Abstract
Myocardin (MYOCD) is a transcriptional co-activator that promotes cardiac or smooth muscle gene programs through its interaction with myocyte-enhancing factor (MEF2) or serum-response factor (SRF). Isoforms of MYOCD with a truncated amino terminus show increased activity when compared with those with the full-length amino terminus, but how this is achieved remains unknown. We identified a rare human sequence variation in MYOCD in a patient with congenital heart disease that resulted in a missense mutation at codon 259 (K259R). This variation created a hypomorphic cardiac isoform with impaired SRF binding and transactivation capacity but did not impair the smooth muscle isoform of MYOCD, which lacks the amino terminus. Consistent with differential effects of the amino terminus on the K259R mutation, we found that the cardiac-specific amino terminus acted in an autoinhibitory fashion to bind MYOCD via specific negatively charged residues and thereby repressed SRF-dependent MYOCD activity. This effect was exaggerated in the MYOCD-K259R mutant. The amino terminus was sufficient to impair MYOCD-dependent fibroblast conversion into smooth muscle cells as well as cardiomyocyte hypertrophy. These findings identify a novel mechanism that regulates levels of MYOCD-dependent activation of the SRF genetic program differentially in cardiac and smooth muscle.
Myocardin (MYOCD)2 was the first recognized member of a family of transcriptional co-activators that binds to serum-response factor (SRF) to activate cardiac or smooth muscle gene programs (1). The two other MYOCD-related transcription factors (MRTF), MRTF-A (also known as MAL, MKL1, or BSAC) and MRTF-B (MKL2), also co-activate SRF, but for unknown reasons, MRTF-B is a weaker activator than either MYOCD or MRTF-A (reviewed in Refs. (2-5). MYOCD, MRTF-A, and MRTF-B are all co-expressed in the developing myocardium early in gestation and are expressed in distinct populations of smooth muscle cells later during development (6-9). In mice, targeted deletion of the SRF interaction domain in MYOCD led to embryonic lethality from cardiovascular defects (6). MYOCD-null mice died midgestation, by embryonic day 10.5, apparently from insufficient smooth muscle specification in the distal aorta and placenta (6). Furthermore, conditional deletion of MYOCD from neural crest resulted in defects in neural crest-derived smooth muscle differentiation (10).
Cardiac and smooth muscle both express MYOCD, but the smooth muscle isoform (MYOCDΔMHD) utilizes an alternate start codon that leads to a partial truncation of the MYOCD homology domain (MHD) (11). The MHD, found in the amino terminus of cardiac MYOCD, contains three RPEL motifs that regulate actin-dependent nuclear localization of MRTF-A/B but not of MYOCD (12). It is thought that actin dynamics do not regulate MYOCD because two of the three RPEL motifs in MYOCD are evolutionarily divergent from the consensus sequence, whereas both MRTFs have three highly conserved RPEL motifs. Although MYOCD is constitutively nuclear, its MHD contains a binding site for myocyte-enhancing factor 2 (MEF2), which activates portions of the cardiac gene program that are distinct from the SRF-dependent program (11). MYOCD truncations that lack the MHD show increased SRF-dependent transactivation both in vitro (1) and in vivo (13), but how this is achieved is unknown.
Multiple transcription factors interact with and synergistically regulate the MYOCD transcriptional complex on DNA. The Jagged/Notch signal transduction pathway is able to inhibit MYOCD-dependent SRF-co-activation via HRT2, a transcriptional repressor, during smooth muscle differentiation (14). Depending on the context, the GATA4/5/6 family of transcription factors is able to either activate or inhibit MYOCD-dependent transactivation (15). Another MYOCD regulator is ELK1, which is necessary for SRF-dependent induction of MYOCD-independent immediate early genes (13). The SRF-binding motifs in MYOCD and ELK1 show strong homology to one another, and ELK1 utilizes this motif to compete with MYOCD for SRF (13). In this way, ELK1 is able to deactivate some MYOCD target genes in favor of growth over differentiation (13). Although these and other transcription factors are known to synergistically regulate MYOCD-dependent activity, it is unknown whether there are mechanisms that act directly on MYOCD to regulate the SRF interaction or how the MHD-dependent decrease in MYOCD activity occurs.
Here, we describe how a rare human MYOCD missense mutation, resulting in a lysine to arginine substitution at codon 259 (K259R), revealed a novel mechanism by which MYOCD activity is regulated. The mutation, which occurred in a subject with thickened pulmonary valves, resulted in a hypomorphic cardiac MYOCD, but did not affect activity of the smooth muscle MYOCD lacking the MHD. In exploring how the MHD influenced the K259R effects, we found that the amino terminus regulated MYOCD activity in an autoinhibitory fashion by binding to MYOCD and disrupting SRF-dependent activation. The MYOCD K259R mutant showed a more pronounced response to the autoinhibitory activity of the MHD. These findings suggest a novel mechanism for regulation of MYOCD activity in cardiac versus smooth muscle.
EXPERIMENTAL PROCEDURES
Plasmid Construction—FLAG-MYOCD truncations, atrial natriuretic factor-luciferase (ANF-Luc), and smooth muscle 22α-luciferase (SM22α-Luc) have been described (1). MHD truncations were created by PCR amplification of MYOCD cDNA with added 5′ end NotI and 3′ end XbaI restriction enzyme sites. K259R, R71R115 → AA, and D130E135 → AA were mutated with the QuikChange II kit (Stratagene), according to the manufacturer's instructions. All mutations were confirmed by sequencing. Primer sequences are available upon request.
Cell Culture and Transfections—Cos-1 cells (American Type Culture Collection) were maintained and passaged as described (16). Transfections were carried out in 12-well dishes unless otherwise indicated. Cells were transfected with 50 ng of each reporter plasmid and 100 ng of each cDNA for 40-44 h and harvested in passive lysis buffer (Promega). Total amounts of DNA were kept constant with corresponding expression vector without a cDNA insert. A CMV-LacZ plasmid was co-transfected for internal transfection efficiency control (1). Western analysis was performed for all transfected proteins to ensure equal expression. Luciferase assays were performed with a PerkinElmer Life Sciences Victor3 plate reader as described (16).
CH310T1/2 cells (American Type Culture Collection) were maintained and passaged as described, except that the plates were coated with 1% gelatin for 30 min before passaging (17). Transfections were performed with FuGENE 6 (Roche Applied Science) according to the manufacturer's instructions. Twenty-four hours after transfection, the cells were switched to Dulbecco's modified Eagle's medium containing 2% horse serum. Seven days later, immunocytochemistry and quantitative real-time (RT) PCR (TaqMan, Applied Biosystems) were performed. For each experiment, 500 ng of each cDNA was used for each well of a six-well plate. A CMV-eGFP plasmid (100 ng, Amaxa) was co-transfected as an internal control for transfection efficiency. Cells were transfected and counted double-blind. Conversion efficiency was calculated as the number of SM-α-actin-positive cells divided by the number of green fluorescent protein-positive cells.
Primary cardiomyocytes were harvested from 0-2-day-old Sprague-Dawley rats as described (18) with minor modifications. The cardiomyocytes were enzymatically separated with 1 μg/ml pancreatin, plated on 0.1% gelatin-coated dishes, and infected 1 day later with lentivirus (pLenti6 Gateway System, Invitrogen) overexpressing protein under control of the EF1 promoter. Twenty-four hours later, phenylephrine (PE) was added to the medium (final concentration, 20 μm.) Forty-eight hours after infection, immunofluorescence and quantitative RT-PCR were performed. Area calculations of the α-actinin-positive cells were made using the Image-Pro 5.0 software; control cells without PE were set at an arbitrary value of 100, and then each condition was set proportionate to the control cells. Cardiomyocyte infection and area calculations were performed double-blind.
Quantitative RT-PCR—Whole RNA was purified with TRI Reagent (Applied Biosystems), treated with DNA-free DNase (Ambion), and synthesized into cDNA with poly(dT) primers (Superscriptase III kit, Invitrogen) according to the manufacturer's instructions. RT-PCR was performed with Applied Biosystems 2× master mix and TaqMan primers (see supplemental Table S1) as described (16). Analysis was performed using the 2(-ΔΔCt) method.
Protein Interaction Assays—Co-immunoprecipitation (Co-IP) and electromobility shift assays (EMSA) were performed as described (1). For Co-IP, 1 μg of FLAG-MYOCD and 100 ng of hemagglutinin-tagged SRF were transfected and harvested 40 h later. For EMSA, protein was translated from 1 μg of each plasmid with the Promega TnT kit. An oligonucleotide containing the c-fos CArG box (1) was 32P-labeled with the Roche Applied Science high prime labeling kit. Quantification of the binding efficiency was made using the ImageJ v1.4 software.
Immunocytochemistry and Western Blot—Immunocytochemistry and Western analysis were performed as described (16). Antibodies were diluted as follows: mouse anti-SM-α-actin (Sigma), 1:1000; mouse anti-α-actinin (Sigma), 1:200; mouse anti-FLAG (M2, Sigma) 1:5000 (Western) or 1:500 (immunocytochemistry); and mouse anti-Myc (9a7, Santa Cruz Biotechnology), 1:1000. Fluorescein isothiocyanate- or TRITC-conjugated goat anti-mouse secondary antibodies were used for immunocytochemistry, and alkaline phosphatase-conjugated goat anti-mouse or goat anti-rabbit antibodies were used for Western analysis (Jackson Immunologicals).
RESULTS
A Unique MYOCD Sequence Variant in a Patient with a Congenital Cardiac Malformation—In a genetic screen of subjects with congenital cardiovascular malformations,3 we found an adenosine-to-guanosine sequence variant in the gene encoding MYOCD (GenBank™ accession number AF532596) that resulted in an arginine substitution for a highly conserved lysine at codon 259 (A776G, supplemental Fig. 1 A). This Hispanic patient had a thickened and stenotic pulmonary valve that obstructed flow of blood from the right ventricle to the pulmonary artery. The frequency of the K259R sequence variation was evaluated in the Dallas Heart Study population of control subjects (19). We found the sequence variation in 1 out of 1137 control individuals, indicating that this is an extremely rare genetic variant. This subject was also Hispanic (out of 212 total Hispanic controls). The valve morphology of the control individual is unknown, and subclinical disease could not be excluded. Sequence alignments show that Lys-259 has been evolutionarily conserved from fruit flies to humans (supplemental Fig. 1B), suggesting that Lys-259 is under selective pressure and may be important for MYOCD function. This residue is also conserved in MRTF-A, but MRTF-B contains an arginine at the corresponding codon (Arg-288, supplemental Fig. 1B). Interestingly, MRTF-B is transcriptionally less active than MYOCD or MRTF-A on most promoters (2).
Cardiac but Not Smooth Muscle Myocardin K259R Is Hypomorphic and Has Reduced SRF Affinity—MYOCD Lys-259 is not contained in the SRF-binding B-box, but does lie within the lysine-rich basic domain that is necessary for SRF binding (Fig. 1A) (1). We therefore hypothesized that K259R might affect MYOCD-dependent promoter activation of SRF-dependent targets (1). Transient co-transfection of the 935-amino acid, cardiac-specific isoform of MYOCD (MYOCD935) with luciferase reporters driven by the SM22α-Luc or ANF-Luc promoter showed that K259R MYOCD935 was hypomorphic when compared with wild type (WT) MYOCD935 (Fig. 1, B and C). The reduced activity was not due to changes in cellular localization, as WT and K259R MYOCD935 both localized to the nucleus (Fig. 1D). We tested whether the reduction in MYOCD activity was due to a corresponding reduction in binding between MYOCD and SRF. In Co-IP assays, we were unable to detect an interaction between MYOCD935 K259R and SRF, but WT MYOCD935 showed normal SRF binding (Fig. 1E). In a more sensitive test involving DNA-protein interactions, EMSA revealed that MYOCD935 K259R could interact with SRF, but this interaction was harder to detect than with WT MYOCD935 (Fig. 1F).
FIGURE 1.
Cardiac myocardin K259R is hypomorphic and has reduced SRF binding affinity. A, schematic of cardiac MYOCD (MYOCD935) and smooth muscle MYOCD (MYOCDΔMHD) showing the locations of known domains and the K259R mutation. K, lysine rich basic domain; B, ELK1-like B-box; QQ, polyglutamine tract; SAP, SAF/Acinus/PIAS domain; LZ, leucine zipper; TA, transactivation. B and C, effect of the K259R mutation on MYOCD935 co-activation of the SM22α-Luc (B) and ANF-Luc (C) promoters. D, immunocytochemistry using FLAG antibody on cells transfected with FLAG-tagged WT or K259R MYOCD935. Nuclear localization is indicated by co-staining with DAPI. E, HA-tagged SRF (HA-SRF) co-immunoprecipitates with WT but not K259R FLAG-tagged MYOCD935. Cell lysates were immunoprecipitated (IP) with anti-FLAG and immunoblotted (IB) with anti-HA antibodies. Inputs are shown. F, EMSA with 32P-labeled c-fos CArG box oligonucleotide (Oligo). SRF impedes oligonucleotide migration, and MYOCD forms a ternary complex with SRF to further impede mobility. K259R MYOCD showed a weaker ability to form the ternary complex. Quantification of band intensities indicates an ∼50% reduction with the mutant form. G, effect of the R288K mutation on MRTF-B co-activation of the SM22α-Luc promoter. Arg-288 corresponds to same codon as Lys-259 in MYOCD. Values are the mean ± S.D. of three experiments in triplicate. Statistical differences were calculated using the Student's t test. *, p < 0.05 versus WT MYOCD.
Since MRTF-B is a weaker activator than MYOCD or MRTF-A, we hypothesized this may be due, in part, to the evolutionarily divergent arginine at codon 288. Mutation of arginine 288 to lysine (R288K), the residue present in MYOCD or MRTF-A, increased MRTF-B-dependent activation of SM22α-Luc nearly 5-fold (Fig. 1G). Although this was still well below the levels achieved with MYOCD or MRTF-A in vitro, it demonstrated that the two basic amino acids are not functionally equivalent in this domain and that an arginine at this residue of the MYOCD family of proteins is sufficient to limit the transactivation potential of MYOCD proteins.
Since the cardiac (MYOCD935) and smooth muscle (MYOCDΔMHD) isoforms also have different levels of activity, we tested whether K259R affected MYOCDΔMHD in a similar fashion to the cardiac isoform. In contrast to what was observed with MYOCD935, K259R MYOCDΔMHD was functionally normal in luciferase reporter assays using the SM22α promoter (Fig. 2A). Unlike with K259R MYOCD935, K259R MYOCDΔMHD appeared to interact with SRF similar to WT MYOCD as assessed by Co-IP (Fig. 2B) and EMSA (Fig. 2C).
FIGURE 2.
Loss of MYOCD MHD rescues K259R effects. A, MYOCDΔMHD and K259R MYOCDΔMHD activated SM22-luciferase similarly. B and C, MYOCD-SRF binding experiments. B, Co-IP showing similar levels of binding of HA-tagged SRF with WT and K259R FLAG-tagged MYOCDΔMHD. Cell lysates were immunoprecipitated (IP) with anti-FLAG and immunoblotted (IB) with anti-HA antibodies. Inputs are shown. C, EMSA of 32P labeled c-fos CArG box oligonucleotide (Oligo). WT and K259R MYOCDΔMHD show similar levels of SRF ternary complex formation (MYOCD + SRF). Values are the mean ± S.D. of three experiments in triplicate.
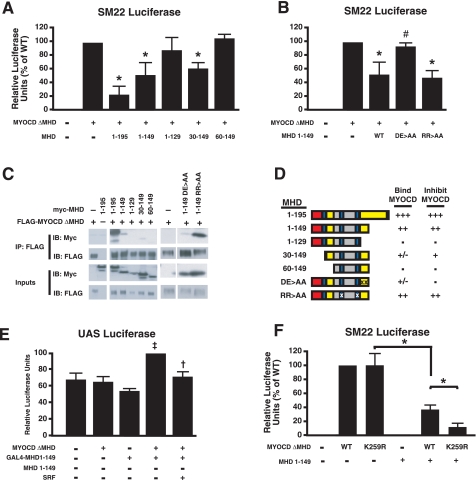
The MYOCD MHD Inhibits MYOCD-dependent Transcription in Vitro by Binding to MYOCD and Blocking Its Interaction with SRF—Because truncating the MHD rescued the K259R hypomorph despite being in a distant domain, we hypothesized that the MHD of MYOCD might normally act as an intramolecular autoinhibitor, consistent with the increased activity of MYOCDΔMHD (20, 21). If this were the case, the MHD, even when separated from the rest of MYOCD, might bind to other domains in MYOCD to mask or allosterically compete for SRF binding. Consistent with this, we found that expression of amino acids 1-195 inhibited transactivation by MYOCDΔMHD on the SRF-dependent reporters described earlier. Amino acids 1-149 also inhibited MYOCD activity in a dose-dependent manner; however, amino acids 1-129 did not (Fig. 3A). Truncation of the MHD amino terminus revealed that amino acids 1-30, but not 30-60, were dispensable for inhibition of MYOCD transactivation (Fig. 3A). Thus, the minimal MYOCD inhibitory domain appeared to lie between amino acids 30 and 149.
FIGURE 3.
The MYOCD MHD acts as an autoinhibitory domain by binding to MYOCD and repressing MYOCD-dependent in vitro activity. A, truncations of MHD to determine minimal inhibitory domain of MYOCD-dependent activation of the SM22 promoter. B, mutation of conserved acidic residues Asp-130 and Glu-135 to alanines (DE → AA) relieved MHD inhibition. Mutation of the RPEL motif by an R72A and R115A change (RR → AA) did not affect MHD inhibition. C, Co-IP of myc-tagged-MHD truncations with FLAG-tagged MYOCDΔMHD to determine minimal binding domain necessary. Cell lysates were immunoprecipitated (IP) with anti-FLAG and immunoblotted (IB) with anti-myc antibodies. Inputs are shown. D, schematic summarizing the ability of MHD truncations to bind to and inhibit MYOCD in vitro. Red box, MEF2-binding motif; blue box, RPEL motifs; yellow box, MHD inhibitory motifs. Xs indicate point mutations. E, mammalian two-hybrid assay between MYOCD and the MHD. MHD-1-149 conjugated to the DNA-binding domain of GAL4 (GAL4-MHD-1-149) and MYOCDΔMHD together activated transcription of the UAS-Luc promoter. MYOCD-dependent activation of GAL4-MHD-1-149 on the UAS-Luc reporter was inhibited by SRF. F, effects of MHD-dependent inhibition on MYOCD transactivation of SM22α-Luc. K259R MYOCDΔMHD responded to MHD-dependent inhibition more than WT MYOCDΔMHD. Values are the mean ± S.D. of three experiments in triplicate. Statistical differences were calculated using the Student's t test. *, p < 0.05 versus MYOCD; #, p < 0.05 versus WT MHD 1-149; †, p < 0.05 versus GAL4-MHD1-149 and MYOCDΔMHD.
The MHD of MRTF-A has been well characterized as an actin-dependent regulator of nuclear localization (12, 21); however, MYOCD is thought to be constitutively nuclear (1, 12). Similar to MRTF-A, MYOCD contains two conserved RPEL motifs in the minimal inhibitory MHD. In previous reports, mutation of the conserved arginines to alanine of the RPEL motif disrupted the MHD-dependent nuclear localization of MRTF-A (12). We found that mutation of the conserved arginines (R71A, R115A) of the two RPEL motifs had no effect on the ability of the MHD to inhibit MYOCD activity, suggesting that the RPEL motifs are not necessary for autoinhibition (Fig. 3B). Another known motif within the MHD is the MEF2-binding site, located in the first 17 amino acids of MYOCD (22). Because amino acids 1-30 were not necessary for inhibition, the MEF2-binding domain may also be dispensable for inhibition of SRF-dependent MYOCD activity (Fig. 3A).
Since the RPEL- and MEF2-binding motif were not necessary and MHD1-129 was insufficient to inhibit MYOCD co-activation, we focused on amino acids 129-149. Sequence analysis identified no recognizable motifs, but revealed two acidic residues that are conserved from humans to amphibians: aspartate 130 (Asp-130) and glutamate 135 (Glu-135, supplemental Fig. 1C). Mutation of these two negatively charged residues to uncharged alanines (DE → AA) disrupted the autoinhibition, effectively mapping some of the essential residues required for inhibition (Fig. 3B).
Next, we investigated whether the MHD was a true autoinhibitory domain that directly binds to MYOCD via intramolecular interactions. In Co-IP experiments, truncated MHD that contained the minimal autoinhibitory domain (30-149) was able to bind to MYOCD, although the 30-149 construct had less affinity than the 1-149 construct (Fig. 3C). Furthermore, mutation of both Asp-130 and Glu-135 disrupted MYOCD-MHD interactions, whereas mutation of the conserved RPEL motifs did not. Autoinhibitory domains generally have to retain intramolecular binding to inhibit protein activity, which may explain why the DE → AA mutations were not inhibitory in the luciferase reporter assays. To seek further evidence of a physical interaction between MYOCD and the MHD, we developed a mammalian two-hybrid assay, with the MHD-1-149 conjugated to the DNA-binding domain of GAL4 (GAL4-MHD). We observed that MYOCD was able to co-activate the UAS-luciferase reporter in the presence of GAL4-MHD (Fig. 3E). Furthermore, SRF was able to inhibit the MYOCD-dependent activation of GAL4-MHD, which suggested that the MHD and SRF could compete for interaction with MYOCD (Fig. 3E).
Since the MHD could compete with SRF for binding to MYOCD and the MHD was necessary for K259R to behave as a hypomorph, we hypothesized that K259R would confer greater autoinhibition when compared with WT MYOCD. Consistent with this hypothesis, K259R MYOCDΔMHD displayed more MHD-dependent inhibition than WT MYOCDΔMHD on the SM22α promoter (Fig. 3F).
The MYOCD MHD Inhibits MYOCD-dependent Smooth Muscle Conversion and Cardiomyocyte Hypertrophy—Full-length MYOCD has a limited capacity to transdifferentiate CH310T1/2 fibroblasts into smooth muscle cells (17). Since truncations of MYOCD lacking the MHD show greater smooth muscle conversion than MYOCD935, we hypothesized that this phenomenon might be explained by MHD autoinhibition. We therefore tested whether introduction of MHD1-149 would repress MYOCD-dependent smooth muscle conversion (Fig. 4, A and B). MYOCDΔMHD converted ∼40% of transfected CH310T1/2 fibroblasts into SM-α-actin-positive cells within 7 days. Consistent with our in vitro reporter assay results, MHD1-149 inhibited the conversion efficiency of MYOCDΔMHD by about 50% (Fig. 4, A and B). Neither the RPEL motifs nor the MEF2-binding domain were necessary, but the two acidic residues (Asp-130, Glu-135) were required for effective inhibition of conversion (Fig. 4B). We next tested the mRNA transcript levels of multiple MYOCD target genes by quantitative RT-PCR to determine whether MHD-dependent inhibition affected genes other than SM-α-actin. MHD-dependent inhibition of MYOCD extended to multiple other direct MYOCD targets such as SM22α, ANF, and smooth muscle calponin (Fig. 4C). These targets were also dependent on Asp-130 and Glu-135.
FIGURE 4.
The MHD of MYOCD inhibits MYOCD-dependent conversion of fibroblasts into smooth muscle cells. A, 10T1/2 fibroblast cells transfected with MYOCD with or without the MHD were subjected to smooth muscle conversion protocol and stained for SM-α-actin as follows: TRITC, red; nuclei, DAPI, blue. Representative pictures are shown. B, double-blind quantification of the number of transfected fibroblasts that converted in to SM-α-actin-positive cells. A panel of MHD truncations was assessed. CMV-GFP was used as an internal control for transfection efficiency. For each conversion condition, experiments were performed three times in duplicate, and green fluorescent protein-positive (GFP +) and TRITC-positive cells were counted in four randomly selected fields. Values are mean ± S.E. (n = 8 fields of cells). C, quantitative RT-PCR for MYOCD-dependent genes in 10T1/2 cells: ANF, Sm22α, and smooth muscle calponin (SMC-Calponin). Co-transfection of WT but not DE → AA MHD inhibited MYOCD-dependent gene expression. Values are the mean ± S.D. of two biological replicates in technical duplicates. Statistical differences were calculated using the Student's t test. *, p < 0.05 versus MYOCD, #, p < 0.05 versus WT MHD 1-149.
We also tested whether the MHD could affect MYOCD activity in cardiomyocytes. MYOCD induces a hypertrophic response in primary cardiomyocytes (18, 23), and a dominant-negative form of MYOCD (DN-MYOCD) inhibits hypertrophy in response to chemicals such as PE. We investigated whether the MHD might mimic the DN-MYOCD effect. Indeed, WT but not DE → AA MHD 1-149 inhibited PE-induced cardiomyocyte hypertrophy, as assessed by measurements of cell surface area (Fig. 5, A and B). This observation suggests that the inhibition of hypertrophy was independent of MEF2 since the DE → AA mutant retains the MEF2-binding domain. The hypertrophic response is normally accompanied by an increase in expression of the ANF, brain natriuretic peptide (BNP (Nppb)), and skeletal muscle α-actin (Sk-α-actin (ACTA1)) transcripts and a decrease in GSK3β expression (18, 23). Consistent with the calculations of surface area, DN-MYOCD and WT MHD inhibited the PE-induced changes in hypertrophic gene expression, and the DE → AA mutant MHD (Fig. 5C) displayed diminished inhibitory activity.
FIGURE 5.
The MYOCD MHD inhibits PE-dependent cardiomyocyte hypertrophy. A and B, MHD inhibition of PE-dependent cardiomyocyte hypertrophy. A, immunocytochemistry of neonatal rat cardiomyocytes subjected to PE (20 μm) for 2 days after being infected for 1 day with lentivirus that overexpresses WT MHD, mutant (DE → AA) MHD, or dominant negative MYOCD (DN-MYOCD). α-Actinin, TRITC, red; nuclei, DAPI, blue. B, double-blind calculation of α-actinin-positive cell surface area of infected cardiomyocytes. Fields of cells in each condition were randomly selected, and area calculations were made using the Image-Pro 5.0 software. Values are mean ± S.E. when compared with the control, which is assigned a value of 100 (n = 80). C, quantitative RT-PCR for hypertrophy-dependent genes: ANF, brain natriuretic peptide (BNP), skeletal muscle α-actin (Sk-α-actin (ACTA1)), and GSK3β. Infection with lentivirus overexpressing DN-MYOCD and WT-MHD but not DE → AA MHD inhibited the hypertrophy-dependent changes in gene expression. Error bars represent S.D. of three biological replicates in technical duplicate. Statistical differences were calculated using the Student's t test. *, p < 0.05 versus PE alone, #, p < 0.05 versus PE with WT MHD.
DISCUSSION
This study shows that MYOCD has the potential for dynamic intramolecular regulation of the SRF-dependent gene program. A rare human mutation, K259R, resulted in attenuated activity of a cardiac isoform of myocardin without affecting a smooth muscle isoform that lacked the amino terminus. This observation led to the discovery that the amino terminus acted as an inhibitor of SRF-dependent myocardin activity. Although neither the MEF2-binding site nor the RPEL motifs in the amino terminus were necessary for this effect, we did find that two evolutionarily conserved acidic residues within the amino terminus were necessary for MHD-dependent inhibition. The K259R mutation was also more responsive to MHD-dependent inhibition, consistent with the notion that the K259R hypomorph is actually a gain of MHD-dependent inhibitory function. MRTF-B, the hypoactive MYOCD family member, showed increased transactivation potential when arginine 288 was mutated to lysine. This suggests that MHD-dependent inhibition may be a general mechanism for regulating the Myocardin family by utilizing the MYOCD Lys-259 and MRTF-B Arg-288 residue.
The differential effects of the human K259R mutation on the function of MYOCD with or without MHD1-149 revealed novel aspects of MYOCD regulation. One interpretation of our finding is that the amino terminus of MYOCD sterically inhibits the SRF-binding domain and that the K259R mutation strengthens this intramolecular interaction between the MHD and the SRF-binding domain. In the smooth muscle form of MYOCD, the autoinhibitory domain (MHD) is absent; therefore, the K259R mutation is inconsequential. This function of the MHD, along with the interaction with MEF2, may explain the differential effect of MYOCD in cardiac and smooth muscle. Interestingly, inclusion of amino acids 149-195 in the MHD constructs increased the binding affinity and inhibitory activity of the MHD upon MYOCD. Of those 46 amino acids 25, or 54%, are conserved to amphibians, and nine of those are acidic (e.g. Asp-130 and Glu-135; data not shown). It is interesting to speculate that a direct interaction occurs between the negatively charged acidic residues of the MHD and the positively charged lysine at 259 or even the entire lysine-rich basic domain. Alternatively, the increased length of the arginine side chain, when compared with lysine, may disrupt MYOCD folding in a way that blocks SRF binding.
In vitro, MYOCD protein is responsive to extracellular hypertrophic stimuli on synthetic promoters, apparently without altered steady state protein levels (18). MYOCD is sumoylated by the PIAS1 ligase and associates with the histone acetyl-transferase p300, which also acetylates proteins (24, 25). Both of these activities are dependent upon lysine residues, and in these instances, arginine is not able to replace the post-translationally modified lysines (24, 25). Therefore, MYOCD may be post-translationally modified at Lys-259 to directly or allosterically disrupt MHD binding to MYOCD. Ultimately, the protein structure of MYOCD will help resolve the precise mechanism of MHD-mediated inhibition of the activity of MYOCD in cardiac tissue.
The patient with the K259R mutation had a thickened pulmonic valve. Although we cannot be certain of the contribution of this mutation, the rarity of the mutation, evolutionary conservation, and the functional consequences would suggest that it is a contributing factor. Outflow tract valves are composed of early outflow tract myocardium, neural crest, and endocardial-derived cells (26). MYOCD-null mutant embryos only survive to embryonic day 10.5, before the formation of cardiac valves, precluding the ability to examine the role of MYOCD in these tissues (6). Selective ablation of the MYOCD gene from neural crest resulted in patent ductus arteriousus but had no effect on valve morphogenesis (10), although MYOCD may play a role in outflow myocardium or endocardium. To determine whether the K259R mutation contributed to the heart disease in the subject or co-segregated with the true disease-causing mutation as part of a haplotype block, further studies will be necessary, including generation of a mouse model of MYOCD K259R. If the knock-in mouse has outflow tract defects, it may provide a valuable tool for understanding valve development and disease. Most genes that are known to play a role in valve development (e.g. vascular endothelial growth factor, nuclear factor of activated T cells 1, and Notch1) are expressed in the endocardium of the prevalve tissue and respond to paracrine signals, such as transforming growth factor-β family members, which are released from the muscle tissue surrounding the valves (27). Since MYOCD is expressed in the myocardium, it may be well situated to induce such paracrine signals (1). Myocardin expression is also induced by transforming growth factor-β, and it acts as a tumor suppressor in precancerous mesenchyme (28). Thus, MYOCD may be involved in guiding early cardiac mesenchymal progenitors into fully differentiated valve tissue, and any reduction in its activity could result in overproduction of undifferentiated valve progenitors and thickened valves.
Supplementary Material
Acknowledgments
We thank E. N. Olson for advice, reagents, and generous help throughout the project and D. Z. Wang, G. Amarasinghe, and A. Muth for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 HL080592 and by University of Texas Southwestern Cardiovascular Training Grant NIH 5-T32-HL007360. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains a supplemental figure and a supplemental table.
This article was selected as a Paper of the Week.
Footnotes
The abbreviations used are: MYOCD, myocardin; MYOCD935, cardiac isoform of MYOCD; MYOCDΔMHD, smooth muscle isoform of MYOCD; MHD, myocardin homology domain; SRF, serum-response factor; MEF2, myocyte-enhancing factor; SM22α, 22-kilodalton smooth muscle protein α (transgel-lin); ANF, atrial natriuretic factor (Nppa); SM-α-actin, smooth muscle α-actin (ACTA2); Co-IP, co-immunoprecipitation; EMSA, electromobility shift assay; MRTF, myocardin-related transcription factor; PE, phenyleph-rine; TRITC, tetramethylrhodamine isothiocyanate; DAPI, 4′,6-diamidino-2-phenylindole; RT-PCR, real-time PCR; WT, wild type; DN, dominant-negative; Luc, luciferase; HA, hemagglutinin.
V. Garg and D. Srivastava, unpublished data.
References
- 1.Wang, D., Chang, P. S., Wang, Z., Sutherland, L., Richardson, J. A., Small, E., Krieg, P. A., and Olson, E. N. (2001) Cell 105 851-862 [DOI] [PubMed] [Google Scholar]
- 2.Wang, D. Z., Li, S., Hockemeyer, D., Sutherland, L., Wang, Z., Schratt, G., Richardson, J. A., Nordheim, A., and Olson, E. N. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 14855-14860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang, D. Z., and Olson, E. N. (2004) Curr. Opin. Genet. Dev. 14 558-566 [DOI] [PubMed] [Google Scholar]
- 4.Cen, B., Selvaraj, A., and Prywes, R. (2004) J. Cell. Biochem. 93 74-82 [DOI] [PubMed] [Google Scholar]
- 5.Parmacek, M. S. (2007) Circ. Res. 100 633-644 [DOI] [PubMed] [Google Scholar]
- 6.Li, S., Wang, D. Z., Wang, Z., Richardson, J. A., and Olson, E. N. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 9366-9370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oh, J., Richardson, J. A., and Olson, E. N. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 15122-15127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li, S., Chang, S., Qi, X., Richardson, J. A., and Olson, E. N. (2006) Mol. Cell. Biol. 26 5797-5808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li, J., Zhu, X., Chen, M., Cheng, L., Zhou, D., Lu, M. M., Du, K., Epstein, J. A., and Parmacek, M. S. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 8916-8921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang, J., Cheng, L., Li, J., Chen, M., Zhou, D., Lu, M. M., Proweller, A., Epstein, J. A., and Parmacek, M. S. (2008) J. Clin. Investig. 118 515-525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Creemers, E. E., Sutherland, L. B., Oh, J., Barbosa, A. C., and Olson, E. N. (2006) Mol. Cell 23 83-96 [DOI] [PubMed] [Google Scholar]
- 12.Miralles, F., Posern, G., Zaromytidou, A. I., and Treisman, R. (2003) Cell 113 329-342 [DOI] [PubMed] [Google Scholar]
- 13.Wang, Z., Wang, D. Z., Hockemeyer, D., McAnally, J., Nordheim, A., and Olson, E. N. (2004) Nature 428 185-189 [DOI] [PubMed] [Google Scholar]
- 14.Proweller, A., Pear, W. S., and Parmacek, M. S. (2005) J. Biol. Chem. 280 8994-9004 [DOI] [PubMed] [Google Scholar]
- 15.Oh, J., Wang, Z., Wang, D., Lien, C., Xing, W., and Olson, E. (2004) Mol. Cell. Biol. 24 8519-8528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao, Y., Ransom, J. F., Li, A., Vedantham, V., von Drehle, M., Muth, A. N., Tsuchihashi, T., McManus, M. T., Schwartz, R. J., and Srivastava, D. (2007) Cell 129 303-317 [DOI] [PubMed] [Google Scholar]
- 17.Wang, Z., Wang, D. Z., Pipes, G. C., and Olson, E. N. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 7129-7134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xing, W., Zhang, T. C., Cao, D., Wang, Z., Antos, C. L., Li, S., Wang, Y., Olson, E. N., and Wang, D. Z. (2006) Circ. Res. 98 1089-1097 [DOI] [PubMed] [Google Scholar]
- 19.Victor, R. G., Haley, R. W., Willett, D. L., Peshock, R. M., Vaeth, P. C., Leonard, D., Basit, M., Cooper, R. S., Iannacchione, V. G., Visscher, W. A., Staab, J. M., and Hobbs, H. H. (2004) Am. J. Cardiol. 93 1473-1480 [DOI] [PubMed] [Google Scholar]
- 20.Peterson, J. R., and Golemis, E. A. (2004) J. Cell. Biochem. 93 68-73 [DOI] [PubMed] [Google Scholar]
- 21.Zaromytidou, A. I., Miralles, F., and Treisman, R. (2006) Mol. Cell. Biol. 26 4134-4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Creemers, E. E., Sutherland, L. B., McAnally, J., Richardson, J. A., and Olson, E. N. (2006) Development (Camb.) 133 4245-4256 [DOI] [PubMed] [Google Scholar]
- 23.Badorff, C., Seeger, F. H., Zeiher, A. M., and Dimmeler, S. (2005) Circ. Res. 97 645-654 [DOI] [PubMed] [Google Scholar]
- 24.Cao, D., Wang, Z., Zhang, C. L., Oh, J., Xing, W., Li, S., Richardson, J. A., Wang, D. Z., and Olson, E. N. (2005) Mol. Cell. Biol. 25 364-376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, J., Li, A., Wang, Z., Feng, X., Olson, E. N., and Schwartz, R. J. (2007) Mol. Cell. Biol. 27 622-632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srivastava, D. (2006) Cell 126 1037-1048 [DOI] [PubMed] [Google Scholar]
- 27.Schroeder, J. A., Jackson, L. F., Lee, D. C., and Camenisch, T. D. (2003) J. Mol. Med. 81 392-403 [DOI] [PubMed] [Google Scholar]
- 28.Milyavsky, M., Shats, I., Cholostoy, A., Brosh, R., Buganim, Y., Weisz, L., Kogan, I., Cohen, M., Shatz, M., Madar, S., Kalo, E., Goldfinger, N., Yuan, J., Ron, S., MacKenzie, K., Eden, A., and Rotter, V. (2007) Cancer Cell 11 133-146 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.