Abstract
To better understand the earliest steps in the assembly of triglyceride (TG)-rich lipoproteins, we compared the biophysical and interfacial properties of two closely related apolipoprotein B (apoB) truncation mutants, one of which contains the complete lipoprotein initiating domain (apoB20.1; residues 1-912), and one of which, by virtue of a 50 amino acid C-terminal truncation, is incapable of forming nascent lipoproteins (apoB19; residues 1-862). Spectroscopic studies detected no major differences in secondary structure, and only minor differences in conformation and thermodynamic stability, between the two truncation mutants. Monolayer studies revealed that both apoB19 and apoB20.1 bound to and penetrated egg phosphatidylcholine (EPC) monolayers; however, the interfacial exclusion pressure of apoB20.1 was higher than apoB19 (25.1 mN/m vs. 22.8 mN/m). Oil drop tensiometry revealed that both proteins bound rapidly to the hydrophobic triolein/water interface, reducing interfacial tension by ∼20 mN/m. However, when triolein drops were first coated with phospholipids (PL), apoB20.1 bound with faster kinetics than apoB19 and also displayed greater interfacial elasticity (26.9 ± 0.8 mN/m vs. 22.9 ± 0.8 mN/m). These data establish that the transition of apoB to assembly competence is accompanied by increases in surface activity and elasticity, but not by significant changes in global structure.
Keywords: lipoprotein assembly, structure and function, fluorescence spectroscopy, circular dichroism spectroscopy, surface chemistry, monolayers, dynamic interfacial activity, very low density lipoproteins
Apolipoprotein B (apoB) is a 4,536 amino acid residue secretory glycoprotein that serves as the major structural component of triglyceride (TG)-rich lipoproteins secreted by the liver (very low density lipoproteins) and intestine (chylomicrons) (1–5). The assembly of apoB-containing lipoproteins is believed to occur in two stages: in the first stage, a precursor apoB-containing emulsion particle is formed in the endoplasmic reticulum (ER) concurrently with apoB translation; in the second stage, these nascent lipoproteins fuse with TG-rich emulsion particles produced in the smooth ER (6–11). Both steps require the activity of microsomal TG transfer protein (MTP), a dedicated ER-localized cofactor that is essential for apoB-containing lipoprotein assembly and secretion (12–14).
The mechanism by which apoB is lipidated during the initial phase of TG-rich lipoprotein assembly is not well understood. It was first proposed that apoB intercalates into the inner leaflet of the ER membrane during its translation and then entrains membrane lipid as it desorbs from the ER membrane (15–18). The observation that apoB becomes associated with the ER membrane immediately after translation (16), and that N-terminal fragments of apoB bind to hydrophobic surfaces (19–24), suggests that apoB possesses avid surface activity, which is an essential requisite of this model. Recently, however, Segrest et al. (25–28) have proposed a different model for the lipidation of apoB that is based upon sequence similarities between apoB and vitellogenin. This model postulates that the N terminal ∼1,000 residues of apoB form two β-sheets that comprise the sides of a lipid-binding cavity held together by a helix-turn-helix motif (27, 29), similar to that noted in the crystal structure of lamprey lipovitellin (LV), the processed form of vitellogenin (30–32). This cavity would then be lipidated by either MTP- or non-MTP-dependent mechanisms (4, 33, 34).
To better understand the structural elements that enable apoB to direct the earliest steps in TG-rich lipoprotein assembly, we have examined the biophysical and interfacial properties of two closely related apoB truncation mutants: apoB20.1 (residues 1–912), which is fully capable of initiating MTP-dependent lipoprotein assembly; and apoB19 (residues 1–862) which, by virtue of a 50 amino acid C-terminal truncation, is incapable of nascent lipoprotein formation (35). We reasoned that the striking difference between the biological behavior of apoB19 and apoB20.1 should be reflected in biophysical properties specific to each model. The data presented here establish that the transition of apoB to assembly competence is accompanied by increased surface activity and interfacial elasticity, but not by significant changes in global structure or molecular stability, properties that are predicted by the membrane intercalation-desorption model (36).
EXPERIMENTAL PROCEDURES
Lipids
Egg phosphatidylcholine (EPC) and triolein were purchased from Sigma-Aldrich. Phospholipid (PL) spreading solutions were prepared by diluting EPC in HPLC grade hexane to a concentration of 1 mg/ml, and stored under argon gas at −20°C. Purity was confirmed by the presence of single bands on thin-layer chromatography. Intralipid was purchased from Baxter Healthcare, and centrifuged in a Ti40.3 rotor (Beckman-Coulter) at 40,000 rpm for 1 h to remove the PL-rich infranatant from the TG-rich emulsion phase (37).
Preparation of apoB truncation mutants
ApoB19 (apoB signal peptide plus mature residues 1–862) and apoB20.1 (apoB signal peptide plus mature residues 1–912) were each engineered to contain a C-terminal His6-tag (35). The 50 amino acid C-terminal peptide present in apoB20.1, but deleted in apoB19, is shown in Fig. 1. Each truncation mutant was expressed in Sf9 cells and purified by nickel-nitrilotriacetic acid-agarose affinity chromatography, as previously described (21, 35, 36). Purified proteins were dialyzed into 10 mM Tris, pH 7.4, 140 mM NaCl, containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, 3 mM EDTA) and 1.5 mM sodium azide, and stored under nitrogen at 4°C. Protein concentration was determined by the bicinchoninic acid method (Pierce). All purified protein preparations displayed single bands upon SDS-polyacrylamide gel electrophoresis and Coomassie blue staining (36). To assure the dissociation of protein multimers, for the spectroscopic studies the purified proteins were incubated for 30 min with 0.2% cholic acid and then dialyzed against phosphate buffered saline (PBS) for 20 h at room temperature (36); for the interfacial activity studies, the proteins were dissolved buffer containing 2.0 M guanidine hydrochloride (GdnHCl).
Fig. 1.
Amino acid sequence of the intervening peptide between apolipoprotein B (apoB)19 and apoB20.1. The bold black box indicates the location of a predicted α-helix. The two light-gray boxes indicate the location of predicted tilted fusogenic peptides.
CD spectroscopy
Circular dichroism (CD) studies were performed using a Jasco J-720 Spectropolarimeter. Spectra of apoB19 and apoB20.1 at 1 μM in PBS were recorded at 25°C from 190 to 250 nm at 0.5 nm intervals using a 1 cm thermostated cell, 1 nm bandwidth, and 2 s time constant. Buffer blanks were digitally subtracted. Thermal denaturation studies were performed by monitoring ellipticity at 222 nm at 2 s intervals as the temperature of the cuvette was increased from 25°C to 75°C at 1°C/min. The enthalpy of denaturation, ΔHD, and thermal denaturation midpoint, Tm, were determined from Van't Hoff plots of ΔG vs. 1/T (38). The entropy of folding, ΔS, was calculated at the denaturation midpoint using the Gibbs-Helmholtz equation (38, 39). Mean residue ellipticity at 222 nm, [Θ]222, was calculated as ([Θ]raw × MRW)/(10 × l × C), where l is the path length in cm and C is the concentration in g/ml, using a mean residue weight (MRW) of 111.9 for apoB19 and 111.6 for apoB20.1. Structural analysis of spectra was performed using the SELCON 3 deconvolution algorithm (40).
Fluorescence spectroscopy
Fluorescence studies were performed using an SLM 8000C spectrofluorometer, as previously described (37, 41). Spectra of 1 μM apoB19 and apoB20.1 in PBS were obtained using a 1 cm cell, an excitation wavelength of 280 nm, a 1 s integration time, and 4 nm slits on both monochromators. Spectra were excitation corrected using a rhodamine quantum counter, and corrected for scatter and Raman emission by digital subtraction of buffer blanks. Denaturation studies were performed by 0.25 M step addition of buffered 6 M GdnHCl. The excitation wavelength dependence of tryptophan emission anisotropy was determined with the instrument in T-format, monochromator A set at 370 nm with 8 nm slits, and a Corning 370 nm bandpass filter on monochromator B (41).
Interfacial activity of apoB truncation mutants at the PL/water interface
Adsorption of apoB19 and apoB20.1 to the PL/water interface was examined using a KSV 5000 Langmuir film balance (KSV Instruments, Helsinki, Finland) (38, 42). EPC monolayers were spread over 5.65 mM Na2HPO4, 3.05 mM NaH2PO4, 80 mM NaCl, pH 7.0 and allowed to stabilize. Surface pressure (Π) at the lipid/water interface was measured with a platinum Wilhelmy plate. Subphase saturating concentrations were determined by injecting increasing amounts of protein in 2.0 M GdnHCl under monolayers spread at 10 mN/m and recording the change in surface tension (ΔΠ). Interfacial exclusion pressure (Πex) was determined by injecting saturating concentrations of protein under EPC monolayers spread at initial pressures (Πi) between 5–25 mN/m and extrapolating plots of ΔΠ versus Πi to ΔΠ = 0.
Dynamic interfacial activity of apoB truncation mutants at the oil/water interface
The interfacial properties of the apoB truncation mutants at the oil/water interface were analyzed using a Tracker® tensiometer (TECLIS-ITConcept, Longessaigne, France). The instrument consists of a computer-controlled syringe that pumps fluids into an optical cuvette; a coherent light source that projects the drop image onto a video chip; and software that digitizes the image and calculates drop area, volume, and interfacial tension (43). Cuvette infusion ports allow circulation of aqueous phase around the oil drop. Triolein drops (10 μl) were formed into a cuvette containing 6 ml of 41.3 mM Tris, pH 7.5, and 25 μg/ml of apoB19 or apoB20.1. The adsorption of protein to the triolein/water interface was recorded as the decrease in interfacial tension (γ) with time. Exponential adsorption rate constants were calculated by log transformation of the initial γ vs. time curves. Interfacial elasticity (ɛ) was determined by sinusoidally oscillating the drop volume by 4 μl at a rate of 6 cycles/min and analyzing the phase shift between drop surface area and surface tension.
Interaction of apoB truncation mutants with PL-coated triolein droplets
Ten μl drops of triolein were formed into a cuvette containing 6 ml of 41.3 mM Tris, pH 7.5 and Intralipid infranatant (250 μg/ml phosphorus) was added (43). When a stable PL monolayer had formed on the drop surface, as evidenced by stabilization of surface tension, excess bulk phase PL was removed by continuous buffer exchange. ApoB truncation mutants (15 μg/ml in 41.3 mM Tris, pH 7.5, 2.0 M GdnHCl) were then injected, and protein adsorption to the PL-coated triolein drop was measured as the time-dependent decrease in γ. Binding rate constants and elasticity measurements were calculated as described above.
RESULTS
Spectroscopic properties of apoB19 and apoB20.1
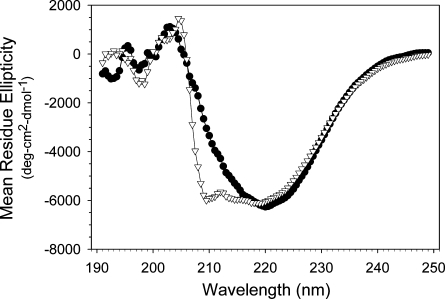
CD spectra of apoB19 and apoB20.1 displayed minima at 222 nm (Fig. 2 and Table 1), characteristic of the presence of α helical structure. Deconvolution analysis of the CD spectra revealed that the extension of apoB19 to form apoB20.1 did not induce a significant change in its secondary structure: apoB19 was predicted to contain 25% α helix, 22% β sheet 22% β turn, and 34% unordered structure and apoB20.1 was predicted to contain 28% α helix, 23% β sheet, 22% β turn, and 31% unordered structure. However, whereas the CD spectrum of apoB20.1 displayed minima at 222 and 208 nm, the CD spectrum of apoB19 displayed only a small shoulder at 208 nm. This CD spectral pattern is characteristic of the presence of coiled-coil structure (44), which suggests that the addition of residues 863–912 may have induced a reorganization of the α helical domains within the apoB20.1 molcule.
Fig. 2.
CD spectra of apoB19 and apoB20.1. Mean residue ellipticity as a function of wavelength was recorded at 25°C at a protein concentration of 1 μM apoB19 (•) or apoB20.1 (▽).
TABLE 1.
Spectroscopic and thermodynamic properties of apoB19 and apoB20.1
| [Θ]222 | Tm | ΔHD | ΔS | λmax | A305/A270 | |
|---|---|---|---|---|---|---|
| deg-cm2-dmol−1 | °C | kcal/mol | cal/mol°K | nm | ||
| ApoB19 | 6,577 | 57.8 | 96.1 | 290.2 | 327.5 ± 1.0 | 1.26 ± 0.04 |
| ApoB20.1 | 6,243 | 56.4 | 71.9 | 218.2 | 328.0 ± 2.0a | 1.30 ± 0.04a |
[Θ]222, mean residue ellipticity at 222 nm; Tm, thermal denaturation midpoint; ΔHD, enthalpy of denaturation; ΔS, entropy of folding; λmax, wavelength of maximum fluorescence emission; A305/A270, tryptophan emission anisotropy ratio.
Means ± SE, n = 3, P = NS by Student's t-test.
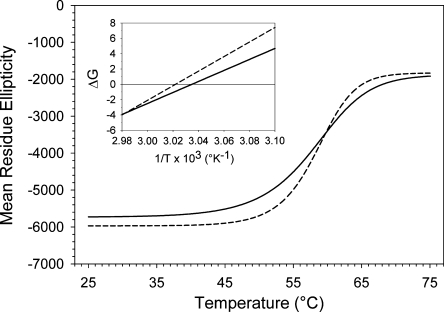
CD-thermal denaturation spectral data were used to calculate thermodynamic stability parameters (Fig. 3; Table 1). For apoB19 the thermal transition midpoint was 57.8°C, the enthalpy of denaturation was 96.1 kcal/mol, and the entropy of unfolding was 290.2 cal/mol°K. For apoB20.1 the thermal denaturation midpoint was 56.4°C, the enthalpy of denaturation was 71.9 kcal/mol, and the entropy of unfolding was 218.2 cal/mol°K. The sigmoidicity of the denaturation curve for apoB20.1 was slightly shallower than that of apoB19, which suggests a loss of intramolecular domain cooperativity in the stabilization of its secondary structure (Fig. 3). Together, these observations suggest that extension of apoB19 to apoB20.1 slightly reduces molecular stability, rather than creating a more thermodynamically stable conformation, as might be expected for completion of a compact, lipid binding cavity.
Fig. 3.
Thermal denaturation of apoB19 and apoB20.1. Mean residue ellipticity at 222 nm of 1 μM solutions of apoB19 (dashed line) and apoB 20.1 (solid line) was sampled at two second intervals as the temperature was raised from 25 to 75°C. The lines are sigmoidal curve fits of the data. Inset: Van't Hoff plots; the solid lines are linear curve fits of the thermal denaturation data. Intercepts of the lines at the X axis yield the reciprocal of the transition temperature midpoint.
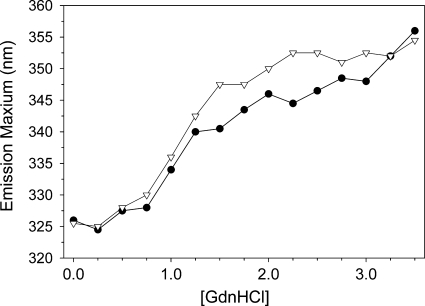
Fluorescence spectroscopy revealed that the emission maxima of apoB19 and apoB20.1 were blue shifted to 327.5 and 328.0 nm, respectively (Table 1), indicating that the three tryptophan residues, present at positions 370, 556, and 694 in both truncation mutants, reside in similar hydrophobic environments. The tryptophan emission anisotropy ratio with excitation at 305 and 270 nm (A305/A270), which provides a measure of intramolecular tyrosine→tryptophan energy transfer efficiency [and is thus in turn a function of the compactness of protein folding (41)] was similar for both truncation mutants (Table 1). Addition of increasing concentrations of GdnHCl to the truncation mutants induced a progressive 30 nm red shift in the tryptophan maximum emission, indicative of protein unfolding (Fig. 4). The concentration of GdnHCl required to induce 50% unfolding was 1.50 M for apoB19 and 1.25 M for apoB20.1. Together these data again suggest that the completion of the lipoprotein initiating domain of apoB is not accompanied by a significant change in native molecular structure, but does slightly decrease its conformational stability.
Fig. 4.
Fluorescence denaturation of apoB19 and apoB20.1. The wavelength of maximum fluorescence of 1 μM solutions of apoB19 (•) and apoB20.1 (▽) was monitored as a function of guanidine hydrochloride (GdnHCl) concentration.
Adsorption of apoB19 and apoB20.1 to EPC monolayers
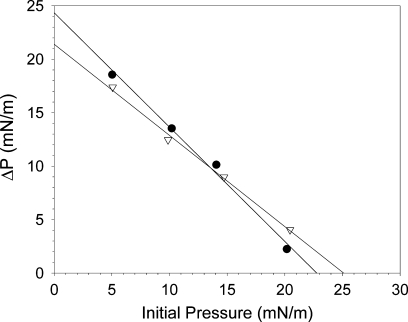
With injection of increasing amounts of apoB19 and apoB20.1 under EPC monolayers spread at 10 mN/m, the increase in surface pressure reached a plateau at a subphase concentrations of 1 × 10−4 g/dl for both proteins (data not shown). To assure saturation of the EPC monolayers, the exclusion pressure experiments were then performed at this subphase concentration (100 μg protein in a 50 ml buffer subphase). Adsorption of apoB19 and apoB20.1 to the monolayers decreased linearly as the initial pressure (Πi) increased (Fig. 5). Extrapolation of the ΔΠ-ΠI curves to zero yielded interfacial exclusion pressures of 22.8 mN/m for apoB19 and 25.1 mN/m for apoB20.1. The higher exclusion pressure of apoB20.1 suggests that it is better able to insert itself into PL monolayers. Of note, these values are lower than exclusion pressures similarly determined for the exchangeable apolipoproteins (apoA-IV, 29 mN/m; apoC-III, 31 mN/m; apoA-I, 33 mN/m; apoC-II, 34 mN/m; and apoA-II, 34 mN/m (42, 45, 46).
Fig. 5.
Interfacial exclusion pressure of apoB19 and apoB20.1. ApoB19 (•) and apoB20.1 (▽) were injected beneath EPC monolayers spread at increasing initial surface pressures, and the resulting change in surface pressure (ΔP) was determined. The solid lines are linear regressions of the data. Extrapolation of the lines to the X axis yields exclusion pressures of 22.8 mN/m for apoB19, and 25.1 mN/m for apoB20.1.
Dynamic interfacial activity of apoB19 and apoB20.1 at the triolein/water interface
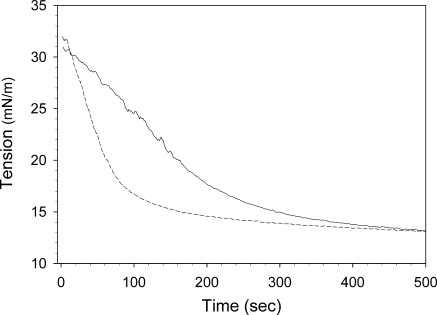
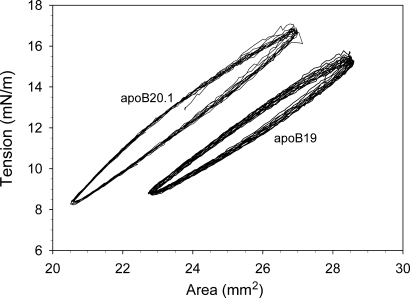
ApoB19 and apoB20.1 bound readily to the triolein/water interface, rapidly decreasing its surface tension from an initial value of 32 mN/m to 11.6 mN/m and 11.1 mN/m, respectively (Fig. 6). The calculated exponential binding rate constants were 3.0 ± 0.2 × 10−3 sec−1 for apoB19, and 1.3 ± 0.2 × 10−3 sec−1 for apoB20.1 (Table 2), which indicates that apoB20.1 interacts more slowly with the oil/water interface, although it ultimately generates the same equilibrium surface tension as apoB19. The interfacial elasticity of apoB19 and apoB20.1 was examined by sinusoidally oscillating the drop volume and recording the change in surface tension as a function of surface area (Fig. 7). The surface area-tension loops for apoB20.1 are shifted to the left relative to apoB19, indicative of a more condensed interfacial conformation. The calculated elasticity of apoB19 was 25.1 ± 0.2 mN/m, whereas the elasticity of apoB20.1 was higher at 26.8 ± 0.1 mN/m (Table 2). These data suggest that addition of only 50 residues to apoB19 confers increased molecular flexibility at hydrophobic interfaces.
Fig. 6.
Dynamic interfacial behavior of apoB19 and apoB20.1 at the oil/water interface. The binding of apoB19 and apoB20.1 to the triolein/water interface was measured using an ITC Tracker oil drop tensiometer. Ten μl drops of triolein were rapidly formed into buffer containing 25 μg/ml apoB19 (dashed line) or apoB20.1 (solid line), and the surface tension was continuously monitored.
TABLE 2.
Dynamic interfacial properties of apoB19 and apoB20.1
| Triolein/water interfacea
|
EPC/triolein/water interfaceb
|
|||||
|---|---|---|---|---|---|---|
| γ | Ka | ɛ | γ | Ka | ɛ | |
| mN/m | (x10−3sec−1) | mN/m | mN/m | (x10−3sec−1) | mN/m | |
| ApoB19 | 11.6 ± 0.2 | 3.0 ± 0.2 | 25.1 ± 0.2 | 11.9 ± 0.5 | 3.5 ± 0.1 | 22.9 ± 0.8c |
| ApoB20.1 | 11.1 ± 0.2 | 1.3 ± 0.2 | 26.8 ± 0.1 | 11.5 ± 0.5 | 4.1 ± 1.0d | 26.9 ± 0.8 |
| P | NS | 0.014 | 0.002 | NS | NS | 0.017 |
Data are means ± SE for 2 to 6 determinations. P values in the third row are B19 versus B20.1 by Student's t-test or Mann-Whitney rank sum test. EPC, egg phosphatidylcholine; γ, final interfacial tension; Ka, exponential binding rate constant; ɛ, interfacial elasticity.
Baseline interfacial tension = 32 mM/m.
Baseline interfacial tension = 22 mM/m.
P = 0.024 versus the elasticity of B19 at the triolein/water interface.
P = 0.040 versus the binding rate constant of B20.1 at the triolein/water interface.
Fig. 7.
Surface tension-area plots of apoB19 and apo 20.1 at the triolein/water interface. Ten μl drops of triolein were formed into buffer containing 25 μg/ml apoB19 or apoB20.1, as indicated. After the tension had stabilized, the drop volume was sinusoidally oscillated at 6 cycles/min and the change in surface tension was continuously monitored as a function of drop surface area.
Dynamic interfacial activity of apoB19 and apoB20.1 at the EPC/triolein/water interface
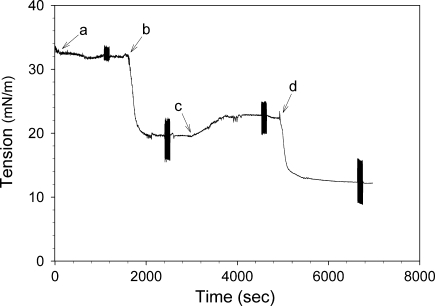
The interfacial behavior of apoB19 and apoB20.1 was further examined using triolein drops coated with an EPC monolayer to simulate interaction with nascent lipoprotein particles in the ER. Triolein drops were formed into buffer (Fig. 8, arrow a) and a micellar EPC solution was injected into the cuvette (arrow b), which caused the interfacial tension to fall to ∼19 mN/m as EPC molecules bound to the triolein/water interface. When unbound PL was washed out of the cuvette by continuous buffer exchange (arrow c), the tension increased to ∼22 mN/m. ApoB19 or apoB20.1 was then injected into the cuvette (arrow d), and the interfacial tension rapidly fell to ∼11 mN/m (Fig. 8 and Table 2). Calculation of exponential binding rate constants revealed that apoB19 bound with a similar rate to naked and EPC-coated triolein drops (3.0 ± 0.2 × 10−3 sec−1 and 3.5 ± 0.1 × 10−3 sec−1, respectively), whereas apoB20.1 bound significantly faster to triolein drops when they were coated with EPC (1.3 ± 0.2 × 10−3 sec−1 vs. 4.1 ± 1.0 × 10−3 sec−1). The elasticity of apoB19 at the EPC/triolein/water interface was significantly lower than at the triolein/water interface, 22.9 ± 0.8 mN/m vs. 25.1 ± 0.2 mN/m, whereas the elasticity of apoB20.1 EPC/triolein/water interface was the same as at the triolein/water interface (Table 2).
Fig. 8.
Dynamic interfacial behavior of apoB19 at the EPC/triolein/water interface. A 10 μl triolein drop was rapidly formed into buffer (arrow a), and was coated with a phospholipid (PL) monolayer by addition of Intralipid (arrow b). Excess unbound PL was washed out of the cuvette by continuous buffer exchange (arrow c). Then 15 μg/ml apoB19 was added to the cuvette (arrow d) and the further decrease in interfacial tension was monitored. The elasticity of the interface at each stage was determined by sinusoidal oscillation of the drop volume (denoted by the rapid vertical displacements) as described under EXPERIMENTAL PROCEDURES. Analyses of apoB20.1 under the same conditions were also performed (36); data not shown.
DISCUSSION
ApoB is a large amphipathic protein that organizes lipids into microemulsion particles within the secretory pathway of lipoprotein producing cells. Two competing models have been proposed to explain how apoB initially acquires lipid: the intercalation/desorption model, which posits that the N-terminal domain of apoB interacts with the ER membrane, thereby sequestering a nidus of neutral lipid (predominately TG) that ultimately desorbs from the membrane as a small, neutral lipid core-containing precursor particle (4); and the LV model, which proposes that the apoB N-terminal domain folds into a globular LV-like lipid binding cavity, which is then filled with a small amount of PL containing a lens of TG (29, 47). As the biophysical characteristics of apoB presumed by each model are distinctive, we examined the structural and interfacial properties of two apoB C-terminal truncation mutants that terminate on either side of a critical functional boundary: apoB20.1, which is assembly-competent; and apoB19, a 50-amino acid truncation mutant of apoB20.1, which is incapable of forming lipoproteins (35).
The structure of the two apoB truncation mutants was analyzed by CD and fluorescence spectroscopy. Both mutants displayed similar secondary structure in their native state, which implies that no major structural reorganization occurs upon completion of the lipoprotein initiating domain. However, the lower transition midpoint, denaturation cooperativity, and enthalpy of denaturation noted for apoB20.1 (Table 1) all suggest that its ordered structure is less stable than apoB19 and thus it is more easily unfolded. In addition, the lower entropy of unfolding for apoB20.1 indicates that it has a greater degree of disorder in its native state. This is further implied by the lower [Θ]222/[Θ]208 ratio for apoB20.1, which is indicative of less coiled-coil structure (44) and disrupted electronic interactions among its aromatic side chains (48). Taken together, these data suggest that the completed lipoprotein initiating domain of apoB adopts a less stable, more loosely folded conformation, predicted by the intercalation-desorption model, rather than a more stable, compact structure predicted by the LV model. Nonetheless, it is also possible that the formation of a lipid-binding cavity could decrease the stability of a truncated apoB in the absence of lipid.
As destabilization of apolipoprotein ordered structure is associated with increased interfacial activity (38), we next examined the behavior of apoB19 and apoB20.1 at the EPC/water, triolein/water, and EPC/triolein/water interfaces. These studies established that apoB19 and apoB20.1 are strongly surface active proteins that can lower the equilibrium surface tension at hydrophobic interfaces to similar levels (Table 2). However, the surface balance studies showed that apoB20.1 has a higher interfacial exclusion pressure for EPC monolayers, consistent with a superior ability to penetrate and/or intercalate into PL membrane interfaces (Fig. 5). It is interesting to note that the absolute reduction in surface tension induced by the apoB truncation mutants was greater with the more hydrophobic triolein/water interface (ΔΠ = ∼21 mN/m), than the EPC/triolein/water interface (ΔΠ = ∼10 mN/m) (Table 2), which further demonstrates that although both apoB mutants bind to PL, they possess a innately higher affinity for the more hydrophobic neutral lipids (i.e., TGs and cholesterol esters) (36). The ability of apoB to lower the surface tension (free energy) of mixed lipid interfaces is a requisite first step in its participation in the membrane reorganization that is central to the intercalation/desorption model.
An unexpected finding was that apoB20.1 bound to the triolein/water interface more slowly than apoB19 (Fig. 6). However, whereas apoB19 bound with similar rates to naked and EPC-coated triolein drops, the binding rate for apoB20.1 increased more than 3-fold when the drops were first coated with EPC, a biophysical model that more closely reflects the mixed lipid surface of a nascent emulsion particle (Table 2). The decrease in surface tension measured by the oil-drop tensiometer reflects a sequence of events: 1) protein diffusion to the triolein/water interface, which is dependent upon molecular weight and solvent viscosity; 2) physical binding and penetration of protein into the interface, which is dependent upon the hydrophobicity of the protein and interface, but can be dramatically altered by the presence of “anchor” domains in the protein; and 3) surface unfolding, which is a function of protein stability. As there is only a 6% difference in molecular weight between apo B19H (96,769 daltons) and apoB20.1 (102,471 daltons), and negligible differences in their molecular hydrophobicity and stability, this interface-dependent kinetic behavior of apoB20.1 may be a function of its superior ability to penetrate PL monolayers, as indicated by its higher interfacial exclusion pressure. In this regard, it is relevant that the 50 amino acid interval between apoB19 and apoB20.1 is predicted to contain two tilted peptides (Fig. 1), a structural motif present in fusogenic proteins that enables them to penetrate deep into the core of lipid bilayers, and thereby induce a structural reorganization that leads to membrane fusion (49).
Interfacial elasticity reflects the ability of surface-active proteins to adapt their conformation to changes in surface geometry. At the triolein/water and EPC/triolein/water interfaces, apoB20.1 displayed greater interfacial elasticity than apoB19 (Table 2). In fact, the elasticity of apo B19 was lower at the surface of the EPC-coated droplet. Although protein elasticity has no direct biological correlate, the facile ability of the lipoprotein initiating domain of apoB to adapt to changes in geometry at mixed lipid interfaces, in combination with its superior binding kinetics, could be critical to its ability to interact with the inner leaflet of the ER membrane, initiate the lipid desorption process (50), and thereafter accommodate the expansion of nascent emulsion particles following membrane desorption.
Taken together, these data establish that extension of apoB19 by only 50 amino acids to form apoB20.1, a fully functional lipoprotein assembly initiating domain, is not accompanied by major changes in its secondary or tertiary structure, but rather, is associated with an increase in its surface activity and interfacial elasticity. These structural and interfacial properties are the hallmarks of surfactant proteins that lower the free energy and increase the stability of hydrophobic interfaces, and are less consistent with completion of a geometrically fixed globular lipid transport protein. Thus, our data are most consistent with a model of lipoprotein assembly in which the surface active initiating domain of apoB interacts with the ER membrane to recruit neutral and polar lipids, and thereafter adapts its conformation and surface activity to promote nascent emulsion particle formation and release into the ER lumen.
Although the biophysical techniques used in this work have been extensively applied to the exchangeable apolipoproteins, the study described herein is the first systematic application of combined spectroscopic and surface chemistry techniques to the analysis of functionally defined forms of apoB. Further such biophysical studies of apoB surface activity and conformational dynamics promise new insights into the underlying mechanisms responsible for nascent lipoprotein formation and lipid transport, processes central to the development of atherosclerotic cardiovascular disease and other chronic diseases, such as metabolic syndrome and obesity.
Abbreviations
apoB, apolipoprotein B
CD, circular dichroism
EPC, egg phosphatidylcholine
ER, endoplasmic reticulum
GdnHCl, guanidine hydrochloride
LV, lipovitellin
MTP, microsomal triglyceride transfer protein
PBS, phosphate buffered saline
PL, phospholipid
TG, triglyceride
Published, JLR Papers in Press, August 18, 2008.
Footnotes
This work was supported by National Institutes of Health grants HL49373 (G.S.S.) and HL30897 (R.B.W.). A.S.L was supported by a predoctoral fellowship from the American Heart Association, Mid-Atlantic Affiliate.
References
- 1.Davidson N. O., and G. S. Shelness. 2000. Apolipoprotein B: mRNA editing, lipoprotein assembly, and presecretory degradation. Annu. Rev. Nutr. 20 169–193. [DOI] [PubMed] [Google Scholar]
- 2.Fisher E. A., and H. N. Ginsberg. 2002. Complexity in the secretory pathway: the assembly and secretion of apolipoprotein B-containing lipoproteins. J. Biol. Chem. 277 17377–17380. [DOI] [PubMed] [Google Scholar]
- 3.Hussain M. M., S. Fatma, X. Y. Pan, and J. Iqbal. 2005. Intestinal lipoprotein assembly. Curr. Opin. Lipidol. 16 281–285. [DOI] [PubMed] [Google Scholar]
- 4.Shelness G. S., and A. S. Ledford. 2005. Evolution and mechanism of apolipoprotein B-containing lipoprotein assembly. Curr. Opin. Lipidol. 16 325–332. [DOI] [PubMed] [Google Scholar]
- 5.Davis R. A., and T. Y. Hui. 2001. 2000 George Lyman Duff memorial lecture—atherosclerosis is a liver disease of the heart. Arterioscler. Thromb. Vasc. Biol. 21 887–898. [DOI] [PubMed] [Google Scholar]
- 6.Alexander C. A., R. L. Hamilton, and R. J. Havel. 1976. Subcellular localization of B apoprotein of plasma lipoproteins in rat liver. J. Cell Biol. 69 241–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rustaeus S., K. Lindberg, P. Stillemark, C. Claesson, L. Asp, T. Larsson, J. Borén, and S. O. Olofsson. 1999. Assembly of very low density lipoprotein: A two-step process of apolipoprotein B core lipidation. J. Nutr. 129 463S–466S. [DOI] [PubMed] [Google Scholar]
- 8.Gusarova V., J. L. Brodsky, and E. A. Fisher. 2003. Apolipoprotein B100 exit from the endoplasmic reticulum (ER) is COPII-dependent, and its lipidation to very low density lipoprotein occurs post-ER. J. Biol. Chem. 278 48051–48058. [DOI] [PubMed] [Google Scholar]
- 9.Gusarova V., J. Seo, M. L. Sullivan, S. C. Watkins, J. L. Brodsky, and E. A. Fisher. 2007. Golgi-associated maturation of very low density lipoproteins involves conformational changes in apolipoprotein B, but is not dependent on apolipoprotein E. J. Biol. Chem. 282 19453–19462. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y., K. Tran, and Z. Yao. 1999. The activity of microsomal triglyceride transfer protein is essential for accumulation of triglyceride within the microsomes in McA-RH7777 cells. J. Biol. Chem. 274 27793–27800. [DOI] [PubMed] [Google Scholar]
- 11.Kilinski A., S. Rustaeus, and J. E. Vance. 2002. Microsomal triglyceride transfer protein is required for lumenal accretion of triacylglycerol not associated with apo B, as well as for apoB lipidation. J. Biol. Chem. 277 31516–31525. [DOI] [PubMed] [Google Scholar]
- 12.Berriot-Varoqueaux N., L. P. Aggerbeck, M. E. Samson-Bouma, and J. R. Wetterau. 2000. The role of the microsomal triglygeride transfer protein in abetalipoproteinemia. Annu. Rev. Nutr. 20 663–697. [DOI] [PubMed] [Google Scholar]
- 13.Wetterau J. R., M. C. Lin, and H. Jamil. 1997. Microsomal triglyceride transfer protein. Biochim. Biophys. Acta. 1345 136–150. [DOI] [PubMed] [Google Scholar]
- 14.Hussain M. M., J. Shi, and P. Dreizen. 2003. Microsomal triglyceride transfer protein and its role in apoB-lipoprotein assembly. J. Lipid Res. 44 22–32. [DOI] [PubMed] [Google Scholar]
- 15.Olofsson S-O., G. Bjursell, K. Boström, P. Carlsson, J. Elovson, A. A. Protter, M. A. Reuben, and G. Bondjers. 1987. Apolipoprotein B: structure, biosynthesis and role in the lipoprotein assembly process. Atherosclerosis. 68 1–17. [DOI] [PubMed] [Google Scholar]
- 16.Pease R. J., G. B. Harrison, and J. Scott. 1991. Cotranslocational insertion of apolipoprotein B into the inner leaflet of the endoplasmic reticulum. Nature. 353 448–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spring D. J., L. W. Chen-Liu, J. E. Chatterton, J. Elovson, and V. N. Schumaker. 1992. Lipoprotein assembly. Apolipoprotein B size determines lipoprotein core circumference. J. Biol. Chem. 267 14839–14845. [PubMed] [Google Scholar]
- 18.Schumaker V. N., M. L. Phillips, and J. E. Chatterton. 1994. Apolipoprotein B and low-density lipoprotein structure: Implications for biosynthesis of triglyceride-rich lipoproteins. Adv. Protein Chem. 45 205–248. [DOI] [PubMed] [Google Scholar]
- 19.Herscovitz H., A. Derksen, M. T. Walsh, C. J. McKnight, D. L. Gantz, M. Hadzopoulou-Cladaras, V. Zannis, C. Curry, and D. M. Small. 2001. The N-terminal 17% of apoB binds tightly and irreversibly to emulsions modeling nascent very low density lipoproteins. J. Lipid Res. 42 51–59. [PubMed] [Google Scholar]
- 20.DeLozier J. A., J. S. Parks, and G. S. Shelness. 2001. Vesicle-binding properties of wild-type and cysteine mutant forms of a1 domain of apolipoprotein B. J. Lipid Res. 42 399–406. [PubMed] [Google Scholar]
- 21.Weinberg R. B., V. R. Cook, J. A. DeLozier, and G. S. Shelness. 2000. Dynamic interfacial properties of human apolipoprotein A-IV and B17 at the air/water and oil/water interface. J. Lipid Res. 41 1419–1427. [PubMed] [Google Scholar]
- 22.Jiang Z. G., D. Gantz, E. Bullitt, and C. J. McKnight. 2006. Defining lipid-interacting domains in the N-terminal region of apolipoprotein B. Biochemistry. 45 11799–11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L., M. T. Walsh, and D. M. Small. 2006. Apolipoprotein B is conformationally flexible but anchored at a triolein/water interface: a possible model for lipoprotein surfaces. Proc. Natl. Acad. Sci. USA. 103 6871–6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang Z. G., M. N. Simon, J. S. Wall, and C. J. McKnight. 2007. Structural analysis of reconstituted lipoproteins containing the N-terminal domain of apolipoprotein B. Biophys. J. 92 4097–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segrest J. P., M. K. Jones, and N. Dashti. 1999. N-terminal domain of apolipoprotein B has structural homology to lipovitellin and microsomal triglyceride transfer protein: a ‘lipid pocket’ model for self-assembly of apoB-containing lipoprotein particles. J. Lipid Res. 40 1401–1416. [PubMed] [Google Scholar]
- 26.Segrest J. P., M. K. Jones, H. De Loof, and N. Dashti. 2001. Structure of apolipoprotein B-100 in low density lipoproteins. J. Lipid Res. 42 1346–1367. [PubMed] [Google Scholar]
- 27.Dashti N., M. Gandhi, X. F. Liu, X. L. Lin, and J. P. Segrest. 2002. The N-terminal 1000 residues of apolipoprotein B associate with mi<!?A3B2 tlsb=-.0w?>crosomal triglyceride transfer protein to create a lipid transfer pocket required for lipoprotein assembly. Biochemistry. 41 6978–6987. [DOI] [PubMed] [Google Scholar]
- 28.Jiang Z., M. Carraway, and C. J. McKnight. 2005. Limited proteolysis and biophysical characterization of the lipovitellin homology region in apolipoprotein B. Biochemistry. 44 1163–1173. [DOI] [PubMed] [Google Scholar]
- 29.Richardson P. E., M. Manchekar, N. Dashti, M. K. Jones, A. Beigneux, S. G. Young, S. C. Harvey, and J. P. Segrest. 2005. Assembly of lipoprotein particles containing apolipoprotein-B: structural model for the nascent lipoprotein particle. Biophys. J. 88 2789–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson T. A., D. G. Levitt, and L. J. Banaszak. 1998. The structural basis of lipid interactions in lipovitellin, a soluble lipoprotein. Structure. 6 895–909. [DOI] [PubMed] [Google Scholar]
- 31.Thompson J. R., and L. J. Banaszak. 2002. Lipid-protein interactions in lipovitellin. Biochemistry. 41 9398–9409. [DOI] [PubMed] [Google Scholar]
- 32.Raag R., K. Appelt, N. H. Xuong, and L. Banaszak. 1988. Structure of the lamprey yolk lipid-protein complex lipovitellin-phosvitin at 2.8 A resolution. J. Mol. Biol. 200 553–569. [DOI] [PubMed] [Google Scholar]
- 33.Sellers J. A., L. Hou, D. R. Schoenberg, S. R. Batistuzzo de Medeiros, W. Wahli, and G. S. Shelness. 2005. Microsomal triglyceride transfer protein promotes the secretion of Xenopus laevis vitellogenin A1. J. Biol. Chem. 280 13902–13905. [DOI] [PubMed] [Google Scholar]
- 34.Dashti N., M. Manchekar, Y. Liu, Z. Sun, and J. P. Segrest. 2007. Microsomal triglyceride transfer protein activity is not required for the initiation of apolipoprotein B-containing lipoprotein assembly in McA-RH7777 cells. J. Biol. Chem. 282 28597–28608. [DOI] [PubMed] [Google Scholar]
- 35.Shelness G. S., L. Hou, A. S. Ledford, J. S. Parks, and R. B. Weinberg. 2003. Identification of the lipoprotein initiating domain of apolipoprotein B. J. Biol. Chem. 278 44702–44707. [DOI] [PubMed] [Google Scholar]
- 36.Ledford A. S., R. B. Weinberg, V. R. Cook, R. R. Hantgan, and G. S. Shelness. 2006. Self-association and lipid binding properties of the lipoprotein initiating domain of apolipoprotein B. J. Biol. Chem. 281 8871–8876. [DOI] [PubMed] [Google Scholar]
- 37.Weinberg R. B., and M. S. Spector. 1985. Structural properties and lipid binding of human apolipoprotein A-IV. J. Biol. Chem. 260 4914–4921. [PubMed] [Google Scholar]
- 38.Weinberg R. B., R. A. Anderson, V. R. Cook, F. Emmanuel, P. <!?A3B2 tlsb=-.01w?>Denefle, A. R. Tall, and A. Steinmetz. 2002. Interfacial exclusion pres<!?A3B2 tlsb=-.01w?>sure determines the ability of apolipoprotein A-IV truncation mutants to activate cholesterol ester transfer protein. J. Biol. Chem. 277 21549–21553. [DOI] [PubMed] [Google Scholar]
- 39.Weinberg R. B., R. A. Anderson, V. R. Cook, F. Emmanuel, P. Denefle, M. Hermann, and A. Steinmetz. 2000. Structure and interfacial properties of chicken apolipoprotein A-IV. J. Lipid Res. 41 1410–1418. [PubMed] [Google Scholar]
- 40.Sreerama N., S. Y. Venyaminov, and R. W. Woody. 1999. Estimation of the number of alpha-helical and beta-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 8 370–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinberg R. B. 1988. Exposure and electronic interaction of tyrosine and tryptophan residues in human apolipoprotein A-IV. Biochemistry. 27 1515–1521. [DOI] [PubMed] [Google Scholar]
- 42.Weinberg R. B., J. A. Ibdah, and M. C. Phillips. 1992. Adsorption of apolipoprotein A-IV to phospholipid monolayers spread at the air/water interface. A model for its labile binding to high density lipoproteins. J. Biol. Chem. 267 8977–8983. [PubMed] [Google Scholar]
- 43.Labourdenne S., N. Gaudry-Rolland, S. Letellier, M. Lin, A. Cagna, G. Esposito, R. Verger, and C. Riviere. 1994. The oil-drop tensiometer: potential applications for studying the kinetics of (phospho)lipase action. Chem. Phys. Lipids. 71 163–173. [Google Scholar]
- 44.Choy N., V. Raussens, and V. Narayanaswami. 2003. Inter-molecular coiled-coil formation in human apolipoprotein E C-terminal domain. J. Mol. Biol. 334 527–539. [DOI] [PubMed] [Google Scholar]
- 45.Krebs K. E., M. C. Phillips, and C. E. Sparks. 1983. A comparison of the surface activities of rat plasma apolipoproteins C–II, C–III-0, C–III-3. Biochim. Biophys. Acta. 751 470–473. [DOI] [PubMed] [Google Scholar]
- 46.Krebs K. E., J. A. Ibdah, and M. C. Phillips. 1988. A comparison of the surface activities of human apolipoproteins A-I and A-II at the air/water interface. Biochim. Biophys. Acta. 959 229–237. [DOI] [PubMed] [Google Scholar]
- 47.Manchekar M., P. E. Richardson, T. M. Forte, G. Datta, J. P. Segrest, and N. Dashti. 2004. Apolipoprotein B-containing lipoprotein particle assembly: lipid capacity of the nascent lipoprotein particle. J. Biol. Chem. 279 39757–39766. [DOI] [PubMed] [Google Scholar]
- 48.Arnold G. E., L. A. Day, and A. K. Dunker. 1992. Tryptophan contributions to the unusual circular dichroism of fd bacteriophage. Biochemistry. 31 7948–7956. [DOI] [PubMed] [Google Scholar]
- 49.Thomas A., and R. Brasseur. 2006. Tilted peptides: the history. Curr. Protein Pept. Sci. 7 523–527. [DOI] [PubMed] [Google Scholar]
- 50.Read J., T. A. Anderson, P. J. Ritchie, B. Vanloo, J. Amey, D. Levitt, M. Rosseneu, J. Scott, and C. C. Shoulders. 2000. A mechanism of membrane neutral lipid acquisition by the microsomal triglyceride transfer protein. J. Biol. Chem. 275 30372–30377. [DOI] [PubMed] [Google Scholar]