populations preceding the HIV epidemic of the mid-to-late twentieth century (Hahn et al. 2000).
For viruses, phylogenetic distance is less important than geography in explaining pathogen community similarity. This is consistent with numerous studies demonstrating wide host ranges in viruses, sometimes spanning multiple taxonomic orders (Taylor et al. 2001; Pedersen et al. 2005; Kuiken et al. 2006). Fast mutation rates and short generation times of viruses, especially RNA viruses (Domingo & Holland 1997), may provide the evolutionary potential for crossing distant host barriers, for example, by rapidly adapting to novel cell surface types (Kuiken et al. 2006). By contrast, host relatedness was the better predictor of protozoan communities. In wild primates, host specificity in this group of pathogens tends to be much greater than for viruses, a possible product of longer generation times and more complex life-history strategies imposing evolutionary constraints on host range expansion (Anderson & May 1991; Pedersen et al. 2005). Host shifts may, therefore, be more frequent among viral pathogens, whereas coinheritance is more common for protozoa. It was not possible to determine the relative importance of geography versus phylogeny in predicting helminth community similarity, although this group demonstrates the highest level of host specificity, and perhaps the most complex life cycles. Helminths are the only group in which transmission by intermediate hosts is common (Pedersen et al. 2005), and this transmission strategy may constrain helminth geographical distributions, as maintenance in a novel host requires coexistence with the appropriate intermediate host.
Our comparison between pathogen taxa assumes that pathogen species delineations are accurate and equivalent between taxonomic groups. For viral pathogens, we followed the International Committee on the Taxonomy of Viruses database (http://www.ncbi.nlm.nih.gov/ICTVdb/Ictv/), and nomenclature for other parasite species followed National Center for Biotechnology Information guidelines. However, species are notoriously difficult to define. Experimental cross-infection studies (e.g. Gilbert & Webb 2007) would provide an empirical test of the relationship between host phylogenetic relatedness and the potential for cross-species infections.
We a priori predicted coinheritance to be the more important determinant of community similarity for pathogens transmitted by direct contact due to the intimate nature of transmission (Lockhart et al. 1996). We therefore expected to find a relatively stronger link with host relatedness for this pathogen subset. Conversely, we predicted pathogens transmitted by indirect contact to demonstrate a relatively stronger correlation with geography, because transmission is decoupled from direct host-to-host contact (Woolhouse et al. 2001). However, the Mantel test provided only weak support for this hypothesis: host relatedness was only marginally the better predictor of community similarity for directly transmitted pathogens, while geographical overlap was either less important or approximately equally as important as relatedness for indirectly transmitted pathogens. Our inconclusive results might be explained by a higher than expected proportion of host shifts among directly transmitted pathogens (Pedersen et al. 2007), which may require repeated close physical contact, only probable when host species are in sympatry. In addition, subtle differences in the relationship between host range and transmission strategy exist within pathogen taxonomic groups (Pedersen et al. 2005). More comprehensive sampling would be required to explore this further.
It is interesting that we do not find a significant association between pathogen sharing and the magnitude of range overlap. It is possible that current range overlap does not correlate with contact rates between hosts because, for example, population densities vary greatly across a species' range or recent range movements have transformed host geographical ranges. Alternatively, pathogen–host biology may be more important in determining the success of the pathogen in the new host following the initial encounter between previously isolated host species. Differences in host ecology might also limit the possibilities for epidemiological contacts across species; for example, interspecific interactions may be low even where the overlap is great, as species may occupy different microhabitats (Bowers & Brown 1982) or have different activity cycles (Kronfeld-Schor & Dayan 2003) and dietary preferences.
Finally, our results show humans to be no more than a naked ape with regard to pathogen sharing. Humans tend to share a greater proportion of pathogens with their closest evolutionary relatives. It was not possible to distinguish between pathogen coinheritance and host shifts from our data; however, many studies have shown cross-species transmission from humans to non-human primates (e.g. poliovirus, syphilis, influenza A, tuberculosis and measles) as well as from non-human primates to humans (e.g. monkey pox, ebola virus, malaria and herpes B; reviewed in Wolfe et al. 2005; Nunn & Altizer 2006). Lastly, we note that HIV-1, perhaps the most devastating recent human EID, was a product of the SIVcpz virus shifting from chimpanzees (Korber et al. 2000), one of our closest living relatives. Here, we considered only pathogen sharing among primate hosts. Wild primates represent a small but not insignificant sample of potential pathogen reservoir species (approx. 75% of zoonotic human EIDs are shared with non-primate reservoirs; Woolhouse & Gowtage-Sequeria 2005). It remains to be evaluated whether the relationship between phylogenetic relatedness and pathogen sharing can be extrapolated across greater evolutionary distances; for example, between non-primate mammals or birds.
In summary, we find primates share more pathogens with their close phylogenetic relatives and geographical neighbours. This is probably a consequence of both pathogen coinheritance and host shifts between primate species. Geographical proximity might provide the opportunity for pathogen transmission to a novel host, especially among viral and directly transmitted pathogens. Greater sharing among close relatives might indicate phylogenetic conservatism of pathogen communities, with pathogens tracking the evolutionary trajectories of their host lineages. Phylogenetic relatedness may also be important in determining whether a pathogen becomes established in a novel host. Close relatives, sharing similar biology and immune responses, will be vulnerable to the same suite of pathogens. Our results provide a critical first step in understanding and predicting the likelihood of future host shifts and disease emergence events.
Acknowledgments
We thank S. Altizer, J. Antonovics, J. L. Gittleman, R. Grenyer, M. E. Hood, A. H. Hurlbert, B. Ledbetter, P. Lindenfors, C. L. Nunn, J. de Roode and H. Wearing for access to data and their comments on the manuscript. This work was in part funded by a grant from Conservation International. A.B.P. was funded by a Royal Society Incoming Research Fellowship. T.J.D. was supported as a postdoctoral associate at the National Center for Ecological Analysis and Synthesis, a center funded by National Science Foundation grant DEB-0072909, and the University of California, Santa Barbara.
Supplementary Material
Table S1. Summary of the results from partial Mantel tests of shared pathogens (βj) against host divergence time and proportional range overlap for each of the pathogen subsets. Table S2. Generalised linear models of shared pathogens (βj) against host divergence time and sympatry for total pathogen community each of the pathogen subsets. Table S3. Regressions of shared pathogens (βj) against host divergence time and sympatry for each of the pathogen subsets including pathogen sample sizes as a covariate in the GLMs
References
- Altizer, S. & Pedersen, A. B. 2008 Evolutionary dynamics of disease threats to biodiversity. In Conservation biology: evolution in action (eds S. Carroll & C. Fox), pp. 268–280. Oxford, UK: Oxford University Press.
- Altizer S, Harvell D, Friedle E. Rapid evolutionary dynamics and threats to biodiversity. Trends Ecol. Evol. 2003;18:589–596. doi:10.1016/j.tree.2003.08.013 [Google Scholar]
- Anderson R.M, May R.M. Oxford University Press; Oxford, UK: 1991. Infectious diseases of humans: dynamics and control. [Google Scholar]
- Antonovics J, Hood M, Partain J. The ecology and genetics of a host shift: Microbotryum as a model system. Am. Nat. 2002;160:S40–S53. doi: 10.1086/342143. doi:10.1086/342143 [DOI] [PubMed] [Google Scholar]
- Barraclough T.G, Hogan J.E, Vogler A.P. Testing whether ecological factors promote cladogenesis in a group of tiger beetles (Coleoptera: Cicindelidae) Proc. R. Soc. B. 1999;266:1061–1067. doi:10.1098/rspb.1999.0744 [Google Scholar]
- Bininda-Emonds O.R.P, et al. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. doi:10.1038/nature05634 [DOI] [PubMed] [Google Scholar]
- Bowers M.A, Brown J.H. Body size and coexistence in desert rodents: chance or community structure. Ecology. 1982;63:391–400. doi:10.2307/1938957 [Google Scholar]
- Charleston M, Robertson D.L. Preferential host switching by primate lentiviruses can account for phylogenetic similarity with the primate phylogeny. Syst. Biol. 2002;51:528–535. doi: 10.1080/10635150290069940. doi:10.1080/10635150290069940 [DOI] [PubMed] [Google Scholar]
- Cleaveland S, Laurenson M.L, Taylor L.H. Disease of humans and their domestic mammals: pathogen characteristics, host range and the risk of emergence. Phil. Trans. R. Soc. B. 2001;356:991–999. doi: 10.1098/rstb.2001.0889. doi:10.1098/rstb.2001.0889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley M.J. Wiley; New York, NY: 2002. Statistical computing: an introduction to data analysis using S-plus. [Google Scholar]
- Daszak P, Berger L, Cunningham A.A, Hyatt A.D, Green D.E, Speare R. Emerging infectious diseases and amphibian population declines. Emerg. Infect. Dis. 1999;5:735–748. doi: 10.3201/eid0506.990601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daszak P, Cunningham A.A, Hyatt A.D. Emerging infectious diseases of wildlife—threats to biodiversity and human health. Science. 2000;287:443–449. doi: 10.1126/science.287.5452.443. doi:10.1126/science.287.5452.443 [DOI] [PubMed] [Google Scholar]
- Davies T.J, Meiri S, Barraclough T.G, Gittleman J.G. Species co-existence and character divergence across carnivores. Ecol. Lett. 2007;10:146–152. doi: 10.1111/j.1461-0248.2006.01005.x. doi:10.1111/j.1461-0248.2006.01005.x [DOI] [PubMed] [Google Scholar]
- Domingo E, Holland J.J. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. doi:10.1146/annurev.micro.51.1.151 [DOI] [PubMed] [Google Scholar]
- Fenton A, Pedersen A.B. Community epidemiology in theory and practice: a conceptual framework for classifying disease threats in human and wild populations. Emerg. Infect. Dis. 2005;11:1815–1821. doi: 10.3201/eid1112.050306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert G.S, Webb C.O. Phylogenetic signal in plant pathogen–host range. Proc. Natl Acad. Sci. USA. 2007;104:4979–4983. doi: 10.1073/pnas.0607968104. doi:10.1073/pnas.0607968104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenyer R, et al. The global distribution and conservation of rare and threatened vertebrates. Nature. 2006;444:93–96. doi: 10.1038/nature05237. doi:10.1038/nature05237 [DOI] [PubMed] [Google Scholar]
- Hahn B.H, Shaw G.M, DeCock K.M, Sharp P.M. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287:607–614. doi: 10.1126/science.287.5453.607. doi:10.1126/science.287.5453.607 [DOI] [PubMed] [Google Scholar]
- Hudson P.J, Rizzoli A, Grenfell B.T, Heesterbeek H, Dobson A.P. Oxford University Press; Oxford, UK: 2001. Ecology of wildlife diseases. [Google Scholar]
- Jaccard P. The distribution of flora in the alpine zone. New Phytol. 1912;11:37–50. doi:10.1111/j.1469-8137.1912.tb05611.x [Google Scholar]
- Kolar C, Lodge D.M. Progress in invasion biology: predicting invaders. Trends Ecol. Evol. 2001;16:199–204. doi: 10.1016/s0169-5347(01)02101-2. doi:10.1016/S0169-5347(01)02101-2 [DOI] [PubMed] [Google Scholar]
- Koleff P, Gaston K.J, Lennon J.J. Measuring beta diversity for presence absence data. J. Anim. Ecol. 2003;72:367–382. doi:10.1046/j.1365-2656.2003.00710.x [Google Scholar]
- Korber B, Muldoon M, Theiler J, Gao F, Gupta R, Lapedes A, Hahn B.H, Wolinsky S, Bhattacharya T. Timing the ancestor of the HIV-1 pandemic strains. Science. 2000;288:1789–1796. doi: 10.1126/science.288.5472.1789. doi:10.1126/science.288.5472.1789 [DOI] [PubMed] [Google Scholar]
- Kronfeld-Schor N, Dayan T. Partitioning of time as an ecological resource. Annu. Rev. Ecol. Syst. 2003;34:153–181. doi:10.1146/annurev.ecolsys.34.011802.132435 [Google Scholar]
- Kuiken T, Holmes E.C, McCauley J, Rimmelzwaan G.F, Williams C.S, Grenfell B.T. Host species barriers to influenza virus infections. Science. 2006;312:394–397. doi: 10.1126/science.1122818. doi:10.1126/science.1122818 [DOI] [PubMed] [Google Scholar]
- Legendre P. Comparison of permutation methods for the partial correlation and partial Mantel tests. J. Stat. Comput. Simul. 2000;67:37–73. doi:10.1080/00949650008812035 [Google Scholar]
- Lindenfors P, Nunn C.L, Jones K.E, Cunningham A.A, Sechrest W, Gittleman J.L. Parasite species richness in carnivores: effects of host body mass, latitude, geographical range and population density. Glob. Ecol. Biogeogr. 2007;16:496–509. doi:10.1111/j.1466-8238.2006.00301.x [Google Scholar]
- Lockhart A.B, Thrall P.H, Antonovics J. The distribution and characteristics of sexually transmitted diseases in animals: ecological and evolutionary implications. Biol. Rev. Camb. Philos. Soc. 1996;71:415–471. doi: 10.1111/j.1469-185x.1996.tb01281.x. doi:10.1111/j.1469-185X.1996.tb01281.x [DOI] [PubMed] [Google Scholar]
- Lockwood J.L, Cassey P, Blackburn T. The role of propagule pressure in explaining species invasion. Trends Ecol. Evol. 2005;20:223–228. doi: 10.1016/j.tree.2005.02.004. doi:10.1016/j.tree.2005.02.004 [DOI] [PubMed] [Google Scholar]
- Nunn C.L, Altizer S.M. The global mammal parasite database: an online resource for infectious disease records in wild primates. Evol. Anthropol. 2005;14:1–2. doi:10.1002/evan.20041 [Google Scholar]
- Nunn C.L, Altizer S.M. Oxford University Press; Oxford, UK: 2006. Infectious diseases in primates: behavior, ecology and evolution. [Google Scholar]
- Nunn C.L, Altizer S, Jones K.E, Sechrest W. Comparative tests of parasites species richness in primates. Am. Nat. 2003;162:597–614. doi: 10.1086/378721. doi:10.1086/378721 [DOI] [PubMed] [Google Scholar]
- Nunn C.L, Altizer S, Sechrest W, Jones K.E, Barton R.A, Gittleman J.L. Parasites and the evolutionary diversification of primate clades. Am. Nat. 2004;164:S90–S103. doi: 10.1086/424608. doi:10.1086/424608 [DOI] [PubMed] [Google Scholar]
- Osterhaus A.D.M.E. Catastrophes after crossing species barriers. Phil. Trans. R. Soc. B. 2001;356:791–793. doi: 10.1098/rstb.2001.0856. doi:10.1098/rstb.2001.0856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page R. University Chicago Press; Chicago, IL: 2003. Tangled trees, phylogeny, cospeciation, and coevolution. [Google Scholar]
- Pedersen A.B, Altizer S, Poss M, Cunningham A.A, Nunn C.L. Patterns of host specificity and transmission among parasites of wild primates. Int. J. Parasitol. 2005;35:647–657. doi: 10.1016/j.ijpara.2005.01.005. doi:10.1016/j.ijpara.2005.01.005 [DOI] [PubMed] [Google Scholar]
- Pedersen A.B, Jones K.E, Nunn C.L, Altizer S.A. Infectious disease and mammalian extinction risk. Conserv. Biol. 2007;21:1269–1279. doi: 10.1111/j.1523-1739.2007.00776.x. doi:10.1111/j.1523-1739.2007.00776.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters M. Cross-species transmissions of simian retroviruses in Africa and risk for human health. Lancet. 2004;363:911–912. doi: 10.1016/S0140-6736(04)15819-4. doi:10.1016/S0140-6736(04)15819-4 [DOI] [PubMed] [Google Scholar]
- Perlman S.J, Jaenike J. Evolution of multiple components of virulence in Drosophila-nematode associations. Evolution. 2003;57:1543–1551. doi: 10.1111/j.0014-3820.2003.tb00362.x. doi:10.1111/j.0014-3820.2003.tb00362.x [DOI] [PubMed] [Google Scholar]
- Pfennig D.W. Effect of predator–prey phylogenetic similarity on the fitness consequences of predation: a trade-off between nutrition and disease? Am. Nat. 2000;155:335–345. doi: 10.1086/303329. doi:10.1086/303329 [DOI] [PubMed] [Google Scholar]
- Ricklefs R.E, Fallon S.M. Diversification and host switching in avian malaria parasites. Proc. R. Soc. B. 2002;269:885–892. doi: 10.1098/rspb.2001.1940. doi:10.1098/rspb.2001.1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelke-Parker M.E, et al. A canine distemper virus epidemic in Serengeti lions (Panthera leo) Nature. 1996;376:441–445. doi: 10.1038/379441a0. doi:10.1038/379441a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B.A. Patterns of association between crucifers and their flower-mimic pathogens: host jumps are more common than coevolution or cospeciation. Evolution. 2001;55:41–53. doi: 10.1111/j.0014-3820.2001.tb01271.x. doi:10.1111/j.0014-3820.2001.tb01271.x [DOI] [PubMed] [Google Scholar]
- Smouse P.E, Long J.C, Sokal R.R. Multiple regression and correlation extensions of the Mantel test of matrix correspondence. Syst. Zool. 1986;35:627–632. doi:10.2307/2413122 [Google Scholar]
- Taylor L.H, Latham S.M, Woolhouse M.E.J. Risk factors for human disease emergences. Phil. Trans. R. Soc. B. 2001;356:983–989. doi: 10.1098/rstb.2001.0888. doi:10.1098/rstb.2001.0888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh P.D, et al. Catastrophic ape decline in western equatorial Africa. Nature. 2003;422:611–614. doi: 10.1038/nature01566. doi:10.1038/nature01566 [DOI] [PubMed] [Google Scholar]
- Webb C.O, Ackerly D.D, McPeek M.A, Donoghue M.J. Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 2002;33:475–505. doi:10.1146/annurev.ecolsys.33.010802.150448 [Google Scholar]
- Wolfe N.D, Daszak P, Kilpatrick A.M, Burke D.S. Bushmeat hunting, deforestation, and prediction of zoonoses emergence. Emerg. Infect. Dis. 2005;11:1822–1827. doi: 10.3201/eid1112.040789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse M.E.J, Gowtage-Sequeria S. Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 2005;11:1842–1847. doi: 10.3201/eid1112.050997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse M.E.J, Taylor L.H, Haydon D.T. Population biology of multi-host pathogens. Science. 2001;292:1109–1112. doi: 10.1126/science.1059026. doi:10.1126/science.1059026 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Summary of the results from partial Mantel tests of shared pathogens (βj) against host divergence time and proportional range overlap for each of the pathogen subsets. Table S2. Generalised linear models of shared pathogens (βj) against host divergence time and sympatry for total pathogen community each of the pathogen subsets. Table S3. Regressions of shared pathogens (βj) against host divergence time and sympatry for each of the pathogen subsets including pathogen sample sizes as a covariate in the GLMs