Summary
A complete mitochondrial (mt) genome sequence was reconstructed from a 38,000-year-old Neandertal individual using 8,341 mtDNA sequences identified among 4.8 Gb of DNA generated from ~0.3 grams of bone. Analysis of the assembled sequence unequivocally establishes that the Neandertal mtDNA falls outside the variation of extant human mtDNAs and allows an estimate of the divergence date between the two mtDNA lineages of 660,000±140,000 years. Of the 13 proteins encoded in the mtDNA, subunit 2 of cytochrome c oxidase of the mitochondrial electron transport chain has experienced the largest number of amino acid substitutions in human ancestors since the separation from Neandertals. There is evidence that purifying selection in the Neandertal mtDNA was reduced compared to other primate lineages suggesting that the effective population size of Neandertals was small.
Introduction
Although it is well established that Neandertals are the hominid form most closely related to present-day humans, their exact relationship with modern humans remains a topic of debate (Hublin and Pääbo, 2006; Soficaru et al., 2006; Harvati et al., 2007). Molecular genetic data first spoke to this issue in 1997 when a 379-base-pair section of the hypervariable I region (HVRI) of the mitochondrial genome (mtDNA) was determined from the Neandertal type specimen found in 1856 in Neander Valley, near Düsseldorf, Germany (Krings et al., 1997). Since then, a total of 15 complete or partial Neandertal HVRI sequences as well as two HVRII sequences (Krings et al., 1999; Krings et al., 2000) have been described. Phylogenetic analyses of these suggest that Neandertal mtDNAs fall outside the variation of modern human mtDNAs. Since the mtDNA genome is maternally inherited without recombination, these results indicate that Neandertals made no lasting contribution to the modern human mtDNA gene pool (Krings et al., 1997; Currat and Excoffier, 2004; Serre et al., 2004).
High-throughput 454 sequencing techniques have recently been applied to ancient DNA (Green et al., 2006; Poinar et al., 2006; Stiller et al., 2006). These methods open new possibilities for the retrieval of ancient DNA that has hitherto relied either on the cloning of random molecules in bacteria (Higuchi et al., 1984; Pääbo, 1985; Noonan et al., 2005; Noonan et al., 2006) or on the PCR amplification of individual DNA sequences of interest (Pääbo and Wilson, 1988; Pääbo et al., 2004). The main benefit of the 454 sequencing technique is the sheer volume of sequence data that make it practical to undertake genome-scale ancient DNA sequencing projects. This is particularly feasible for mitochondrial genomes (Gilbert et al., 2007), given their smaller size relative to the nuclear genome and their abundance in cells, where typically several hundred mtDNAs per nuclear genome exist.
454 sequence data from ancient DNA have also allowed an increased understanding of DNA diagenesis, i.e. how DNA is modified during deposition in a burial context. In particular, they have allowed a quantitative model of how DNA degradation and chemical modification occurs and how the effects of these processes interact with the molecular manipulations used to generate sequencing libraries (Briggs et al., 2007). Notably, although it was previously known that a high rate of cytosine deamination occurs in ancient DNA (Hofreiter et al., 2001) it has become clear that this is particularly prevalent in the ends of the ancient molecules presumably because these are often single-stranded (Briggs et al., 2007). Deamination of cytosine residues results in uracil residues that are read as thymine by DNA polymerases, leading to a high rate of C to T transitions. A high rate of G to A transitions observed near the 3’-ends of sequence reads is thought to be caused by deaminated cytosine residues on the complementary strands used as templates during the fill-in reaction to create blunt ends when sequencing libraries are constructed (Briggs et al., 2007).
By 454 sequencing, we have generated 34.9-fold coverage of the Neandertal mtDNA genome from a Neandertal bone (Vindija bone 33.16) excavated in 1980 from Vindija Cave, Croatia (Malez, 1982). It has been dated to 38,310+/−2,130 years before present (Serre et al., 2004). Previously, the mtDNA HVRI sequence of this bone has been determined (Serre et al., 2004) as well as 2,414 bp of mtDNA sequences by 454 sequencing (Green et al., 2006). Here, we present its complete mtDNA sequence as well as the insights it allows into recent human and Neandertal mtDNA evolution.
Results
DNA sequence determination
Three DNA extracts, each from 100–200 mg of a Neandertal bone (Vindija 33.16) were prepared in our clean room facility where several precautions against DNA contamination are implemented (Experimental Procedures). These include complete separation from other parts of the laboratories, direct delivery of all equipment and reagents to the facility, positive pressure generated with filtered air that excludes particles larger than 0.2 µm, and UV irradiation and bleach treatment of all surfaces. The bone surface was removed prior to extraction. However, the interior of bones are also often contaminated with modern human DNA, presumably due to past washing and other treatments of Neandertal bones. Thus, we analyzed each extract for contamination by extant human mtDNA by PCR using primers flanking positions in the HVRI that distinguish extant humans from Neandertals (Green et al., 2006) and amplify both types of mtDNA with similar efficiencies. Following amplification, we cloned the PCR product and sequenced 103–112 clones to determine the ratio of Neandertal to extant human mtDNA. The contamination rate in the three extracts ranged from 0% to 0.9% (Supp. Fig. 1).
From these extracts, we generated a total of nine 454 libraries in the clean room facility using 454-adapters with a Neandertal-specific sequence key that is unique to this project and thus unequivocally identifies each sequence determined as derived from the extract of a Neandertal bone (Briggs et al., 2007). This allows detection of any contamination that may be introduced in subsequent handling and sequencing steps outside the clean room. To maximize the library and sequence yield, we incorporate two modifications to the standard 454 protocol that reduce the need to perform titration runs of libraries (Meyer et al., 2008) and allows more molecules to be retrieved during library preparation (Maricic and Pääbo, unpublished results). From these libraries we generated a total of 39 million sequence reads by 147 runs on the GS FLX sequencing platform. Bases were called using the standard 454 signal threshold and filtering criteria with minor modifications tailored for short, ancient sequence reads (Meyer et al., 2008)
Neandertal sequences were identified within each run as described previously (Green et al., 2006) with the chief criterion being sequence similarity to a primate genome. MtDNA sequences were identified from these using further criteria (see Experimental Procedures) including similarity to the human mtDNA at least as great as to any nuclear DNA sequence. While the total fraction of sequences that are identified as Neandertal varied across libraries and library pools, the ratio of putative Neandertal nuclear DNA sequences to mtDNA sequences varied little among these libraries and averages 171. This corresponds to approximately 2,100 mtDNA molecules per cell. In total, 8,341 mtDNA sequences were identified. They are of an average length of 69.4 bp (standard deviation=26.4), with the shortest fragment identified being 30 bp (limited by the length cut-off in the analysis pipeline) and the longest fragment being 278 bp (limited by the flow cycles performed on the GS FLX instrument).
MtDNA genome assembly
Ancient DNA sequences present a challenge for DNA sequence assembly since they are typically short and exhibit high rates of nucleotide misincorporation. A further complication is that pyrosequencing (Ronaghi et al., 1998), for example as performed on the GS FLX platform, calls long polymers of the same base with reduced accuracy. With these issues in mind, we designed an assembly procedure for ancient DNA. In short, each sequence identified as mtDNA was aligned over its entire length to the human reference mtDNA sequence (UCSC build hg18). These alignments were then merged and each alignment column was examined to determine the majority base, yielding an assembled mtDNA sequence. Homopolymer lengths at positions where the reference human carries ≥5 identical bases were determined by analysis of the raw signal distributions as described in Supp. Data.
Following this, some problematic regions remained in the assembly. These include four regions of a total length of 20 bp where no sequence coverage existed, eight other regions amounting to a total of 117 bp covered by only single reads, nine positions where no majority base existed due to low coverage, and 31 homopolymers for which the data were not sufficient to determine their length. These regions were amplified by PCR from a Vindija 33.16 bone extract in two-step multiplex PCRs (Krause et al., 2006), cloned and sequenced by Sanger technology in order to complete the assembly (Experimental Procedures).
We then reapplied our mtDNA fragment detection pipeline to all Neandertal DNA sequences determined from the bone but compared them to the assembled Neandertal mtDNA instead of the reference human mtDNA. This resulted in the detection of an additional 721 mtDNA sequences that were initially missed since they have a higher similarity to the human nuclear genome than human mtDNA and thus could only be identified using the assembled Neandertal mtDNA. Interestingly, 522 of these sequences were similar to a single nuclear mtDNA insertion on chromosome 1 (hg18 position 554,327–560,165). In total, 8,341 mtDNA fragments totaling 578,733 nt yielded a final assembly of 16,565 nt where no position has less than 9-fold sequence coverage.
Estimates of contamination
In order to estimate the level of human contamination among the mtDNA sequences determined from these libraries, we realigned the individual sequences to the assembled mtDNA and used a number of differences between the putative Neandertal and human mtDNAs as diagnostic markers. We then examined each alignment to determine if it contained a position that allowed it to be classified as of either Neandertal or extant human origin.
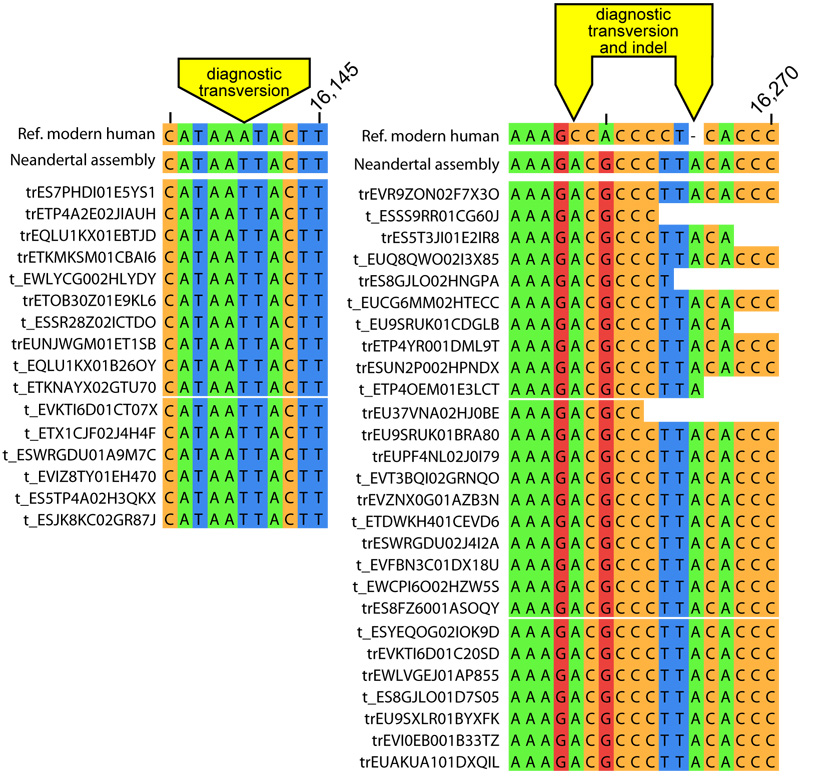
First, we used three human-Neandertal differences in the HVR1: an A to T and a C to A transversion and an insertion of an A where all Neandertal sequences determined to date, including Vindija 33.16 (Serre et al., 2004), differ from all 1,865 contemporary human HVRI sequences available in the mtDB database (Ingman and Gyllensten, 2006). Forty three sequences overlapped these HVRI positions and all were of the Neandertal type (Fig. 1). Second, we used four transversions between the putative Neandertal mtDNA and human mtDNAs that occur outside the HVR1 and are not adjacent to homopolymers and not polymorphic among the 1,865 human mtDNA sequences. Among 192 sequences that overlapped these positions, 186 carried the base of the Neandertal assembly (Supp. Fig. 2). Of the 6 sequences that did not, five were cases where the observed base was T, the assembly base was C, and the modern human base was G or A. Thus, these are likely to be the result of deaminated cytosine residues that are common in ancient DNA (Hofreiter et al., 2001; Briggs et al., 2007; Brotherton et al., 2007). The alternative that these are contamination by a previously unknown modern human mtDNA sequence is unlikely because of the large number of modern human sequences available. One single sequence matches the human base at a diagnostic position. Interestingly, this sequence shows features commonly associated with ancient DNA (Supp. Fig. 2) in that it begins with a C to T mismatch to the assembly, ends with two G to A mismatches, and is 65 nucleotides long and thus relatively short. In spite of this, it may represent a contaminant since modern DNA sequences retrieved from ancient specimens can carry such features (Sampietro et al., 2006). In order to estimate contamination levels, we thus disregard the 5 C to T mismatches as uninformative and count the last sequence as a contaminant. This yields a total of 229 out of 230 sequences carrying Neandertal diagnostic positions and an estimate of the contamination rate of 0.4% with a 95% confidence interval of 0.01% to 2.4%
Figure 1.
Sequences overlapping HVRI positions carrying diagnostic differences between Neandertal and extant humans. All 43 sequences overlapping the three diagnostic positions are shown.
In order to maximize our power to detect contamination, we also used all positions in the Neandertal assembly that differ from extant humans and for which at least 99% of a panel of 311 worldwide modern human mtDNA sequences do not differ, ignoring positions that could be due to cytosine deamination. One hundred thirty-three positions fulfilled these criteria and allowed 1,963 fragments to be classified. Nine were of extant human origin yielding a contamination estimate of 0.5% (95% confidence interval 0.21% to 0.87%)
We conclude that so few of the mtDNA sequences determined derive from extant humans that they will not compromise the assembly which has a 35-fold average coverage. The assembled mtDNA sequence therefore represents a reliable reconstruction of the mtDNA that this Neandertal individual carried when alive.
MtDNA sequence analyses
Alignment of the 16,565 nt Neandertal mtDNA to the 16,568 nt human revised Cambridge reference mtDNA sequence (rCRS) (Andrews et al., 1999) revealed 206 differences (195 transitions and 11 transversions). In the non-coding control region, the Neandertal sequence contains a deletion of four base pairs (CACA) at rCRS position 514 and a previously known insertion of one base pair (Serre et al., 2004) following position 16,263. The 13 protein-coding genes, the 22 tRNA genes and two rRNA genes in the Neandertal mtDNA lack notable structural differences when compared to the human and chimpanzee mtDNAs (Supp. Table 2).
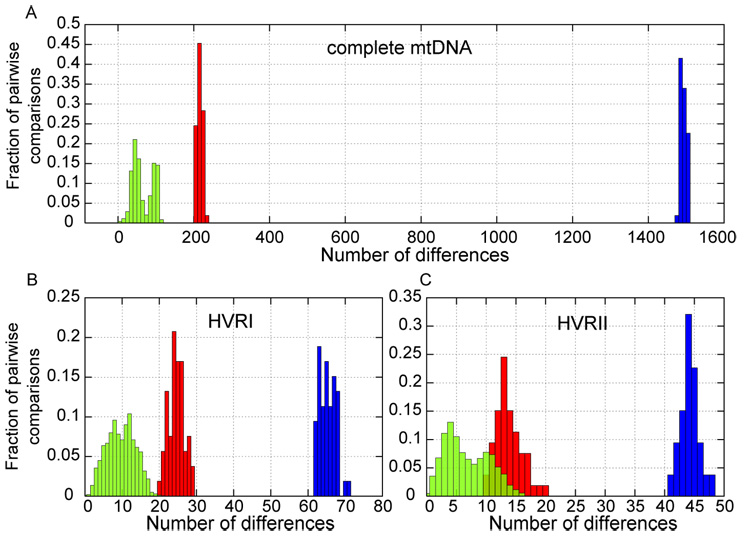
Figure 2A shows the distribution of pairwise sequence differences among 53 humans from around the world (Ingman et al., 2000), between these and the Neandertal, and between the modern humans and the chimpanzee mtDNAs. Among the humans, the number of differences ranges from 2 to 118 and is bimodal. The peak with a mode around 99 differences contains comparisons that involve at least one member of deep clades containing sub-Saharan African mtDNAs. The peak with a mode around 44 differences involves comparisons between individuals outside these clades. By contrast, the number of differences between human mtDNAs and the Neandertal mtDNA ranges from 201 to 234 and is unimodal. Thus, the Neandertal mtDNA falls outside the variation of extant humans even when nucleotide differences, uncorrected for multiple substitutions, are counted. Notably, when the comparisons are restricted to only the HVRI or HVRII, the distributions of pairwise differences among extant humans and between humans and the Neandertal overlap illustrating that multiple substitutions complicate the reconstruction of mtDNA relationships when only these regions are studied (Figure 2B, C).
Figure 2.
Distribution of pairwise mtDNA sequence differences among 53 humans (green), between humans and the Neandertal mtDNA (red), and between humans and chimpanzee (blue) in (A) the complete mtDNA; (B) the HVRI (Neandertal pos. 16,044–16,411); and (C) the HVRII (pos. 57–372).
MtDNA divergence
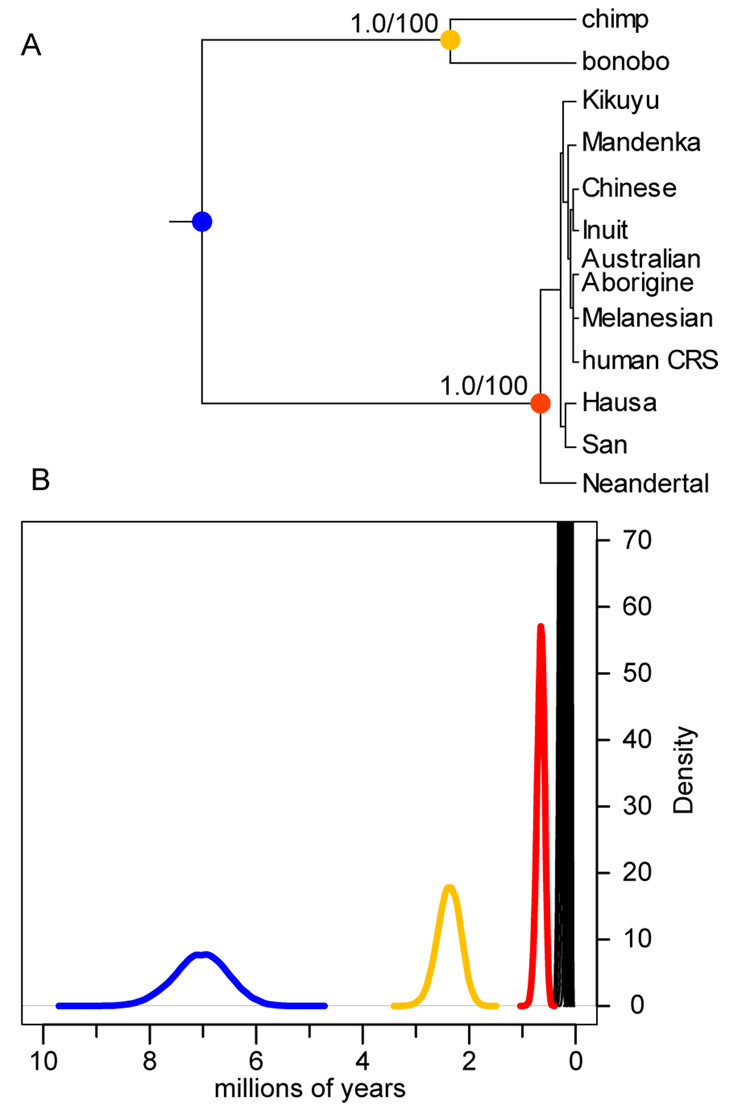
To further explore the evolutionary relationship between Neandertal and extant human mtDNAs, we estimated a phylogenetic tree using the complete mtDNA of Neandertal, 53 human mtDNAs, the human rCRS, and chimpanzee and bonobo mtDNAs as outgroups. Neighbor-joining, maximum likelihood, parsimony, and a Bayesian approach (Supp. Data) all indicated that the 54 extant humans formed a monophyletic group to the exclusion of the Neandertal with complete support for each tree-building measure (100% bootstrap support; 1.0 posterior probability). Unsurprisingly, this topology is also found when analysis is restricted to various subsets of the sequence such as protein-coding sequences and RNA-coding sequences (Supp. Data).
In order to estimate the date of the divergence of the Neandertal and extant human mtDNAs, we compared the Neandertal mtDNA to 10 divergent human mtDNAs (Fig. 3A) and tested if we could reject a molecular clock assuming the topology of the Bayesian tree (Supp. Data). This was not the case indicating no significant heterogeneity in evolutionary rates among the sequences. To allow divergences of mtDNA sequences to be transformed to calendar years, we assumed - based on the fossil record - that humans and chimpanzees diverged between six (Galik et al., 2004) to eight million years ago (Brunet et al., 2002; Lebatard et al., 2008). This results in an estimate of the mean divergence time between Neandertal and extant human mtDNAs of 660,000 with a 95% credibility interval of 520,000 to 800,000 years ago (Fig. 3B) (see also Supp. Data) which overlaps with previous estimates based on HVRI and II sequences (Krings et al., 1997; Krings et al., 1999). Since the width of the divergence credibility interval increases almost linearly with the posterior mean of the divergence estimate (Supp. Fig. 5), further sequence data would be unlikely to decrease the width of the credibility interval (Yang and Rannala, 2006). However, if the estimated date of the divergence between humans and chimpanzees or current assumptions about how the mtDNA evolves would be incorrect, the estimates in calendar years of the divergence of the Neandertal and human mtDNAs would need to be revised.
Figure 3.
Phylogenetic tree and divergence time estimate of mtDNA sequences. (A) Bayesian phylogenetic tree of complete mtDNA sequences of the Neandertal, 10 extant humans, one chimpanzee, and one bonobo. Identical topologies for the Neandertal and chimpanzee/bonobo split are produced by each tree-building methods. The Bayesian posterior probability and the bootstrap support values are shown for two internal nodes. (B) Posterior distribution of divergence times at each internal node using a 6–8 Mya for the ape/hominid divergence (blue node). The extant human divergences are shown in black, the Neandertal/human divergence in red, the chimpanzee/bonobo divergence in yellow, and the ape/hominid in blue.
Mitochondrial protein evolution
Next we estimated the substitutions that occurred in each of the 13 mtDNA protein-coding genes on the Neandertal and the human lineages using parsimony with the chimpanzee as an outgroup (Table 1). The total number of silent substitutions assigned to the Neandertal lineage, 44, is lower than the number assigned to the human lineage, 57. An apparent shortening of the Neandertal lineage relative to the human branch is also observed when all substitutions in the mtDNA are analyzed (Supp. Fig. 3). This dearth of Neandertal lineage differences is not clearly concentrated in any particular region of the mtDNA (Supp. Fig. 4A) or class of substitutions (Supp. Fig. 4B,C) and is not statistically significant (Supp. Data). It may reflect stochastic variation in the amount of substitutions on the two lineages, perhaps in combination with the fact that the Neandertal mtDNA is ~38,000 years older than the human mtDNA sequence. However, for amino acid substitutions, we observe a similar number on the Neandertal (20) and human (18) lineages. Thus, relative to silent substitutions more amino acid replacements occurred on the Neandertal lineage.
Table 1.
Number of synonymous and non-synonymous substitutions in each protein coding mtDNA gene assigned to the Neandertal or extant human lineage by parsimony using the chimpanzee as an outgroup.
| Gene | Neandertal synonymous | Neandertal non-synonymous | Extant human synonymous | Extant human non-synonymous |
|---|---|---|---|---|
| ND1 | 4 | 2 | 5 | 2 |
| ND2 | 6 | 1 | 3 | 1 |
| COX1 | 8 | 0 | 8 | 0 |
| COX2 | 0 | 0 | 3 | 4 |
| ATP8 | 2 | 1 | 3 | 0 |
| ATP6 | 3 | 3 | 1 | 2 |
| COX3 | 1 | 1 | 3 | 1 |
| ND3 | 1 | 1 | 5 | 1 |
| ND4L | 3 | 1 | 1 | 0 |
| ND4 | 5 | 0 | 9 | 0 |
| ND5 | 7 | 5 | 6 | 4 |
| ND6 | 1 | 1 | 1 | 0 |
| CYTB | 3 | 4 | 9 | 3 |
| Total | 44 | 20 | 57 | 18 |
To explicitly test for an increase in amino acid replacements on the Neandertal lineage, we evaluated the evolution of the 12 protein-coding genes (excluding ND6, see Supp. Data) across seven primates (human, Neandertal, chimpanzee, bonobo, gorilla, orangutan, and baboon) using a maximum likelihood framework. The ratio of amino acid replacements per replacement site (dN) to silent substitutions per silent site (dS) can reveal the influence of natural selection on protein evolution with low dN/dS (≪1) indicating purifying selection and high dN/dS (>1) indicating positive selection. Overall, dN/dS was quite low across the entire phylogeny (0.051–0.087; Supp. Table 3), consistent with strong purifying selection on mitochondrial proteins in primates (Hasegawa et al., 1998). However, dN/dS varied significantly among branches in the phylogeny (all P<0.0001; Supp. Table 3). While lineage-specific estimates of dN/dS were sensitive to both the underlying model of sequence evolution and the number of outgroups considered (see Supp. Fig. 6 and Supp. Table 3), we consistently observed the highest dN/dS on the Neandertal lineage and this value was significantly higher than background rates in all comparisons (all P < 0.01; Supp. Table 3).
To determine if any of the 13 proteins encoded in the mtDNA show an unusual pattern of evolution in humans since the divergence from the Neandertal, we contrasted the ratio of nucleotide polymorphisms in the 54 humans to fixed differences to Neandertal at synonymous and non-synonymous sites (Table 2). Under a standard neutral model the ratio of diversity to divergence should be the same for both classes of sites (McDonald and Kreitman, 1991). Whereas this is the case for 12 of the protein-coding genes, COX2 has an excess of amino acid divergence (P=0.021), consistent with the action of positive directional selection. However, when corrected for multiple tests of 13 genes, this yields only suggestive evidence for adaptive evolution of COX2 (Bonferroni α =0.0038).
Table 2.
Polymorphism within humans and divergence to Neandertal at synonymous and nonsynonymoussites in protein-coding mtDNA genes. The neutrality index value (Rand and Kann, 1996) is the ratio of polymorphism to divergence at non-synonymous sites versus this ratio atsynonymous sites. Values smaller than 1 indicate an excess of non-synonymous divergence. Values greater than 1 indicate excess non-synonymous polymorphism within humans.
| Gene | Fixed synonymous differences | Human synonymous polymorphic sites | Fixed non-syn. differences | Human non-syn. polymorphic sites | Neutrality index | P-value uncorrected (Fisher’s exact test) |
|---|---|---|---|---|---|---|
| ND1 | 7 | 19 | 2 | 8 | 1.47 | 1 |
| ND2 | 7 | 30 | 1 | 13 | 3.03 | 0.419 |
| COX1 | 13 | 40 | 0 | 8 | - | 0.184 |
| COX2 | 2 | 19 | 4 | 3 | 0.08 | 0.021 |
| ATP8 | 4 | 6 | 1 | 2 | 1.33 | 1 |
| ATP6 | 2 | 22 | 2 | 9 | 0.41 | 0.575 |
| COX3 | 3 | 25 | 2 | 4 | 0.24 | 0.205 |
| ND3 | 5 | 10 | 1 | 3 | 1.50 | 1 |
| ND4L | 2 | 9 | 1 | 1 | 0.22 | 0.423 |
| ND4 | 7 | 43 | 0 | 12 | - | 0.328 |
| ND5 | 11 | 51 | 7 | 23 | 0.71 | 0.58 |
| ND6 | 1 | 20 | 0 | 5 | - | 1 |
| CYTB | 10 | 26 | 5 | 17 | 1.31 | 0.764 |
COX2 has experienced four amino acid substitutions on the human mtDNA lineage after its divergence from the Neandertal lineage about 660,000 years ago. In order to see if any of these amino acids vary among humans today, we analyzed human mtDNA sequences in mtDB (Ingman and Gyllensten, 2006). At one of the four positions (rCRS pos. 7,868), one of 2,704 individuals carries the same base as the Neandertal. Because mtDNA is inherited without recombination and because the Neandertal mtDNA falls outside the variation of modern human mtDNA, this single modern human observation represents a reversion to the ancestral state seen in Neandertals and chimpanzees. Thus, these four amino acid substitutions occurred in the relatively short period after the divergence of Neandertal and extant human mtDNAs and before the most recent common ancestor of current human mtDNAs. The observation of four non-synonymous substitutions on the modern human lineage and no amino acid changes on the Neandertal lineage stands in contrast to the overall trend of more non-synonymous evolution in Neandertal protein-coding genes (Table 1) and deserves consideration.
Subunit 2 of cytochrome c oxidase
COX2 encodes subunit 2 of the cytochrome c oxidase, or complex IV, of the inner mitochondrial membrane. Complex IV catalyzes the reduction of molecular oxygen to water using electrons from cytochrome c and protons from within the mitochondrial matrix. By acting as a proton pump (Belevich et al., 2006) it maintains a proton gradient across the mitochondrial inner membrane which drives the phosphorylation of ADP to ATP (Mitchell, 1966; Reid et al., 1966). It has previously been noted that genes encoding various components of the electron transport chain evolve quickly in primates (Doan et al., 2004; Grossman et al., 2004).
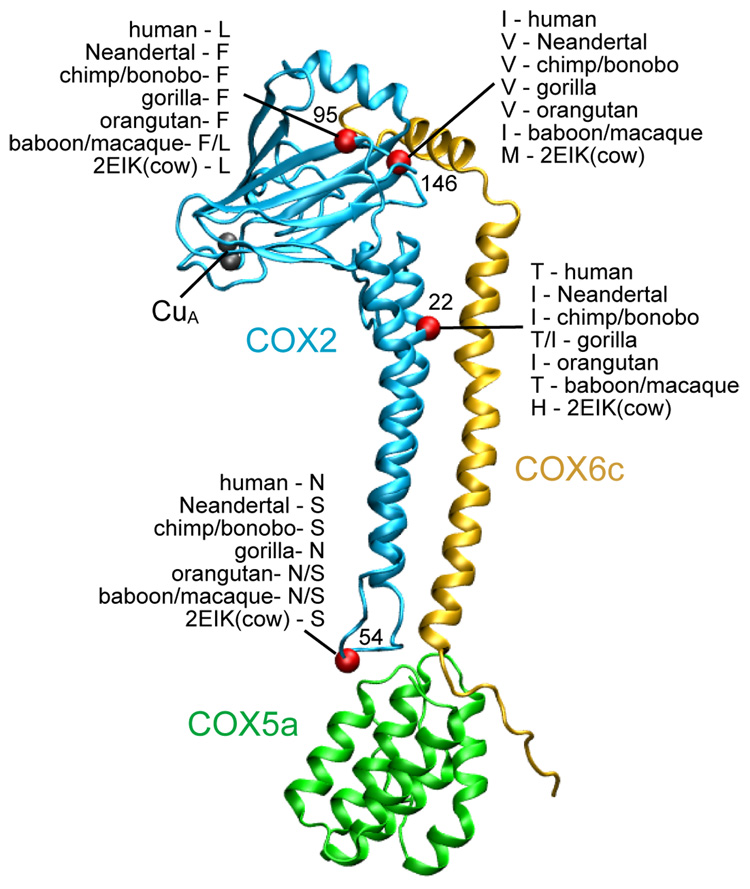
To determine what functional effects, if any, these amino acid substitutions may have, we examined the crystal structure of the bovine cytochrome c oxidase complex (Figure 4) (Muramoto et al., 2007) All four substitutions are spatially far from functionally critical parts of the protein such as the cytochrome c binding area and the bimetallic CuA center that passes electrons from cytochrome c to COX1. However, three of them interact with the nuclear encoded COX5a and COX6c subunits, which are thought to perform regulatory functions. It has been noted that COX5a evolves fast in primates (Uddin et al., 2008). However, the two COX5a sites that differ between humans and chimpanzees are not in close proximity to COX2. It may also be of relevance that two of the four amino acid residues that are specific to modern humans relative to Neandertals are variable among apes and all four are variable when Old World monkeys are considered (Figure 4).
Figure 4.
COX2 protein sequence differences between Neandertal and modern humans in structural context. The amino acid positions of the four differences are shown in red. The copper center of COX2 (CuA) is also shown. The amino acid at each position in some primate and the cow sequences (from which the structure (PDB identifier:2EIK) is derived) are indicated.
DNA diagenesis
The 8,341 mtDNA sequences comprising the Neandertal mtDNA assembly allow us to gauge a number of factors of relevance to the state of preservation of mtDNA in this late Pleistocene individual. These estimates have an advantage over previous ones (e.g. Briggs et al. 2007) that were based on inferences from alignments to human and chimpanzee genome sequences and were thus potentially affected by alignment uncertainties or uncertain inferences about evolutionary processes.
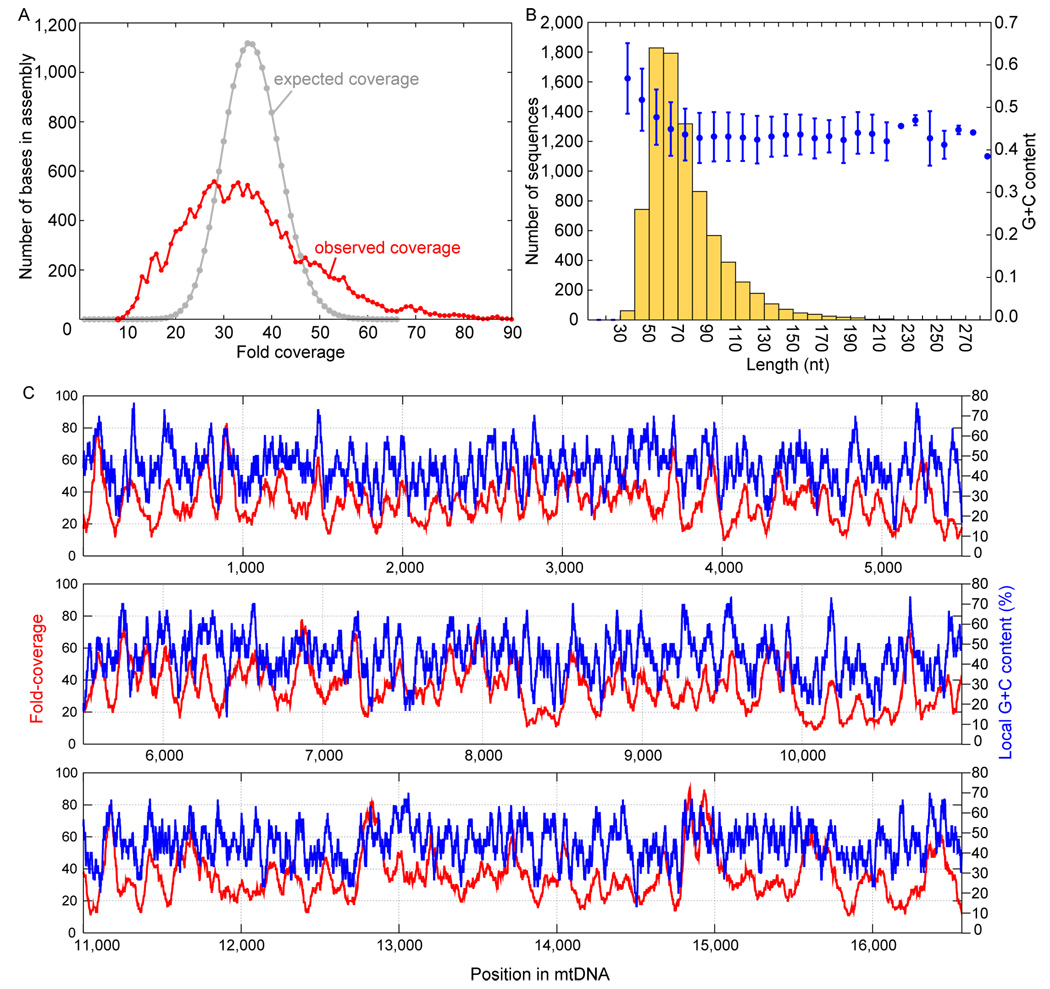
One feature of interest is that the retrieval of DNA across the mtDNA genome is more uneven than expected under a model of random sampling (Lander and Waterman, 1988) (Figure 5A). The variation in DNA retrieval is largely explained by a positive correlation (r=0.49) between GC content and the number of recovered fragments (Figure 5C). Furthermore, for shorter fragments, GC content is negatively correlated with length (Figure 5B). This may be due to denaturation of short, AT-rich fragments during library preparation.
Figure 5.
Sequence coverage and base composition of the Neandertal mtDNA.(A) The expected (grey) and observed (red) distribution of sequence depths in the Neandertal mtDNA assembly at 34.9 fold over-all coverage. (B) The length distribution of sequences (yellow) and, for each length bin, the mean and standard deviation of GC content (blue). (C) G+C content within a sliding window 30 bp 5’ and 3’ of each position (blue) and the observed coverage (red) at each position.
Because the GS FLX read length of about 250 nucleotides is much longer than the average sequence length of 69 nucleotides, fragmentation points can be inferred for nearly all sequences. As previously reported (Briggs et al., 2007), purines are over-represented before, and pyrimidines after, fragmentation points (Supp. Table 4). This is consistent with that depurination in ancient DNA which would induce strand breaks. When dinucleotides made up of the bases immediately 5’ and 3’ of fragmentation points are analyzed (Supp. Table 4), the most over-represented bases are G 5’ of breaks and T 3’ of breaks, where breaks occur ~2.9 times more often than would be expected if fragmentation was random and ~2.1 times more often than predicted by the individual contributions of G and T separately. Further investigations are necessary to understand the biochemical basis for this.
When we compare the observed rates of nucleotide misincorporations to maximum likelihood estimates for the expected numbers of misincorporations based on a model we previously developed (Briggs et al., 2007), we find the model fits the overall data well (Supp. Fig. 7). However, longer fragments have higher misincorporation rates (and by inference higher deamination rates), and shorter fragments have lower misincorporation rates than predicted by the model. This could be due to a reduced power to detect and align fragments that are both short and contain nucleotide misincorporations. Alternatively, it could represent a difference between shorter and longer fragments, for example if longer fragments have, on average, longer single-stranded overhanging ends.
Discussion
Neandertal genetic history
The complete Neandertal mtDNA genome confirms and extends previous insights into the genetic history of Neandertals. Firstly, it confirms that the Neandertal mtDNA falls outside the variation of extant human mtDNA variation. Second, it shows that the Neandertal mtDNA diverged from the extant human mtDNA lineage on the order of 660,000 years ago. Thus, the most recent common ancestor of human and Neandertal mtDNA lived more than two or three times as long ago as the most recent common ancestor of extant human mtDNAs.
A striking observation from the analysis of the 13 protein-coding genes in the mtDNA is that the ratio of non-synonymous to synonymous evolutionary rates is significantly higher on the Neandertal lineage. One plausible explanation is that Neandertals had a smaller effective population size and thus less effective purifying selection than humans. Previous studies have reported that mtDNA dN/dS ratios tend to be higher within than between species of great apes, including extant humans (Nachman et al., 1996; Templeton, 1996; Hasegawa et al., 1998; Kivisild et al., 2006). These results suggest that slightly deleterious amino acid variants segregate within populations and that differences in the intensity of purifying selection may affect mtDNA dN/dS ratios. Previous estimates based on mean pairwise differences (MPD) within the mtDNA HVRI suggested that Neandertals (MPD=5.5) had an effective population size similar to modern Europeans (MPD=4.0) or Asians (MPD=6.3), but lower than in modern Africans (MPD=8.1) (Krause et al., 2007b). Recent population genetic analyses have revealed a higher mtDNA amino acid substitution rate (Elson et al., 2004) and relatively more deleterious autosomal nuclear variants (Lohmueller et al., 2008) in Europeans than in Africans, presumably due to the smaller effective population size of Europeans. Thus, it seems plausible that Neandertals had a long-term effective population size smaller than that of modern humans. Population reductions caused by recurrent glaciations in Eurasia during Neandertals’ ~400,000 years of existence may have contributed to this. Further work will reveal if a small effective population size is compatible with the extent of nucleotide diversity seen in the Neandertal nuclear genome.
A tantalizing finding is that when all substitutions in the mtDNA are analyzed, the Neandertal mtDNA lineage is shorter than the human lineage by approximately 20% (Supp. Fig. 3B). It is tempting to speculate that this is due to the fact that the Neandertal mtDNA is ~38,000 years older than the extant human mtDNA sequences to which it is compared. However, under the assumption that the evolutionary dates are reasonably accurate, the reduction in length is about three times as large as would be expected if it was entirely due to the age of the fossil. It should be noted that for small evolutionary distances such as these, there is a large stochastic component to phylogenetic branch length. Thus, although the evolutionary dates are clearly dependent on many tenuous assumptions, it seems reasonable to assume that the majority of the discrepancy in length between the Neandertal and extant human mtDNA lineages is due to stochastic differences in the amounts of substitutions that have come to fixation on the two lineages.
Another interesting observation is that COX2 stands out among proteins encoded in the mitochondrial genome as having experienced four amino acid substitutions on the modern human mtDNA lineage. Further work is warranted to elucidate the functional consequences of these amino acid substitutions. However, all these substitutions are in regions of the protein that based on the crystal structure do not have any obvious function, and they are variable among primates. Hence, they may represent either minor adaptive advantages, perhaps of regulatory relevance, or have no significant functional consequences for mitochondrial function. Unless other evidence for their importance becomes available, we see no need to invoke positive selection to account for the evolution of COX2 on the human lineage.
DNA sequence authenticity
Two features of ancient DNA in general and Neandertal DNA in particular may cause errors in nucleotide sequences in excess of what is expected from modern DNA. First, chemical modification in the ancient DNA may cause nucleotide misincorporations, and, second, the unintended presence in the experiments of DNA from extant humans may be mistaken for Neandertal DNA. In the case of the Neandertal mtDNA genome sequences presented here, high average coverage of the random sequence reads in combination with amplification and sequencing of positions where coverage is low or where longer nucleotide homopolymers may cause base calling problems make us confident that the error rates from both these sources are low. Thus, the amount of data now accumulated for the Neandertal mtDNA allows extrapolations to what will be required to arrive at a reasonably reliable nuclear DNA sequences from the Neandertal.
With respect to nucleotide misincorporations, the current data show that errors causing a transition occur at positions carrying either a C or a G base in ~2.7% of sequence reads. By contrast, all other errors occur in less than 0.1% of reads (Supp. Fig. 8). Thus, C to T and G to A misincorporations are the predominant sequencing errors that need to be considered in terms of an ancient genome assembly. Since these misincorporations are drastically accumulated towards the 5’ and 3’ ends of molecules (Supp. Fig. 7), respectively, and since there is a tendency for strand breaks in the ancient DNA to occur 3’ of purines and in particular at GT dinucleotides (Supp. Table 4), this might conceivably result in an accumulation of misincorporations at certain nucleotide positions where even multiple retrieval of the same sequence may then not always allow the correct base to be determined. However, since GT dinucleotides cause only an approximately 3-fold increase in the likelihood of a break compared to random positions in the genome, already rather low sequence coverage makes this effect unlikely to cause errors in the assembled sequence. In fact, although it has been argued that C to T misincorporations are accumulated at particular sites in ancient mtDNA molecules (Gilbert et al., 2003), no positions in the mtDNA assembly have a mixture of observed bases that would confuse assembly (Supp. Fig. 9) and higher relative amounts of mismatches correlate with lower sequence coverage, suggesting that stochastic events among few recovered sequences rather than hotspots for DNA modifications are the basis of these observations. For example, the least supported position of the Neandertal mtDNA assembly (pos. 5,476) is supported by 8 C observations and 3 T observations (Supp. Fig. 9). Thus, we conclude that nucleotide misincorporations will not impede the determination of reliable Pleistocene genome sequences provided that reasonable sequence coverage is achieved.
Contamination with extant human DNA is the other dominant source of erroneous Neandertal sequences. Given the high coverage and the fact that the best estimate of the contamination rate here is 0.5% (with an upper 95% confidence limit of 0.87%), we do not expect contamination to affect the mtDNA sequence assembly to any appreciable level. Under the assumption that the Neandertal mtDNA sequence is reliable, it is a useful tool for gauging contamination when sequencing the Neandertal nuclear genome. Previously, assays to determine contamination within Neandertal fossil extracts were limited to the HVRI which carry few positions where extant humans differ from Neandertals. By contrast, the complete Neandertal mtDNA now offers 133 such positions. This enables a reliable estimation of mtDNA contamination by analyzing sequence reads from 454 libraries rather than by PCR-based assays of the DNA extracts. For example, when we do this in a small preliminary data set initially published from this fossil (Green et al., 2006), 10 of 10 sequences are classified as Neandertal. However, in further unpublished sequencing runs from that library, 8 out of 75 diagnostic sequences derive from extant human mtDNA, suggesting a contamination rate of ~11% (C.I. 4.7–20%). This is in agreement with the suggestion (Wall and Kim, 2007) that contamination occurred in that experiment. That library was constructed outside our clean room facility and before the introduction of the Neandertal-specific key which is crucial for the detection of contamination by other 454 libraries and was therefore not used for the subsequent Neandertal genome sequencing project (Briggs et al., 2007). However, with the help of the mtDNA presented here, such levels of contamination are now easily detectable from single 454 sequencing runs. Nevertheless, since the ratios of nuclear to mtDNA may differ between endogenous and contaminating DNA, it is of importance to identify nuclear DNA sequences diagnostic for Neandertals. Data collected from Vindija bone 33.16 now begin to make this possible (Krause et al., 2007a).
Outlook for nuclear genome sequencing
Irrespective of the fact that the amino acid substitutions in the human COX2 protein may not be of functional significance, they illustrate the power of Neandertal DNA sequences for finding accumulations of recent evolutionary changes in human genes. A genome-wide analysis of human and Neandertal genes holds the promise to identify many such cases for nuclear genes. However, whereas the mtDNAs of extant humans are monophyletic with respect to Neandertals, this is not expected to be the case for most nuclear genes (Pääbo, 1999). This fact will be helpful for the identification of recent positive selection in humans. If, for example, a completed selective sweep affected a gene in human ancestors after their separation from their common ancestors with Neandertals, extant human diversity in that region of the genome would coalesce to the exclusion of Neandertal alleles. Consequently, the combination of the accumulation of recent human substitutions in coding genes or conserved sequence elements along with reduced human diversity that excludes Neandertals may be a hallmark for genes and genomic regions that have been important in the emergence of fully modern humans.
Technically, the Neandertal mtDNA presented here is a useful forerunner for the sequencing of the Neandertal nuclear genome. First, the mtDNA assembly and the sequences of which it is comprised can be used to tune the parameters of models of DNA diagenesis. These parameters, in turn, can then be used to properly detect and align ancient Neandertal DNA sequence to the human genome. This will be particularly crucial before multifold coverage of the Neandertal nuclear genome is achieved. Second, the high coverage allows a first tentative estimate of how deeply a Neandertal genome would need to be sequenced in order to arrive at a reasonable accuracy of the sequence given the biased retrieval of sequences (Fig. 5), the misincorporation patterns (Supp. Fig. 8) and other features of the DNA extracted from Neandertal bones. By resampling our sequences to simulate different depths of coverage, we find that to achieve an error-rate of one in ten thousand, 12-fold coverage would be required (Supp. Fig. 10). Although all factors that effect ancient DNA error rates are unlikely to be identical between mitochondrial and nuclear DNA, for example homopolymer lengths and DNA methylation, this lends us confidence that a reliable Neandertal genome sequence will be achievable.
Experimental Procedures
DNA extraction
DNA was extracted as described (Serre et al., 2004). After the first DNA extraction from 100–200 mg of bone a second DNA extraction (“re-extract”) was carried out from the remaining bone pellet as described (Rohland and Hofreiter, 2007a; Rohland and Hofreiter, 2007b) except that incubation was over night at 56°C. One extract of the Vindija 33.16 bone (Vi80 [33.16] 120402, Supp. Fig. 1) as well as the two re-extracts (Vi80 [33.16] 120402re, Vi80 [33.16] 031201re, Supp. Fig. 1) were used for 454 library preparation and high throughput sequencing. For PCR verification of the initial sequence assembly, DNA was extracted from 90 mg of the bone as described (Rohland and Hofreiter, 2007b).
Assembly of the mtDNA sequence
MtDNA sequences were identified using megablast (Zhang et al., 2000) and was required to be ≥30 nucleotides long and show ≥90% identity to the reference human mtDNA (GI: 17981852) or a version of this sequence carrying the Vindija HVRI. MtDNA alignments, which had to be at least as high-scoring as the best alignment against the human nuclear genome, were merged and regions where the assembly was problematic were amplified by PCR. While nuclear insertions of mtDNA could in theory confound the assembly, this is unlikely for several reasons (see Supp. Data). The Neandertal mtDNA sequence is available under accession number AM948965.
Evolutionary analyses
We aligned Neandertal, human, ape and monkey mtDNAs (identifiers in Supp. Data) and estimated trees using neighbor-joining, maximum parsimony, maximum likelihood and Bayesian approaches. Support for nodes was assessed using bootstrap replicates and a likelihood ratio test was used to test the clock assumption. To date mtDNA divergences, we estimated the posterior distribution of divergence times assuming that chimpanzees and humans diverged 6–8 million years ago. For protein analyses, we concatenated 12 genes, excluding ND6 and regions with overlapping reading frames and codons with insertion-deletion variation, and performed McDonald-Kreitman tests (McDonald and Kreitman, 1991)
Supplementary Material
Acknowledgments
We thank the Croatian Academy of Sciences and Arts, the Berlin-Brandenburg Academy of Sciences and the Presidential Innovation Fund of the Max Plank Society for making this work possible. We are indebted to Mark Stoneking, Weiwei Zhai, John Huelsenbeck, Jérôme Fuchs, Mario Mörl, Nick Patterson, David Reich and Timothy White for helpful discussions. We would also like to thank Bruce Rannala and Ziheng Yang for help with mcmctree, James Hudson Bullard for help with figures, and Christine Green for editing. AS, PFLJ, and MS are supported by NIH grant R01-GM40282. PLFJ is also supported by a Chang-Lin Tien scholarship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- Belevich I, Verkhovsky MI, Wikstrom M. Proton-coupled electron transfer drives the proton pump of cytochrome c oxidase. Nature. 2006;440:829–832. doi: 10.1038/nature04619. [DOI] [PubMed] [Google Scholar]
- Briggs AW, Stenzel U, Johnson PL, Green RE, Kelso J, Prufer K, Meyer M, Krause J, Ronan MT, Lachmann M, Pääbo S. Patterns of damage in genomic DNA sequences from a Neandertal. Proc Natl Acad Sci U S A. 2007;104:14616–14621. doi: 10.1073/pnas.0704665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotherton P, Endicott P, Sanchez JJ, Beaumont M, Barnett R, Austin J, Cooper A. Novel high-resolution characterization of ancient DNA reveals C > U-type base modification events as the sole cause of post mortem miscoding lesions. Nucleic Acids Res. 2007;35:5717–5728. doi: 10.1093/nar/gkm588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet M, Guy F, Pilbeam D, Mackaye HT, Likius A, Ahounta D, Beauvilain A, Blondel C, Bocherens H, Boisserie JR, et al. A new hominid from the Upper Miocene of Chad, Central Africa. Nature. 2002;418:145–151. doi: 10.1038/nature00879. [DOI] [PubMed] [Google Scholar]
- Currat M, Excoffier L. Modern humans did not admix with Neanderthals during their range expansion into Europe. PLoS Biol. 2004;2:e421. doi: 10.1371/journal.pbio.0020421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan JW, Schmidt TR, Wildman DE, Uddin M, Goldberg A, Huttemann M, Goodman M, Weiss ML, Grossman LI. Coadaptive evolution in cytochrome c oxidase: 9 of 13 subunits show accelerated rates of nonsynonymous substitution in anthropoid primates. Mol Phylogenet Evol. 2004;33:944–950. doi: 10.1016/j.ympev.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson JL, Turnbull DM, Howell N. Comparative genomics and the evolution of human mitochondrial DNA: assessing the effects of selection. Am J Hum Genet. 2004;74:229–238. doi: 10.1086/381505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galik K, Senut B, Pickford M, Gommery D, Treil J, Kuperavage AJ, Eckhardt RB. External and internal morphology of the BAR 1002'00 Orrorin tugenensis femur. Science. 2004;305:1450–1453. doi: 10.1126/science.1098807. [DOI] [PubMed] [Google Scholar]
- Gilbert MT, Tomsho LP, Rendulic S, Packard M, Drautz DI, Sher A, Tikhonov A, Dalen L, Kuznetsova T, Kosintsev P, et al. Whole-genome shotgun sequencing of mitochondria from ancient hair shafts. Science. 2007;317:1927–1930. doi: 10.1126/science.1146971. [DOI] [PubMed] [Google Scholar]
- Gilbert MT, Willerslev E, Hansen AJ, Barnes I, Rudbeck L, Lynnerup N, Cooper A. Distribution patterns of postmortem damage in human mitochondrial DNA. Am J Hum Genet. 2003;72:32–47. doi: 10.1086/345378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Krause J, Ptak SE, Briggs AW, Ronan MT, Simons JF, Du L, Egholm M, Rothberg JM, Paunovic M, Pääbo S. Analysis of one million base pairs of Neanderthal DNA. Nature. 2006;444:330–336. doi: 10.1038/nature05336. [DOI] [PubMed] [Google Scholar]
- Grossman LI, Wildman DE, Schmidt TR, Goodman M. Accelerated evolution of the electron transport chain in anthropoid primates. Trends Genet. 2004;20:578–585. doi: 10.1016/j.tig.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Harvati K, Gunz P, Grigorescu D. Cioclovina (Romania): affinities of an early modern European. J Hum Evol. 2007;53:732–746. doi: 10.1016/j.jhevol.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Cao Y, Yang Z. Preponderance of slightly deleterious polymorphism in mitochondrial DNA: nonsynonymous/synonymous rate ratio is much higher within species than between species. Mol Biol Evol. 1998;15:1499–1505. doi: 10.1093/oxfordjournals.molbev.a025877. [DOI] [PubMed] [Google Scholar]
- Higuchi R, Bowman B, Freiberger M, Ryder OA, Wilson AC. DNA sequences from the quagga, an extinct member of the horse family. Nature. 1984;312:282–284. doi: 10.1038/312282a0. [DOI] [PubMed] [Google Scholar]
- Hofreiter M, Jaenicke V, Serre D, Haeseler Av A, Pääbo S. DNA sequences from multiple amplifications reveal artifacts induced by cytosine deamination in ancient DNA. Nucleic Acids Res. 2001;29:4793–4799. doi: 10.1093/nar/29.23.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hublin JJ, Pääbo S. Neandertals. Curr Biol. 2006;16:R113–R114. doi: 10.1016/j.cub.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Ingman M, Gyllensten U. mtDB: Human Mitochondrial Genome Database, a resource for population genetics and medical sciences. Nucleic Acids Res. 2006;34:D749–D751. doi: 10.1093/nar/gkj010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingman M, Kaessmann H, Pääbo S, Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- Kivisild T, Shen P, Wall DP, Do B, Sung R, Davis K, Passarino G, Underhill PA, Scharfe C, Torroni A, et al. The role of selection in the evolution of human mitochondrial genomes. Genetics. 2006;172:373–387. doi: 10.1534/genetics.105.043901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause J, Dear PH, Pollack JL, Slatkin M, Spriggs H, Barnes I, Lister AM, Ebersberger I, Pääbo S, Hofreiter M. Multiplex amplification of the mammoth mitochondrial genome and the evolution of Elephantidae. Nature. 2006;439:724–727. doi: 10.1038/nature04432. [DOI] [PubMed] [Google Scholar]
- Krause J, Lalueza-Fox C, Orlando L, Enard W, Green RE, Burbano HA, Hublin JJ, Hanni C, Fortea J, de la Rasilla M, et al. The derived FOXP2 variant of modern humans was shared with Neandertals. Curr Biol. 2007a;17:1908–1912. doi: 10.1016/j.cub.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Krause J, Orlando L, Serre D, Viola B, Prufer K, Richards MP, Hublin JJ, Hanni C, Derevianko AP, Pääbo S. Neanderthals in central Asia and Siberia. Nature. 2007b;449:902–904. doi: 10.1038/nature06193. [DOI] [PubMed] [Google Scholar]
- Krings M, Capelli C, Tschentscher F, Geisert H, Meyer S, von Haeseler A, Grossschmidt K, Possnert G, Paunovic M, Pääbo S. A view of Neandertal genetic diversity. Nat Genet. 2000;26:144–146. doi: 10.1038/79855. [DOI] [PubMed] [Google Scholar]
- Krings M, Geisert H, Schmitz RW, Krainitzki H, Pääbo S. DNA sequence of the mitochondrial hypervariable region II from the neandertal type specimen. Proc Natl Acad Sci U S A. 1999;96:5581–5585. doi: 10.1073/pnas.96.10.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krings M, Stone A, Schmitz RW, Krainitzki H, Stoneking M, Pääbo S. Neandertal DNA sequences and the origin of modern humans. Cell. 1997;90:19–30. doi: 10.1016/s0092-8674(00)80310-4. [DOI] [PubMed] [Google Scholar]
- Lander ES, Waterman MS. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics. 1988;2:231–239. doi: 10.1016/0888-7543(88)90007-9. [DOI] [PubMed] [Google Scholar]
- Lebatard AE, Bourles DL, Duringer P, Jolivet M, Braucher R, Carcaillet J, Schuster M, Arnaud N, Monie P, Lihoreau F, et al. Cosmogenic nuclide dating of Sahelanthropus tchadensis and Australopithecus bahrelghazali: Mio-Pliocene hominids from Chad. Proc Natl Acad Sci U S A. 2008;105:3226–3231. doi: 10.1073/pnas.0708015105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller KE, Indap AR, Schmidt S, Boyko AR, Hernandez RD, Hubisz MJ, Sninsky JJ, White TJ, Sunyaev SR, Nielsen R, et al. Proportionally more deleterious genetic variation in European than in African populations. Nature. 2008;451:994–997. doi: 10.1038/nature06611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malez M, Ullrich H. Neuere plaäanthropologische Untersuchungen am Material aus der Höhle Vindija. Palaeontologia Jugoslavia. 1982;29:1–44. [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Meyer M, Briggs AW, Maricic T, Hober B, Hoffner B, Krause J, Weihmann A, Pääbo S, Hofreiter M. From micrograms to picograms: quantitative PCR reduces the material demands of high-throughput sequencing. Nucleic Acids Res. 2008;36:e5. doi: 10.1093/nar/gkm1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. Biol Rev Camb Philos Soc. 1966;41:445–502. doi: 10.1111/j.1469-185x.1966.tb01501.x. [DOI] [PubMed] [Google Scholar]
- Muramoto K, Hirata K, Shinzawa-Itoh K, Yoko-o S, Yamashita E, Aoyama H, Tsukihara T, Yoshikawa S. A histidine residue acting as a controlling site for dioxygen reduction and proton pumping by cytochrome c oxidase. Proc Natl Acad Sci U S A. 2007;104:7881–7886. doi: 10.1073/pnas.0610031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman MW, Brown WM, Stoneking M, Aquadro CF. Nonneutral mitochondrial DNA variation in humans and chimpanzees. Genetics. 1996;142:953–963. doi: 10.1093/genetics/142.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan JP, Coop G, Kudaravalli S, Smith D, Krause J, Alessi J, Chen F, Platt D, Pääbo S, Pritchard JK, Rubin EM. Sequencing and analysis of Neanderthal genomic DNA. Science. 2006;314:1113–1118. doi: 10.1126/science.1131412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan JP, Hofreiter M, Smith D, Priest JR, Rohland N, Rabeder G, Krause J, Detter JC, Pääbo S, Rubin EM. Genomic sequencing of Pleistocene cave bears. Science. 2005;309:597–599. doi: 10.1126/science.1113485. [DOI] [PubMed] [Google Scholar]
- Pääbo S. Molecular cloning of Ancient Egyptian mummy DNA. Nature. 1985;314:644–645. doi: 10.1038/314644a0. [DOI] [PubMed] [Google Scholar]
- Pääbo S. Human evolution. Trends Cell Biol. 1999;9:M13–M16. [PubMed] [Google Scholar]
- Pääbo S, Poinar H, Serre D, Jaenicke-Despres V, Hebler J, Rohland N, Kuch M, Krause J, Vigilant L, Hofreiter M. Genetic analyses from ancient DNA. Annu Rev Genet. 2004;38:645–679. doi: 10.1146/annurev.genet.37.110801.143214. [DOI] [PubMed] [Google Scholar]
- Pääbo S, Wilson AC. Polymerase chain reaction reveals cloning artefacts. Nature. 1988;334:387–388. doi: 10.1038/334387b0. [DOI] [PubMed] [Google Scholar]
- Poinar HN, Schwarz C, Qi J, Shapiro B, Macphee RD, Buigues B, Tikhonov A, Huson DH, Tomsho LP, Auch A, et al. Metagenomics to paleogenomics: large-scale sequencing of mammoth DNA. Science. 2006;311:392–394. doi: 10.1126/science.1123360. [DOI] [PubMed] [Google Scholar]
- Rand DM, Kann LM. Excess amino acid polymorphism in mitochondrial DNA: contrasts among genes from Drosophila, mice, and humans. Mol Biol Evol. 1996;13:735–748. doi: 10.1093/oxfordjournals.molbev.a025634. [DOI] [PubMed] [Google Scholar]
- Reid RA, Moyle J, Mitchell P. Synthesis of adenosine triphosphate by a protonmotive force in rat liver mitochondria. Nature. 1966;212:257–258. doi: 10.1038/212257a0. [DOI] [PubMed] [Google Scholar]
- Rohland N, Hofreiter M. Ancient DNA extraction from bones and teeth. Nat Protoc. 2007a;2:1756–1762. doi: 10.1038/nprot.2007.247. [DOI] [PubMed] [Google Scholar]
- Rohland N, Hofreiter M. Comparison and optimization of ancient DNA extraction. Biotechniques. 2007b;42:343–352. doi: 10.2144/000112383. [DOI] [PubMed] [Google Scholar]
- Ronaghi M, Uhlen M, Nyren P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363–365. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Sampietro ML, Gilbert MT, Lao O, Caramelli D, Lari M, Bertranpetit J, Lalueza-Fox C. Tracking down human contamination in ancient human teeth. Mol Biol Evol. 2006;23:1801–1807. doi: 10.1093/molbev/msl047. [DOI] [PubMed] [Google Scholar]
- Serre D, Langaney A, Chech M, Teschler-Nicola M, Paunovic M, Mennecier P, Hofreiter M, Possnert G, Pääbo S. No evidence of Neandertal mtDNA contribution to early modern humans. PLoS Biol. 2004;2:E57. doi: 10.1371/journal.pbio.0020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soficaru A, Dobos A, Trinkaus E. Early modern humans from the Pestera Muierii, Baia de Fier, Romania. Proc Natl Acad Sci U S A. 2006;103:17196–17201. doi: 10.1073/pnas.0608443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stiller M, Green RE, Ronan M, Simons JF, Du L, He W, Egholm M, Rothberg JM, Keates SG, Ovodov ND, et al. Patterns of nucleotide misincorporations during enzymatic amplification and direct large-scale sequencing of ancient DNA. Proc Natl Acad Sci U S A. 2006;103:13578–13584. doi: 10.1073/pnas.0605327103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. In: PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Associates S, editor. Sunderland, MA: Sinauer Associates; 2003. [Google Scholar]
- Templeton AR. Contingency tests of neutrality using intra/interspecific gene trees: the rejection of neutrality for the evolution of the mitochondrial cytochrome oxidase II gene in the hominoid primates. Genetics. 1996;144:1263–1270. doi: 10.1093/genetics/144.3.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M, Opazo JC, Wildman DE, Sherwood CC, Hof PR, Goodman M, Grossman LI. Molecular evolution of the cytochrome c oxidase subunit 5A gene in primates. BMC Evol Biol. 2008;8:8. doi: 10.1186/1471-2148-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Kim SK. Inconsistencies in Neanderthal genomic DNA sequences. PLoS Genet. 2007;3:1862–1866. doi: 10.1371/journal.pgen.0030175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Hasegawa M. Models of amino acid substitution and applications to mitochondrial protein evolution. Mol Biol Evol. 1998;15:1600–1611. doi: 10.1093/oxfordjournals.molbev.a025888. [DOI] [PubMed] [Google Scholar]
- Yang Z, Rannala B. Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds. Mol Biol Evol. 2006;23:212–226. doi: 10.1093/molbev/msj024. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.