Abstract
SALMs are a family of five adhesion molecules whose expression is largely restricted to the CNS. Initial reports showed that SALM1 functions in neurite outgrowth while SALM2 is involved in synapse formation. To investigate the function of SALMs in detail, we asked if all five are involved in neurite outgrowth. Expression of epitope-tagged proteins in cultured hippocampal neurons showed that SALMs are distributed throughout neurons, including axons, dendrites, and growth cones. Over-expression of each SALM resulted in enhanced neurite outgrowth, but with different phenotypes. Neurite outgrowth could be reduced by applying antibodies targeting the extracellular leucine rich regions of SALMs and with RNAi. Through over-expression of deletion constructs, we found that the C-terminal PDZ binding domains of SALMs 1–3 are required for most aspects of neurite outgrowth. In addition, by using a chimera of SALMs 2 and 4, we found that the N-terminus is also involved in neurite outgrowth.
Introduction
Neurite outgrowth is a fundamental event in the development and maintenance of synaptic connections in the nervous system. Through highly regulated mechanisms, young neurons undergo axonal/dendritic polarization, and subsequent outgrowth of these neurites is essential to the establishment of synaptic connections that lead to brain function (da Silva and Dotti, 2002). Cell adhesion molecules (CAMs) are a diverse class of proteins that function in neurite outgrowth, synaptic development and maintenance, and cell adhesion at synaptic and non-synaptic sites (Craig and Banker, 1994; Dalva et al., 2007). Several CAMs are enriched at growth cones and are required for normal neurite outgrowth. For example, neural cell adhesion molecule (NCAM), N-cadherin, and L1-CAMs have been shown to regulate neurite outgrowth through various mechanisms, including changes in intracellular calcium levels, associations with cytoskeletal proteins at growth cones, and the activation of FGFR and MAPK signaling cascades (Doherty et al., 2000; Francavilla et al., 2007; Meiri et al., 1998; Utton et al., 2001). In humans, mutations in L1-CAMs lead to various neurological disorders, including hydrocephalus and MASA (mental retardation, aphasia, shuffling gait, and adducted thumbs) syndrome, and expression of constructs encoding L1 with these known mutations leads to deficits in neurite outgrowth (Moulding et al., 2000). While a wealth of information implicates CAMs in neurite outgrowth, the mechanism is highly complex and not completely understood.
Synaptic adhesion-like molecules (SALMs) are a family of CAMs that is largely restricted to the CNS and is involved in neurite outgrowth and synapse formation (Ko et al., 2006; Morimura et al., 2006; Wang et al., 2006). SALMs are also present in the adult where they may play a role in synaptic maintenance and other cellular interactions. Five family members have been identified: SALMs 1–5 (Ko et al., 2006; Morimura et al., 2006; Wang et al., 2006). The domain structure of SALMs includes extracellular leucine-rich repeats (LRR), an immunoglobulin C2-like domain (IgC2), a fibronectin type III (FN3) domain, a transmembrane (TM) region, and a PDZ-BD (PSD-95, Discs-large, ZO-1, binding domain; absent in SALMs 4 and 5). This domain structure is homologous with that of various related CAMs that function in outgrowth, including AMIGO, LINGO, NGL-1, and FLRT proteins (Chen et al., 2006).
Over-expression of SALM1 in young (4 days in vitro, DIV4) primary hippocampal cultures promotes an increase in neurite outgrowth (Wang et al., 2006), while alterations in SALM2 expression affects synapse formation and may play a role in regulating the balance of excitatory and inhibitory synapses (Ko et al., 2006). Therefore, individual SALMs may have a range of different functions. Alternatively, all SALMs may have multiple roles and function in neurite outgrowth and synapse formation in developing animals, as well as maintenance of synapses in adults. To investigate these possibilities, we have studied the role of all SALMs in neurite outgrowth by using a combination of over-expression, RNAi-mediated knock-down of expression, and blocking of function with antibodies to extracellular domains. Our results show that all SALMs promote neurite outgrowth, but with various phenotypes.
Results
Distribution of SALMs in neurons
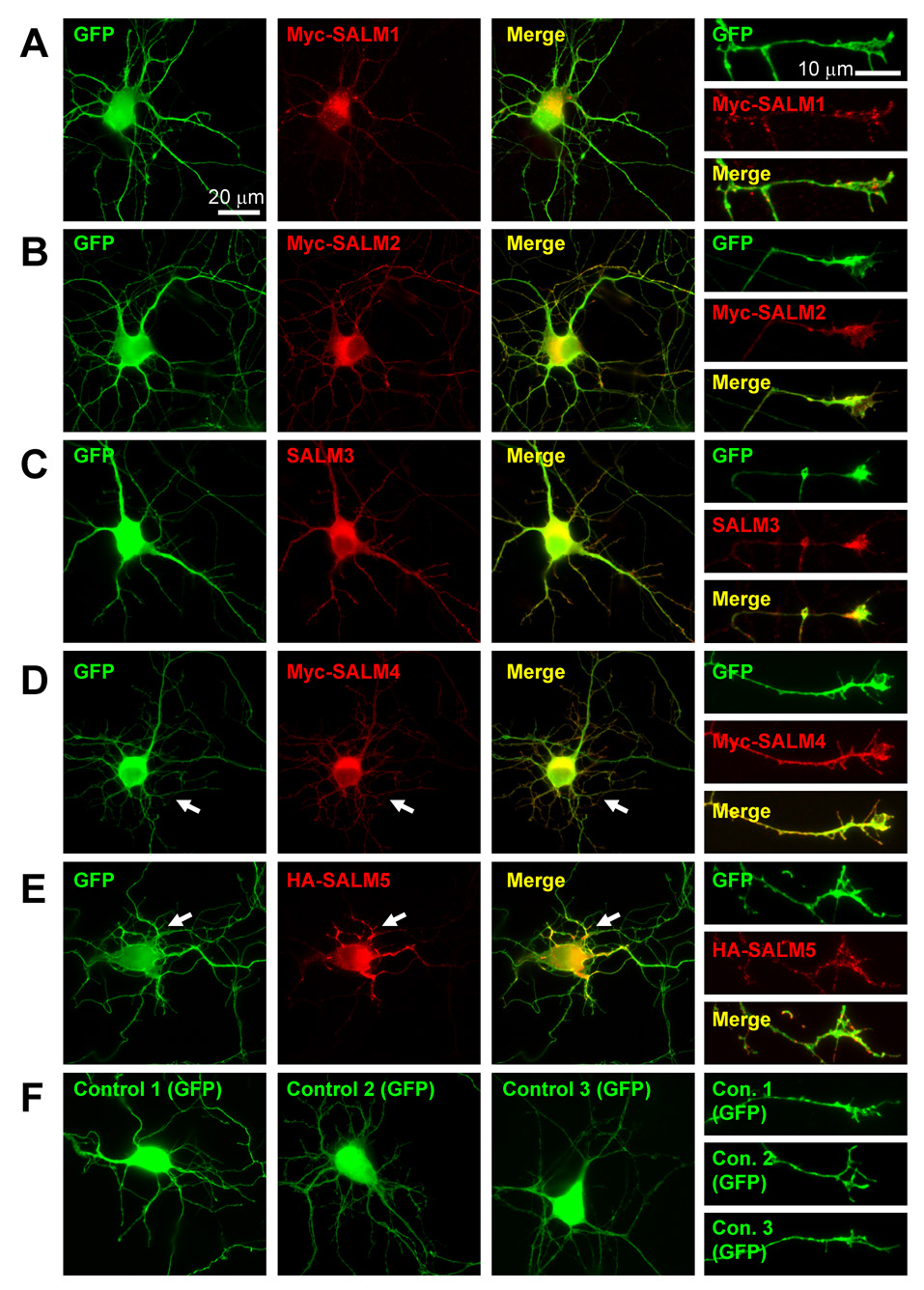
SALM1 and SALM2 localize to both axons and dendrites (Ko et al., 2006; Wang et al., 2006). Additionally, SALM1 co-localizes with NMDA receptors (Wang et al., 2006), while SALM2 co-localizes with both pre- and post-synaptic proteins at excitatory synapses in mature neurons (Ko et al., 2006). To understand the roles of SALMs in neurite outgrowth, we began by characterizing the cellular localization and morphological effects of overexpressed SALMs early in neuronal development. Young primary hippocampal neurons (DIV4) were co-transfected with GFP and myc-SALM1, myc-SALM2, untagged SALM3, myc-SALM4, or HA-SALM5 cDNA constructs. Neurons transfected with GFP and pcDNA 3.1+ empty vector were used as a control, and immunocytochemistry was performed 48 hours after transfection. Transfected SALM constructs over-expressed their respective proteins by about 300%, as compared to endogenous SALM levels (data not shown). Over-expressed SALMs are localized throughout the cell in the soma, axons, dendrites, and growth cones (Fig. 1) with a largely diffuse pattern. However, punctate staining is present and is particularly apparent when staining is restricted to SALMs present on the cell surface (supplemental Fig. 1). Therefore, SALMs are present in intracellular pools represented by the diffuse staining as well as on the surface where they appear more clustered.
Fig. 1. Localization and morphological characteristics of transfected SALM proteins.
DIV4 primary hippocampal cultures were co-transfected with SALM and GFP cDNAs. Neurons co-transfected with pcDNA3.1+ (empty vector) and GFP were used for control. Immunostaining was performed 48 hours later and the SALM localization and cell morphology were examined. Transfected SALMs localize throughout the cell body, axons, and dendrites. Representative examples of neurons transfected with SALMs 1–5 are shown in A–E, respectively. Scale bars, 20 µm. Transfected SALMs are enriched at growth cones (A–E, fourth column). Scale bars, 10 µm. Transfected SALM4 and SALM5, which do not contain PDZ-BDs, often exhibit distinct phenotypes as compared to SALMs 1–3. D, Myc-SALM4-transfected cells show a large increase in the number of short primary processes (arrows). HA-SALM5-transfected cells often display an increase in the number of primary processes crossing/overlapping with each other. E, Transfected SALM5 accumulates at these crossing points (arrows). Representative examples of GFP and pcDNA3.1+ co-transfected (control) cell body and growth cone morphology are shown in F.
The various SALMs qualitatively appear to have distinct effects on cell morphology. For example, SALM4-transfected neurons often had a dramatic increase in the number of shorter primary neurites protruding from the cell body (Fig. 1D, arrows). These shorter neurites appeared to be dendritic, as MAP2 immunostaining localized at these neurites (data not shown). Many SALM5-transfected neurons showed another unique phenotype. The primary neurites often overlapped and appeared to adhere to one another at regions proximal to the cell body. SALM5 accumulated at these points of neurite adhesion (Fig. 1E, arrows). These various changes in cell morphology of SALM-transfected neurons imply that individual SALMs may have different functions in the CNS.
SALMs promote neurite outgrowth
To further extend our initial qualitative observations indicating that SALMs may affect neurite outgrowth, we performed a detailed computer-assisted analysis of neuronal morphology using Metamorph Neurite Outgrowth Module (v7.0r3). The parameters tested included total neurite outgrowth (defined as the total skeletonized pixel area in µm), mean process length, number of primary neurites extending from the cell body, and number of total neurite branches. Primary hippocampal cultures were transfected at DIV4, a time when significant neurite outgrowth takes place (Dotti et al., 1988). Cultures were co-transfected with GFP (to visualize the entirety of each cell) and myc-SALM1, myc-SALM2, SALM3, myc-SALM4, or HA-SALM5 cDNA. Neurons were fixed and immunostained for GFP and SALM proteins 48 hours after transfection (DIV6); GFP staining was used to quantify neurite outgrowth and SALM staining served to verify SALM expression.
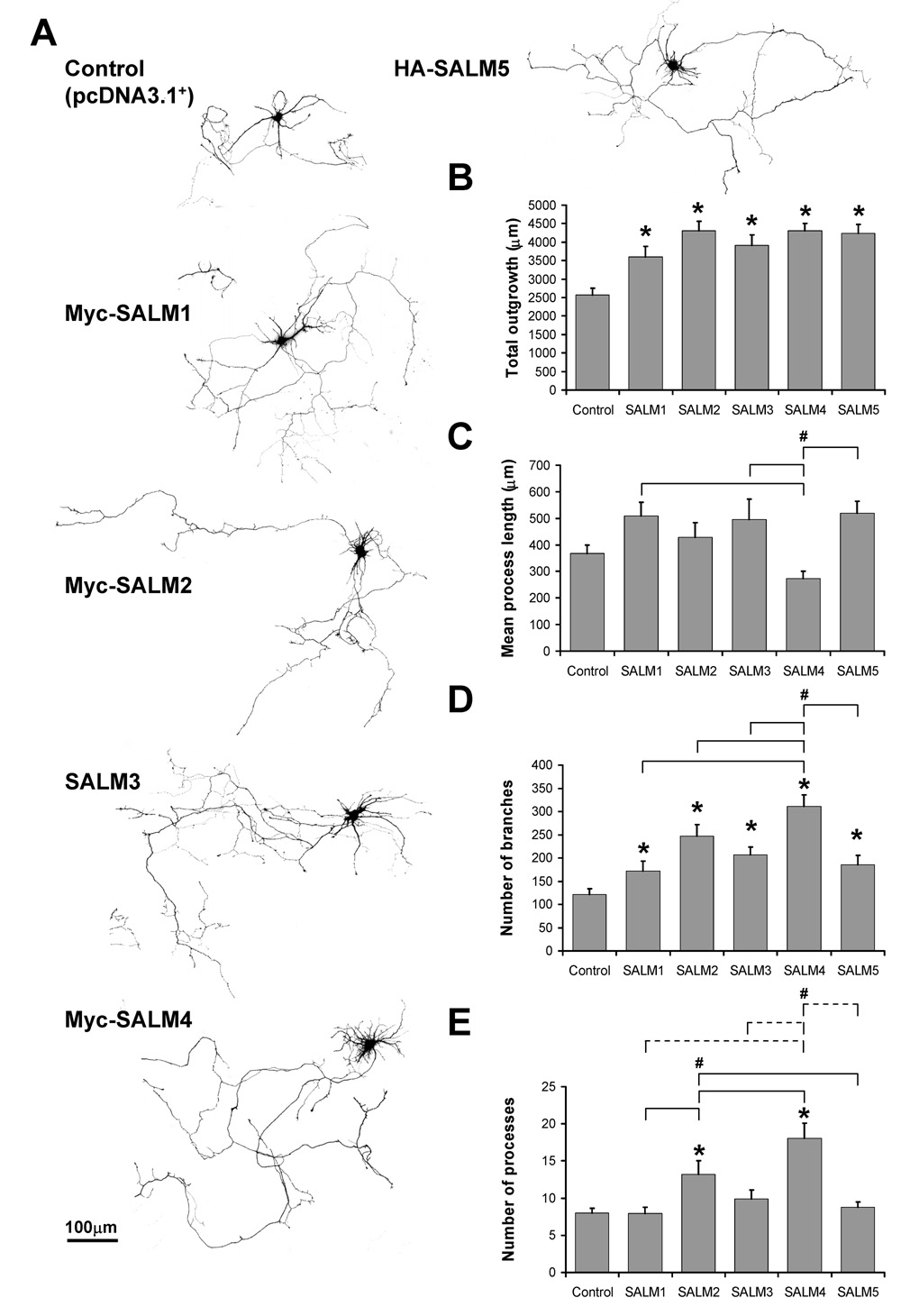
Our results showed that SALMs promote neurite outgrowth with various phenotypes. SALMs 1–5 all promoted significant increases in total outgrowth and number of branches, as compared to the control (Fig. 2B and D, respectively). Additionally, SALM4 promoted more branching than other SALMs (Fig. 2D). SALM2 promoted an increase in the number of primary processes, as compared to the control, SALM1, and SALM5 (Fig. 2E). Consistent with the dramatic increase in short primary neurites described earlier, the number of processes in SALM4-transfected cells more than doubled, as compared to control and other SALM-transfected neurons, including those transfected with SALM2 (Fig. 2E). The mean process length of SALM4-transfected neurons was significantly less than that of SALM1, SALM3, and SALM5-transfected neurons (Fig. 2C). Together, this increase in short primary neurites in SALM4 is a visually distinctive phenotype as compared to the other SALMs. Thus, SALMs promote neurite outgrowth with various distinct phenotypes, and the major outgrowth parameters that SALMs 1–5 modify are total outgrowth and neurite branching.
Fig. 2. SALMs promote neurite outgrowth.
DIV4 primary hippocampal neurons were co-transfected with GFP and myc-SALM1, myc-SALM2, SALM3, myc-SALM4, HA-SALM5, or pcDNA 3.1+ vector (control). Immunostaining was performed 48 hours later, and neurite outgrowth was analyzed using Metamorph Neurite Outgrowth software (v7.0r3). Analysis was based on the transfected GFP signal. A, Representative examples of transfected neurons. B, Quantifications of neurite outgrowth: all five SALMs promote increases in total outgrowth, as compared to control, but do not promote increases in mean process length (C). The mean process length of myc-SALM4-transfected neurons is less than that of neurons transfected with myc-SALM1, SALM3, and HA-SALM5. D, All five SALMs promote increases in the number of branches, as compared to control. Additionally, myc-SALM4 promotes increases number of branches as compared to the other SALMs. E, Myc-SALM2 promotes increases in the number of processes, as compared to control, myc-SALM1, and HA-SALM5. Myc-SALM4 promotes increases in processes as compared to myc-SALM1, myc-SALM2, SALM3, HA-SALM5 (dotted lines) and control (n=15–17, values shown are mean +/− SEM, and analyzed by one-way ANOVA, * represents significance with respect to control, # represents significance with respect to other conditions, p<0.05). Scale bar, 100 µm.
SALMs promote axonal and dendritic outgrowth
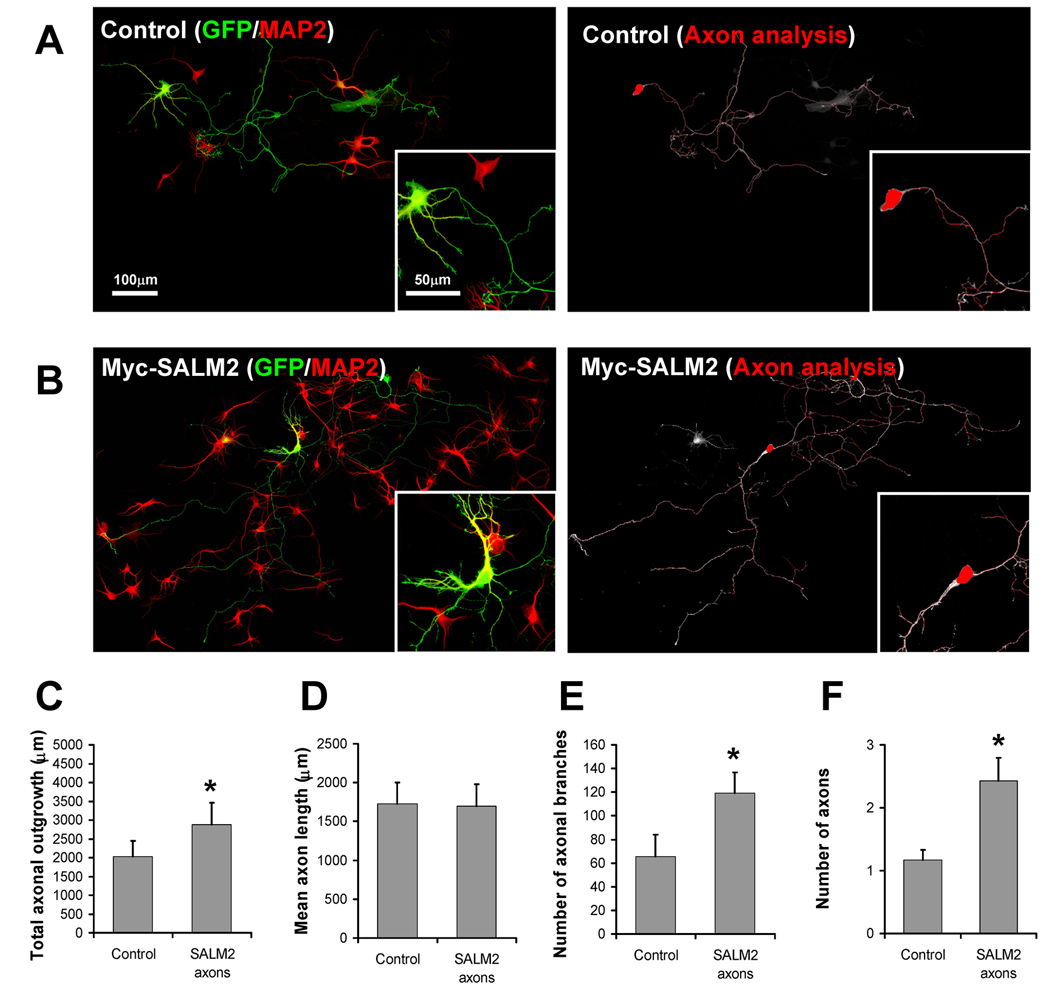
The increase in the number of MAP2-positive processes extending from the cell body, particularly evident with SALM4, indicates a change in dendrite growth. In order to determine if the SALM-mediated increases in number of processes were axonal as well, we co-transfected DIV4 neurons with SALM2 and GFP, immunostained for MAP2, and imaged the cells as described earlier. Since more than 90% of all GFP-transfected cells also expressed SALM2 (data not shown), we used GFP expression as a positive indicator of SALM2 transfection in these experiments. Along with MAP2 staining, the transfected cells were analyzed through the morphological criteria described in the methods to distinguish axons from dendrites (Fig. 3A and 3B, top panels). Once axons and dendrites were identified, the dendrites were digitally removed from the cell and the resulting axon-only image was analyzed for neurite outgrowth (Fig. 3A and 3B: bottom panels). Our results showed a 75% increase in total axonal outgrowth (3543 ± 371.42 µm for SALM2, 2029 ± 419.03 for control) (Fig. 3C), 108% increase in axon number (2.43 ± 0.37 axons for SALM2, 1.17 ± 0.17 for control) (Fig. 3F), and an 82% increase of axonal branches for SALM2 (119 ± 17.57 branches for SALM2, 65 ± 18.37 for control) (Fig. 3E), as compared to control axons, showing that axon growth is also affected by SALM expression. Analysis of the non-axonal processes also showed increases in outgrowth with SALM2 expression. These results showed that the SALMs affect both axon and dendrite outgrowth, although the effects may vary with individual SALMs.
Fig. 3. SALM2 promotes axonal outgrowth.
DIV4 primary hippocampal neurons were co-transfected with GFP and myc-SALM2 or pcDNA 3.1+ empty vector (control). Representative examples of control and myc-SALM2 transfected cells used for analysis are shown in A and B, respectively. Axons and dendrites were identified by morphological criteria and the dendritic marker MAP2 (A and B, left). Dendrites were digitally separated from the cell body, and the resulting axon-only images were analyzed for neurite outgrowth (A and B, right). Scale bars for composite images and insets 100 µm and 50 µm, respectively. Myc-SALM2 promotes an increase in total axonal outgrowth (C), number of axonal branches (E), and number of axons (F), as compared to controls. D, Myc-SALM2 does not promote increases in mean axon length (n=7, values shown are mean +/− SEM, and analyzed by unpaired students t-test, *p<0.05).
Application of antibodies directed to the extracellular LRR region of SALMs inhibits neurite outgrowth
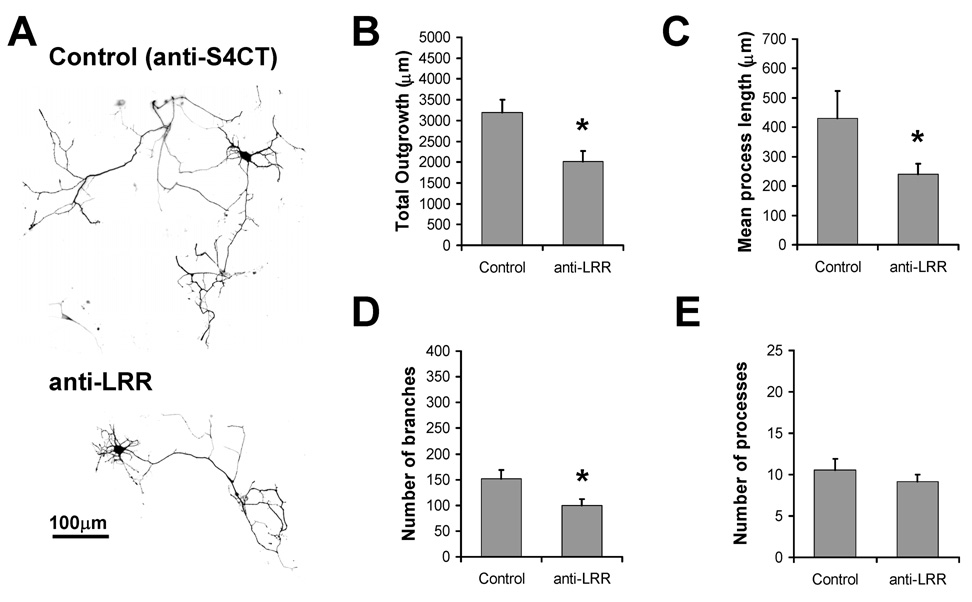
While over-expression experiments suggest a role for SALMs in neurite outgrowth, they do not address a role for endogenous SALMs. Previous studies have demonstrated the efficacy of applying antibodies directed to extracellular domains of endogenous transmembrane proteins, including cell adhesion molecules like cadherin, L1-CAM, and NCAM, to block their activity (Garcia-Castro et al., 2000; Lindner et al., 1983; Muller et al., 1996; Tang et al., 1998). The extracellular domains of SALMs contain several functional regions of potential protein-protein interaction, including six highly conserved LRR regions, a FN3 domain, and an IgC2-like domain. Recently, Seabold et al. (2008) showed that application of polyclonal antibodies that bind to the LRR of SALMs (anti-LRR) inhibited SALM4 trans interactions in transfected heterologous cells. Directed to amino acids 38–297 of SALM2 (a region of high similarity among the SALMs), anti-LRR has been shown to interact with the extracellular domains of all SALMs expressed in heterologous cells (Seabold et al., 2008). To test the ability of anti-LRR application to inhibit SALM-mediated outgrowth, DIV4 primary hippocampal neurons were transfected with GFP and treated by bath application with anti-LRR. To control for variability due to the antibody application technique, parallel cultures were treated with antibodies directed to the intracellular C-terminus of SALM4 (anti-S4CT). Neurons were fixed and analyzed for neurite outgrowth 48 hours after antibody application. Representative examples of treated neurons are shown in Fig. 4A. Anti-LRR-treated neurons showed a significant reduction in total outgrowth, mean process length, and neurite branches (Fig. 4B, C, and D). There was no significant change in the number of processes (Fig. 4E). While these results are in accordance with increases in neurite outgrowth and branches promoted by the over-expression of SALMs, we cannot rule out the possibility that anti-LRR antibodies interact with a variety of other endogenous proteins present on the surface of the neurons which have LRR domains, and that these interactions contribute to the changes in neurite outgrowth.
Fig. 4. Application of antibodies directed to the extracellular LRR region of SALMs inhibits neurite outgrowth.
DIV4 primary hippocampal neurons were transfected with GFP and treated by bath application with antibodies directed to the LRR region of SALM2 (anti-LRR) or to the C-terminus of SALM4 (anti-S4CT), as control. Immunostaining was performed 48 hours later, and neurite outgrowth was analyzed using Metamorph. A, Representative examples of transfected/treated neurons. Neurons treated with anti-LRR showed a significant decrease in total outgrowth (B), mean process length (C) and number of branches (D), as compared to the control condition. Anti-LRR treatment did not have an effect on the number of processes (E). (n=15, values shown are mean +/− SEM, and analyzed by unpaired students t-test, *p<0.05. Scale bar, 100 µm.)
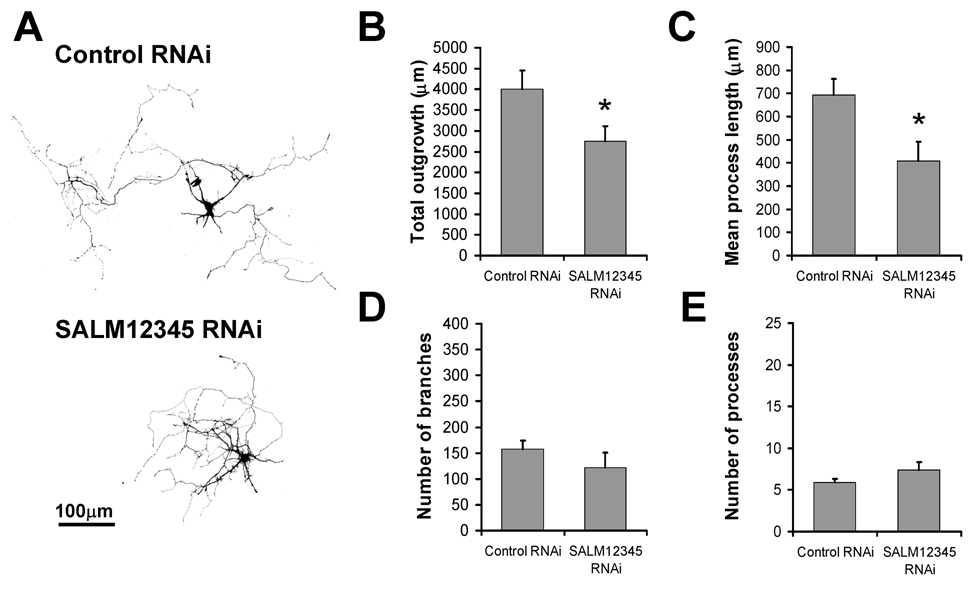
RNAi knock-down of SALM expression reduces neurite outgrowth
To further examine the function of SALMs in mediating neurite outgrowth, individual RNAi plasmid constructs were generated for SALMs 1–5 using pcDNA™6.2-GW/EmGFP-miR micro RNAi expression vectors, which co-express GFP. To test the efficacy of these RNAi constructs, each one was co-transfected with its respective SALM cDNA into neurons at DIV2. Over-expressed SALM levels were analyzed at DIV6 by fluorescence immunocytochemistry of transfected neurons and compared to those transfected with control RNAi. Co-transfection of each RNAi construct with the respective SALM cDNA resulted in significant reduction of SALM over-expression (supplemental Fig. 2), with decreases in SALM over-expression of 28%, 75%, 60%, 61%, and 43% for SALMs 1–5, respectively.
We initially transfected neurons with individual SALM RNAi constructs and measured neurite outgrowth. While there was a trend indicating a decrease in outgrowth and process length with the individual RNAi constructs (data not shown), the changes were not statistically significant. Since all SALMs promote neurite outgrowth and multiple SALMs are likely co-expressed in neurons (Morimura et al., 2006), loss of one SALM protein may have only a slight effect on outgrowth. Therefore, we examined the effects of knocking down all five SALMs on neurite outgrowth. DIV2 neurons were quintuple transfected with SALMs 1–5 RNAi constructs, and analyzed DIV6. GFP-positive RNAi-transfected cells showed a decrease in total outgrowth and mean process length (Fig. 5B and C, respectively), with no change in the number of branches or processes (Fig. 5D and E, respectively). These results indicate that cumulative knockdown of endogenous SALMs 1–5 expression effectively inhibits neurite outgrowth.
Fig. 5. RNAi knock-down of SALM expression reduces neurite outgrowth.
Individual RNAi plasmid constructs were designed and generated for SALMs 1–5 (Invitrogen custom services). To knockdown the expression of endogenous SALM proteins, DIV2 neurons were quintuple transfected with SALMs 1–5 RNAi constructs (SALM12345 RNAi), and analyzed four days later (DIV6). A, Representative examples of transfected cells. GFP-positive RNAi-transfected cells showed a significant decrease in total outgrowth and mean process length (B and C, respectively). The number of branches and processes were not significantly different, compared to control (D and E, respectively). (n=10, values shown are mean +/− SEM, and analyzed by unpaired students t-test, *p<0.05. Scale bar, 100 µm.)
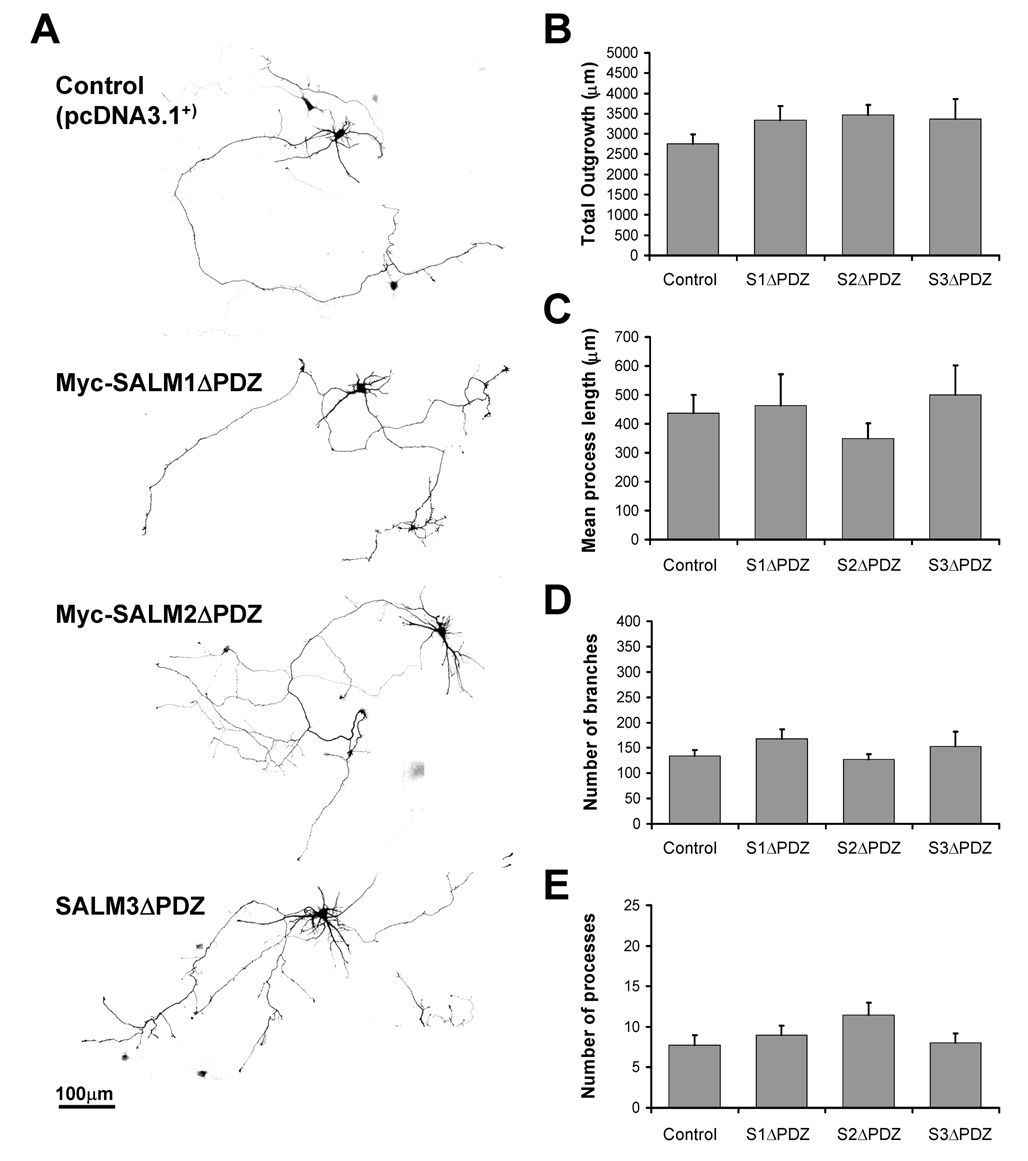
Neurite outgrowth is mediated by the PDZ binding domains of SALMs
PDZ interactions between CAMs and PDZ domain-containing proteins are critical for a variety of mechanisms, including synapse formation and regulation, as well as neurite outgrowth (for review, see Dalva et al., 2007). For example, neurexin and neuroligin complexes regulate synapse formation through direct interactions with the PDZ proteins PSD-95 and CASK (Irie et al., 1997) and have been implicated in mediating neurite outgrowth (Grifman et al., 1998). SALMs 4 and 5 differ from SALMs 1–3 in that they do not have a PDZ-BD, and they have visually distinctive phenotypes that may be related to the lack of PDZ interactions (SALM4 promotes a dramatic increase in the number of short primary neurites, and SALM5 often induces primary neurites to overlap and apparently adhere to each other). To examine the roles of PDZ interactions in SALM-mediated neurite outgrowth, we generated mutant cDNA constructs of SALMs 1, 2, and 3 in which the distal C-terminal 4 (for SALMs 1 and 3) or 7 (for SALM2) amino acids corresponding to the PDZ-BD were deleted (SALM1ΔPDZ, SALM2ΔPDZ, and SALM3ΔPDZ, respectively). The SALMΔPDZ constructs were individually co-transfected with GFP into primary neurons at DIV4 and analyzed for neurite outgrowth. Deleting the PDZ-BD greatly attenuated the neurite outgrowth facilitated by the exogenous full-length SALMs 1–3. As seen in Fig. 6B–E, the amounts of total outgrowth, mean process length, neurite branches, and number of processes were all similar to that of control. These results indicate that the PDZ-BD functions in neurite branching, which was the primary parameter of outgrowth seen by transfecting the full-length SALMs. Thus, the mechanism of SALM-mediated neurite outgrowth for this subset of SALMs likely involves interactions with PDZ domain-containing proteins.
Fig. 6. Neurite outgrowth is mediated by the PDZ domains of SALMs.
Primary hippocampal neurons (DIV4) were co-transfected with GFP and myc-SALM1ΔPDZ, myc-SALM2ΔPDZ, SALM3ΔPDZ, or pcDNA 3.1+ vector (control). Immunostaining was performed 48 hours later, and neurite outgrowth was analyzed using Metamorph. A, Representative examples of transfected neurons. Transfection of myc-SALM1ΔPDZ, myc-SALM2ΔPDZ, or SALM3ΔPDZ do not promote increases in total outgrowth (B), mean process length (C), number of branches (D), or the number of processes (E). (n=10, values shown are mean +/− SEM, and analyzed by one-way ANOVA, *p<0.05. Scale bar, 100 µm.)
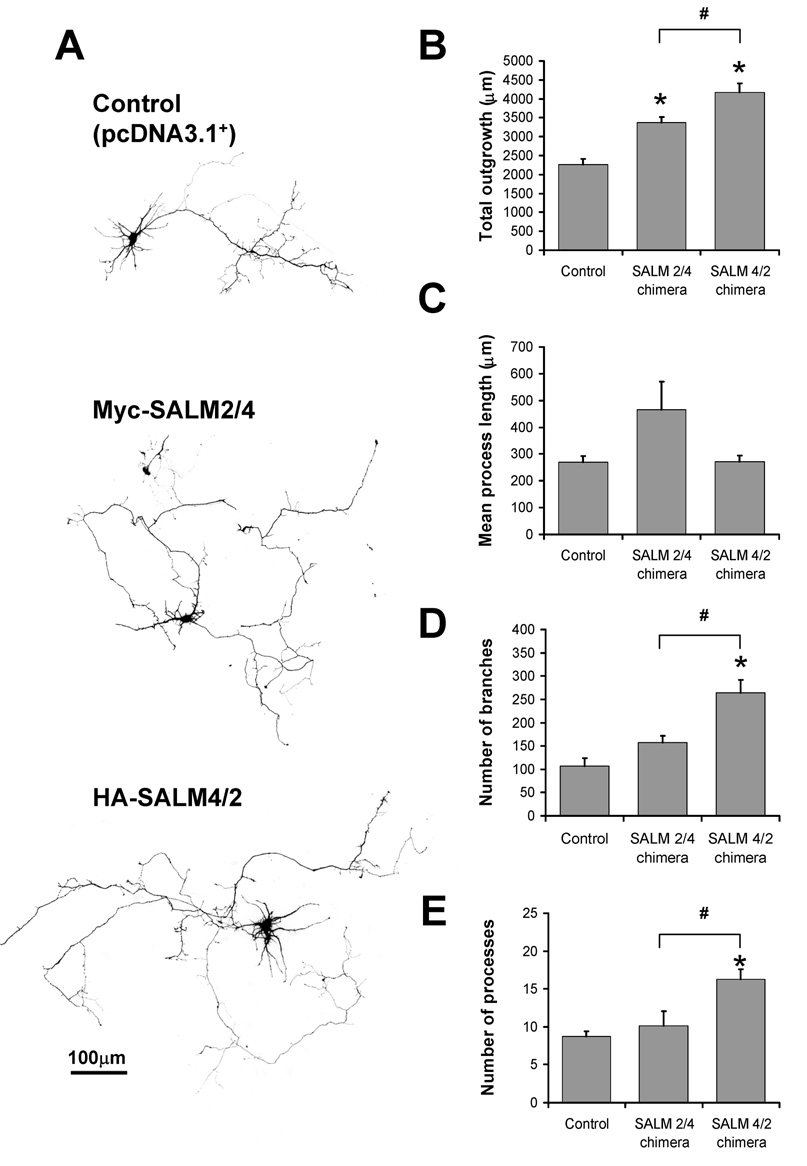
Neurite outgrowth phenotype is determined by both the N-and C-termini
The individual SALMs display distinct phenotypes in promoting neurite outgrowth. The most visually apparent and unique phenotype is that of SALM4, which promotes dramatic increases in the number of short, primary processes extending from the cell body. By examining and replacing the domains of SALM4, we can investigate which domains are involved in regulating various facets of neurite outgrowth. The N-termini of the SALMs contain various protein-protein interaction domains that are highly homologous among the family members, while the C-termini show considerable sequence variability among the family members. Additionally, SALMs 1–3 have C-terminal PDZ-BDs that contribute to neurite outgrowth, while SALMs 4–5 do not. Therefore, it is conceivable that either the N- or C-terminal regions, or both, may contribute various functions in the mechanism of SALM-mediated neurite outgrowth, and that these functions may be unique to regions of the individual SALMs. To examine the domains involved in neurite outgrowth, we generated chimera constructs with the N-termini and C-termini of SALM2 and SALM4. Myc-SALM2/4 contains the N-terminus of SALM2 and the C-terminus of SALM4, while HA-SALM4/2 contains the N-terminus of SALM4 and the C-terminus of SALM2.
When transfected into DIV4 cultures, both chimera constructs promoted a significant increase in total outgrowth (Fig. 7B), which is consistent with both SALM2 and SALM4 enhancing outgrowth (Fig 2). However, the most apparent phenotype was that of HA-SALM4/2-transfected cells, as they additionally promoted an increase in total outgrowth compared to Myc-SALM2/4 (Fig 7B). HA-SALM4/2 also promoted an increase in process number, as compared to both control and myc-SALM2/4, similar to the phenotype seen with transfected SALM4 (Fig 2). HA-SALM4/2 promoted a statistically significant increase in branching, as compared to control and myc-SALM2/4 (Fig. 7D). Increases in mean process length were not statistically significant (Fig 7C), as was also seen for both SALM2 and SALM4 (Fig 2).
Fig. 7. Neurite outgrowth phenotype is determined by both the N-and C-termini of SALMs.
We generated SALM2/SALM4 chimera constructs in which the N-termini and C-termini of SALM2 and SALM4 were switched. Myc-SALM2/4 contains the N-terminus of SALM2 and the C-terminus of SALM4, while HA-SALM4/2 contains the N-terminus of SALM4 and the C-terminus of SALM2. Chimera constructs were individually co-transfected with GFP at DIV4 and analyzed 48 hours later. A, Representative examples of pcDNA3.1+ (control) and chimera-transfected cells. Both Myc-SALM2/4 and HA-SALM4/2 exhibited increases in total outgrowth (B), but not mean process length (C), as compared to control. HA-SALM4/2 promoted increases in number of branches (D) and process number (E), as compared to both control and Myc-SALM2/4. The HA-SALM4/2-mediated increase in process number is similar in phenotype to the increase seen with SALM4, indicating that the N-terminus of SALM4 confers this property (n=10, values shown are mean +/− SEM, and analyzed by one-way ANOVA, * represents significance with respect to control, # represents significance with respect to other conditions, p<0.05. Scale bar, 100 µm).
These results suggest that both the N and C termini play a role in neurite outgrowth. The most prominent result was that SALM4/2 chimera showed a significant increase in the number of processes, similar to that of SALM4, suggesting that the N-terminus plays a role in outgrowth. Total outgrowth was increased for both chimeras, which is similar to results seen with the over-expression of SALM2 and SALM4, as both increase outgrowth. However, SALM2ΔPDZ over-expression did not increase outgrowth (Fig 6), and the SALM2/4 chimera also lacks the PDZ-BD found in SALM2. This result may suggest that the SALM4 C-terminus contains another domain with a function similar to that of the PDZ-BD. On the other hand, while both SALM2 and SALM4 increased branching, only the SALM4/2 chimera increased branching. This supports our finding that the PDZ-BD of SALM2 is required for branching. Therefore, the mechanism of SALM-mediated outgrowth may involve an intricate interplay between multiple functional regions on each individual molecule, including the N-terminal domains, the C-terminus (including the PDZ-binding domains for SALMs 1–3), and conserved regions common to the various SALMs.
Discussion
Here we show that all five SALMs promote neurite outgrowth when over-expressed in young primary hippocampal cultures, with various phenotypes. Application of antibodies directed to the extracellular domain, or knockdown of endogenous SALMs using RNAi in hippocampal cultures, inhibits neurite outgrowth. The nature of the phenotype from over-expression is determined largely by the extracellular domain, although the PDZ-BD is required for most aspects of neurite outgrowth of SALMs 1–3. SALMs 4 and 5, which lack the PDZ-BD, also effectively promote neurite outgrowth, suggesting that their intracellular C-termini either function somewhat differently from SALMs 1–3 or that they have alternative interacting mechanisms to substitute for the PDZ interaction.
The extracellular domain structure of the SALMs is homologous with that of a variety of related proteins that regulate neurite outgrowth, including AMIGO, NGL, LINGO, FLRT, NLRR, and PAL protein families (Chen et al., 2006). Similar to the SALMs, these proteins contain extracellular LRRs and variations in the presence and number of Ig-like and FN3 domains. Like SALMs, the AMIGO family members contain six LRRs and promote neurite outgrowth in hippocampal neurons via their extracellular region (Kuja-Panula et al., 2003). In contrast, LINGO proteins contain twelve LRRs and negatively regulate neurite outgrowth and regeneration in brain and spinal cord by enhancing myelin-mediated inhibition and participating in the Nogo/OMgp/MAG-NgR signaling pathway (Mi et al., 2004). Therefore, the SALMs join a list of proteins with a general domain structure (X number of LRR +/− IgC2 +/− FN3) that regulate neurite outgrowth. However, the nature of the outgrowth is variable, likely reflecting specific combinations of the protein-interaction domains, interactions with extracellular binding partners, and intracellular signaling mechanisms.
The extracellular N-termini of SALMs are involved in neurite outgrowth
Several factors could play a role in the differential effects of SALM over-expression on neurite outgrowth. For example, a different distribution of the SALMs within the neuron may lead to selective outgrowth of axons or dendrites. To address this question, we expressed epitope-tagged constructs, and found that all five SALMs are distributed throughout the neuron, including in the growth cone. These results indicate that distribution alone is unlikely to be responsible for the different effects of the individual SALMs.
Alternatively, the differential outgrowth effects of the SALMs could be due to the different properties of interactions with other proteins through their extracellular domains. Our results with the SALM2 and SALM4 chimeras show that the N-terminal regions of SALMs play a major role in regulating neurite outgrowth. This is not unexpected since the extracellular regions of related proteins, such as AMIGO (Kuja-Panula et al., 2003), have been shown to mediate neurite outgrowth. While the N-termini are highly conserved among the SALMs, there are regions of high variability. Additionally, sequence analysis reveals that there is a potential N-linked glycosylation site in the first LRR region of SALMs 1–3, but not SALMs 4 and 5 (data not shown). Glycosylation is a highly regulated post-translational modification which influences a wide variety of functional consequences including cell adhesion, signal transduction, and the trafficking of proteins to the cell surface (for review, see Scheiffele and Fullekrug, 2000).
Our findings showing that the N-termini play a role in neurite outgrowth suggest that the N-termini of individual SALMs may interact with different binding partners or with the same binding partners, but with different affinities. The mechanisms by which the SALMs may interact or signal through extracellular domains has not been extensively studied. However, Seabold et al. (2008) recently reported that certain SALMs form trans associations in heterologous cells. SALMs 4 and 5 form homomeric, but not heteromeric, trans associations. Therefore, these interactions may play a role in determining SALM 4 and 5 neurite outgrowth phenotypes. Multiple mechanisms remain to be investigated for the SALMs. In addition to direct interactions through extracellular domains of molecules on the cell surface, secreted and cleaved fragments of adhesion molecules can function in neurite outgrowth. For example, NCAM is proteolytically cleaved by the metalloproteases TACE, and the inhibition of this cleavage inhibits NCAM-mediated neurite outgrowth (Kalus et al., 2006). A similar mechanism may be in play for the SALMs.
SALM-mediated neurite outgrowth involves PDZ domain interactions
A distinguishing feature of the SALMs is the presence of a PDZ-BD at the C-terminus of SALMs 1–3. PDZ interactions mediate a variety of essential processes in the CNS, including the clustering and trafficking of ion channels and facilitation of signaling at the synapse (El-Husseini et al., 2000; Kim and Sheng, 2004). PDZ proteins are also involved in neurite outgrowth (Charych et al., 2006; Hoogenraad et al., 2005). GRIP1, an AMPA receptor-associated multi-PDZ domain protein, mediates dendritic formation and outgrowth by regulating EphB receptor trafficking (Hoogenraad et al., 2005). Firestein and colleagues have shown that PSD-95 regulates dendritic branching in an activity-independent manner (Charych et al., 2006). Overexpression of PSD-95 decreases dendritic branching in immature neurons, while knocking down PSD-95 increases it (Charych et al., 2006). Previous studies (Ko et al., 2006; Wang et al., 2006) showed that SALMs 1 and 2 interact with PSD-95 and other MAGUKs (membrane associated guanylate kinases), and that this interaction is required to recruit PSD-95 to presumed synaptic locations. Therefore, it is possible that interactions between SALMs and MAGUKs may be important for the proper regulation of early neuronal development. Our results show that the PDZ-BDs (in SALMs 1–3) and the N-termini of the SALMs are involved in neurite outgrowth. Other adhesion molecules mediate outgrowth-related mechanisms through PDZ interactions. NrCAM (an L1-CAM) binds to MAGUKs and this association regulates trafficking to the membrane (Davey et al., 2005; Dirks et al., 2006). Neuroligin-1 (NGL) contains LRR and Ig-like regions and also has been implicated in regulating neurite outgrowth (Grifman et al., 1998). The synaptic localization of NLG is mediated by PDZ-interactions (Irie et al., 1997), and NLG regulates the balance of excitatory and inhibitory synaptic contacts through interactions with the third PDZ domain of PSD-95 (Song et al., 1999). Previous studies (Ko et al., 2006; Wang et al., 2006) showed that SALMs 1 and 2 interact with members of the MAGUKs and that this interaction is required to recruit PSD-95 to presumed synaptic locations.
The PDZ-BD has significantly different roles in trafficking of the individual SALMs; in heterologous cells and neurons, SALM1ΔPDZ is excluded from the cell surface (Wang et al., 2006). SALM2ΔPDZ, however, is expressed on the cell surface throughout the neuron (data not shown). The requirement of the PDZ-BD for normal function of SALMs 1–3 suggests that either the PDZ interaction is required for the clustering of the SALM molecule, or that the PDZ protein is required to organize the SALM C-termini with other molecules associated with signaling. The slight, but not statistically significant, increase in total outgrowth of SALMs 1–3-ΔPDZ, would be consistent with the interpretation that the PDZ interaction is required for efficient coupling to intracellular signaling cascades; without it, only a minor effect remained, likely due to random associations. Therefore, SALMs may mediate neurite outgrowth through PDZ-mediated second messenger signaling cascades, which are disrupted in these mutant constructs. Among these, MAPK cascades and FGFR cascades at growth cones have been characterized in mediating neurite outgrowth in response to stimuli from CAMs such as L1, N-cadherin, and NCAM (Doherty et al., 2000). SALMs 4 and 5 do not contain PDZ-BDs, but over-expression still effectively promotes neurite outgrowth. A likely explanation is that they contain another motif(s) in their C-termini that substitute for the PDZ-BD, but these remain to be identified.
Dual functions for SALMs
The results of the present study, which indicate that all SALMs can enhance neurite outgrowth, agrees with previous studies showing that SALM1 helps recruit proteins to the synapse (Wang et al., 2006) and that SALM2 plays a role in determining the number of excitatory synapses (Ko et al., 2006). Together these studies suggest that SALMs participate in at least two functions, neurite outgrowth and synapse formation. Although not yet demonstrated functionally, their presence in mature synapses suggests a role in synapse maintenance in adults. This dual role is not without precedent among adhesion molecules, as the cadherins are critical to the development of the CNS and have roles in neurite outgrowth and synapse formation (for review, see Redies, 2000). N-cadherin mediates calcium-dependent cell adhesion and has been demonstrated to promote neurite outgrowth through various methods including over-expression studies (Matsunaga et al., 1988), function-blocking antibodies (Bixby et al., 1987), and as substrates for neuronal culture (Bixby and Zhang, 1990). N-cadherins also localize to growth cones, and mediate growth cone migration (Letourneau et al., 1990). In mature neurons, N-cadherins are components of NMDA receptor complexes (Husi et al., 2000), and function in synaptic plasticity and long term potentiation (LTP) (Tang et al., 1998), and in the formation and maintenance of synapses (Fannon and Colman, 1996). L1-CAMs also are involved in various mechanisms during development including, neurite outgrowth, growth cone motility, and axonal fasciculation (Hortsch, 2000; Kamiguchi and Lemmon, 1997, 1998). L1-CAMs mediate neurite outgrowth in developing neurons through direct interactions with ankrynB (Nishimura et al., 2003), and via MAP kinase signaling (Whittard et al., 2006). L1-CAMs are also important in LTP, synapse alignment, and synapse formation (Godenschwege et al., 2006; Triana-Baltzer et al., 2006). This suggests that some of the processes involved in synapse formation and neurite outgrowth are shared, but raises questions about how these two processes are differentially regulated. For example, what determines the proportions of SALMs dedicated to synapse formation compared to neurite outgrowth, or what developmental change signals the switch in functions. We initially hypothesized that the PDZ interaction would not be relevant to neurite outgrowth, but our results showed that the PDZ-BD is required for neurite outgrowth. Therefore, the PDZ-BD cannot be used to differentiate between the two processes, unless different PDZ interactions regulate outgrowth earlier in development, while others regulate synapse formation.
Experimental Methods
Antibodies
Anti-Myc monoclonal (hybridoma purchased from ATCC, Manassas, VA, clone #9E10) and anti-hemagglutinin (HA) monoclonal (Covance, Denver, PA, clone #16B12) primary antibodies were used at a 1:1000 dilution. Anti-GFP polyclonal (Chemicon, Billerica, Massachusetts) was used at 1:2000. SALM antisera were produced in rabbits (Covance) as described in Seabold et al. (2008). Briefly, for detection of transfected SALM3, polyclonal antibodies were generated using a peptide directed to the C-terminus of SALM3, (amino acids 621–636: [NH2]-CRGVGGSAERLEESVV-[COOH]). For detection of transfected SALM3ΔPDZ, polyclonal antibodies were generated using a peptide directed to the N-terminus of SALM3 (amino acids 377–389: [NH2]-TSAEGGRPGPSDI-[COOH]. For function-blocking antibody experiments, an antibody that recognizes the LRR regions of the SALMs was generated by expressing the SALM2 LRR region (residues 38–297) as a glutathione S-transferase (GST) fusion protein, and produced in rabbits (Covance).
cDNA constructs
Cloning and epitope-tagging of SALM cDNA constructs was conducted as previously described (Seabold et al., 2008; Wang et al., 2006). The Myc tag was inserted into the sequence of SALM1 before residue 21, the SALM2 Myc tag was inserted before residue 33, the SALM4 Myc tag was inserted before residue 24, and the SALM5 HA-tag was inserted before residue 18. Non-tagged SALM3 cDNA was utilized in our experiments due to difficulties in detecting epitope-tagged SALM3 through immunocytochemistry.
To generate SALM2 and SALM3 constructs lacking the PDZ-BD, stop codons were introduced by site-directed mutagenesis (Stratagene, La Jolla, CA) at W759 for SALM2(Δ7) and E633 for SALM3(Δ7). The GFP construct (pEGFP-N2) was purchased from Clontech. Chimeras of SALM2 and SALM4 were made by creating a BamHI site after the transmembrane domain of SALM2 and SALM4. A single amino acid was changed in the HA-SALM2 (D574S) and the Myc-SALM4 (G566S) constructs for subcloning purposes. The C-terminal region of each construct was then excised and exchanged by utilizing the BamHI and EcoRI sites from the multiple cloning site in the pcDNA3.1+ vector.
Primary hippocampal cultures and transfections
Primary hippocampal neuronal cultures were prepared as previously described (Sans et al., 2005; Wang et al., 2006). Briefly, embryonic day 18 (E18) hippocampi from Sprague-Dawley rats (Harlan, Indianapolis, IN) were dissected and dissociated with trypsin EDTA (Invitrogen, Carlsbad, California). Neurons were plated onto poly-ornithine/ fibronectin-coated coverslips in 2% fetal bovine serum/Neurobasal medium (Invitrogen) in six-well culture plates. Fifty percent of the culture media was changed to Neurobasal media plus B27 (Invitrogen) 72 hours after plating. All animal procedures were done in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (National Institutes of Health publication 85–23) under National Institute on Deafness and Other Communication Disorders protocol 1167-07. Transfections of DIV4 hippocampal cultures were performed using a calcium phosphate method (Clontech, Mountain View, CA) with modifications. Briefly, Neurobasal/B27 culture medium was replaced with 5 ml DMEM one hour prior to transfection. One to five micrograms of cDNA was mixed with 2 M calcium solution, and added to the same volume of 2X Hepes buffered saline. The plasmid cDNA/calcium solution was incubated for 20 min at room temperature, and then applied to the neurons for 20 min. Neurons were washed twice with DMEM and cultured in the original Neurobasal/B27 medium at 37°C, 5% CO2. Control cells were co-transfected with empty vector (pcDNA 3.1+) and GFP (Green Fluorescent Protein construct) in each experimental group. Forty-eight hours after transfection (DIV6), neurons were fixed and processed for immunostaining. For neurite outgrowth experiments using RNAi plasmids, neurons were transfected at DIV2 and fixed for immunocytochemistry at DIV6 to allow sufficient time for knockdown of endogenous proteins.
Immunocytochemistry
Immunocytochemistry was performed as previously described (Sans et al., 2005). Neurons were fixed with 4% paraformaldehyde (PFA), washed with phosphate buffered saline (PBS) and permeabilized with 0.25% Triton-X/PBS. Neurons were then blocked with 10% normal goat serum (NGS)/PBS/0.1% Triton X-100 for one hour, and then incubated with primary antibodies at room temperature in 3% NGS/PBS/0.1% Triton X-100 for one hour. Neurons were washed and incubated with AlexaFluor 488 or 555 secondary antibodies (Molecular Probes, Carlsbad, California) for 30 minutes, washed and mounted on slides using Prolong Antifade Gold (Invitrogen). For surface staining experiments, live neurons were incubated with primary antibodies for 30 minutes at 4 °C, fixed with 4% PFA for 20 minutes, blocked with 10% NGS/PBS, and incubated with secondary antibodies for 30 minutes.
RNAi constructs
RNAi constructs were designed and synthesized using the Invitrogen miR RNAi custom synthesis service. Double-stranded oligos were synthesized and inserted into pcDNA6.2GW/EmGFP-miR vector, which utilizes an shRNA designed to have an RNAi effect in the context of micro RNA (miRNA) expression. The sequence region targeted for RNAi in SALM1 was 5’-TGC TGT ACT GAA GTC CAT CAA CTC ATG TTT TGG CCA CTG ACT GAC ATG AGT TGG GAC TTC AGTA. The sequence region targeted for SALM2 was 5’-TGC TGA GAA ATA GCG GTC AGT GAG ATG TTT TGG CCA CTG ACT GAC ATC TCA CTC CGC TAT TTCT. The sequence region targeted for SALM3 was 5’-TGC TGA AGA GTT GCC AAC CAG CCT GTG TTT TGG CCA CTG ACT GAC ACA GGC TGT GGC AAC TCTT. The sequence region targeted for SALM4 was 5’-TGC TGA AAT GCA GCA GGC CTG TCA TGG TTT TGG CCA CTG ACT GAC CAT GAC AGC TGC TGC ATTT, and the region targeted for SALM5 was 5’-TGC TGA AAT CTG ACA GAC ACA ACG CTG TTT TGG CCA CTG ACT GAC AGC GTT GTC TGT CAG ATTT. Control cells were transfected with RNAi plasmids containing an insert predicted to not target any known human, mouse, or rat gene (pcDNA 6.2-GW/EmGFP–miR-neg2): TGC TGT ATT GCG TCT GTA CAC TCA CCG TTT TGG CCA CTG ACT GAC GGT GAG TGC AGA CGC AATA.
Image acquisition and neurite outgrowth analysis
Images were taken using a Nikon E1000M microscope equipped with a CCD camera using a Plan Fluor 20x (0.5 NA) dry or Plan Apo 60x (1.4 NA) oil-immersion objective. For neurite outgrowth experiments, only transfected neurons without overlapping neurites from adjacent GFP-transfected cells were selected for analysis. All experiments were performed in triplicate. Partial images were systematically acquired for sections of each cell using the 20x objective until the entire cell was imaged. Partial images were digitally overlaid into a single composite image using Adobe Photoshop 7.0 (Adobe, San Jose, CA). Reconstructed images exhibiting the cell in its entirety were analyzed using Metamorph Neurite Outgrowth Module v7.0r3 (Molecular Devices, Sunnyvale, CA). If visual inspection of the resulting trace was incorrect, settings for each cell were individually adjusted until the resulting trace accurately represented that of the neuron. For SALM2 axonal measurements, dendrites were identified by MAP2 immunostaining and through morphological criteria. As described in Kaech and Banker (2006): dendrites were identified as processes that emerged gradually from the cell body, tapered with distance, had a radial orientation, and had a length of 200–300 µm. Axons were identified as processes that were are thinner at their origin, exhibited less taper, often turned at 90 degree angles, and extended over millimeters. Dendrites from reconstructed images were digitally severed from the cell body using Adobe Photoshop 7.0, and the resulting images were analyzed using Metamorph. Statistical analysis was performed using GraphPad Prism 4.0 (GraphPad Software Inc., San Diego, CA). Statistical significance between two groups were determined with a two-tailed, paired Student's t-test, while significance among multiple groups was determined with an analysis of variance (ANOVA), followed by the post-hoc Newman-Keuls multiple-comparison test. Differences were considered significant at p < 0.05. All values are reported as mean ± SEM. For signal intensity studies, images were taken at consistent exposure times and contrast values, and imported to Metamorph for analysis of pixel intensity of transfected and non-transfected cells.
Supplementary Material
DIV4 primary hippocampal neurons were co-transfected with GFP and myc-SALM1, myc-SALM2, myc-SALM4, or HA-SALM5. Surface labeling with myc or HA antibodies was performed 48 hours after transfection. A, Representative examples of myc-SALM2 surface labeling are shown. Scale bar, 20 µm. B, SALM surface expression is punctate (arrows/inset), and accumulated at growth cones. Scale bar, 10 µm.
Individual RNAi plasmid constructs were designed and generated for SALMs 1–5 using pcDNA™6.2-GW/EmGFP-miR micro RNAi expression vectors, which co-expresses GFP. DIV4 primary hippocampal neurons were co-transfected individually with myc-SALM1, myc-SALM2, SALM3, myc-SALM4, or HA-SALM5 cDNA and their respective SALM RNAi plasmid (e.g. myc-SALM1cDNA and SALM1 RNAi), or scramble RNAi negative control (e.g. myc-SALM1 cDNA and scramble RNAi). Expression levels of transfected myc-SALM1 with scramble RNAi or SALM1 RNAi are shown in rows A1 and A2, respectively. Expression levels of SALM2-SALM5 with scramble RNAi (GFP) or SALM RNAi (GFP) are shown in rows B–E, respectively. Scale bar, 20 µm. For the characterization of each SALM RNAi construct, images were taken and processed with identical exposure times and contrast levels. Expression of transfected SALMs was reduced by 28%, 75%, 60%, 61%, and 43% for SALMs 1–5, respectively (n=5).
Acknowledgements
We are grateful to Dr. Kai Chang, Dr. Ya-Xian Wang, and Ms. Linna Ge for excellent technical assistance. We would like to thank Dr. Ronald Petralia, Dr. Rana Al-Hallaq, and members of the Wenthold lab for invaluable discussions and suggestions on the preparation of this manuscript. This work was supported by the National Institute on Deafness and Other Communication Disorders (NIDCD) Intramural Research Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bixby JL, Pratt RS, Lilien J, Reichardt LF. Neurite outgrowth on muscle cell surfaces involves extracellular matrix receptors as well as Ca2+-dependent and -independent cell adhesion molecules. Proc Natl Acad Sci U S A. 1987;84:2555–2559. doi: 10.1073/pnas.84.8.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bixby JL, Zhang R. Purified N-cadherin is a potent substrate for the rapid induction of neurite outgrowth. J Cell Biol. 1990;110:1253–1260. doi: 10.1083/jcb.110.4.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charych EI, Akum BF, Goldberg JS, Jornsten RJ, Rongo C, Zheng JQ, Firestein BL. Activity-independent regulation of dendrite patterning by postsynaptic density protein PSD-95. J Neurosci. 2006;26:10164–10176. doi: 10.1523/JNEUROSCI.2379-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Aulia S, Li L, Tang BL. AMIGO and friends: an emerging family of brain-enriched, neuronal growth modulating, type I transmembrane proteins with leucine-rich repeats (LRR) and cell adhesion molecule motifs. Brain Res Rev. 2006;51:265–274. doi: 10.1016/j.brainresrev.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Craig AM, Banker G. Neuronal polarity. Annu Rev Neurosci. 1994;17:267–310. doi: 10.1146/annurev.ne.17.030194.001411. [DOI] [PubMed] [Google Scholar]
- da Silva JS, Dotti CG. Breaking the neuronal sphere: regulation of the actin cytoskeleton in neuritogenesis. Nat Rev Neurosci. 2002;3:694–704. doi: 10.1038/nrn918. [DOI] [PubMed] [Google Scholar]
- Dalva MB, McClelland AC, Kayser MS. Cell adhesion molecules: signalling functions at the synapse. Nat Rev Neurosci. 2007;8:206–220. doi: 10.1038/nrn2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey F, Hill M, Falk J, Sans N, Gunn-Moore FJ. Synapse associated protein 102 is a novel binding partner to the cytoplasmic terminus of neurone-glial related cell adhesion molecule. J Neurochem. 2005;94:1243–1253. doi: 10.1111/j.1471-4159.2005.03271.x. [DOI] [PubMed] [Google Scholar]
- Dirks P, Thomas U, Montag D. The cytoplasmic domain of NrCAM binds to PDZ domains of synapse-associated proteins SAP90/PSD95 and SAP97. Eur J Neurosci. 2006;24:25–31. doi: 10.1111/j.1460-9568.2006.04899.x. [DOI] [PubMed] [Google Scholar]
- Doherty P, Williams G, Williams EJ. CAMs and axonal growth: a critical evaluation of the role of calcium and the MAPK cascade. Mol Cell Neurosci. 2000;16:283–295. doi: 10.1006/mcne.2000.0907. [DOI] [PubMed] [Google Scholar]
- Dotti CG, Sullivan CA, Banker GA. The establishment of polarity by hippocampal neurons in culture. J Neurosci. 1988;8:1454–1468. doi: 10.1523/JNEUROSCI.08-04-01454.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Husseini AE, Schnell E, Chetkovich DM, Nicoll RA, Bredt DS. PSD-95 involvement in maturation of excitatory synapses. Science. 2000;290:1364–1368. [PubMed] [Google Scholar]
- Fannon AM, Colman DR. A model for central synaptic junctional complex formation based on the differential adhesive specificities of the cadherins. Neuron. 1996;17:423–434. doi: 10.1016/s0896-6273(00)80175-0. [DOI] [PubMed] [Google Scholar]
- Francavilla C, Loeffler S, Piccini D, Kren A, Christofori G, Cavallaro U. Neural cell adhesion molecule regulates the cellular response to fibroblast growth factor. J Cell Sci. 2007;120:4388–4394. doi: 10.1242/jcs.010744. [DOI] [PubMed] [Google Scholar]
- Garcia-Castro MI, Vielmetter E, Bronner-Fraser M. N-Cadherin, a cell adhesion molecule involved in establishment of embryonic left-right asymmetry. Science. 2000;288:1047–1051. doi: 10.1126/science.288.5468.1047. [DOI] [PubMed] [Google Scholar]
- Godenschwege TA, Kristiansen LV, Uthaman SB, Hortsch M, Murphey RK. A conserved role for Drosophila Neuroglian and human L1-CAM in central-synapse formation. Curr Biol. 2006;16:12–23. doi: 10.1016/j.cub.2005.11.062. [DOI] [PubMed] [Google Scholar]
- Grifman M, Galyam N, Seidman S, Soreq H. Functional redundancy of acetylcholinesterase and neuroligin in mammalian neuritogenesis. Proc Natl Acad Sci U S A. 1998;95:13935–13940. doi: 10.1073/pnas.95.23.13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenraad CC, Milstein AD, Ethell IM, Henkemeyer M, Sheng M. GRIP1 controls dendrite morphogenesis by regulating EphB receptor trafficking. Nat Neurosci. 2005;8:906–915. doi: 10.1038/nn1487. [DOI] [PubMed] [Google Scholar]
- Hortsch M. Structural and functional evolution of the L1 family: are four adhesion molecules better than one? Mol Cell Neurosci. 2000;15:1–10. doi: 10.1006/mcne.1999.0809. [DOI] [PubMed] [Google Scholar]
- Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci. 2000;6:661–669. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- Irie M, Hata Y, Takeuchi M, Ichtchenko K, Toyoda A, Hirao K, Takai Y, Rosahl TW, Sudhof TC. Binding of neuroligins to PSD-95. Science. 1997;277:1511–1515. doi: 10.1126/science.277.5331.1511. [DOI] [PubMed] [Google Scholar]
- Kaech S, Banker G. Culturing hippocampal neurons. Nat Protoc. 2006;1:2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- Kalus I, Bormann U, Mzoughi M, Schachner M, Kleene R. Proteolytic cleavage of the neural cell adhesion molecule by ADAM17/TACE is involved in neurite outgrowth. J Neurochem. 2006;98:78–88. doi: 10.1111/j.1471-4159.2006.03847.x. [DOI] [PubMed] [Google Scholar]
- Kamiguchi H, Lemmon V. Neural cell adhesion molecule L1: signaling pathways and growth cone motility. J Neurosci Res. 1997;49:1–8. doi: 10.1002/(sici)1097-4547(19970701)49:1<1::aid-jnr1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Kamiguchi H, Lemmon V. A neuronal form of the cell adhesion molecule L1 contains a tyrosine-based signal required for sorting to the axonal growth cone. J Neurosci. 1998;18:3749–3756. doi: 10.1523/JNEUROSCI.18-10-03749.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- Ko J, Kim S, Chung HS, Kim K, Han K, Kim H, Jun H, Kaang BK, Kim E. SALM synaptic cell adhesion-like molecules regulate the differentiation of excitatory synapses. Neuron. 2006;50:233–245. doi: 10.1016/j.neuron.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Kuja-Panula J, Kiiltomaki M, Yamashiro T, Rouhiainen A, Rauvala H. AMIGO, a transmembrane protein implicated in axon tract development, defines a novel protein family with leucine-rich repeats. J Cell Biol. 2003;160:963–973. doi: 10.1083/jcb.200209074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letourneau PC, Shattuck TA, Roche FK, Takeichi M, Lemmon V. Nerve growth cone migration onto Schwann cells involves the calcium-dependent adhesion molecule, N-cadherin. Dev Biol. 1990;138:430–442. doi: 10.1016/0012-1606(90)90209-2. [DOI] [PubMed] [Google Scholar]
- Lindner J, Rathjen FG, Schachner M. L1 mono- and polyclonal antibodies modify cell migration in early postnatal mouse cerebellum. Nature. 1983;305:427–430. doi: 10.1038/305427a0. [DOI] [PubMed] [Google Scholar]
- Matsunaga M, Hatta K, Nagafuchi A, Takeichi M. Guidance of optic nerve fibres by N-cadherin adhesion molecules. Nature. 1988;334:62–64. doi: 10.1038/334062a0. [DOI] [PubMed] [Google Scholar]
- Meiri KF, Saffell JL, Walsh FS, Doherty P. Neurite outgrowth stimulated by neural cell adhesion molecules requires growth-associated protein-43 (GAP-43) function and is associated with GAP-43 phosphorylation in growth cones. J Neurosci. 1998;18:10429–10437. doi: 10.1523/JNEUROSCI.18-24-10429.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Lee X, Shao Z, Thill G, Ji B, Relton J, Levesque M, Allaire N, Perrin S, Sands B, Crowell T, Cate RL, McCoy JM, Pepinsky RB. LINGO-1 is a component of the Nogo-66 receptor/p75 signaling complex. Nat Neurosci. 2004;7:221–228. doi: 10.1038/nn1188. [DOI] [PubMed] [Google Scholar]
- Morimura N, Inoue T, Katayama K, Aruga J. Comparative analysis of structure, expression and PSD95-binding capacity of Lrfn, a novel family of neuronal transmembrane proteins. Gene. 2006;380:72–83. doi: 10.1016/j.gene.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Moulding HD, Martuza RL, Rabkin SD. Clinical mutations in the L1 neural cell adhesion molecule affect cell-surface expression. J Neurosci. 2000;20:5696–5702. doi: 10.1523/JNEUROSCI.20-15-05696.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller D, Wang C, Skibo G, Toni N, Cremer H, Calaora V, Rougon G, Kiss JZ. PSA-NCAM is required for activity-induced synaptic plasticity. Neuron. 1996;17:413–422. doi: 10.1016/s0896-6273(00)80174-9. [DOI] [PubMed] [Google Scholar]
- Nishimura K, Yoshihara F, Tojima T, Ooashi N, Yoon W, Mikoshiba K, Bennett V, Kamiguchi H. L1-dependent neuritogenesis involves ankyrinB that mediates L1-CAM coupling with retrograde actin flow. J Cell Biol. 2003;163:1077–1088. doi: 10.1083/jcb.200303060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redies C. Cadherins in the central nervous system. Prog Neurobiol. 2000;61:611–648. doi: 10.1016/s0301-0082(99)00070-2. [DOI] [PubMed] [Google Scholar]
- Sans N, Wang PY, Du Q, Petralia RS, Wang YX, Nakka S, Blumer JB, Macara IG, Wenthold RJ. mPins modulates PSD-95 and SAP102 trafficking and influences NMDA receptor surface expression. Nat Cell Biol. 2005;7:1179–1190. doi: 10.1038/ncb1325. [DOI] [PubMed] [Google Scholar]
- Scheiffele P, Fullekrug J. Glycosylation and protein transport. Essays Biochem. 2000;36:27–35. doi: 10.1042/bse0360027. [DOI] [PubMed] [Google Scholar]
- Seabold GK, Wang PY, Chang K, Wang CY, Wang YX, Petralia RS, Wenthold RJ. The salm family of adhesion-like molecules forms heteromeric and homomeric complexes. J Biol Chem. 2008 doi: 10.1074/jbc.M709456200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JY, Ichtchenko K, Sudhof TC, Brose N. Neuroligin 1 is a postsynaptic cell-adhesion molecule of excitatory synapses. Proc Natl Acad Sci U S A. 1999;96:1100–1105. doi: 10.1073/pnas.96.3.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Hung CP, Schuman EM. A role for the cadherin family of cell adhesion molecules in hippocampal long-term potentiation. Neuron. 1998;20:1165–1175. doi: 10.1016/s0896-6273(00)80497-3. [DOI] [PubMed] [Google Scholar]
- Triana-Baltzer GB, Liu Z, Berg DK. Pre- and postsynaptic actions of L1-CAM in nicotinic pathways. Mol Cell Neurosci. 2006;33:214–226. doi: 10.1016/j.mcn.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Utton MA, Eickholt B, Howell FV, Wallis J, Doherty P. Soluble N-cadherin stimulates fibroblast growth factor receptor dependent neurite outgrowth and N-cadherin and the fibroblast growth factor receptor co-cluster in cells. J Neurochem. 2001;76:1421–1430. doi: 10.1046/j.1471-4159.2001.00140.x. [DOI] [PubMed] [Google Scholar]
- Wang CY, Chang K, Petralia RS, Wang YX, Seabold GK, Wenthold RJ. A novel family of adhesion-like molecules that interacts with the NMDA receptor. J Neurosci. 2006;26:2174–2183. doi: 10.1523/JNEUROSCI.3799-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittard JD, Sakurai T, Cassella MR, Gazdoiu M, Felsenfeld DP. MAP kinase pathway-dependent phosphorylation of the L1-CAM ankyrin binding site regulates neuronal growth. Mol Biol Cell. 2006;17:2696–2706. doi: 10.1091/mbc.E06-01-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DIV4 primary hippocampal neurons were co-transfected with GFP and myc-SALM1, myc-SALM2, myc-SALM4, or HA-SALM5. Surface labeling with myc or HA antibodies was performed 48 hours after transfection. A, Representative examples of myc-SALM2 surface labeling are shown. Scale bar, 20 µm. B, SALM surface expression is punctate (arrows/inset), and accumulated at growth cones. Scale bar, 10 µm.
Individual RNAi plasmid constructs were designed and generated for SALMs 1–5 using pcDNA™6.2-GW/EmGFP-miR micro RNAi expression vectors, which co-expresses GFP. DIV4 primary hippocampal neurons were co-transfected individually with myc-SALM1, myc-SALM2, SALM3, myc-SALM4, or HA-SALM5 cDNA and their respective SALM RNAi plasmid (e.g. myc-SALM1cDNA and SALM1 RNAi), or scramble RNAi negative control (e.g. myc-SALM1 cDNA and scramble RNAi). Expression levels of transfected myc-SALM1 with scramble RNAi or SALM1 RNAi are shown in rows A1 and A2, respectively. Expression levels of SALM2-SALM5 with scramble RNAi (GFP) or SALM RNAi (GFP) are shown in rows B–E, respectively. Scale bar, 20 µm. For the characterization of each SALM RNAi construct, images were taken and processed with identical exposure times and contrast levels. Expression of transfected SALMs was reduced by 28%, 75%, 60%, 61%, and 43% for SALMs 1–5, respectively (n=5).