Abstract
To evaluate the effect of genetic background on high-density lipoprotein cholesterol (HDL) levels in Soat1−/− mice, we backcrossed sterol O-acyltransferase 1 (Soat1)−/− mice, originally reported to have elevated HDL levels, to C57BL/6 mice and constructed a congenic strain with only a small region (3.3Mb) of 129 alleles, specifically excluding the nearby apolipoprotein A-II (Apoa2) gene from 129. HDL levels in these Soat1−/− mice were no different from C57BL/6, indicating that the passenger gene Apoa2 caused the previously reported elevation of HDL in these Soat1−/− mice. Because many knockouts are made in strain 129 and then subsequently backcrossed into C57BL/6, it is important to identify quantitative trait loci (QTL) that differ between 129 and C57BL/6 so that one can guard against effects ascribed to a knockout but really caused by a passenger gene from 129. To provide such data, we generated 528 F2 progeny from an intercross of 129S1/SvImJ and C57BL/6 and measured HDL concentrations in F2 animals first fed chow and then atherogenic diet. A genome wide scan using 508 single-nucleotide polymorphisms (SNPs) identified 19 QTL, 2 of which were male specific and 2 were female specific. Using comparative genomics and haplotype analysis, we narrowed QTL on chromosomes 3, 5, 8, 17, and 18 to 0.5, 6.3, 2.6, 1.1, and 0.6 Mb, respectively. These data will serve as a reference for any effort to test the impact of candidate genes on HDL using a knockout strategy.
Keywords: Soat1, QTL, comparative genomics, haplotype
Plasma high-density lipoprotein cholesterol (HDL) is a quantitative trait determined by the interactions of multiple genes and environmental factors. Because elevated plasma HDL is protective for cardiovascular disease (1), there has been considerable interest in understanding genetic factors contributing to variations in HDL levels. One method to determine whether a candidate quantitative trait loci (QTL) gene truly affects the phenotype is to construct a mouse line with a deficiency of the gene. Gene-targeted mice are usually initially created on one of the many substrains of 129 mice, and the null mutation subsequently transferred into C57BL/6 (B6). Thus, unless separated by recombinations, the region flanking the targeted gene from 129 will be carried with the targeted gene (2). This can lead to incorrect conclusions if allelic differences between 129 and B6 affect HDL. The HDL levels of the targeted mice might be attributable to the effects of the targeted gene when the difference is really caused by flanking linked loci. This is exemplified by the observations described in this paper that elevated HDL levels in Soat1 knockout (KO) mice were due to a passenger allele of Apoa2 from strain 129 rather than the null mutation. Soat1 encodes sterol O-acyltransferase (acyl-CoA: cholesterol acyltransferase, ACAT) 1, which is an intracellular enzyme that catalyzes the formation of cholesteryl esters from cholesterol and fatty acyl-CoA (3).
The problem of misinterpreting the cause of an altered phenotype in a knockout mouse is reduced if one has a detailed map of the QTL differences between the strain in which the knockout was constructed and the strain it was backcrossed into, usually 129 and B6. To obtain such a map, we generated a new B6 × 129 F2 cross and identified the genetic loci contributing to the difference in HDL levels between B6 and 129 mice. Although we previously crossed B6 and 129 strains to identify the genetic loci influencing HDL (4), only females fed an atherogenic diet were used for the purpose of finding genetic loci controlling atherosclerosis. The cross we now generated is improved in several ways: the cross is considerably larger (528 compared with 294 mice) for better resolution, both sexes were included allowing us to test the influence of sex, HDL was measured in chow and atherogenic diet fed mice, and more dense genotyping was used (508 SNPs compared with 111 Mit markers). We also used bioinformatics tool to quickly narrow some of the QTL.
MATERIALS AND METHODS
Mice and diets
C57BL/6J (B6), 129S1/SvImJ (129), and B6.129 Soat1−/− (stock 003322) mice were obtained from The Jackson Laboratory (Bar Harbor, ME). Mice were housed in a climate-controlled facility with a 14 h:10 h light-dark cycle. After weaning, mice were fed a chow diet containing 19% protein and 6% fat (LabDiet, 5K52, PMI Nutritional International, Bentwood, MO) and had free access to food and water. For certain experiments, mice were fed an atherogenic diet at 8 weeks of age containing 15% (w/w) dairy fat, 1% cholesterol, and 0.5% cholic acid as described previously (5). This study was conducted in conformity with the Public Health Service Policy on Humane Care and Use of Laboratory Animals. Experiments were approved by the Institutional Animal Care and Use Committee of The Jackson Laboratory.
To map QTLs, B6 females were mated to 129 males to produce the F1 progeny, which were intercrossed to produce 528 F2 progeny (269 females and 259 males) as reported previously (6). To reduce the size of the 129 region in the B6.129 Soat1−/− mice, a N11 male mouse was mated to female B6 mice to produce N12 progeny, and one N12 male offspring was backcrossed again to B6, and so on until N15. At each generation the male progeny were genotyped and the male carrying the smallest region around the Soat1 gene was selected to be the parent of the next generation. At N15, we intercrossed the heterozygous KO mice to generate homozygous Soat1−/− mice.
Measuring plasma HDL concentrations
Blood samples were obtained at 8 weeks of age from chow-fed mice and at 16 weeks of age from mice fed the atherogenic diet. Before blood was collected, animals were fasted for 4 h in the morning. Blood was collected from the retro-orbital sinus in tubes containing EDTA, centrifuged at 9,000 rpm for 5 min. Plasma was frozen at −20°C until assay. We measured plasma HDL concentrations using an enzymatic reagent kit (# 650207, Beckman Coulter, Fullerton, CA) according to manufacturer's recommendations on the Synchron CX Delta System (Beckman Coulter).
DNA isolation and genotyping
DNA was isolated from the tail tips by using genomic DNA purification kit (Gentra Systems, Minneapolis, MN). For Soat1−/− mice, the progeny were genotyped using markers listed in supplementary Table I. For the B6 × 129 progeny, genotyping was performed using a mouse SNP array with 508 SNPs polymorphic between B6 and 129 as described previously (6). For mapping purposes, the megabase (Mb) position of each SNP was ascertained from the NCBI mouse genome build 36.
QTL analysis
QTL mapping for HDL was performed using R/qtl (7) version 1.07–12 for Windows. It is an add-on package for the freely available and widely used statistical language/software R (http://www.rqtl.rog). As described previously (6, 8), a three-step QTL analysis was conducted to search first for main effects, second for pair-wise gene interactions, and finally to integrate all main and interacting QTL phenotype associations into a multiple regression. In single locus scans, the sex was first included as an additive covariate in the genome scan to account for overall differences in phenotypes between the sexes. A second set of scans included an interaction between sex and the putative QTL at each locus to identify sex-specific QTL. The difference in LOD scores (ΔLOD) between these two scans constitutes a test for sex-specific effects. We applied a significance threshold of LOD > 2.0 corresponding to P < 0.01, based on the 2 degree of freedom (df) chi-square distribution of the log likelihood ratio, for QTL-by-sex interactions (9). QTL were deemed significant if they either met or exceeded the 95% genome-wide adjusted threshold, which was assessed by 1,000-permutation analysis for each trait; they were deemed suggestive if they either met or exceeded the 37% genome-wide adjusted threshold but were not significant. QTL confidence intervals were calculated according to posterior probability (10). In the second stage, we performed a search for significant pairs of loci using a simultaneous search strategy. Each pair of loci in the genome was assessed for both additive and epistatic effects on the phenotype values. The primary purpose of the pairwise analysis is to identify potential epistatic interactions. The method employed was implemented as scantwo function in R/qtl using the algorithm described previously (10). We refined the multiple-QTL model at the third stage with fitqtl function in R/qtl using a backward elimination approach. We combined all significant and suggestive QTL and interactions in a multiple regression model. QTLs may be removed if they fail to achieve significance (at P < 0.01) after the effects of other QTLs have been taken into account. This is a conservative approach, as only those QTLs that were detected in the genome scans and retained in the final model are reported.
To assess whether there were separate QTL on the same chromosome, we conditioned the trait value on one of the markers and computed the LOD score at the second marker using the residual values and vice versa. The LOD score difference between 2-QTL and 1-QTL (ΔLOD) was used to judge whether there were two separate QTL on the same chromosome, the significant ΔLOD is 1.99 (P < 0.05) (11).
Statistical analysis
HDL values are given as the mean ± SD. One-way ANOVA was used for determining whether the mean phenotype values of progeny with different genotypes at a specific marker were significantly different. Data were analyzed using GraphPad Prism (Windows v5.00; GraphPad Software, San Diego, CA).
Comparative genomics and haplotype analysis
Homologous chromosomal regions between mouse and human were found at http://www.informatics.jax.org/reports/homologymap/mouse_human.shtml. When human and mouse QTL were located in the homologous locations, we assumed they were caused by the same gene and reduced the mouse QTL region to that homologous to the human QTL (12). If a confidence interval was not provided in the original report, we used a 1-LOD score drop from the QTL peak; if no chromosome LOD score plot was provided, we used a confidence interval of 20 Mb on either side of the reported peak.
The reduced region from comparative genomics was further restricted by haplotype analysis (12). In the first step, we examined a dense SNP map and excluded those genomic regions within the QTL where 129 and B6 had an identical SNP pattern. Such regions are likely to be identical by descent (IBD) and do not contain ancestral polymorphisms. This step assumes that the mutation causing the QTL is ancestral rather than recent. Such an assumption is correct most of the time with two strains, and when the QTL has been found in a second cross using different strains, it is highly likely that the mutation is ancestral. The second step of haplotype analysis compares haplotypes of additional strains if the QTL has been found previously in a cross with different strains. This step assumes that QTL found in the same location in different crosses are nevertheless caused by the same QTL gene. The haplotype analysis was performed by using SNPs of all the strains that were parents of the QTL crosses in current and previous studies. We found those regions where the strains carrying the allele that increased the trait were identical but differed from the strains carrying the allele that decreased the trait. For example, QTL on Chr 3, we searched the SNPs in regions where the high-allele strains B6 and NZB had identical SNPs, low-allele strains 129, C3H and SM had identical SNPs, and these SNPs differed between the high and low alleles. SNPs were downloaded from the Mouse Phenome Database (www.phenome.org).
RESULTS
Comparison of HDL levels in Soat1−/− mice with and without Apoa2 Val61 allele from strain 129
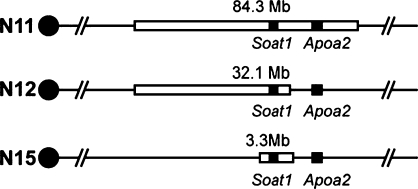
HDL cholesterol is elevated in Soat1−/− mice both in the original report with homozygous mice in a mixed genetic background (83 ± 19 mg/dl in female Soat1−/− vs. 48 ± 12 mg/dl in female controls) (13) and after these mice had been backcrossed for 11 generations (N11) to C57BL/6 by The Jackson Laboratory (Table 1). However, Soat1, located at 158.4Mb on chromosome 1, is just 15 Mb upstream of a major QTL gene that affects HDL (14), namely Apoa2 (apolipoprotein A2) located at 173.1Mb. Because Soat1 and Apoa2 are only 15 Mb apart and because the 129 allele (Val61) of Apoa2 leads to higher HDL (14), the increased HDL in Soat1−/− mice could be caused by the Apoa2 gene, by the Soat1 knockout, or both. We genotyped N11 mice using markers spanning the region from 102.3Mb to 186.6Mb (NCBI build 36) (see supplementary Table I), which includes Soat1 and Apoa2. The N11 Soat1−/− mice carried 129 alleles for the entire genotyped region (Fig. 1), indicating that the Apoa2 allele from strain 129 was carried as a passenger gene in the Soat1−/− congenic strain. To determine whether the elevated HDL in the knockout was caused by the knockout itself or by the Apoa2 Val61 allele, we backcrossed male N11 mice with female B6 mice for four more generations, at each generation selecting a male parent for the next generation that contained a crossover narrowing the amount of 129 genome carried by the Soat1−/+ mouse. At N15, we had reduced the 129 region to 3.3 Mb between 158.3Mb and 161.6Mb (Fig. 1), eliminating the Apoa2 Val61 allele and considerable other genomic DNA derived from 129. These mice were intercrossed to generate homozygous Soat1−/− mice with Apoa2 from B6. Comparing these knockout mice to B6 mice showed no difference in HDL in 8-week-old mice fed chow (Table 1). We concluded that Soat1 deficiency did not influence HDL levels and that the increased HDL levels previously reported in Soat1−/− mice (13) were caused by the passenger 129 allele of Apoa2. Because many knockouts are made in strain 129 and then subsequently backcrossed into B6, it becomes increasingly important to identify QTLs that differ between 129 and B6 so that one can guard against effects ascribed to a knockout but really caused by a passenger gene from 129.
TABLE 1.
Plasma HDL concentrations (mg/dl) in Soat1−/− mice
| Soat1 genotype | Female | Male |
|---|---|---|
| −/− (N11) | 89 ± 6a (n = 10) | 107 ± 5a (n = 10) |
| −/− (N15) | 61 ± 7 (n = 11) | 74 ± 8 (n = 13) |
| +/+ | 63 ± 5 (n = 20) | 73 ± 6 (n = 20) |
HDL, high-density lipoprotein cholesterol; Soat1, sterol O-acyltransferase 1. Data are presented as the means ± SD.
P < 0.01 vs. controls.
Fig. 1.
Separating sterol O-acyltransferase 1 (Soat1) and apolipoprotein A-II (Apoa2). The blank box represents 129 alleles; the black line represents B6 allele. Black boxes show location for Soat1 and Apoa2.
QTL analysis of strains B6 and 129 for genetic factors affecting plasma HDL concentrations
Plasma HDL levels for the parental, (B6 × 129) F1, and 528 F2 mice are summarized in Table 2. HDL levels were significantly higher in 129 than in B6 mice, and higher in males than in females in both strains. In F1 mice, HDL levels were intermediate and significantly different (P < 0.01 by ANOVA) from those of both parental strains. In strain 129, HDL levels increased in mice fed an atherogenic diet by 80% in females and 45% in males. HDL levels in the F2 populations were distributed normally.
TABLE 2.
Plasma HDL concentrations in B6, 129, F1, and F2, progeny
| Chow diet
|
Atherogenic diet
|
|||
|---|---|---|---|---|
| Female | Male | Female | Male | |
| B6 (n = 10) | 56 ± 7 | 70 ± 7 | 64 ± 5 | 95 ± 6 |
| 129 (n = 10) | 93 ± 11a | 128 ± 7a | 167 ± 12a | 185 ± 14a |
| F1 (n = 10) | 86 ± 7b | 107 ± 7c | 110 ± 9c | 127 ± 10c |
| F2 (n = 528)d | 83 ± 21 | 99 ± 20 | 130 ± 40 | 160 ± 50 |
Data are presented as the means ± SD.
P < 0.01 vs. B6.
P < 0.01 from B6 but not significantly different from 129.
P < 0.01 vs. B6 and 129.
Because it is the distribution, not the mean, among the F2 population that is most important for detecting genetic linkage to a phenotype, we did not test for significant differences between F2 progeny and either the parental strains or F1 progeny.
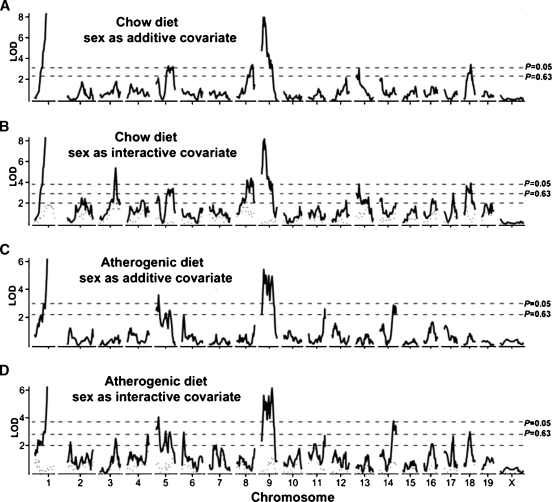
To account for overall average HDL differences between the sexes, we performed single-locus genome scans with sex as an additive covariate (Fig. 2A, C). To identify sex-specific QTL, we carried out a second set of single-locus genome scans but with sex as an interactive covariate (Fig. 2B, D) as previously described (6, 8). The difference in LOD (ΔLOD) between the scans with sex as an additive or interactive covariate constitutes a test for QTL-by-sex interaction as shown by the dotted line in Fig. 2B, D. A ΔLOD greater than two between the sex-additive and sex-interactive LOD scores indicates a locus at which the QTL differed between the sexes. Details of the QTL including significance levels (suggestive, P < 0.63; significant, P < 0.05; defined by 1,000 permutation tests), peak marker loci, LOD scores, and 95% confidence intervals (CIs) are summarized in Table 3. We named any QTL that were significant in our cross or suggestive but were found previously.
Fig. 2.
Genome-wide scans for high-density lipoprotein cholesterol (HDL). A: Chow-fed mice with sex as additive covariate. B: Chow-fed mice with sex as interactive covariate. C: Atherogenic diet-fed mice with sex as additive covariate. D: Atherogenic diet-fed mice with sex as interactive covariate. The horizontal dashed lines represents the suggestive (P = 0.63) and significant (P = 0.05) genome-wide levels as determined by 1,000 permutation tests. The dotted curves at the bottom of B and D depict the Δ logarithm of the odds ratio (LOD) scores between scans A and B, and C and D, respectively. ΔLOD > 2.0 (denoted by the lower horizontal dashed line in B and D) indicates a quantitative trait loci (QTL) that differ significantly between sexes (P < 0.05).
TABLE 3.
QTL identified by single or pairwise genome-wide scan of F2 mice
| LODb
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| QTLa | Chr | Peak Mb | 95% CI | Chow | Ath | Nearest marker | High allele | Human QTL homologous regionc | Coincident QTLd |
| Hdlq15e | 1 | 173.4 | 170–175 | 68.6 | 30 | rs31507136 | 129 | 1q25–23 | B6x129 |
| Hdlq21f | 3 | 124.8 | 117–129 | 5.4 | 2.5 | rs13477384 | B6 | 1p31–21 | B6xC3H |
| NZBxSM | |||||||||
| Hdlq52 | 5 | 23.4 | 7–33 | 2.1 | 3.6 | rs13478142 | 129 | 7q21–31.2 | NZBxSM |
| PERAxI | |||||||||
| Hdlq1 | 5 | 91.4 | 79–127 | 3.3 | 2.5 | rs13478390 | 129 | 4p1, 4q1, 1p22, 22q1, 12q24 | B6xNZB/CAST |
| NZBxSM | |||||||||
| Hdlq53 | 6 | 15.6 | 6–26 | — | 2.9 | rs13478640 | 129 | 7q31–35 | B6xSPRET |
| Hdlq44g | 8 | 79.7 | 71–90 | 4.3 | — | rs13479840 | B6 | 8p23–22, 22q11–12, 4q28–31 | B6xA/J |
| Hdlq16 | 8 | 107.7 | 94–121 | 3.4 | — | rs30731821 | B6 | 16q21–24 | B6xCAST |
| Hdlq17 | 9 | 35.1 | 27–43 | 8.0 | 5.4 | rs13480142 | 129 | 11q22–24 | RIIIx129 |
| B6xCAST | |||||||||
| Hdlq54 | 9 | 91 | 60–95 | 3.5 | 4.8 | rs3693091 | 129 | 15q21–24, 6q14, 6p12, 3q24 | |
| 11 | 115.4 | 110–117 | — | 2.6 | rs13481245 | 129 | — | ||
| 13 | 23.7 | 4–52 | 3.0 | — | rs8267056 | 129 | — | ||
| Hdlq55 | 14 | 100 | 94–117 | — | 2.9 | rs6179144 | 129 | — | MRLxSJL |
| Hdlq56f | 17 | 63.6 | 38–86 | 2.9 | 2.7 | rs13483068 | B6 | 6p12.3–q12, 3p25, 2q14 | NZBxSM |
| Hdlq57g | 18 | 22.9 | 10–36 | 3.8 | — | rs13483251 | 129 | 18q12, 2q21 | NZBxSM |
| Hdlq47 | 18 | 52.4 | 36–63 | 3.4 | — | rs3722312 | 129 | — | B6xA/J |
| B6xC3H | |||||||||
| Hdlq58h | 7 | 64 | — | — | rs3719301 | — | 15q1–2 | B6xC3H | |
| Hdlq18h | 12 | 11.5 | — | — | rs3717933 | — | 7q2–3, 7p21–15 | RIIIx129 | |
| Hdlq59h | 19 | 47.2 | — | — | rs3023496 | — | 10q23–25 | NZBxRF | |
| Hdlq60h | X | 124 | — | — | rs6205221 | — | |||
Quantitative trait loci (QTL) were named if they were significant or if they were suggestive but confirmed QTL reported previously. They were given the same name if the crosses identifying them shared at least one common parental strain and a new name if the crosses identifying them involved no common strains.
Logarithm of the odds ratio (LOD) scores of the analysis with sex as additive covariate are given with the exception of sex-specific QTL. Suggestive and significant LODs are 2.3/3.1 for chow diet and 2.2/3.0 for atherogenic die using sex as additive covariate. Significant LOD scores are bolded.
Human homologous region of the mouse QTLs were retrieved from Genome Orthology Map at http://www.informatics.jax.org/reports/homologymap/mouse_human.shtml.
Mouse HDL QTL were reviewed recently by Rollins et al (15). Strains in bold carry the allele for high HDL.
Apoa2 is the underlying gene.
Male specific QTL.
Female specific QTL.
Interacting QTL are selected from pairwise gene-interaction search.
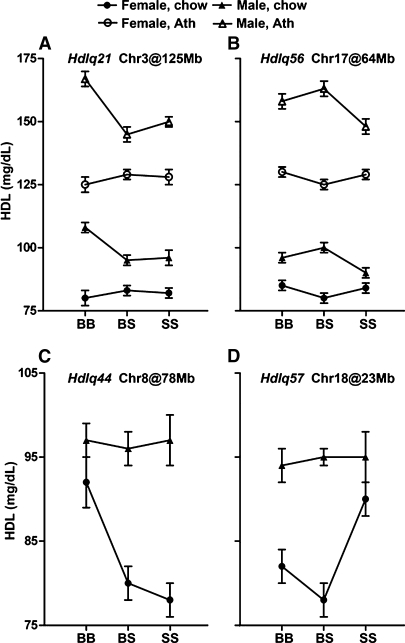
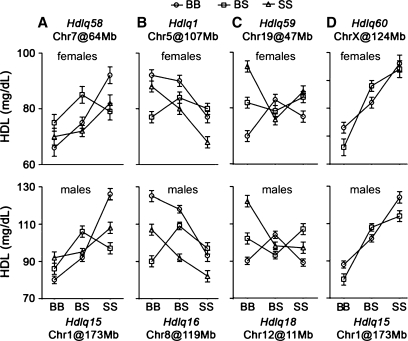
Our analysis revealed 10 significant and 5 suggestive HDL QTLs distributed on Chrs 1, 3, 5, 6, 8, 9, 11, 13, 14, 17, and 18 (Table 3). QTL on Chrs 8, 13, and 18 had reduced significance on the atherogenic diet compared with chow conditions, while QTL on Chrs 6, 11, and 14 were specific for the atherogenic diet. We found strong evidence that 3 significant QTLs named Hdlq21 (Chr 3 at Mb 124.8), Hdlq44 (Chr 8 at Mb 79.7), and Hdlq57 (Chr 18 at Mb 22.9), and 1 suggestive QTL (Hdlq56 on Chr 17 at Mb 63.6) had sex-specific effects. Whereas Hdlq21 (Fig. 3A) and Hdlq56 (Fig. 3B) affected plasma HDL levels only in males, Hdlq44 (Fig. 3C) and Hdlq57 (Fig. 3D) affected only females.
Fig. 3.
Allele effects for the sex-specific QTL. Hdlq21 (A) and Hdlq56 (B) found in males fed either diet, and Hdlq44 (C) and Hdlq57 (D) found in females fed a chow diet. Chromosome number and the QTL position in Mb are given for each QTL. Homozygosity for B6 alleles is represented by BB, homozygosity for 129 alleles is represented by SS, and heterozygosity at a locus is represented by BS. Y axes show mean values of HDL; error bars represent SD.
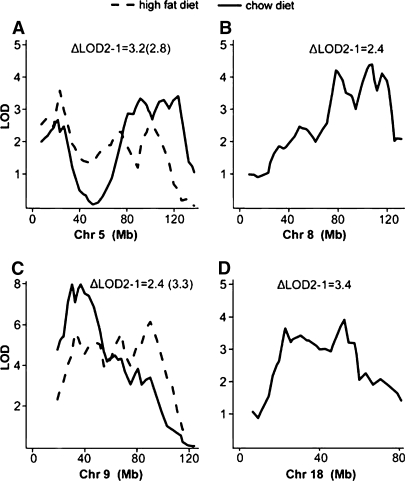
The shapes of the LOD curves for Chr 5, 8, 9, and 18 in the genome scans suggested that they each represented more than one QTL (Fig. 4). To resolve these QTLs, we computed maximum LOD scores for models with 1, 2, and 3 QTLs and compared the ΔLOD between the models (a significant ΔLOD is >1.99). The evidence indicated at least 2 QTLs each for Chrs 5 and 9 (Fig. 4A, C) for both diets and sexes. To test for multiple QTL on Chrs 8 and 18 for mice fed the chow diet, we allowed the QTLs to have sex interaction. We found evidence (ΔLOD2-1 are 2.4 and 3.4 for Chrs 8 and 18, respectively) for 2 QTL (Fig. 4B, D), with 1 QTL interacting with sex on each chromosome.
Fig. 4.
Linked QTL for HDL were identified on Chrs 5, 8, 9, and 18. Multilocus scans were carried out on a per-chromosome basis. The LOD score difference between one- and two-QTL models (ΔLOD2-1) was used as a test statistics to judge whether there were two separate QTL on the same chromosome. Threshold LOD differences were computed by permutation tests: ΔLOD = 1.99 (P < 0.05), ΔLOD = 1.83 (P < 0.10). ΔLOD for chow is listed first; the ΔLOD for atherogenic diet is in brackets. The solid line represents the scan from mice fed chow diet, and the dashed line represents the scan from mice fed atherogenic diet. Loci on Chrs 8 and 18 were detected only in females fed chow. A: Chr 5; B: Chr 8; C: Chr 9; D: Chr 18.
Pairwise genome scans with sex as an additive covariate revealed three significant epistatic interactions in mice fed a chow diet (Fig. 5A–C). In the first, homozygous 129 alleles at Hdlq15 (Chr 1) interacted with B6 homozygozity at Hdlq58 (Chr 7) to significantly increase HDL levels (Fig. 5A). In the second, Hdlq1 on Chr 5 interacted with homozygous 129 alleles at Hdlq16 on Chr 8 to significantly decrease HDL levels (Fig. 5B). In the third, although neither Hdlq18 on Chr12 nor Hdlq59 on Chr19 affected HDL levels by themselves, homozygotes for B6 alleles at both loci contributed significantly to decreased HDL levels (Fig. 5C). In mice fed an atherogenic diet, Hdlq15 on Chr 1 and Hdlq60 on X significantly interacted to increase HDL levels (Fig. 5D).
Fig. 5.
The effects of gene interactions detected by the pairwise genome scan. The upper panel of each interaction is from females, the lower one is from males. Homozygosity for B6 alleles is represented by BB, homozygosity for 129 alleles represented by SS, and heterozygosity is represented by BS. Y axes show mean values of HDL; error bars represent SD.
Finally, to evaluate the relative contributions of each QTL when considered together, we combined all significant, suggestive, and interacting QTL in a multiple regression model, eliminating terms that did not meet the P = 0.01 significance level based on the multiple regression F test (Table 4). In addition to these QTL, sex contributes significantly to the multilocus model, accounting for 18.4% of the total HDL variance in chow-fed mice. These models confirmed the importance of the QTL-by-sex and QTL-by-QTL interactions. The proportions of the total HDL variance accounted for in the F2 population fed the chow or atherogenic diet are 71.1% and 49.7%, respectively (Table 4).
TABLE 4.
Multiple regression models for loci involved in HDL cholesterol levels on chow and atherogenic diet
| Chr@Mb | dfa | %varianceb | F value | P value | ||||
|---|---|---|---|---|---|---|---|---|
| Chow diet | ||||||||
| Sex | 3 | 18.4 | 95.0 | <2 × 10−16 | ||||
| Chr1@173.4 | 6 | 33.0 | 85.2 | <2 × 10−16 | ||||
| Chr3@124.8 | 4 | 2.5 | 9.7 | 1.6 × 10−7 | ||||
| Chr5@107.4 | 6 | 2.8 | 7.2 | 2.5 × 10−7 | ||||
| Chr7@64 | 6 | 1.9 | 5.0 | 6.9 × 10−5 | ||||
| Chr8@115.7 | 6 | 1.5 | 3.6 | 0.0008 | ||||
| Chr9@36.8 | 2 | 2.9 | 22.5 | 4.9 × 10−10 | ||||
| Chr12@11.5 | 6 | 1.4 | 3.6 | 0.0016 | ||||
| Chr13@23.7 | 2 | 0.9 | 6.9 | 0.0011 | ||||
| Chr18@52.48 | 2 | 0.7 | 5.8 | 0.0034 | ||||
| Chr19@47.2 | 6 | 1.6 | 4.2 | 0.0004 | ||||
| Chr1@173.4:Chr7@64 | 4 | 1.1 | 4.2 | 0.0026 | ||||
| Chr5@107.4:Chr8@115.7 | 4 | 0.9 | 3.7 | 0.0053 | ||||
| Chr12@11.5:Chr19@47.2 | 4 | 1.3 | 5.2 | 0.0004 | ||||
| Chr3@124.8:sex | 2 | 0.6 | 4.7 | 0.0091 | ||||
| Total | 35 | 71.1 | ||||||
| Atherogenic diet | ||||||||
| Chr1@173.4 | 8 | 23.5 | 25.4 | <2 × 10−16 | ||||
| Chr5@91.4 | 2 | 1.4 | 5.9 | 3.0 × 10−3 | ||||
| Chr6@15.6 | 2 | 1.7 | 7.2 | 8.8 × 10−4 | ||||
| Chr9@35.1 | 2 | 1.2 | 5.0 | 7.5 × 10−3 | ||||
| Chr9@91.0 | 2 | 2.3 | 9.8 | 6.7 × 10−5 | ||||
| Chr11@115.4 | 2 | 2.3 | 9.9 | 6.2 × 10−5 | ||||
| Chr14@100 | 2 | 1.3 | 5.7 | 3.5 × 10−3 | ||||
| ChrX@124 | 9 | 14.8 | 14.2 | <2 × 10−16 | ||||
| Chr1@173.4: ChrX@124 | 6 | 3.2 | 4.5 | 1.8 × 10−4 | ||||
| Total | 21 | 49.7 | ||||||
df, degree of freedom.
%variance indicates the percentage of the total F2 phenotypic variance associated with each marker.
Narrowing QTL using bioinformatics tools
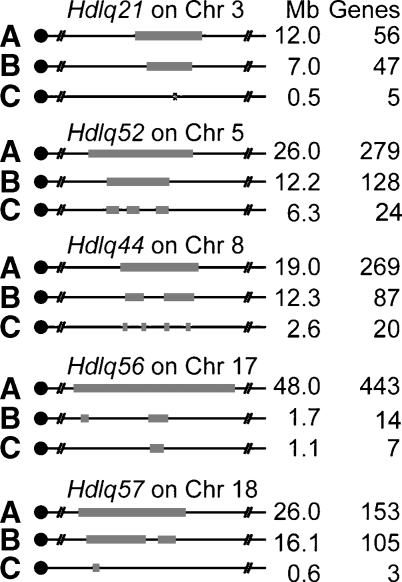
Most QTL identified in this study had been identified in human homologous regions and in other mouse crosses (Table 3), allowing us to narrow the QTL and reduce the number of candidate genes in each by comparative genomics, haplotyping, and other bioinformatic tools (12). Rodent and human QTLs for the same trait often map to homologous genomic locations (6, 8, 15–17). If homologous QTLs have the same underlying gene, a correct assumption in many cases (18–20), comparing mouse-human homology maps may reduce a QTL region in both species to the interval for which they are homologous. This comparative genomics step narrowed the Chr 3 QTL from 12 to 7 Mb, the Chr 5 QTL from 26 to 12.2 Mb, the Chr 8 QTL from 19 to 12.3 Mb, the Chr 17 QTL from 48 to 1.7 Mb, and the Chr 18 QTL from 26 to 16.1 Mb (Fig. 6, step B).
Fig. 6.
Narrowing QTL by bioinformatics. A: The 95% confidence interval (CI) and the number of gene in each QTL identified in cross B6 × 129; B: Comparative genomics. QTLs were narrowed by homology with human HDL QTL. Hdlq21 was homologous with human 1p31.3–21.3 (Mb 93–100); Hdlq52 was homologous with human 7q21–31.2 (Mb 81.1–84.6, Mb 86.3–92, Mb 102–105); Hdlq44 was homologous with human 8p23–22 (Mb 19–20), 22q11.2–q12.3 (Mb 32–34.3) and 4q28.3–q31.2 (Mb 141–150); Hdlq56 was homologous with human 6p12.3–q12 (Mb 49.5–49.9), 3p25 (Mb 52.9–53.2), and 2q13 (Mb 107.2–108.2); and Hdlq57 was homologous with human 18q12.1–22.2 (Mb 18–33, Mb 37–39) and 2q14.3–23.3 (Mb 127.5–128.4). C: Haplotype analysis. Each QTL was reduced by analyzing the haplotypes of the strains involved in the QTL crosses (Table 3) (15).
Because approximately 97% of genetic variation among inbred mouse strains is ancestral (21), regions that are identical by descent between the parental strains of a cross can be inferred by shared SNP patterns. Such regions are unlikely to contain QTL genes, especially if the QTLs have been found in multiple crosses. Thus, we compared the haplotypes based on SNPs of the parental strains for each cross pair (Table 3), and eliminated from the QTL those regions for which the strains were identical. Based on the assumption that the same gene causes the QTL in the different crosses, we then further reduced the QTL by comparing SNPs to identify the genomic regions shared among high-allele strains but different from low-allele strains. Haplotype analysis further narrowed the Chr 3 QTL to an 0.5 Mb, 5-gene region, the Chr 5 QTL to a 6.3 Mb, 24-gene region, the Chr 8 QTL to a 2.57 Mb, 20-gene region, the Chr 17 QTL to a 1.1 Mb, 7-gene region, and the Chr 18 QTL to an 0.6 Mb, 3-gene region (Fig. 6, step C, list of genes in supplementary Table II).
DISCUSSION
Knockout mice are usually created by injecting genetically engineered 129 embryonic stem cells into B6 host blastocysts, and the resulting knockout mice are widely used to study gene function. This approach is effective to evaluate candidate genes that affect monogenic diseases such as Lesch-Nyhan syndrome (22) and cystic fibrosis (23–25). However, when this approach used to evaluate candidate genes for complex traits such as HDL levels, precautions should be taken to minimize the impact of genetic background and passenger genes, which may complicate the analysis and lead to incorrect conclusions. This problem is exemplified by the example of Soat1−/− mice as reported in this paper. In the original study (13), Soat1 KO mice, carried on a mixed 129 and B6 background, were reported to have elevated HDL levels, an effect ascribed to the Soat1 deficiency. Even after The Jackson Laboratory backcrossed the Soat1 KO mice to B6 mice for 10 generations to provide the knockout on a clean genetic background, the knockout still had elevated HDL levels. Nevertheless, we carried out additional backcrosses to reduce the 129 region around Soat1 and to specifically eliminate the 129 allele of Apoa2, which we knew elevated HDL (14). The resulting Soat1−/− mice had HDL levels that did not differ from the B6 control, indicating that the original elevation in HDL most likely resulted from the Apoa2 passenger gene.
This example points to the need for a very good QTL map between B6 and 129 when interpreting data from knockout mice. Although this is not the first QTL cross between these two strains, it is the only cross that has both sexes, both chow and atherogenic diet, and has more mice and more markers than previous crosses. Knowing the location of all QTL affecting HDL between these strains is important in at least two ways. First, it is critical for assessing results from any studies using B6, 129 KO mice to test HDL candidate genes. If the gene for which a knockout is made maps to a region already containing an HDL QTL, one must be careful to determine whether any HDL difference is caused by the gene deficiency or by another QTL gene in the region. Second, the location of these QTL will provide important information that can be used if the KO mouse is backcrossed to B6. Generally, the null mutant mouse is backcrossed to B6 for 10 generations, a process that requires more than 2 years. However, this process can be shortened considerably by eliminating the 129 alleles from all known QTL. To do so, the backcross progeny are tested for the SNPs flanking each QTL interval identified in this study (provided in supplemental Table III); selecting a mouse for the next generation that is homozygous B6 for these regions ensures that no passenger genes from 129 affecting HDL are present in the final congenic. This eliminates the need to perform a whole genome screen and decreases the number of backcross generations required, thus decreasing both the cost and the time.
The findings in our present and previous (26) studies also prove the importance of taking gender into account in analyzing HDL QTL: two loci (Hdlq21 and Hdlq56) had effects only in male mice and two (Hdlq44 and Hdlq57) had effects only in female mice. The strong gender effects were also seen in the analysis of the multiple-QTL models. On chow diet, sex contributes significantly to the multilocus model, accounting for 18.4% of the total HDL variance (71.1%). Although sex is not as an independent term contributing to the variance of HDL values on atherogenic diet, the QTL on the X chromosome accounts for 14.8% of the total HDL variance (49.7%) in the multilocus model (Table 4).
The results of the present study differ from our previous cross using the same parental strains (4). In addition to the QTL observed in the previous cross, we identified 12 additional QTL because this study used both sexes, two diets, and had greater statistical power due to the larger number of mice. However, we did fail to confirm a significant QTL on Chr 12 at 20 cM (47 Mb) found previously. We think the most likely explanation is the length of time the atherogenic diet was fed: 14 weeks in the previous study and only 8 weeks in this study. In this study we did find a QTL on Chr 12 at 11 Mb interacting with the QTL on Chr 19 (Fig. 5C), but this seems too far from the one located in the previous study to be the same.
The QTL found on Chrs 1, 3, 5, 6, 8, 9, 17, 18, and 19 are located in regions homologous to human QTL for HDL. Rodent and human QTL for the same trait often map to homologous genomic locations (6, 8, 15–17), suggesting that the same genes underlie the homologous QTL in both species. In fact, there are many examples where concordant QTL are caused by the same genes in mouse and human (18–20, 27), including mutations in Apoa2, which are associated with plasma HDL levels in mice (14) and humans (28). For concordant QTL, comparison of the mouse-human homology maps may reduce the QTL region to that shared in both species. Although the use of comparative genomics is helpful in narrowing QTL, care should be taken that these regions are adequately covered by markers. Furthermore, it is possible that genes with similar function are clustered in the genome; if clustering is present, then one gene could have mutated in humans and another nearby one mutated in mouse, so one must remember that the candidates must be adequately tested in both species.
Because a majority of the genetic variation among inbred mouse strains is ancestral (21), regions that are IBD between the parental strains of a cross can be inferred using SNP genotype data (29). Such regions are unlikely to contain QTL genes especially if the QTL has been found in multiple crosses. Therefore, it is possible to narrow the QTL by excluding genetic segments that are inferred to be IBD between parental strains of the crosses by haplotype analysis. The B6 and 129 strains are well genotyped, which facilitates the identification of any polymorphism causing amino-acid changes between strains. Using this approach, we have significantly narrowed five of the QTL we identified and provided the reduced list of genes in supplemental Table II. For Hdlq44 on Chr 8, one gene on the short list is Lpl, lipoprotein lipase, which is known to regulate plasma HDL levels in mice and humans.
The interaction between genetic loci may give clues to identify the candidate genes. We found that Hdlq15 on Chr 1 and Hdlq58 on Chr 7 interact with each other. The gene underlying Hdlq15 has been identified as Apoa2 (14). A candidate gene for Hdlq58 is Nr2f2 (nuclear receptor subfamily 2, group F, member 2, at 70.2Mb), which was found to regulate the transcription of Apoa2 (30), and its polymorphisms were found to be significantly associated with HDL levels in a recent genome-wide association study (31).
In summary, our study demonstrated the problem of passenger genes in knockout mice and provided a detailed genetic map of the genetic loci controlling HDL levels that differ in B6 and 129 mice. This knowledge is critical and must be considered when B6/129 KO mice are used to study gene functions in regulating HDL levels. Furthermore, we narrowed some of these HDL QTL by using comparative genomics and haplotype analysis, which will facilitate the identification of the QTL genes.
Supplementary Material
Acknowledgments
The authors thank Harry Whitmore for excellent technical assistance.
Abbreviations
Apoa2, apolipoprotein A-II
HDL, high-density lipoprotein cholesterol
IBD, identical by descent
KO, knockout
LOD, logarithm of the odds ratio
QTL, quantitative trait loci
Soat1, sterol O-acyltransferase 1
SNP, single-nucleotide polymorphism
Published, JLR Papers in Press, September 4, 2008.
Footnotes
This work was funded by the US National Institutes of Health grants to BP (HL 81162, HL74086, and HL77796), the American Heart Association grant 0725905T to ZS and by the Novartis Institutes for Biomedical Research.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three tables.
References
- 1.Brewer H. B., Jr. 2004. High-density lipoproteins: a new potential therapeutic target for the prevention of cardiovascular disease. Arterioscler. Thromb. Vasc. Biol. 24 387–391. [DOI] [PubMed] [Google Scholar]
- 2.Lusis A. J., J. Yu, and S. S. Wang. 2007. The problem of passenger genes in transgenic mice. Arterioscler. Thromb. Vasc. Biol. 27 2100–2103. [DOI] [PubMed] [Google Scholar]
- 3.Buhman K. F., M. Accad, and R. V. Farese. 2000. Mammalian acyl-CoA:cholesterol acyltransferases. Biochim. Biophys. Acta. 1529 142–154. [DOI] [PubMed] [Google Scholar]
- 4.Ishimori N., R. Li, P. M. Kelmenson, R. Korstanje, K. A. Walsh, G. A. Churchill, K. Forsman-Semb, and B. Paigen. 2004. Quantitative trait loci analysis for plasma HDL-cholesterol concentrations and atherosclerosis susceptibility between inbred mouse strains C57BL/6J and 129S1/SvImJ. Arterioscler. Thromb. Vasc. Biol. 24 161–166. [DOI] [PubMed] [Google Scholar]
- 5.Nishina P. M., J. Verstuyft, and B. Paigen. 1990. Synthetic low and high fat diets for the study of atherosclerosis in the mouse. J. Lipid Res. 31 859–869. [PubMed] [Google Scholar]
- 6.Su Z., R. Korstanje, S. W. Tsaih, and B. Paigen. 2008. Candidate genes for obesity revealed from a C57BL/6J x 129S1/SvImJ intercross. Int. J. Obes. 32 1180–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broman K. W., H. Wu, S. Sen, and G. A. Churchill. 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 19 889–890. [DOI] [PubMed] [Google Scholar]
- 8.Su Z., S. W. Tsaih, J. Szatkiewicz, Y. Shen, and B. Paigen. 2008. Candidate genes for plasma triglyceride, FFA, and glucose revealed from an intercross between inbred mouse strains NZB/B1NJ and NZW/LacJ. J. Lipid Res. 49 1500–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Solberg L. C., A. E. Baum, N. Ahmadiyeh, K. Shimomura, R. Li, F. W. Turek, G. A. Churchill, J. S. Takahashi, and E. E. Redei. 2004. Sex- and lineage-specific inheritance of depression-like behavior in the rat. Mamm. Genome. 15 648–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sen S., and G. A. Churchill. 2001. A statistical framework for quantitative trait mapping. Genetics. 159 371–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X., I. Le Roy, E. Nicodeme, R. Li, R. Wagner, C. Petros, G. A. Churchill, S. Harris, A. Darvasi, J. Kirilovsky, et al. 2003. Using advanced intercross lines for high-resolution mapping of HDL cholesterol quantitative trait loci. Genome Res. 13 1654–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiPetrillo K., X. Wang, I. M. Stylianou, and B. Paigen. 2005. Bioinformatics toolbox for narrowing rodent quantitative trait loci. Trends Genet. 21 683–692. [DOI] [PubMed] [Google Scholar]
- 13.Meiner V. L., S. Cases, H. M. Myers, E. R. Sande, S. Bellosta, M. Schambelan, R. E. Pitas, J. McGuire, J. Herz, and R. V. Farese, Jr. 1996. Disruption of the acyl-CoA:cholesterol acyltransferase gene in mice: evidence suggesting multiple cholesterol esterification enzymes in mammals. Proc. Natl. Acad. Sci. USA. 93 14041–14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X., R. Korstanje, D. Higgins, and B. Paigen. 2004. Haplotype analysis in multiple crosses to identify a QTL gene. Genome Res. 14 1767–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rollins J., Y. Chen, B. Paigen, and X. Wang. 2006. In search of new targets for plasma high-density lipoprotein cholesterol levels: promise of human-mouse comparative genomics. Trends Cardiovasc. Med. 16 220–234. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y., J. Rollins, B. Paigen, and X. Wang. 2007. Genetic and genomic insights into the molecular basis of atherosclerosis. Cell Metab. 6 164–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt C., N. P. Gonzaludo, S. Strunk, S. Dahm, J. Schuchhardt, F. Kleinjung, S. Wuschke, H. G. Joost, and H. Al-Hasani. 2008. A meta-analysis of QTL for diabetes-related traits in rodents. Physiol. Genomics. 34 42–53. [DOI] [PubMed] [Google Scholar]
- 18.Stoll M., A. E. Kwitek-Black, A. W. Cowley, Jr., E. L. Harris, S. B. Harrap, J. E. Krieger, M. P. Printz, A. P. Provoost, J. Sassard, and H. J. Jacob. 2000. New target regions for human hypertension via comparative genomics. Genome Res. 10 473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hillebrandt S., H. E. Wasmuth, R. Weiskirchen, C. Hellerbrand, H. Keppeler, A. Werth, R. Schirin-Sokhan, G. Wilkens, A. Geier, J. Lorenzen, et al. 2005. Complement factor 5 is a quantitative trait gene that modifies liver fibrogenesis in mice and humans. Nat. Genet. 37 835–843. [DOI] [PubMed] [Google Scholar]
- 20.Wang X., M. Ria, P. M. Kelmenson, P. Eriksson, D. C. Higgins, A. Samnegard, C. Petros, J. Rollins, A. M. Bennet, B. Wiman, et al. 2005. Positional identification of TNFSF4, encoding OX40 ligand, as a gene that influences atherosclerosis susceptibility. Nat. Genet. 37 365–372. [DOI] [PubMed] [Google Scholar]
- 21.Frazer K. A., C. M. Wade, D. A. Hinds, N. Patil, D. R. Cox, and M. J. Daly. 2004. Segmental phylogenetic relationships of inbred mouse strains revealed by fine-scale analysis of sequence variation across 4.6 mb of mouse genome. Genome Res. 14 1493–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seegmiller J. E., F. M. Rosenbloom, and W. N. Kelley. 1967. Enzyme defect associated with a sex-linked human neurological disorder and excessive purine synthesis. Science. 155 1682–1684. [DOI] [PubMed] [Google Scholar]
- 23.Kerem B., J. M. Rommens, J. A. Buchanan, D. Markiewicz, T. K. Cox, A. Chakravarti, M. Buchwald, and L. C. Tsui. 1989. Identification of the cystic fibrosis gene: genetic analysis. Science. 245 1073–1080. [DOI] [PubMed] [Google Scholar]
- 24.Riordan J. R., J. M. Rommens, B. Kerem, N. Alon, R. Rozmahel, Z. Grzelczak, J. Zielenski, S. Lok, N. Plavsic, J. L. Chou, et al. 1989. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 245 1066–1073. [DOI] [PubMed] [Google Scholar]
- 25.Rommens J. M., M. C. Iannuzzi, B. Kerem, M. L. Drumm, G. Melmer, M. Dean, R. Rozmahel, J. L. Cole, D. Kennedy, N. Hidaka, et al. 1989. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science. 245 1059–1065. [DOI] [PubMed] [Google Scholar]
- 26.Korstanje R., R. Li, T. Howard, P. Kelmenson, J. Marshall, B. Paigen, and G. Churchill. 2004. Influence of sex and diet on quantitative trait loci for HDL cholesterol levels in an SM/J by NZB/BlNJ intercross population. J. Lipid Res. 45 881–888. [DOI] [PubMed] [Google Scholar]
- 27.Ruivenkamp C. A., T. van Wezel, C. Zanon, A. P. Stassen, C. Vlcek, T. Csikos, A. M. Klous, N. Tripodis, A. Perrakis, L. Boerrigter, et al. 2002. Ptprj is a candidate for the mouse colon-cancer susceptibility locus Scc1 and is frequently deleted in human cancers. Nat. Genet. 31 295–300. [DOI] [PubMed] [Google Scholar]
- 28.Blanco-Vaca F., J. C. Escola-Gil, J. M. Martin-Campos, and J. Julve. 2001. Role of apoA-II in lipid metabolism and atherosclerosis: advances in the study of an enigmatic protein. J. Lipid Res. 42 1727–1739. [PubMed] [Google Scholar]
- 29.Wiltshire T., M. T. Pletcher, S. Batalov, S. W. Barnes, L. M. Tarantino, M. P. Cooke, H. Wu, K. Smylie, A. Santrosyan, N. G. Copeland, et al. 2003. Genome-wide single-nucleotide polymorphism analysis defines haplotype patterns in mouse. Proc. Natl. Acad. Sci. USA. 100 3380–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ladias J. A., M. Hadzopoulou-Cladaras, D. Kardassis, P. Cardot, J. Cheng, V. Zannis, and C. Cladaras. 1992. Transcriptional regulation of human apolipoprotein genes ApoB, ApoCIII, and ApoAII by members of the steroid hormone receptor superfamily HNF-4, ARP-1, EAR-2, and EAR-3. J. Biol. Chem. 267 15849–15860. [PubMed] [Google Scholar]
- 31.Saxena R., B. F. Voight, V. Lyssenko, N. P. Burtt, P. I. de Bakker, H. Chen, J. J. Roix, S. Kathiresan, J. N. Hirschhorn, M. J. Daly, et al. 2007. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 316 1331–1336. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.