Abstract
S6K1 (p70S6K) is a serine kinase downstream from Akt in the insulin signaling pathway that is involved in negative feedback regulation of insulin action. S6K1 is also activated by TNF-α, a pro-inflammatory cytokine. However, its role remains to be characterized. In the current study, we elucidated a mechanism for S6K1 to mediate TNF-α-induced insulin resistance in adipocytes and hepatocytes. S6K1 was phosphorylated at Thr-389 in response to TNF-α. This led to phosphorylation of IRS-1 by S6K1 at multiple serine residues including Ser-270, Ser-307, Ser-636, and Ser-1101 in human IRS-1 (Ser-265, Ser-302, Ser-632, and Ser-1097, in rodent IRS-1). Direct phosphorylation of these sites by S6K1 was observed in an in vitro kinase assay using purified IRS-1 and S6K1. Phosphorylation of all these serines was increased in the adipose tissue of obese mice. RNAi knockdown demonstrated an important role for S6K1 in mediating TNF-α-induced IRS-1 inhibition that led to impaired insulin-stimulated glucose uptake in adipocytes. A point mutant of IRS-1 (S270A) impaired association of IRS-1 with S6K1 resulting in diminished phosphorylation of IRS-1 at three other S6K1 phosphorylation sites (Ser-307, Ser-636, and Ser-1101). Expression of a dominant negative S6K1 mutant prevented TNF-induced Ser-270 phosphorylation and IRS-1 protein degradation. Moreover, in IKK2 (but not IKK1)-null cells, TNF-α treatment did not result in Thr-389 phosphorylation of S6K1. We present a new mechanism for TNF-α to induce insulin resistance that involves activation of S6K by an IKK2-dependent pathway. S6K directly phosphorylates IRS-1 on multiple serine residues to inhibit insulin signaling.
TNF-α is a pro-inflammatory cytokine implicated in development of insulin resistance in obesity (1, 2). Several signaling pathways may play important roles in tumor necrosis factor (TNF)2-induced insulin resistance (3–7). Serine phosphorylation of IRS-1 at various residues by distinct kinases may be a common mechanism by which TNF-α impairs insulin-induced glucose uptake. For example, JNK (5, 8), IKK (6), ERK (4, 5, 9, 10), PKCζ (11–13), PKCθ (14, 15), Akt (16, 17), GSK-3 (18–20), IRAK (21), and mTOR (17, 22) may all participate in insulin resistance by mediating serine phosphorylation of IRS-1. S6K1 (ribosomal protein S6 kinase 1, p70S6K, S6K in this study) was reported to inhibit IRS-1 function through induction of IRS-1 degradation (22). Recent studies demonstrate that S6K mediates mTOR signaling to phosphorylate IRS-1 at serine residues (rodent/human) including Ser-302/307 (23), Ser-307/312 (24), Ser-632/636 (17), and Ser-1097/Ser-1101 (25). S6K knock-out mice are protected against diet-induced insulin resistance, and this phenotype is associated with reduced phosphorylation of IRS-1 Ser-636 (Ser-632 in rodent) (26). However, it remains possible that S6K phosphorylates IRS-1 at additional sites. Additionally, direct phosphorylation of IRS-1 by S6K has not been previously demonstrated in a kinase assay.
S6K/IRS-1 interaction is involved in regulation of insulin sensitivity by amino acids and insulin (27–30). However, the role of S6K in TNF-induced insulin resistance is not clear. Direct evidence supporting a mechanism for S6K1 to mediate TNF-α-induced insulin resistance is lacking. In obese mice, S6K activity is significantly elevated in liver, adipose tissue, and skeletal muscle (30, 31) and this may contribute to insulin resistance. However, the molecular mechanisms underlying the elevated S6K activity are not clear in obesity. Two studies suggest that S6K may be activated by TNF-α (7, 17). However, the molecular mechanism by which TNF-α activates mTOR/S6K is controversial. In an initial study, Ozes et al. (17) reported that the PI3K/Akt/mTOR pathway mediated the TNF-α signal. In a more recent study, Lee et al. (32) demonstrated that S6K can be activated by TNF-α through a PI3K-independent pathway. It remains to be determined which pathway mediates the S6K activation by TNF-α.
Given the potentially important role of S6K and TNF-α in the control of insulin sensitivity, it is of interest to determine if S6K mediates TNF signaling related to insulin resistance. It is also important to determine the relationship between S6K and other serine kinases that are activated by TNF-α for insulin resistance. The association of IRS-1 Ser-636 phosphorylation with TNF-α treatment suggests a potential mechanism for S6K to mediate TNF signals for insulin resistance (7, 17).
In the present study, we conducted a systematic analysis of the role of S6K in mediating TNF-α-induced insulin resistance. Our data suggest that TNF-α activates S6K through IKK2, and that S6K directly phosphorylates IRS-1 at four serine residues including Ser-265/270, Ser-302/307, Ser-632/636, and Ser-1097/1101 in rodents/humans. The Ser-265/270 is required for S6K to phosphorylate IRS-1 at other three serines.
MATERIALS AND METHODS
Animals—Male C57BL/6J-Lepob, and C57BL/6J mice were purchased from Jackson Laboratory (Bar Harbor, ME) at 5 weeks of age and used in the study according to an animal protocol approved by the institutional animal care and use committee. Mice were housed in a regular cage at 4 mice/cage with free access to water and standard chow unless noted. Epididymal fat from mice fasted overnight were collected, frozen in liquid nitrogen, and stored at –70 °C until further analysis. All procedures were performed in accordance with National Institutes of Health guidelines for the care and use of animals.
Cells and Reagents—Cell lines including mouse NIH-3T3 (CRL-1658) and human embryonic kidney (HEK) 293 (CRL-1573) were purchased from the American Type Culture Collection (ATCC). IKK wild-type, IKK1, or IKK2 knock-out cell lines were described in a previous study (6). The H4IIE cell line stably transfected with Flag-IRS1 wild type was a gift from Dr. Richard A. Roth at Stanford University Medical School, Stanford, CA 94305-5174 (33). All cells were maintained in Dulbecco's modified Eagle's culture medium supplemented with 10% fetal calf serum. Phospho-IRS-1 (Ser-312/307) antibody (07-247) was from Upstate Biotechnology (Lake Placid, NY). Antibodies to phospho-Ser-307/302 (2384) and phospho-Ser-1101/1097 (2385) in IRS-1, phospho-Thr-308 (9275) and phospho-Ser-473 (9271) in Akt, phospho-Thr-389 (9205) in p70S6 were obtained from Cell Signaling (Beverly, MA). Antibodies to phospho-Ser-270 (sc-17192) in IRS-1, IRS-1 (sc-7200), and IκBα (sc-371) were from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies to S6K (ab9366), phospho-Ser-636/632 of IRS-1 (ab47764) and β-actin (ab6276) were from Abcam (Cambridge, UK). Rapamycin (A-275), LY294002 (ST420), and SP600125 (EI-305) were acquired from Biomol (Plymouth Meeting, PA). 15-Deoxyprostaglandin J2 (15dPGJ2, 538927), PD98059 (513000), and SB203580 (203580) were purchased from Calbiochem. Wortmannin (W-1628), Type II collagenase (C6885), and TNF-α (T6674) were from Sigma. Purified p70S6 kinase (T412E), active IKK2 (IKKβ), and PKCθ were obtained from Upstate Biotechnology (Lake Placid, NY).
Generation of Adenovirus—Adenovirus carrying a dominant negative S6K1 (S6K-DN) was constructed using ViraPower Adenoviral Expression System (K4930-00), which was from Invitrogen (Carlsbad, CA). Briefly, S6K1-DN cDNA with HA tag was inserted into TOPO pENTR vector (K2400-20) and was recombined into the adenovirus expression plasmid pAd/CMV/V5-DEST. The pAd/CMV/V5-DEST plasmid with S6K1 cDNA was digested with the PacI endonuclease and transfected with 293A cells for production of adenovirus. The medium supernatant containing adenovirus was collected 3 days later and titrated according to the manufacturer's instructions.
Immunoblotting—Adipose tissue was homogenized in cold lysis buffer followed by sonication (6). Lysis buffer contains 1% Nonidet P-40, 50 mm Hepes, pH 7.6, 250 mm NaCl, 10% glycerol, 1 mm EDTA, 20 mm β-glycerophosphate, 1 mm sodium orthovanadate, 1 mm sodium metabisulfite, 1 mm benzamidine hydrochloride, 10 μg/ml leupeptin, 20 μg/ml aprotinin, 1 mm phenylmethylsulfonyl fluoride. Cultured cells were kept in serum-free media overnight and treated with various reagents as indicated. After treatment, whole cell lysates were made in lysis buffer with sonication, and the supernatant was used for immunoblotting after centrifugation at 10,000 × g for 10 min at 4 °C. Total protein (100 μg) in 50 μl of reducing sample buffer was used for immunoblotting as described previously (34). Immunoblots were quantified using a scanning densitometer in conjunction with NIH ImageJ software. Signals were normalized to loading controls.
Plasmids and Transfection—Expression vectors for HA-tagged IRS-1 wild type and IRS-1 S270A mutant were constructed in pCIS2 expression vector as described (11). Plasmids for HA-S6K1 WT (8984) and HA-S6K1 dominant negative (8985) were obtained from Addgene (Cambridge, MA) (35). The plasmids for HA-IKK2 WT, HA-IKK2 kinase dead mutant, and GST-IRS-1 were described previously (6). Constructs were expressed in HEK293 cells by transient transfection using Lipofectamine. Purified GST-IRS-1 was described previously (6).
Immunoprecipitation—Immunoprecipitation was carried out using whole cell lysates (400 μg of total protein), 2–4 μgof antibody, and 20 μl of protein A- or protein G-Sepharose beads (Amersham Biosciences). After treatment, cell lysates were prepared by sonication in cell lysis buffer. IP was conducted by incubating the whole cell lysate with antibody for 3–4 h at 4 °C. The immune complex was washed five times in cell lysis buffer before being used for immunoblotting or kinase assays.
Kinase Assay—For each in vitro kinase assay, purified GST-IRS-1 or HA-IRS-1 protein was used as substrate. The proteins were diluted in kinase assay buffer (20 mm Hepes, pH 7.6, 20 mm MgCl2, 20 mm glycerophosphate, 1 mm dithiothreitol, 10 μm ATP, 1 mm EDTA, 1 mm sodium orthovanadate, 0.4 mm phenylmethylsulfonyl fluoride, 20 mm creatine phosphate). The kinase assay was conducted at 37 °C for 30 min in 20 μl of kinase assay buffer and 0.2 μg (2 μl) of kinase, such as S6K1, IKK2 (IKKβ), or PKCθ. The product was resolved by 8% SDS-PAGE and immunoblotted with phosphospecific IRS-1 antibodies.
RNA Interference—Mouse S6K-specific shRNA was expressed in retroviral silencing plasmids. The shRNA plasmids were made under a contract service agreement with Origene (Rockville, MD). Expression vectors for four independent shRNA were cotransfected into NIH-3T3 cells using Lipofectamine 2000 (Invitrogen). Empty vector was used as a negative control. 60 h after transfection, cells were kept in serum-free medium overnight and then stimulated without or with TNF-α or insulin for 30 min.
Primary Adipocytes—Preadipocytes were isolated from epididymal fat pads of C57BL/6J mice as described elsewhere (36). The tissue was digested with Type II collagenase, and single cells were plated in a 100-mm flask in normal culture medium. After 24 h, cells attached to the flask were used as preadipocytes. The preadipocytes were plated into a 6-well plates and differentiated into adipocytes in standard adipogenic mixture (5 μg/ml insulin, 0.5 mm isobutylmethylxanthine, and 10 μm dexamethasone). Mature adipocytes were used at day 9 of differentiation.
Insulin-induced Glucose Uptake—3T3-L1 preadipocytes (5 × 105/well) were differentiated into adipocytes in a 12-well plate, and used in the glucose uptake assay as described elsewhere (15). In experiments using dominant negative mutant of S6K, the differentiated 3T3-L1 cells were infected by the S6K-DN adenovirus for 24 h and glucose uptake was examined after an additional 24 h.
Statistical Analysis—All experiments were repeated independently at least three times with consistent results. For most figures with immunoblots, a representative blot is shown for each experiment along with a bar graph representing the mean ± S.E. of multiple independent experiments determined by densitometric analysis normalized to appropriate controls. Student's t test or one-way analysis of variance was used as appropriate in statistical analyses of the data. p < 0.05 was considered to indicate statistical significance.
RESULTS
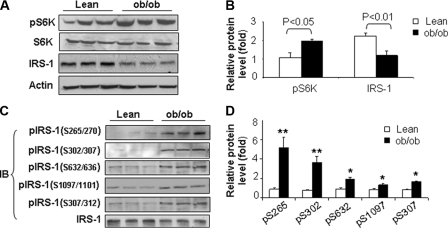
Association between Increased S6K Phosphorylation and Serine Phosphorylation of IRS-1 in ob/ob Mice—We hypothesized that S6K1 participates in the TNF-α-induced insulin resistance of obesity. Therefore, we first examined the relationship between S6K1 phosphorylation at Thr-389 (a proxy for S6K1 activity) and serine phosphorylation of IRS-1 in adipose tissue from ob/ob mice (Fig. 1). As expected, phosphorylation of S6K1 was significantly elevated in adipose tissue from ob/ob mice when compared with samples from lean control mice (Fig. 1, A and B). Expression levels for S6K1 protein were comparable between lean and ob/ob mice. However, expression of IRS-1 protein was decreased in adipose tissue from ob/ob mice (Fig. 1, A and B).
FIGURE 1.
Obesity is associated with increased phosphorylation of S6K at Thr-389 and IRS-1 at Ser-270. A, S6K phosphorylation is increased in adipose tissue of ob/ob mice. Lean control mice and ob/ob mice were fasted overnight before being sacrificed (n = 3 for each group). Whole cell lysates were made from epididymal fat and used for immunoblotting experiments. Samples were immunoblotted with antibodies against S6K, phospho-S6K (pThr-389), IRS-1, and actin. B, quantification of S6K phosphorylation and IRS-1 protein level in adipose tissue of lean and obese mice. C, IRS-1 serine phosphorylation in adipose tissue. IRS-1 protein was immunoprecipitated (IP) from whole cell lysates using anti-IRS-1 antibody. Samples were immunoblotted with phosphospecific IRS-1 antibodies to pSer-265/270, pSer-302/307, pSer-307/312, pSer-632/636, pSer-1097/1101 in the rodent/human IRS-1 protein. Each abbreviation for serines stands for the two corresponding serines in rodent (Ser-265) and human (Ser-270) IRS-1. The abbreviations are used in the same way in rest of study unless specified. D, quantification of IRS-1 phosphorylation in adipose tissue of lean and obese mice. The phosphosignal at each serine site was normalized using the non-phosphosignal. The mean signal of three mice was obtained for the lean mice. The signal in the obese mice was obtained in the same way. An average was obtained from three experiments. The signal from lean mice was used as a basic unit to determine the fold-change in the obese group. All experiments presented in this figure were repeated independently three times with consistent results. Each data point represents mean ± S.E. of three independent experiments (n = 3). *, p < 0.05. **, p < 0.001.
In several independent studies, S6K is reported to be involved with phosphorylation of IRS-1 at multiple serine residues including Ser-302/307, Ser-632/636, Ser-1097/1101, and Ser-307/312 in the mouse/human IRS-1 proteins (17, 23–26). Each abbreviation for serines stands for the two corresponding serines in rodent (Ser-302) and human (Ser-307) IRS-1. The abbreviations are used in the same way in rest of our study unless specified. These previously published studies did not compare all of these sites in the same experimental preparation. Therefore, using phosphospecific antibodies, we demonstrated that serine phosphorylation at all of these sites was elevated in IRS-1 in adipose tissue from ob/ob mice when compared with samples from lean control mice (Fig. 1, C and D). In addition, we evaluated serine phosphorylation of Ser-265/270 (Fig. 1, C and D) because the amino acid sequence surrounding this site is homologous to known S6K phosphorylation sites (Fig. 3A). Interestingly, Ser-265/270 phosphorylation was also increased in the samples from ob/ob mice. Thus, increased S6K phosphorylation in adipose tissue from ob/ob mice is associated with increased phosphorylation of IRS-1 at multiple serine residues as well as diminished protein expression of IRS-1. This raises the possibility that S6K may be directly phosphorylating IRS-1 at these 5 serine residues resulting in accelerated degradation of IRS-1.
FIGURE 3.
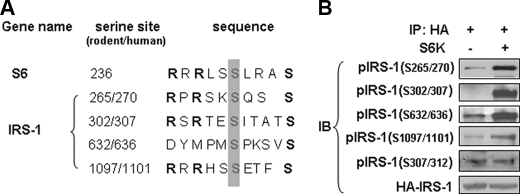
S6K1 directly phosphorylates IRS-1 at multiple serine sites in vitro. A, putative S6K phosphorylation sites in IRS-1. Amino acid sequence of S6K phosphorylation site in the 40S ribosomal protein S6 was used to generate a consensus S6K-specific sequence template for analysis of IRS-1. We identified four candidate S6K-specific phosphorylation motifs in IRS-1. Conserved arginine and serine are shown in bold, and putative serine phosphorylation sites for S6K are shown in gray box. B, in vitro kinase assay used purified S6K1 kinase and HA-IRS-1 as a substrate. HA-IRS-1 (human IRS-1) was expressed in HEK293 cells and isolated by immunoprecipitation with anti-HA antibody. Samples were resolved using SDS-PAGE and immunoblotted with phosphospecific antibodies against IRS-1.
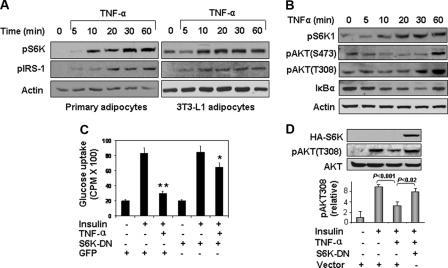
Role of S6K in Mediating TNF-induced Insulin Resistance—To evaluate the potential role of S6K in mediating TNF-α-induced insulin resistance, we first examined the ability of TNF-α to stimulate phosphorylation of S6K at Thr-389 and IRS-1 at Ser-265/270 (Fig. 2A). In both primary adipocytes and 3T3-L1 adipocytes, TNF-α stimulated a time-dependent increase in phosphorylation of S6K at Thr-389 and IRS-1 at Ser-265/270. Similarly in rat hepatoma cells (H4IIE), S6K1 phosphorylation was induced by TNF-α as early as 10 min (Fig. 2B). Akt phosphorylation at Thr-308 and Ser-473 was also increased in response to TNF-α in a time-dependent manner that seemed some 20–60 min later than the S6K phosphorylation. This difference in time course between S6K1 and Akt activation in response to TNF-α suggests that S6K1 activation may be independent of Akt. However, it should be noted that the sensitivity of the phospho-antibodies used against different proteins may not be similar. Thus, firm conclusions regarding relative time courses of phosphorylation for different proteins are not warranted based on these data alone. In these experiments, we also assessed IkBα degradation as a control for TNF-α activity (Fig. 2B).
FIGURE 2.
Role of S6K to mediate TNF-α-induced insulin resistance. A, TNF-α treatment (10 ng/ml) causes time-dependent increase in phosphorylation of S6K (Thr-389) and IRS-1 (Ser-265/270) in primary and 3T3-L1 adipocytes. B, time course of S6K phosphorylation in response to TNF-α treatment in rat hepatoma cells (H4IIE). Cells were serum-starved overnight and then treated with TNF-α (10 ng/ml) for the indicated times. S6K phosphorylation at Thr-389 was determined by immunoblotting with a phosphospecific antibody. Immunoblotting with antibodies against IkBa and phospho-Akt (T308, S473) were used as positive controls for TNF-α activity. C, insulin-stimulated glucose uptake was examined in 3T3-L1 adipocytes. 3T3-L1 adipocytes were infected by S6K-DN adenovirus. In control cells, 3T3-L1 adipocytes were infected with GFP adenovirus. The glucose uptake assay was performed after TNF-α pretreatment for 4 h. Each bar represents mean ± S.E. of three independent experiments. *, p < 0.05 and **, p < 0.001. D, S6K-DN blocked TNF-inhibition of insulin-stimulated phosphorylation of Akt (Thr-308) in 3T3-L1 cells. The cells were pretreated with TNF-α for 4 h before addition for insulin (100 nm for 15 min). Results from three independent experiments were quantified by scanning densitometry and plotted as mean ± S.E. in the bar graph underneath the immunoblot.
We next evaluated insulin-stimulated glucose uptake in 3T3-L1 adipocytes without or with pretreatment with TNF-α in the absence or presence of expression of a dominant negative mutant of S6K (S6K-DN) (Fig. 2C). As expected, insulin-stimulated glucose uptake was significantly impaired when cells were pretreated with TNF-α. Importantly, the effect of TNF-α to cause this insulin resistance was substantially blunted in cells expressing S6K-DN. Moreover, insulin-stimulated phosphorylation of Akt (Thr-308), a key mediator of insulin-stimulated glucose uptake (37), was also significantly diminished by TNF-α pretreatment (p < 0.001) and this effect of TNF-α was significantly abrogated by expression of S6K-DN (p < 0.02) (Fig. 2D). Taken together, these results suggest that S6K plays a key role in mediating impairment in metabolic insulin signaling caused by TNF-α that contributes to insulin resistance.
Identification of IRS-1 Ser-265/270 as a Novel Phosphorylation Site for S6K1—To identify novel serine phosphorylation sites on IRS-1 for S6K1, we compared an amino acid motif surrounding the phosphorylation site on S6, an authentic substrate of S6K1, with the whole IRS-1 amino acid sequence (Fig. 3A). The S6 motif is characterized by the sequence RXRXXS, in which the distal serine residue is the kinase target. We identified similar motifs in four of the IRS-1 phosphoserine sites evaluated in our present study (Ser-265/270, Ser-302/307, Ser-632/636, and Ser-1097/1101). This motif was not present for Ser-307/312 (Fig. 3A).
We next evaluated the ability of S6K1 to directly phosphorylate IRS-1 by conducting in vitro kinase assays with purified S6K1 and recombinant HA-tagged IRS-1 (human IRS-1) immunoprecipitated from transfected 293 cells (Fig. 3B). Phosphorylation of IRS-1 was detected with phosphospecific antibodies as in Fig. 1B. Using this method, we found that S6K1 directly phosphorylated Ser-265/270, Ser-302/307, Ser-632/636, and Ser-1097/1101 in IRS-1. Interestingly, specific phosphorylation of IRS-1 at Ser-307/312 by S6K1 was not observed. These results are consistent with our sequence analysis (Fig. 3A) and support the specificity of our in vitro kinase assay. Thus, we have identified Ser-265/270 as a novel S6K1 target in the IRS-1 protein. Our observations are unlikely to result from nonspecific interaction between S6K1 and IRS-1 since Ser-307/312 phosphorylation was not induced by S6K under identical assay conditions.
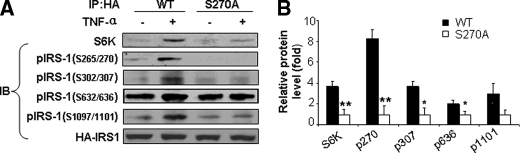
Regulatory Role for Phosphorylation of IRS-1 at Ser-265/270 by S6K1—Serine phosphorylation of IRS-1 at multiple sites has been implicated in inhibition of insulin signaling. It is possible that phosphorylation of IRS-1 at one serine residue may regulate the ability of S6K1 to phosphorylate other sites on IRS-1. To evaluate this possibility, we substituted Ser-270 with alanine in the human IRS-1 protein (S270A mutant). This serine site was selected for further investigation because it represents a novel phosphorylation target for S6K that has not previously been investigated. The HA-tagged IRS-1 mutant (S270A) was expressed in HEK293 cells (Fig. 4). In control cells, the wild type HA-IRS-1 was expressed. TNF-α induced association between S6K1 and wild-type IRS-1 in the control cells (as determined by co-immunoprecipitation). By contrast, this association was significantly reduced by the S270A mutation in IRS-1 although it was still detectable. Moreover, the TNF-α-stimulated increase in phosphorylation of HA-IRS-1 at Ser-270 and the other three S6K serine phosphorylation sites (Ser-302/307, Ser-632/636, and Ser-1097/1101) was also significantly reduced in IRS-1 with the S270A mutation (Fig. 4, A and B). These data suggest that phosphorylation of IRS-1 at Ser-265/270 is critical for regulating the interaction of IRS-1 with S6K1 in response to TNF-α and for phosphorylation of other serine residues in IRS-1 by S6K1.
FIGURE 4.
Phosphorylation of Ser-270 regulates ability of S6K to interactive with IRS-1 and phosphorylate other phosphorylation sites on IRS-1. A, HEK293 cells were transfected with either HA-tagged wild-type IRS-1, or IRS-1 S270A mutant. After overnight serum-starvation, transfected cells were treated without or with TNF-α (10 ng/ml, 30 min) to induce S6K activation. After immunoprecipitation (IP) with anti-HA antibody, IRS-1/S6K association was determined by immunoblotting with S6K antibody. Phosphorylation of HA-IRS-1 wild-type and S270A mutant was determined by immunoblotting with phosphospecific antibodies against pSer-270, pSer-307, pSer-636, and pSer-1101 in the human IRS-1. B, quantification of immunoblots. Each bar represents the mean ± S.E. of three independent experiments. *, p < 0.05 and **, p < 0.001.
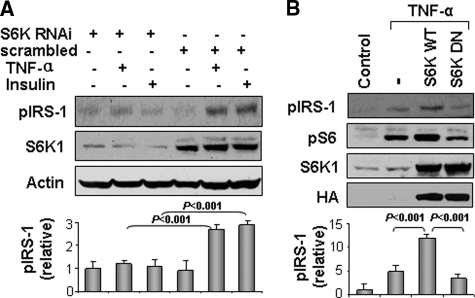
S6K1 Is Required for TNF-α-mediated Phosphorylation of IRS-1 at Ser-265/270—To determine the role of S6K in the TNF-α signaling pathway related to IRS-1 phosphorylation, we blocked S6K1 function in cells using two distinct strategies: RNAi-mediated gene knockdown of S6K1 and expression of a dominant negative mutant of S6K1. In cells expressing RNAi targeting S6K1, the S6K1 protein was reduced by 90% (Fig. 5A). Under these conditions, IRS-1 phosphorylation at Ser-265/270 in response to TNF-α or insulin treatment was significantly reduced when compared with control cells infected with scrambled RNAi (p < 0.001) (Fig. 5A). Similarly, when S6K1 dominant negative mutant was expressed, TNF-α-stimulated phosphorylation of IRS-1 at Ser-265/270 was significantly inhibited (when compared with control cells overexpressing wild-type S6K1) (p < 0.001) (Fig. 5B). In these experiments, phosphorylation of S6 (classical substrate of S6K1) was used as a control for S6K activity to demonstrate that the dominant negative mutant of S6K was inhibiting endogenous S6K1 function. Taken together, these data suggest that S6K1 is required for the TNF-induced IRS-1 phosphorylation at Ser-265/270.
FIGURE 5.
S6K1 is required for TNF-α-stimulated phosphorylation of IRS-1 at Ser-270. A, IRS-1 phosphorylation at Ser-265/270 in response to TNF-α or insulin with S6K knockdown. NIH3T3 cells were infected with retrovirus vector expressing RNAi to knockdown S6K or control scrambled RNAi. After serum-starvation, the NIH3T3 cells were stimulated with TNF-α or insulin (100 nm) for 30 min and Ser-265 (Ser-270) phosphorylation was examined in IRS-1. B, HEK293 cells were transiently transfected with wild-type S6K (S6K-WT) or dominant genitive S6K (S6K-DN). Ser-265 (Ser-270) phosphorylation was examined in IRS-1 after TNF-α treatment. For both A and B, results from at least three independent experiments were quantified by scanning densitometry and plotted as mean ± S.E. in the bar graph underneath the immunoblots.
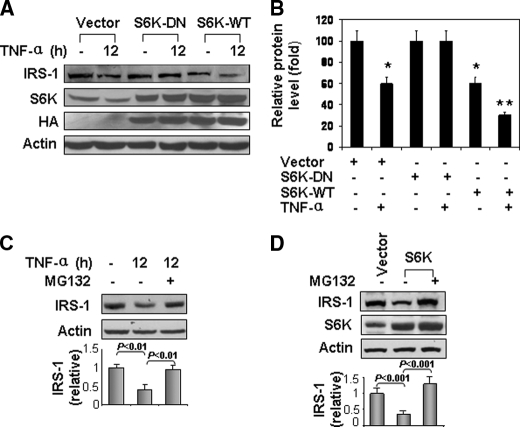
Role of S6K1 in IRS-1 Degradation in Response to TNF-α Treatment—Serine phosphorylation leads to degradation of IRS-1 protein. To investigate potential mechanisms underlying the reduction in IRS-1 in adipose tissue of ob/ob mice (Fig. 1A), we examined the role of S6K1 in IRS-1 degradation in cells. We reasoned that S6K1-mediaetd serine phosphorylation may promote IRS-1 protein degradation. To test this possibility, IRS-1 degradation was examined in cells where S6K1 activity was inhibited by expression of S6K-DN in HEK293 cells (Fig. 6A). In control cells transfected with empty vector, IRS-1 protein was significantly reduced by ∼40% after TNF-α treatment. A similar reduction was observed in cells overexpressing wild-type S6K1. This reduction was further increased by TNF-treatment resulting in ∼70% loss in total IRS-1 protein (Fig. 6B). By contrast, in cells expressing S6K-DN, TNF-α treatment did not result in any significant reduction in IRS-1 expression. Moreover, IRS-1 degradation induced by TNF-α or S6K overexpression was substantially blocked in the presence of the proteasome inhibitor MG132 (Fig. 6, C and D). Taken together, these data strongly suggest that S6K is required for TNF-induced IRS-1 degradation.
FIGURE 6.
S6K1 plays a role in TNF-induced IRS-1 degradation. A, HEK293 cells were transiently transfected with expression vectors for wild type S6K (S6K-WT) or its dominant negative mutant (S6K-DN). After serum starvation, cells were treated without or with TNF-α (10 ng/ml) for 12 h. Protein level of IRS-1, and S6K1 were assessed by immunoblotting. B, quantification of immunoblots in A. Each bar represents mean ± S.E. of three independent experiments. *, p < 0.05. **, p < 0.001. C, proteasome inhibitor MG132 blocked TNF-α-induced degradation of IRS-1. After serum starvation, HEK293 cells were pretreated with MG132 (15 μm) for 30 min, and then stimulated with TNF-α (10 ng/ml) for 12 h. D, HEK 293 cells were transiently transfected with expression vector for S6K. After 36 h, cells were treated with MG132 (15 μm) for 12 h. For both C and D, results from three independent experiments were quantified by scanning densitometry and plotted as mean ± S.E. in the bar graph underneath the immunoblots.
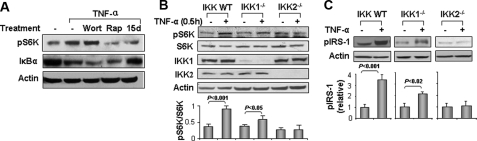
Activation of S6K by TNF-α Requires IKK2—The mechanism regulating S6K1 activation by TNF-α is controversial. One early study (17) suggests a role for PI3K. More recently, the IKK2 pathway has been implicated (32). Both of these prior studies agree on the requirement of mTOR for S6K1 activation. To examine these pathways in our experimental system, we evaluated S6K1 phosphorylation in response to TNF-α treatment without or with inhibition of PI3K or IKK2 using chemical inhibitors (Fig. 7). TNF-induced S6K1 phosphorylation was not substantially influenced by pretreatment of cells with Wortmannin (PI3K inhibitor). However, 15dPGJ2 (IKK2 inhibitor) and rapamycin (mTOR inhibitor) both markedly inhibited the effect of TNF-α to stimulate phosphorylation of S6K (Fig. 7A).
FIGURE 7.
Role of IKK2 in S6K phosphorylation in response to TNF-α treatment. A, role of mTOR and IKK in S6K activation by TNF-α. H4IIE cells were pretreated with PI3K inhibitor Wortmannin (Wort, 100 nm), mTOR inhibitor Rapamycin (Rap, 200 nm) or IKK inhibitor 15dPGJ2 (15d, 5 μm) and then treated with TNF-α. Cell lysates were immunoblotted with antibodies against IκBα or pS6K1 (Thr-389). B, role of IKK2 in activation of S6K by TNF-α. Embryonic fibroblasts from IKK wild type, IKK2–/– and IKK1–/– mice were used to determine the role of IKK in S6K phosphorylation in response to TNF-α. Phosphorylation of S6K was determined after TNF treatment for 30 min. The signal was quantified and normalized with non-phospho-S6K. C, IRS-1 phosphorylation at Ser-265/270 was examined in IKK-null MEFs without or with treatment with TNF-α (10 ng/ml, 30 min). The signal was quantified and normalized with non-phospho-IRS-1. For both B and C, results from at least three independent experiments were quantified by scanning densitometry and plotted as mean ± S.E. in the bar graph underneath the immunoblots.
To further evaluate the role of IKK2, we examined S6K1 phosphorylation in IKK2-null (IKK2–/–) MEF cells (Fig. 7B). In the control cells (wild type or IKK1-null (IKK1–/–) MEFs), S6K phosphorylation was significantly induced by TNF-α (p < 0.001 and 0.05, respectively). By contrast, in IKK2–/– cells, TNF-α was unable to induce phosphorylation of S6K1. Interestingly, basal phosphorylation of S6K1 in IKK2-null cells was higher than that in control cells. This basal increase may be due to signaling unrelated to TNF-α. S6K protein was expressed at comparable levels in all three lines of MEFs (Fig. 7B). Taken together, our results suggest that IKK2 is required for S6K activation by TNF-α.
Role of IKK2 in IRS-1 Phosphorylation—Because IKK2 is required for TNF-α to induce S6K1 phosphorylation, we next tested whether IKK2 is necessary for IRS-1 phosphorylation at the S6K1 serine phosphorylation site Ser-265/270 (Fig. 7C). In the control cells (wild type, and IKK1–/– MEFs), TNF-α significantly induced IRS-1 phosphorylation at Ser-265/270 (p < 0.001 and p < 0.02, respectively). By contrast, no increase in IRS-1 phosphorylation at Ser-265/270 was observed in the IKK2-null cells in response to TNF-α treatment. Thus, in the absence of IKK2, TNF-α is not able to enhance phosphorylation of IRS-1 Ser-265/270 (most likely because of an inability to activate S6K1).
DISCUSSION
Data from our study suggest that S6K represents a signal integration point for TNF-α and insulin. S6K is activated downstream from IRS-1/PI3K/Akt/mTOR in the insulin signaling pathway to regulate protein synthesis (38). It also mediates mTOR signaling in the negative feedback regulation of the insulin signal transduction pathway by phosphorylation of IRS-1 that is upstream of PI3K (27, 39–41). The S6K-IRS1 interaction represents a molecular mechanism by which insulin, other hormones, and amino acids induce insulin resistance (30, 42). In the current study, we demonstrated an important role for S6K to mediate TNF-α signaling in the induction of insulin resistance. This role is supported by data from RNAi-mediated S6K knockdown, and S6K inhibition by expression of a dominant negative mutant. In these experiments, TNF-α was unable to inhibit insulin-induced glucose uptake or Akt activation (indicated by Thr-308 phosphorylation).
In our experimental system, TNF-α induces S6K activation via an Akt-independent mechanism that requires IKK2. In TNF-treated cells, S6K phosphorylation occurred somewhat earlier than Akt phosphorylation. However, it should be noted that the sensitivity of the phospho-antibodies used against different proteins may not be similar. Thus, firm conclusions regarding relative time courses of phosphorylation for different proteins are not warranted based on these data alone. Inhibition of PI3K by Wortmannin did not block the ability of TNF-α to induce S6K phosphorylation. Importantly, using IKK2-null cells, we showed that IKK2 was required for TNF-α activation of S6K and subsequent inhibition of IRS-1 function. Recently, IKK2 was reported to activate mTOR by inhibition of TSC1 (an inhibitor of the mTOR complex) (32, 43). This suggests that IKK2-mTOR interaction is important in the regulation of tumor angiogenesis. Our study establishes the role of this pathway in regulation of energy metabolism. We demonstrated that the IKK2-S6K pathway mediated TNF-α signaling to negatively regulate insulin action. The molecular mechanism involves S6K phosphorylation of IRS-1 at four serines residues: Ser-265/270, Ser-302/307, Ser-632/636, and Ser-1097/1101 (rodent/human). This modification of IRS-1 leads to degradation of IRS-1 protein and insulin resistance. These conclusions are supported by our experiments using the proteasome inhibitor MG132 (Fig. 6, C and D).
It was previously reported that PI3K/AKT/mTOR signaling is required for TNF-α inhibition of IRS-1 function (17). In that study, TNF-α is used at 250 ng/ml (10 nm) to activate the PI3K/AKT pathway (17). LY 294002 was used to block PI3K activity while kinase dead Akt was used to block Akt activity to suggest that activation PI3K/Akt is required for mTOR activation in the TNF-α signaling pathway. Our current study is consistent with this early report in that we found mTOR is required for TNF-α-induced insulin resistance. However, our data also suggest that PI3K/Akt is not required for TNF-α-mediated activation of mTOR/S6K. Instead, IKK2 is required for mTOR/S6K activation. This discrepancy may be related to differences in experimental systems, including dosage of TNF-α (10 ng/ml in our study), choice of PI3K inhibitor, and choice of cell models. In the current study, Wortmannin was used to inhibit PI3K. It is known that LY 294002 inhibits PI3K as well as IKK/NF-κB activity (44). In the current study, IKK2 knock-out cells were used to strongly support the role of IKK2 in the S6K activation.
It was previously reported that S6K may be involved in phosphorylation of four serine sites in IRS-1. These are Ser-302/307 (23), Ser-307/312 (24), Ser-632/636 (17), Ser-1097/1101 (25) in rodent/human IRS-1 protein. However, evidence that S6K directly phosphorylates these residues was missing because the kinase/substrate relationship was not tested using in vitro kinase assays previously. In the current study, three of the four sites (except Ser-307/312) were confirmed in our in vitro kinase assay. Moreover, we identified Ser-265/270 as a novel S6K target. Ser-307/312 of IRS-1 was suggested as a S6K target serine (24). This is not supported by our data from the kinases assay. In kinase assay, we confirmed that this site is directly phosphorylated by IKK2 or PKCθ, not by S6K (Fig. S1). This result is in line with observations about IKK2 and PKCs in the regulation of IRS-1 phosphorylation (6, 45–48). Ser-1097/1101 site was previously reported as a PKCθ substrate (14). In a recent study, this site was suggested to be an S6K target (25). This was confirmed in the kinase assays for S6K in the current study. The fact that we observed phosphorylation of Ser-307/312 in intact cells (Fig. 1C) but not in our in vitro kinase assay (Fig. 3B) may be explained by the fact that Ser-307/312 is phosphorylated by IKK2 or JNK1 in intact cells whereas, IKK2 and JNK1 are not present in our in vitro kinase assays.
We identified a novel interaction among phospho-serine sites in the control of IRS-1 function. It is generally believed that phosphorylation of multiple serine sites in IRS-1 are required for substantial inhibition of IRS-1 function (49). Human IRS-1 has more than 50 serine/threonine residues that are potential targets for post-translational modification by phosphorylation (50). We hypothesized that there may be a regulatory hierarchy among the serine residues in IRS-1 protein. Previously, there was no direct evidence addressing this possibility. The S6K-IRS1 interaction provides an excellent model to test this possibility. There are four S6K target serines in IRS-1. To determine if one serine influences the other three serines in IRS-1 phosphorylation by S6K, we employed a S270A mutant of human IRS-1. Ser-270 in IRS-1 was first reported as a potential target of Akt in 1999 (16). Here, using in vitro kinase assay, we demonstrate, for the first time, that this Ser-270 is directly phosphorylated by S6K. To investigate the role of this novel S6K target site on IRS-1 in more depth, we constructed a point mutant with substitution of alanine for serine at position 270 in human IRS-1. The other three S6K target sites that we found were previously identified and reported by others in the literature. Thus, we did not investigate further these phosphorylation sites as we preferred to focus on our novel findings in more depth. Interestingly, it turned out that Ser-270 appeared to regulate phosphorylation at the other 3 sites by S6K. That is, this mutant had reduced of phosphorylation at the other three serines (Ser-302/307, Ser-632/636, and Ser-1097/1101 in rodent/human IRS-1). This is likely due to reduced association between S6K-IRS1 as indicated by the substantially reduced S6K in the IP product of the S270A mutant IRS-1. Thus, our data suggest that among the four S6K target serines we identified in IRS-1, Ser-270/267 is required for S6K-IRS1 interaction and phosphorylation of the other three S6K-specific serine phosphorylation sites.
Our data suggest that IKK2 may inhibit IRS-1 function through both direct and indirect mechanisms. In an earlier study, IKK2 was shown to phosphorylate IRS-1 directly at Ser-307/312 in mouse/human IRS-1 protein (6). This mechanism was supported by data from an in vitro kinase assay in the earlier study (6), and was confirmed in the current study. In addition, we now provide evidence for an indirect mechanism by which IKK2 inhibits IRS-1 function. In this novel mechanism, IKK2 acts through activation of the mTOR/S6K1 pathway to promote IRS-1 phosphorylation at four additional serine sites, which are themselves not directly phosphorylated by IKK2. Thus, the current study extends our knowledge about molecular mechanisms of IKK2 action in the regulation of insulin signaling.
In summary, we demonstrated that S6K may link TNF-α to the negative feedback loop of insulin signaling that inhibits insulin action. In this new mechanism, IKK2 mediates the action of TNF-α to activate mTOR/S6K. S6K then directly phosphorylates IRS-1 at four serine residues. IKK2 may promote IRS-1 serine phosphorylation directly at Ser-307/312, and indirectly at Ser-265/270, Ser-302/307, Ser-632/636, and Ser-1097/1101 through S6K. All of the kinase-specific serine phosphorylation sites in IRS-1 identified in our study were demonstrated directly with in vitro kinase assays. We conclude that Ser-265/270 is a novel S6K target that is required for substantial interaction between S6K and IRS-1. This interaction is required for S6K phosphorylation of the other three S6K-specific serine phosphorylation sites in IRS-1. Our study provides new insights into the molecular mechanism of insulin resistance induced by pro-inflammatory cytokines such as TNF-α that may be relevant to obesity and other metabolic diseases.
This work was supported, in whole or in part, by National Institutes of Health Intramural Research Program (NCCAM) (to M. J. Q.), NIDDK CNRU Grant 1P30 DK072476 (Genomic Core), and the NIH Grant DK68036 and ADA Research Award 7-07-RA-189 (to J. Y.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: TNF, tumor necrosis factor; HA, hemagglutinin; DN, dominant negative.
References
- 1.Hotamisligil, G. S. (1999) Exp. Clin. Endocrinol. Diabetes 107 119–125 [DOI] [PubMed] [Google Scholar]
- 2.Peraldi, P., and Spiegelman, B. (1998) Mol. Cell Biochem. 182 169–175 [PubMed] [Google Scholar]
- 3.Peraldi, P., Xu, M., and Spiegelman, B. M. (1997) J. Clin. Investig. 100 1863–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engelman, J. A., Berg, A. H., Lewis, R. Y., Lisanti, M. P., and Scherer, P. E. (2000) Mol. Endocrinol. 14 1557–1569 [DOI] [PubMed] [Google Scholar]
- 5.Rui, L., Aguirre, V., Kim, J. K., Shulman, G. I., Lee, A., Corbould, A., Dunaif, A., and White, M. F. (2001) J. Clin. Investig. 107 181–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao, Z., Hwang, D., Bataille, F., Lefevre, M., York, D., Quon, M. J., and Ye, J. (2002) J. Biol. Chem. 277 48115–48121 [DOI] [PubMed] [Google Scholar]
- 7.Gao, Z., Zuberi, A., Quon, M., Dong, Z., and Ye, J. (2003) J. Biol. Chem. 278 24944–24950 [DOI] [PubMed] [Google Scholar]
- 8.Aguirre, V., Uchida, T., Yenush, L., Davis, R., and White, M. F. (2000) J. Biol. Chem. 275 9047–9054 [DOI] [PubMed] [Google Scholar]
- 9.De Fea, K., and Roth, R. A. (1997) J. Biol. Chem. 272 31400–31406 [DOI] [PubMed] [Google Scholar]
- 10.De Fea, K., and Roth, R. A. (1997) Biochemistry 36 12939–12947 [DOI] [PubMed] [Google Scholar]
- 11.Ravichandran, L. V., Esposito, D. L., Chen, J., and Quon, M. J. (2001) J. Biol. Chem. 276 3543–3549 [DOI] [PubMed] [Google Scholar]
- 12.Bourbon, N. A., Sandirasegarane, L., and Kester, M. (2002) J. Biol. Chem. 277 3286–3292 [DOI] [PubMed] [Google Scholar]
- 13.Liu, Y. F., Paz, K., Herschkovitz, A., Alt, A., Tennenbaum, T., Sampson, S. R., Ohba, M., Kuroki, T., LeRoith, D., and Zick, Y. (2001) J. Biol. Chem. 276 14459–14465 [DOI] [PubMed] [Google Scholar]
- 14.Li, Y., Soos, T. J., Li, X., Wu, J., DeGennaro, M., Sun, X., Littman, D. R., Birnbaum, M. J., and Polakiewicz, R. D. (2004) J. Biol. Chem. 279 45304–45307 [DOI] [PubMed] [Google Scholar]
- 15.Gao, Z., Zhang, X., Zuberi, A., Hwang, D., Quon, M. J., Lefevre, M., and Ye, J. (2004) Mol. Endocrinol. 18 2024–2034 [DOI] [PubMed] [Google Scholar]
- 16.Paz, K., Liu, Y. F., Shorer, H., Hemi, R., LeRoith, D., Quan, M., Kanety, H., Seger, R., and Zick, Y. (1999) J. Biol. Chem. 274 28816–28822 [DOI] [PubMed] [Google Scholar]
- 17.Ozes, O. N., Akca, H., Mayo, L. D., Gustin, J. A., Maehama, T., Dixon, J. E., and Donner, D. B. (2001) Proc. Natl. Acad. Sci. U. S. A 98 4640–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eldar-Finkelman, H., and Krebs, E. G. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 9660–9664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liberman, Z., and Eldar-Finkelman, H. (2005) J. Biol. Chem. 280 4422–4428 [DOI] [PubMed] [Google Scholar]
- 20.Ilouz, R., Kowalsman, N., Eisenstein, M., and Eldar-Finkelman, H. (2006) J. Biol. Chem. 281 30621–30630 [DOI] [PubMed] [Google Scholar]
- 21.Kim, J. A., Yeh, D. C., Ver, M., Li, Y., Carranza, A., Conrads, T. P., Veenstra, T. D., Harrington, M. A., and Quon, M. J. (2005) J. Biol. Chem. 280 23173–23183 [DOI] [PubMed] [Google Scholar]
- 22.Haruta, T., Uno, T., Kawahara, J., Takano, A., Egawa, K., Sharma, P. M., Olefsky, J. M., and Kobayashi, M. (2000) Mol. Endocrinol. 14 783–794 [DOI] [PubMed] [Google Scholar]
- 23.Harrington, L. S., Findlay, G. M., Gray, A., Tolkacheva, T., Wigfield, S., Rebholz, H., Barnett, J., Leslie, N. R., Cheng, S., Shepherd, P. R., Gout, I., Downes, C. P., and Lamb, R. F. (2004) J. Cell Biol. 166 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlson, C. J., White, M. F., and Rondinone, C. M. (2004) Biochem. Biophys. Res. Commun. 316 533–539 [DOI] [PubMed] [Google Scholar]
- 25.Tremblay, F., Brule, S., Hee Um, S., Li, Y., Masuda, K., Roden, M., Sun, X. J., Krebs, M., Polakiewicz, R. D., Thomas, G., and Marette, A. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 14056–14061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Um, S. H., Frigerio, F., Watanabe, M., Picard, F., Joaquin, M., Sticker, M., Fumagalli, S., Allegrini, P. R., Kozma, S. C., Auwerx, J., and Thomas, G. (2004) Nature 431 200–205 [DOI] [PubMed] [Google Scholar]
- 27.Tremblay, F., and Marette, A. (2001) J. Biol. Chem. 276 38052–38060 [DOI] [PubMed] [Google Scholar]
- 28.Tremblay, F., Krebs, M., Dombrowski, L., Brehm, A., Bernroider, E., Roth, E., Nowotny, P., Waldhausl, W., Marette, A., and Roden, M. (2005) Diabetes 54 2674–2684 [DOI] [PubMed] [Google Scholar]
- 29.Giraud, J., Leshan, R. L., Lee, Y.-H., and White, M. F. (2004) J. Biol. Chem. 279 3447–3454 [DOI] [PubMed] [Google Scholar]
- 30.Um, S. H., D'Alessio, D., and Thomas, G. (2006) Cell Metab. 3 393–402 [DOI] [PubMed] [Google Scholar]
- 31.Khamzina, L., Veilleux, A., Bergeron, S., and Marette, A. (2005) Endocrinology 146 1473–1481 [DOI] [PubMed] [Google Scholar]
- 32.Lee, D.-F., Kuo, H.-P., Chen, C.-T., Hsu, J.-M., Chou, C.-K., Wei, Y., Sun, H.-L., Li, L.-Y., Ping, B., Huang, W.-C., He, X., Hung, J.-Y., Lai, C.-C., Ding, Q., Su, J.-L., Yang, J.-Y., Sahin, A. A., Hortobagyi, G. N., Tsai, F.-J., Tsai, C.-H., and Hung, M.-C. (2007) Cell 130 440–455 [DOI] [PubMed] [Google Scholar]
- 33.Greene, M. W., Sakaue, H., Wang, L., Alessi, D. R., and Roth, R. A. (2003) J. Biol. Chem. 278 8199–8211 [DOI] [PubMed] [Google Scholar]
- 34.Gao, Z., He, Q., Peng, B., Chiao, P. J., and Ye, J. (2006) J. Biol. Chem. 281 4540–4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schalm, S. S., and Blenis, J. (2002) Curr. Biol. 12 632–639 [DOI] [PubMed] [Google Scholar]
- 36.Ye, J., Gao, Z., Yin, J., and He, H. (2007) Am. J. Physiol. Endocrinol. Metab. 293 E1118–E1128 [DOI] [PubMed] [Google Scholar]
- 37.Cong, L. N., Chen, H., Li, Y., Zhou, L., McGibbon, M. A., Taylor, S. I., and Quon, M. J. (1997) Mol. Endocrinol. 11 1881–1890 [DOI] [PubMed] [Google Scholar]
- 38.Ye, J. (2007) Endocr. Metab. Immune Disord Drug Targets 7 65–74 [DOI] [PubMed] [Google Scholar]
- 39.Li, J., DeFea, K., and Roth, R. A. (1999) J. Biol. Chem. 274 9351–9356 [DOI] [PubMed] [Google Scholar]
- 40.Takano, A., Usui, I., Haruta, T., Kawahara, J., Uno, T., Iwata, M., and Kobayashi, M. (2001) Mol. Cell. Biol. 21 5050–5062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ueno, M., Carvalheira, J. B., Tambascia, R. C., Bezerra, R. M., Amaral, M. E., Carneiro, E. M., Folli, F., Franchini, K. G., and Saad, M. J. (2005) Diabetologia 48 506–518 [DOI] [PubMed] [Google Scholar]
- 42.Patti, M. E., and Kahn, B. B. (2004) Nat. Med. 10 1049–1050 [DOI] [PubMed] [Google Scholar]
- 43.Dan, H. C., Adli, M., and Baldwin, A. S. (2007) Cancer Res. 67 6263–6269 [DOI] [PubMed] [Google Scholar]
- 44.Xie, P., Browning, D. D., Hay, N., Mackman, N., and Ye, R. D. (2000) J. Biol. Chem. 275 24907–24914 [DOI] [PubMed] [Google Scholar]
- 45.Yuan, M., Konstantopoulos, N., Lee, J., Hansen, L., Li, Z. W., Karin, M., and Shoelson, S. E. (2001) Science 293 1673–1677 [DOI] [PubMed] [Google Scholar]
- 46.Nakajima, K., Yamauchi, K., Shigematsu, S., Ikeo, S., Komatsu, M., Aizawa, T., and Hashizume, K. (2000) J. Biol. Chem. 275 20880–20886 [DOI] [PubMed] [Google Scholar]
- 47.Dey, D., Mukherjee, M., Basu, D., Datta, M., Roy, S. S., Bandyopadhyay, A., and Bhattacharya, S. (2005) Cell Physiol Biochem. 16 217–228 [DOI] [PubMed] [Google Scholar]
- 48.Jiang, G., Dallas-Yang, Q., Liu, F., Moller, D. E., and Zhang, B. B. (2002) J. Biol. Chem. 278 180–186 [DOI] [PubMed] [Google Scholar]
- 49.White, M. F. (2002) Am. J. Physiol. Endocrinol. Metab. 283 E413–E422 [DOI] [PubMed] [Google Scholar]
- 50.Araki, E., Sun, X. J., Haag, B. L., 3rd, Chuang, L. M., Zhang, Y., Yang-Feng, T. L., White, M. F., and Kahn, C. R. (1993) Diabetes 42 1041–1054 [DOI] [PubMed] [Google Scholar]