Abstract
Membrane-type 1 matrix metalloproteinase (MT1-MMP; MMP-14) drives fundamental physiological and pathological processes, due to its ability to process a broad spectrum of substrates. Because subtle changes in its activity can produce profound physiological effects, MT1-MMP is tightly regulated. Currently, many aspects of this regulation remain to be elucidated. It has recently been discovered that O-linked glycosylation defines the substrate spectrum of MT1-MMP. We hypothesized that a mutual interdependency exists between MT1-MMP trafficking and glycosylation. Lectin precipitation, metabolic labeling, enzymatic deglycosylation, and site-directed mutagenesis studies demonstrate that the LL572 motif in the cytoplasmic tail of MT1-MMP influences the composition of the complex O-linked carbohydrates attached to the hinge region of the protein. This influence appears to be independent from major effects on cell surface trafficking. MT1-MMP undergoes extensive processing after its synthesis. The origins and the molecular characters of its multiple forms are incompletely understood. Here, we develop and present a model for the sequential, post-translational processing of MT1-MMP that defines stages in the post-synthetic pathway pursued by the protein.
Proteases regulate processes as diverse as apoptosis, blood clotting, angiogenesis, embryogenesis, and inflammation. Furthermore, proteolytic activity shapes the most distinctive and devastating feature of invasive cancers: their ability to break the boundaries of intact tissue structures and form colonies at a distance from their origin (1, 2). Proteolytic activity constitutes a final common pathway in the development of all invasive neoplasms (2–5). Other characteristics of cancers, their limitless replicative potential, sustained angiogenesis, self sufficiency in growth signals, and the ability to evade apoptosis, are also affected by proteolytic activity, if not entirely dependent upon it (2, 5–7). Matrix metalloproteinases (MMPs),2 a family of zinc-binding proteases, have long been associated with metastasis and tumor cell invasion. Elevated levels of MMPs have been verified in almost all human cancers (7, 8).
Mechanisms that confine proteolytic activity in the pericellular microenvironment have been found to be essential regulators of tumor microecology and are prerequisites for tumor cell invasion (9). The most obvious mediators of such localized proteolytic activity are the transmembrane- and glycosylphosphatidylinositol-anchored membrane type (MT) MMPs (10, 11). MT1-MMP (MMP-14) is considered the prototype of these membrane-tethered proteases due to its broad spectrum proteolytic capabilities and prominent involvement in cancer spreading (12–14).
Because MMPs have broadly overlapping substrate targets, most MMP knockout mice display no dramatic phenotypes (4, 15). So far, only the loss of MT1-MMP has been demonstrated to be incompatible with normal postnatal development. MT1-MMP-deficient mice display a variety of bone and joint disorders, including dwarfism, osteopenia, and arthritis (15).
The majority of MMPs and most other proteases are secreted as inactive pro-enzymes that undergo limited proteolysis for their activation. MT1-MMP is activated intracellularly and has the potential to activate other proteases such as MMP-2 on the cell surface. This makes it an important trigger of proteolytic cascades. In general, the activity of MT1-MMP is closely controlled post-translationally by its activation, inactivation, various endogenous inhibitors, oligomerization, protein trafficking, and glycosylation (4). It is this last mechanism that has most recently received major attention (4, 16, 17), because it was demonstrated that post-translational modification of the hinge domain by O-glycosylation specifically modifies a subset of activities of MT1-MMP (17). Unglycosylated mutants retained the ability to degrade collagen-I and induce autocleavage but lost the important ability to activate pro-MMP-2 (18, 19).
The information about the molecular and cellular processes that regulate the O-linked glycosylation of proteins is limited. Consequently, no cellular mechanisms that are involved in the generation of distinctive glycosylation species of MT1-MMP have been identified until now. We hypothesized in this context that a mutual interdependency exists between MT1-MMP trafficking and glycosylation. The function for the cytoplasmic tail of MT1-MMP in determining this protein's subcellular trafficking has been well established (for example see Refs. 20–22). Here, we demonstrate that the cytoplasmic tail's dileucine motif LL572 also influences the O-glycosylation pattern of MT1-MMP. This influence appears to be independent of major effects on MT1-MMP distribution, as demonstrated by metabolic pulse-chase experiments and assessments of steady-state surface expression levels. In addition, a comprehensive model for the sequential, post-translational processing of MT1-MMP is elaborated and presented.
EXPERIMENTAL PROCEDURES
General Reagents—Unless otherwise noted, all reagents were purchased from Sigma-Aldrich.
Sequence Analysis of MT1-MMP—The amino acid sequence of MT1-MMP was obtained from Swiss-Prot (P50281). Protein alignment and identification of domains were performed on the T-coffee server (www.ch.embnet.org/software/Tcoffee.html). The prediction of O- and N-glycosylation sites was performed by analyzing the full-length sequences on the NetOGlyc 3.1 (www.cbs.dtu.dk/services/NetOGlyc) and NetNGlyc 1.0 (www.cbs.dtu.dk/services/NetNGlyc) server (Center for Biological Sequence Analysis, Technical University of Denmark, Lyngby, Denmark) (23). The prediction of N-glycosylation sites was based on identification of Asn-Xaa-Ser/Thr consensus sequences.
DNA Constructs—cDNAs of FLAG-tagged (FL), human MT1-MMP constructs were kindly provided by the laboratory of Prof. Motoharu Seiki (Institute of Medical Science, University of Tokyo, Tokyo, Japan) (24). A hemagglutinin (HA) tag (YPYDVPDYA) was introduced between residues Gly501 and Gly502 in the stalk (linker-2) region of all FL-tagged MT1-MMP constructs (Fig. 1a) by a two-step overlapping PCR strategy, followed by subcloning XhoI/FseI digestion fragments into the MT1-MMP FL constructs. In short, the first step of the PCR yielded two fragments from separate PCR reactions that covered the complete coding sequence of WT MT1-MMP, with the HA tag in the overlapping region. In the second step, both fragments were combined and served as templates to generate the complete cDNA carrying the HA tag. The outer primers in this strategy were 5′-TAGTAGATGTCTCCCGCCCCAAGACCCCCCCG-3′ and 5′-GATGATTCAGACCTTGTCCAGCAGGGAACGC-3′. The inner primers were 5′-AGCGTAGTCTGGGACGTCGTATGGGTATCCCGATGGGCAGCCCATCC-3′ and 5′-TACCCATACGACGTCCCAGACTACGCTGGCCGGCCGGATGAGGGGACTGAGG-3′. Prior to XhoI and FseI digestion, the final PCR product was gel-purified and cloned into the pCRIITopo (Invitrogen, Carlsbad, CA) vector according to the manufacturer's instructions. Point mutations were introduced by PCR with site-directed mutagenesis using mismatch primers. The primers were 5′-GGTGGCTGTGCACGCGCTGGGCC-3′ and 5′-GGCCCAGCGCGTGCACAGCCACC-3′ for E240A; 5′-ACCCCCAGGCGAGCGGCCTACTGCCAGCGTTCC-3′ and 5′-GGAACGCTGGCAGTAGGCCGCTCGCCTGGGGGT-3′ for LL571/572AA (LeuLeu571/572 to Ala); 5′-GCGACTGCTCGCCTGCCAGCGTTCC-3′ and 5′-GGAACGCTGGCAGGCGAGCAGTCGC-3′ for Y573A; 5′-CGACTGCTCTACGCCCAGCGTTCCC-3′ and 5′-GGGAACGCTGGGCGTAGAGCAGTCG-3′ for C574A; 5′-TTCCCCGCTAAGATGCCCCCTCAACCCAGGGCTGCTGCTCGGCC-3′ and 5′-GGCCGAGCAGCAGCCCTGGGTTGAGGGGGCATCTTAGCGGGGAA-3′ for CHO-4 (Thr291/Thr299/Thr300/Ser301 to Ala); 5′-GCTGCTGCTCGGCCTGCTGTTCC-3′ and 3′-GGAACAGCAGGCCGAGCAGCAGC-5′ for CHO-5 (Thr291/Thr299/Thr300/Ser301/Ser304 to Ala). The CHO-4 construct served as the template for the CHO-5 construct. Correctness of all DNA constructs was confirmed by automated fluorescent sequencing.
FIGURE 1.
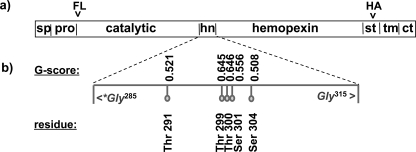
Schematic domain model of MT1-MMP and tag localization. a, MT1-MMP has a distinct domain structure. In the constructs used for the present studies, the FLAG epitope tag (FL) was inserted between the furin cleavage site and the N terminus of the catalytic domain (ct). After the autocleavage of MT1-MMP and shedding of the catalytic domain the FLAG tag is thus released into the extracellular media. To ensure the capacity to detect the remaining membrane-associated portion of the cleaved protein, a HA tag (HA) was introduced in the stalk domain (st) that links the hemopexin domain to the transmembrane domain. This tag localization enables identification of most membrane-tethered MT1-MMP metabolites. b, glycosylation analysis. The protein sequence of MT1-MMP was analyzed with the NetNGlyc 1.0 Server and NetOGlyc 3.1 server for potential O- and N-linked glycosylation sites. No N-glycosylated residues were found. The prediction of O-glycosylated residues is based upon a trained neural network approach. If the score of the best general predictor, the G-score, is above the threshold of 0.5 then the residue is predicted to be glycosylated. All putative glycosylation sites are in the hinge domain (hn) of the protease (sp = signal peptide, pro = pro-domain, tm = transmembrane domain, ct = cytoplasmic tail).
Tissue Culture—Cell culture media and reagents were purchased from Invitrogen. COS cells were grown in flasks under standard conditions (37 °C and 5% CO2) in minimal essential medium supplemented with l-glutamine (GlutaMAX), 10% fetal calf serum, and penicillin/streptomycin.
Procedures for cDNA Transfection and Sample Preparation—Cells were seeded in 6-well plates and transfected at 85–90% confluency 24 h after seeding. Lipofectamine2000 (Invitrogen) was used for transfections according to the manufacturer's instructions. Unless otherwise noted, cells were lysed 24 h after transfection according to the following protocol. Cells were washed twice with ice-cold PBS (137 mm NaCl, 2.7 mm KCl, 1.4 mm KH2PO4, 10 mm Na2HPO4, 1 mm CaCl2, 1 mm MgCl2, pH 7.4) before the addition of 1 ml of lysis buffer (50 mm Tris-HCl, 150 mm NaCl, supplemented with 1% Triton X-100, pH 7.5) per well, containing a 2× final concentration of complete protease inhibitor mixture tablets with EDTA (Roche Diagnostics, Penzberg, Germany). Cells were scraped from the wells and rocked on ice for 45 min. Cells lysates were cleared by centrifugation (17,000 × g, 30 min, 4 °C). 10-μl aliquots of cell lysates were mixed with reducing, 6× SDS-PAGE sample buffer (350 mm Tris-HCl, 10% (w/v) SDS, 30% (v/v) glycerol, 600 mm dithiothreitol, 0.01% (w/v) bromphenol blue; pH 6.8) and heated (10 min and 56 °C).
SDS-PAGE, Western Blot Analysis, and Antibodies—Standard protocols for Western blot analysis have been described elsewhere (25). Unless otherwise noted, Western blots were blocked with 5% dry milk in TBS-T and incubated overnight with a mouse anti-β actin monoclonal antibody (mAb) (ab6276, Abcam Inc., Cambridge, MA; 1:4000), anti-HA mAb (MMS-101R, Covance, Berkeley, CA; 1:1000), anti-MT1-MMP hinge region rAb (AB815; Chemicon, Temecula, CA; 1:2500) or anti-FLAG (M2) mAb (F1804; Sigma; 1:500).
Lectin Precipitation Assay—Cells were transfected with double tagged MT1-MMP constructs and lysed according to the protocol described above. For lectin precipitation, 900 μl of cell lysate were preincubated with 25 μg of biotinylated wheat germ agglutinin (WGA) or biotinylated, succinylated wheat germ agglutinin (sWGA) (Vector Laboratories, Burlingame, CA) for 4 h at 4 °C before 60 μl of a washed 50% slurry of streptavidin conjugated beads (Pierce Biotechnology, Rockford, IL) were added. After overnight incubation at 4 °C on a rotating platform, beads were washed five times in 1% Triton X-100 lysis buffer. Proteins were released by adding 2× sample buffer and heating (10 min and 56 °C). Blots of WGA precipitates were incubated with anti-HA mAb. Western blots of lysate samples were incubated with anti-HA, anti-FL, and anti-β-actin mAb.
Direct Metabolic Labeling of GalNAc—The basic principle and chemistry of the metabolic in vivo labeling of O-linked glycosides by bioorthogonal chemical tags have been described elsewhere in detail (26–28). The method is based on the metabolic incorporation of an azide modified sugar, here 1,3,4,6-tetra-O-acetyl-N-azidoacetyl-α,β-d-galactosamine (Ac4-GalNAz), into protein glycan structures. The glycoproteins which have incorporated the modified sugar are detected by the specific reaction of the azide modified sugar with an alkyne. In brief, COS cells were transfected with double tagged MT1-MMP constructs and incubated for 48 h under tissue culture conditions at a 40 μm final concentration of Ac4-GalNAz (C33365, Invitrogen). Cells were lysed according to the protocol described above, and cell lysates were immunoprecipitated with anti-HA mAb (compare below). Proteins were released by boiling (5 min at 95 °C) the protein A beads (Pierce) in 50 mm Tris-HCl, pH 8.5, containing 1% SDS. Covalent binding of glycoside incorporated Ac4-GalNAz to 5-carboxytetramethylrhodamine (TAMRA) alkyne (C33370, Invitrogen) followed by protein precipitation by methanol and chloroform was performed according to the manufacturer's instructions. Proteins were separated by SDS-PAGE gel electrophoresis. TAMRA fluorescence was detected in the polyacrylamide gels with a UV-light source and a charge-coupled device camera system with an ethidium bromide filter set (Gel iX Imager, Intas Science Imaging Instruments GmbH, Goettingen, Germany).
Cell-surface Biotinylation—Protocols and standard procedures for cell-surface protein biotinylation have been described previously (29). In brief, manipulations were performed on a rocking platform at 4 °C with ice-cold reagents and buffers. Cell-surface biotinylation was carried out 24 h after transfection of cells in 6 well with double tagged MT1-MMP constructs. Cells were washed twice with PBS and once with biotinylation buffer (10 mm triethanolamine, 2 mm CaCl2, 125 mm NaCl, pH 8.9). 1 ml of biotinylation buffer supplemented with 2.5 mm sulfo-NHS-SS-biotin (EZ-Link, Pierce) was added twice per well for incubations of 25 min each. Cells were washed twice for 5 min with PBS supplemented with 100 mm glycine and once with PBS before they were lysed. Biotinylated proteins were captured with streptavidin beads (see protocol for the lectin precipitation assay).
Immunoprecipitation—Cell lysates and immunoprecipitates were prepared according to the procedures described for the lectin precipitation assay. For immunoprecipitations, cells were transfected with MT1-MMP constructs. 15μg of anti-HA mAb (MMS-101R, Covance, Berkeley, CA) were added to the cleared lysates. Immobilized protein A beads (Pierce) were washed, blocked with 3% bovine serum albumin in lysis buffer for 4 h and washed again prior to the addition of 50 μl of a 50% slurry of these beads to each preincubated lysate-Ab mix.
Enzymatic Desialylation—Enzymatic desialylation with sialidase A (GK80040, Prozyme Inc., San Leandro, CA) was performed according to the manufacturer's instructions. In short, transfected cells were lysed, and 10 μl of lysate was mixed and incubated with 3 μl of 5× reaction buffer and 2 μl of sialidase A (activity > 5 units/ml) or solvent control for 12 h at 37 °C. The reaction was stopped by addition of 6× reducing sample buffer and heating (10 min for 56 °C). Samples were immunoblotted with anti-HA mAb.
Metabolic Labeling and Pulse-chase Experiments—Experiments were performed according to the procedures described elsewhere (30). In short, COS cells were transfected with double tagged MT1-MMP constructs. 24 h after transfection, COS cells were washed twice with PBS and incubated at 37 °C for 40 min in depletion media (serum, methionine, and cysteine-free minimal essential medium) before pulse labeling with 150μCi of l-[35S]methionine/cysteine in 1 ml of depletion medium per 35-mm dish for 25 min at 37 °C. Cells were placed on ice after 30 min and washed twice with ice-cold chase media (minimal essential medium supplemented with 10% fetal bovine serum and 10 mm l-methionine and 10 mm l-cysteine, pH 7.4, at 5% CO2). Cells were then switched back to 37 °C and incubated with chase media. After the time intervals indicated in Fig. 5, individual dishes were placed on ice, washed with ice-cold PBS, and lysed. Cell lysates were immunoprecipitated with anti-HA mAb. Lysates of untransfected cells and lysates of transfected cells without addition of precipitating Ab were used as controls (Fig. 5).
FIGURE 5.
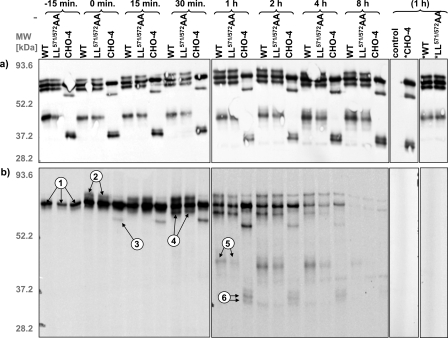
Pulse-chase analysis of MT1-MMP. COS cells were transfected with HA-tagged MT1-MMP constructs, metabolically labeled with l-[S35]methionine for 25 min, and chased for the indicated intervals. a, comparable expression levels of MT1-MMP constructs were confirmed by immunoblotting with monoclonal anti-HA Ab. b, immunoprecipitates of MT1-MMP with monoclonal anti-HA Ab were analyzed by SDS-PAGE and autoradiography. The combination of the results from the metabolic pulse-chase with the glycosylation analysis of MT1-MMP by site-directed mutagenesis and enzymatic desialylation (compare Fig. 2, b and c) allow the assignment of the protein bands to distinct processing steps. In brief, these steps include: 1, synthesis of the protein; 2, glycosylation (absent in CHO); 3 and 4, activation with loss of the pro-domain; and 5 and 6, generation of a membrane-tethered, autocatalytic cleavage product (see “Results” and “Discussion” for further details) (control = untransfected COS; * = transfected controls without precipitating Ab).
RESULTS
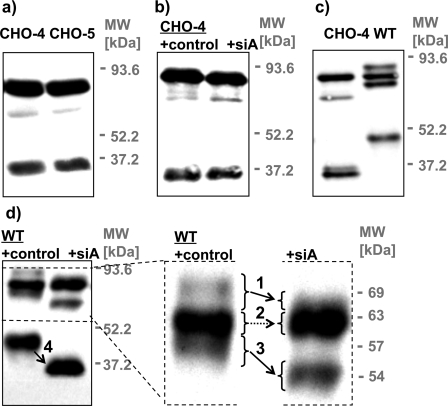
Sequence and Glycosylation Analysis of MT1-MMP—To generate a glycosylation-defective CHO mutant of MT1-MMP that could be used as an appropriate control for lectin precipitation assays, ezymatic dessialylation, and pulse-chase experiments, the glycosylated residues of MT1-MMP first had to be identified. Analysis of potential N-glycosylation sites in MT1-MMP with the NetNGlyc server software revealed the absence of the canonical Asn-Xaa-Ser/Thr N-glycosylation sequence, indicating that this protein cannot be subject to this post-translational modification. The prediction of O-glycosylated residues is less straightforward, because no consensus recognition sequence is known for O-glycosyltransferases (23). Five residues were predicted by the trained neural network of the NetOGlyc server as potential sites for O-glycosylation (Fig. 1b). All residues are within the small hinge domain, which connects the large catalytic and hemopexin domains. The prediction identifies 76% of glycosylated and 93% of non-glycosylated residues correctly (23). Consequently, the possibility of a false-positive prediction of a glycosylated residue is higher than missing one. If the G-score (the score from the best general predictor) of a residue is above the threshold of 0.5, it is predicted to be glycosylated. The confidence of the prediction increases with the score. Five residues of MT1-MMP received scores above 0.5 (Thr291, Thr299, Thr300, Ser301, and Ser304), however, one of these residues (Ser304) had a G-score of only 0.508 (Fig. 1b). To examine the potential glycosylation status of these residues two mutant constructs, CHO-4 and CHO-5, were prepared. CHO-5 includes all of the mutations present in CHO-4 (T291A, T299A, T300A, S301A) as well as the mutation S304A (Fig. 1b). Because the electrophoretic mobilities of the MT1-MMP putative glycosylation mutants CHO-4 and CHO-5 did not differ from one another (Fig. 2a) we conclude that Ser304 is not glycosylated. Furthermore, although enzymatic deglycosylation produces a dramatic shift in the electrophoretic mobility of wild-type MT1-MMP (Fig. 2d), no such shift was observed after the enzymatic deglycosylation of CHO-4 (Fig. 2b), demonstrating that the mutations incorporated into CHO-4 are sufficient to abrogate O-glycosylation of the MT1-MMP protein. Accordingly, CHO-4 was used for all subsequent experiments.
FIGURE 2.
Mutagenic elimination of glycosylation sites and enzymatic deglycosylation analysis of MT1-MMP. COS cells were transfected with MT1-MMP WT or constructs in which putative glycosylation sites were removed by mutagenesis (CHO). Cells were lysed 24 h after the transfection. Cell lysates were immunoblotted with monoclonal anti-HA Ab. a, mutants that are deficient in four (CHO-4 not including Ser304) or five (CHO-5 including Ser304) out of five putative O-glycosylation sites (compare Fig. 1b) showed no difference in electrophoretic mobility. b and d, cell lysates were incubated with sialidase A (siA) or solvent (control) prior to immunoblotting with monoclonal anti-HA Ab. b, CHO-4 displayed no shift in apparent molecular mass after sialidase A treatment, confirming that no additional sugars are attached at residue Ser304. c, expression of CHO-4 yields distinct bands of 63, 53, and two bands of ∼34 kDa, whereas expression of WT MT1-MMP results in the generation of bands of 69, 63, 57, and 44 kDa. d, glycosylated processed forms of MT1-MMP were in addition identified by a shift in their apparent molecular mass after sialidase A treatment (arrows 1, 3, and 4). This analysis confirms that the 63-kDa band (no shift, arrow 2) represents the only unglycosylated form of the WT protein.
Expression of MT1-MMP and Glycosylation Analysis of Its Metabolites—The extensive post-translational processing of MT1-MMP produces a diversity of discrete metabolites. It was critical for our study to distinguish the protein's glycosylated and unglycosylated species. The location of the HA tag close in the protein's sequence to its single transmembrane-spanning segment permits the detection of all membrane-tethered metabolites. Western blot analysis (with an anti-HA Ab) displays a distinct expression pattern of HA-tagged CHO-4 and WT MT1-MMP expressed in COS cells. Both proteins comprise several metabolites, of which they share only one with an apparent molecular mass of 63 kDa (Fig. 2c). Besides the common 63-kDa protein, expression of the CHO-mutant generates additional bands of 53 and two bands of ∼34 kDa, whereas the WT protein's other metabolites have apparent molecular masses of ∼69, 57, and 44 kDa (Fig. 2, c and d). Because enzymatic deglycosylation is expected to alter only the electrophoretic mobility of glycosylated proteins, we conclude that any of the multiple forms of MT1-MMP that exhibit a change in apparent molecular mass after sialidase A treatment possess sialic acid-containing glycosylation. Based upon this criterion, all of the MT1-MMP forms (Fig. 2d, arrows 1, 3, and 4) detected except for the 63-kDa protein (Fig. 2d, arrow 2) are glycosylated. Accordingly, we suggest that the 63-kDa protein is the biosynthetic precursor from which all of the glycosylated protein species must be derived. This hypothesis is explored further in the pulse-chase analysis presented below and in Fig. 5.
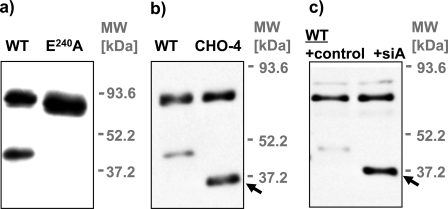
Autocleavage of MT1-MMP—Expression of the wild-type MT1-MMP protein in COS cells results in the appearance of a glycosylated 44-kDa metabolite that is likely to be the product of autocatalytic cleavage (Fig. 2, c and d). The catalytically inactive MT1-MMP construct E240A was used to rule out a possible role of endogenously expressed COS cell proteases in this proteolytic cleavage of MT1-MMP (Fig. 3a). Because expression of E240A did not result in the generation of a membrane-tethered 44-kDa product, we conclude that this product is generated by self-proteolysis.
FIGURE 3.
MT1-MMP autocleavage. a, COS cells were transfected with WT MT1-MMP and the catalytically inactive MT1-MMP mutant E240A. Cells were lysed 24 h after the transfection, and lysates were immunoblotted with monoclonal anti-HA antibody. E240A served as a negative control for the cleavage of MT1-MMP by endogenous COS cell proteases. Generation of the 44-kDa form in COS cells is clearly attributable solely to the self-cleavage of MT1-MMP. b, COS cells were transfected with WT MT1-MMP and the glycosylation site mutant CHO-4. Cells were lysed 24 h after the transfection. Cell lysates were immunoblotted with the polyclonal anti-hinge domain Ab815. c, COS cells were transfected with WT MT1-MMP and lysed 24 h after the transfection. Cell lysates were incubated with sialidase A (siA) or solvent (control) and immunoblotted with the polyclonal anti-hinge domain Ab815 (arrowheads: membrane-tethered self-cleavage product of MT1-MMP).
It has been suggested that glycosylation exerts a major effect on the self-proteolysis of MT1-MMP (31). This conclusion was mainly based on the observation that the prevalence of the 44-kDa autocleavage product increased after inhibition of O-glycosylation in cells by a synthetic inhibitor (31). In our hands, the MT1-MMP WT and the glycosylation-defective mutant CHO-4 proteins yielded similar amounts of autocleavage product, when the anti-HA-Ab was used for detection (Figs. 2c,4e, and 6a). In contrast, results obtained utilizing the anti-hinge domain Ab (AB815, Chemicon), which has been used in previous studies for immunoblotting, suggested enhanced generation of self-cleavage product in CHO-4 compared with WT (arrow, Fig. 3b). Interestingly, enzymatic desialylation of MT1-MMP WT in vitro also considerably increased the detection of the autocleavage product with the anti-hinge domain Ab (arrow, Fig. 3c). Because MT1-MMP activity was blocked during the in vitro desialylation, this increase cannot be attributed to enhanced autocleavage induced by desialylation. Instead, we conclude that the affinity of the anti-hinge region antibody varies dramatically with the glycosylation status of this domain.
FIGURE 4.
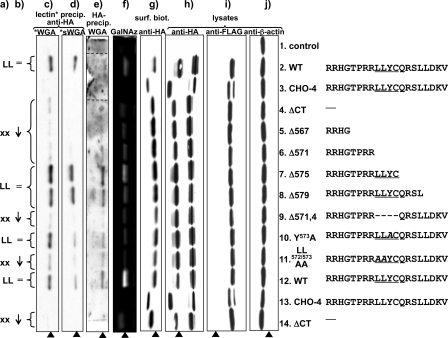
Analysis of MT1-MMP constructs. COS cells were transfected with double (FLAG and HA)-tagged MT1-MMP constructs. Compare Fig. 1a for localization of the tags within the protein. a, presence (LL) or absence (xx) of the dileucine motif LL572 in the sequence of the cytoplasmic tail of the corresponding MT1-MMP protein construct. b, degree of glycosylation as detected by lectin binding and incorporation of azide modified galactosamine in comparison to the wild-type MT1-MMP protein. c and d, Western blot analysis of biotinylated wheat germ agglutinin (WGA)(c) or biotinylated, succinylated wheat germ agglutinin (sWGA)(d) precipitates. WGA binds GlcNAc and sialic acids, whereas sWGA binds only to GlcNAc. MT1-MMP was immunoblotted with monoclonal anti-HA Ab. e, MT1-MMP was precipitated with monoclonal anti-HA antibody, and Western blots were probed with biotinylated WGA and streptavidin-conjugated horseradish peroxidase (lanes 2 and 3 originate from the same blot, but their positions were shifted in the preparation of the figure to maintain the alignment of lanes throughout the entire panel). f, O-linked glycosides in MT1-MMP constructs were metabolically labeled in live COS cells with azide modified d-galactosamine (GalNAz) as a bioorthogonal tag. GalNAz was detected by covalent linking to TAMRA alkyne and imaging of TAMRA fluorescence in polyacrylamide gels after SDS-page analysis of proteins. g, transfected COS were cell-surface biotinylated. Biotinylated proteins were pulled down with streptavidin beads, and MT-MMP was detected with anti-HA Ab. As a control for protein expression and efficiency of protein loading and cell lysis, cell lysates were immunoblotted with monoclonal anti-HA Ab (h), monoclonal anti-FLAG Ab (i), and anti b-actin Ab (j)(arrowhead: O-glycosylated MT1-MMP self-cleavage product).
FIGURE 6.
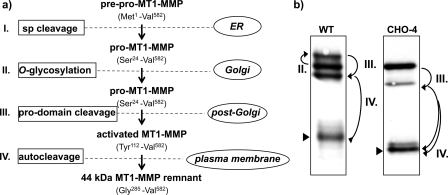
Sequential processing of MT1-MMP. a, schematic model based on data from enzymatic desialylation, site-directed mutagenesis, and metabolic pulse-chase experiments of MT1-MMP. b, Western blot analysis of MT1-MMP WT and its carbohydrate-deficient CHO-4 mutant expressed in COS. I, The 23-amino acid signal peptide (sp) is removed upon synthesis in the endoplasmic reticulum to yield the inactive, unglycosylated proform of MT1-MMP. II, O-glycosylation of the hinge domain in the Golgi apparatus is mandatory if glycosylation sites are present, but not a prerequisite for further processing, as demonstrated by the CHO-4 mutant. III, MT1-MMP is activated by the removal of its pro-domain by pro-protein convertase activity of furin or furin-like enzyme activity. IV, autocleavage causes shedding of the catalytic domain and results in the generation of a membrane-tethered remnant with the hinge domain at its N terminus (arrowhead).
Glycosylation Analysis of MT1-MMP Mutants—To investigate a possible connection of the glycosylation of MT1-MMP and its trafficking, the effects of alterations introduced into MT1-MMP's cytoplasmic tail motifs were investigated by lectin precipitation and metabolic labeling with Ac4-Gal-NAz. Western blot analysis of total cell lysates using the anti-HA and anti-β-actin antibodies was employed to ensure that any differences in lectin binding were not due to unequal expression levels of the MT1-MMP constructs, or to variations in cell lysis efficiency or in the total protein loaded per lane (Fig. 4, h–j). Fig. 4i depicts the results of Western blotting using an antibody directed against the FLAG tag epitope, which is inserted at the N terminus of the full-length MT1-MMP protein (Fig. 1), and in Fig. 4j the blot is probed with an antibody directed against actin, which serves as a loading control. The data presented in this figure demonstrate that all of the constructs whose behaviors will be analyzed in subsequent figures are expressed at similar levels in transfected COS cells.
The black arrowhead in Fig. 4 indicates the position of the 44 kDa MT1-MMP self cleavage product. As demonstrated by desialylation, this metabolite is O-glycosylated and no unglycosylated species of this protein species can be observed (Fig. 2 d). Differences in metabolic labeling of O-linked glycosides and lectin precipitability were particularly apparent through direct comparison of the 44 kDa autocleavage fragment. For reasons of clarity and figure composition only this band is displayed in Fig. 4 c to g. Major differences were detected in the susceptibility of the various constructs to lectin precipitation Fig. 4c and metabolic labeling by GalNAz (Fig. f). The CHO-4 mutant showed only minimal interaction with WGA (Fig. 4c) and sWGA (Fig. 4d) and no metabolic labeling by GalNAz (Fig. 4f), consistent with the conclusion that all relevant glycosylation sites are abrogated in this construct.
The differences in lectin precipitability and metabolic incorporation of GalNAz indicate that the presence of the dileucine LL572 motif has a major impact on the glycosylation of MT1-MMP (Fig. 4, a–f). The truncation constructs ΔCT, Δ567, and Δ571 incorporate less GalNAz and are less precipitated by WGA and sWGA as compared with the full-length WT protein. In contrast, Δ575 and Δ579 display GalNAz labeling and susceptibility to lectin precipitation that is comparable to that of the WT MT1-MMP protein (Fig. 4, c, d, and f). The Δ571 and Δ575 mutants differ from one another in length and in the presence of the 571LLYC574 sequence. In interpreting the patterns detected with these constructs, the different precipitation behaviors and metabolic GalNAz labeling of the Δ579 and Δ571–4 constructs are instructive. These constructs have the same total length but differ from one another in the presence of a defined subset of amino acids, namely the 571LLYC574 sequence. Furthermore, the cytoplasmic tail's second dileucine motif LL579 is disrupted in Δ579 and intact in Δ571–4. Conversely, LL572 is intact in Δ579 and absent in Δ571–4. Therefore, these data suggest that the LL572 motif modulates the glycosylation status of the MT1-MMP polypeptide.
To further specify the function of 571LLYC574, the dileucine and tyrosine motifs embodied within this sequence were each individually disrupted by alanine mutagenesis. The LL571/572AA mutation results in a pronounced reduction in metabolic GalNAz integration and susceptibility to WGA and sWGA pull down as compared with that observed with the WT protein and the Y573A and C574A mutants (Fig. 4, c, d, and f).
In principle, the lectin precipitation of MT1-MMP could be indirect and thus the differing susceptibilities of the constructs to lectin precipitation could be explained by differences in their affinity for another glycosylated protein that is pulled down by WGA and sWGA. As an additional control, and to test this possibility the preceding experiment was performed in reverse. HA-tagged MT1-MMP constructs were precipitated first with anti-HA Ab. GlcNAc- or sialic acid-modified proteins on the blot were then visualized by blotting with WGA (Fig. 4e). This approach is hampered by the fact that WGA binds to any protein on the blot containing this modification, including the IgG used in the immunoprecipitations. Hence, Western blots were blocked with an optimized concentration of 2.5% bovine serum albumin, which served as a compromise allowing effective blocking of nonspecific protein binding to the blot without resulting in excessive binding of WGA to the bovine serum albumin blocking agent. Although this compromise results in a relatively high background signal, the specific bands corresponding to the 44-kDa autocleavage product can be readily identified (arrow, Fig. 4e). The data from this experiment are essentially consistent with the results from WGA precipitations. The clones that possess the LL572 motif in their C-terminal tails are O-glycosylated in the hinge domain.
Surface Biotinylation Studies—Previous studies of the cytoplasmic tail of MT1-MMP have focused primarily on its influence on the trafficking and subcellular localization of the protein. We wondered, therefore, whether there is a connection between steady-state surface expression and the glycosylation of MT1-MMP. No such association was observed (Fig. 4g). The distinct behavior manifest by the 571LLYC574 construct with respect to its susceptibility to WGA and sWGA precipitation did not extend to alterations in the extent of its presence at the plasma membrane. The cell-surface expression levels of the 571LLYC574-omitting construct Δ571–4 and the LL571/572AA mutant are equivalent to that of WT. The surface expression level of Δ579 was increased as compared with WT and in particular as compared with that of the Δ571–4 protein. As noted above, the cytoplasmic tail's LL579 motif is disrupted in Δ579 and intact in Δ571–4, whereas LL572 is absent in Δ571–4 and present in Δ579. The interruption or removal of LL579 (ΔCT, Δ567, Δ571, Δ575, and Δ579) but not LL572 (LL571/572AA, Δ571–4) resulted in general in the enhanced steady-state surface expression of MT1-MMP.
Metabolic Labeling and Processing of MT1-MMP—A metabolic pulse-chase experiment was performed to investigate the sequential order in which the various forms of the MT1-MMP protein appear following synthesis and to explore possible differences in the processing of the LL572 and CHO-4 mutant in comparison to the full-length wild-type protein. Western blot analysis demonstrated equal and constant total expression of the various MT1-MMP constructs during the entire pulse-chase protocol (Fig. 5a). The first band that became apparent after 10 min of 35S pulse labeling incubation has a molecular mass of 63 kDa (Fig. 5b, arrows 1). This band represents the only common metabolite of the WT protein and CHO-4 (Fig. 2c). It is unglycosylated, because it has the same electrophoretic mobility in all three constructs, including CHO-4, and it did not shift after sialidase A treatment of MT1-MMP WT (Fig. 2d, arrow 2). The band's molecular mass of 63 kDa matches the calculated molecular mass of pro-MT1-MMP after the loss of its 23-amino acid signal peptide (32). Pro-MT1-MMP rapidly underwent an increase in molecular mass in LL572 and WT but not in CHO-4 (Fig. 5, arrows 2), which we interpret to reflect glycosylation of the protein (Fig. 6). This higher molecular mass band (apparent molecular mass of ∼69 kDa) must be sialylated, because its electrophoretic mobility was altered after incubation with sialidase A (Fig. 2d, arrow 1). This form is also absent in the CHO mutant (Fig. 2c). In the same post-synthetic time interval, CHO-4 was processed to a 53-kDa fragment (Fig. 5b, arrow 3) that corresponds to the calculated molecular mass of active MT1-MMP without the pro-domain. Activation of MT1-MMP has previously been demonstrated to occur intracellularly by limited proteolysis between Arg111 and Tyr112, respectively, its pro- and catalytic domain, by furin or furin-like enzymes (Fig. 6) (10, 33). Concurrently or with a short delay, WT and LL571/572AA underwent the same apparent cleavage, resulting in a 57-kDa protein (Fig. 5b, arrows 4). The 57-kDa fragment is glycosylated, as demonstrated by its behavior in SDS-PAGE following desialylation (Fig. 2d, arrow 3), and must therefore be a product of the corresponding higher molecular mass band, but not the unglycosylated 63-kDa pro-enzyme (Fig. 5b). It corresponds essentially to the 53-kDa fragment found in CHO-4 (Fig. 2c). WT and LL571/572AA underwent autocleavage to a 44-kDa fragment (Figs. 5b, arrows 5, and 6), whereas CHO-4 produced at least two fragments of ∼34 kDa (Figs. 5, arrows 6, and 6). Again, the 44-kDa autocleavage product was glycosylated and sialylated (Fig. 2d, arrow 4). It was, therefore, a direct cleavage product of the activated, “mature” 57-kDa protein (Fig. 6). No fragments matching the weights of the unglycosylated autocleavage products of CHO-4 were observed with WT, LL571/572AA, or with any cytoplasmic tail mutant. We conclude, therefore, that glycosylation precedes autocleavage but is not a prerequisite for further MT1-MMP processing. CHO-4 cleavage products match exactly the predicted molecular mass of previously described MT1-MMP fragments, as calculated from their amino acid sequences (summarized in Fig. 6) (32, 34–36). As noted above, the higher resolution achieved with the large SDS-PAGE gels used for separating proteins in the pulse-chase experiments revealed that the self-proteolysis product of the CHO-4 construct consists actually of at least two distinct subspecies of different molecular weights (Fig. 5, arrows 6).
DISCUSSION
The pathways by which MMPs regulate development, homeostasis, and disease are not fully understood. Insight into their function in diverse biological processes will require the elucidation of their own intricate regulation. A more thorough knowledge of the mechanisms involved in this complex regulation may lead to the development of specific therapeutic interventions that modify proteolytic activities involved in distinct processes by interfering with their regulation rather than by blocking them globally. Here, we present a new model for the metabolic processing of MT1-MMP and demonstrate that the presence of the dileucine motif LL572 in its cytoplasmic tail modulates the processing of the protein as revealed by metabolic labeling of O-linked glycosides by azide modified d-galactosamine and WGA and sWGA precipitation.
WGA is a lectin that is positively charged at a pH in the physiological range (37). It binds glycoconjugates containing N-acetylglucosamine (GlcNAc) and sialic acids, whereas its succinylated form is negatively charged at a physiological pH and fails to interact with sialic acids (17, 37). Sialic acids are acidic monosaccharides that are typically located at the terminal positions of glycoconjugates (38, 39). The sialic acid family comprises currently over 50 naturally occurring members that display a considerable structural and biochemical diversity (39, 40). Because sWGA does not bind sialic acids and the precipitation of MT1-MMP by WGA and sWGA displays an essentially similar pattern, we conclude that the dileucine LL572 motif's effect cannot be limited to the terminal residues of the MT1-MMP sugar complexes. Nonetheless, changes in the affinity of an anti-hinge domain antibody after sialidase A treatment of MT1-MMP in vitro emphasize the functional relevance of sialic acids for the antigenic presentation of the hinge region (Fig. 3, b and c). This effect becomes particularly apparent in the 44-kDa autocleavage product.
It had previously been suggested that O-glycosylation serves to stringently regulate the self-cleavage of MT1-MMP (31). The data presented in this report indicate, however, that apparent glycosylation-induced effects on autocleavage are actually attributable in large measure to the effects of glycosylation on the ability of the antibody used in those studies to detect the cleavage product. Our data demonstrate that glycosylation does indeed mask autocleavage sites within the hinge domain of MT1-MMP, and the absence of glycosylation enables self-cleavage of these alternate cleavage sites. Glycosylation has little effect, however, on the total extent of self-proteolysis. The preferential autocleavage site for MT1-MMP has previously been identified to reside between Gly284 and Gly285 (32, 34, 35). Recent evidence indicated that mutation of the preferential autocleavage site Gly284/Gly285 enforced downstream cleavage between Gln296 and Ser304 (41). Strikingly, this stretch includes three out of four potentially glycosylated residues (Fig. 1b). In the past, the low sequence homology of hinge domains among MMPs has been exploited for the generation of specific antibodies. Because glycosylation appears to be a common feature of hinge domains (data not shown), our data suggest that the results obtained with such probes need to be interpreted with caution.
The cytoplasmic tail of MT1-MMP has generally been credited as the primary determinant of the trafficking pattern pursued by this protein (20, 24, 42). The results from metabolic pulse-chase and cell surface biotinylation experiments presented here indicate that the effect of LL572 on the O-glycosylation pattern of MT1-MMP is likely to be independent of changes in trafficking and steady-state surface expression. Previously, the LLY573 sequence has been demonstrated to bind the μ2 subunit of adaptor protein-2 in a yeast two-hybrid assay (24). Membrane protein interaction with adaptor protein-2 is known to induce internalization via clathrin-coated pits. However, the precise binding motif had not been fully defined, because both tyrosine and dileucine motifs are potential adaptor protein-2 binding sites (43–46). Our results indicate that the mutation of the dileucine motif LL572 had no effect on the expression level of MT1-MMP at the cell surface, whereas mutation of the other dileucine motif LL579 increased the population of MT1-MMP resident at the cell surface (24).
Most notably, a number of studies have linked dileucine motifs to a function in protein folding that is independent from cell surface trafficking. For instance, the dileucine motif at the C terminus of the human multidrug resistance P-glycoprotein (Pgb or ABCB1) has been demonstrated to be essential for the correct folding of this protein (47). Mutation of the dileucine motif caused the expression of an immature protein. Although the underlying mechanism responsible for this effect is still unclear, it appears that the dileucine motif did not function as a plasma membrane targeting signal. Furthermore, dileucine motifs in the C terminus of the vasopressin V2 receptor and ATP-sensitive potassium channels (KATP) have been shown to be required for the exit from the endoplasmic reticulum (48, 49). In polarized epithelial cells, such as Madin-Darby canine kidney cells, a dileucine motif has been proven to be necessary and sufficient to target E-cadherin to the basolateral cell surface (50). Mutation of the motif caused aberrant apical expression of E-cadherin. Still, the basic mechanisms that define these effects of dileucine motifs have not yet been delineated. In our case, MT1-MMP with a mutated or interrupted dileucine LL572 is delivered to the cell surface, but displays a glycosylation pattern that is distinct from that of the wild-type protein. This effect could be attributed, for example, to perturbation of an interaction with a chaperone that depends on the dileucine motif, resulting as a consequence in incorrect protein folding. This might hamper downstream trafficking to or within the Golgi apparatus or protein-protein interactions that ultimately modulate the O-glycosylation pattern.
Differences in the nature and extent of pro-MMP-2 activation and substrate degradation have been extensively described for cytoplasmic tail truncation mutants of MT1-MMP, and have been interpreted in relation to the putative effects of tail mutations on MT1-MMP cell surface trafficking and expression (20, 24). Our data suggest that these functional differences may also be attributable in part to the novel influence that the cytoplasmic tail exerts upon the extent and composition of the O-glycosylation of MT1-MMP, as glycosylation has been shown to be a significant determinant of the substrate profile of MT1-MMP (17). Conversely, effects of glycosylation on microdomain trafficking have been discussed, because glycans contain signals that are responsible for the partitioning of glycoproteins into subcellular domains (51, 52). Interestingly, truncation of the cytoplasmic tail of MT1-MMP has been shown to induce aberrant and persistent inclusion of this protein into lipid rafts (53). On a functional level, the diversity of MT1-MMP glycosylation subtypes found in different cell lines may reflect the involvement of these proteins in regulating an equally diverse array of biological processes (17).
To the best of our knowledge, a cytoplasmic tail motif has not previously been implicated in the regulation of the O-glycosylation pattern of a protein. Because protein-coupled oligosaccharides are not primary gene products but are instead the product of post-translational modifications, our understanding of the factors that regulate their composition and assembly remains somewhat rudimentary (54, 55). Hence, the unique regulation of the glycosylation pattern of MT1-MMP by a dileucine motif in this protein's cytoplasmic tail sheds light on a novel mechanism involved in the O-linked glycosylation and post-translational modification of MT1-MMP in particular and in the maturation of transmembrane proteins in general.
Acknowledgments
We thank Kaisa Lehti (Biomedicum, University of Helsinki, Helsinki, Finland) and Naohiko Koshikawa and Motoharu Seiki (Institute of Medical Science, University of Tokyo, Tokyo, Japan) for providing us with MT1-MMP cDNAs.
This work was supported, in whole or in part, by National Institutes of Health Grants DK17433 and DK072614 (to M. J. C.). This work was also supported by the Deutsche Forschungsgemeinschaft (Grants LU 854/2-1 and LU 854/3-1) and the FRONTIER program of the Excellence Initiative at Heidelberg University (to T. L.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: MMP, matrix metalloproteinase; MT, membrane type; Ac4-GalNAz, 1,3,4,6-tetra-O-acetyl-N-azidoacetyl-α,β-d-galactosamine; CHO, Chinese hamster ovary; CHO-4, glycosylation-deficient mutant of MT1-MMP; FL, FLAG tag; HA, hemagglutinin; mAb, monoclonal antibody; rAb, rabbit antibody; siA, sialidase A; sWGA, biotinylated succinylated wheat germ agglutinin; TAMRA, 5-carboxytetramethylrhodamine; WGA, biotinylated wheat germ agglutinin; WT, wild type; PBS, phosphate-buffered saline.
References
- 1.Ludwig, T., Ossig, R., Graessel, S., Wilhelmi, M., Oberleithner, H., and Schneider, S. W. (2002) Am. J. Physiol. 283 F319–F327 [DOI] [PubMed] [Google Scholar]
- 2.Ludwig, T. (2005) BioEssays 27 1181–1191 [DOI] [PubMed] [Google Scholar]
- 3.Ludwig, T., Puttmann, S., Bertram, H., Tatenhorst, L., Paulus, W., Oberleithner, H., and Senner, V. (2005) J. Cell. Physiol. 202 690–697 [DOI] [PubMed] [Google Scholar]
- 4.Ludwig, T. (2006) in Nanomaterials for Cancer Diagnosis (Kumar, C., ed) pp. 377–412, John Wiley & Sons, Ltd., New York
- 5.Mohamed, M. M., and Sloane, B. F. (2006) Nat. Rev. Cancer 6 764–775 [DOI] [PubMed] [Google Scholar]
- 6.Hanahan, D., and Weinberg, R. A. (2000) Cell 100 57–70 [DOI] [PubMed] [Google Scholar]
- 7.Egeblad, M., and Werb, Z. (2002) Nat. Rev. Cancer 2 161–174 [DOI] [PubMed] [Google Scholar]
- 8.Sternlicht, M. D., and Werb, Z. (2001) Annu. Rev. Cell Dev. Biol. 17 463–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kusick, S., Bertram, H., Oberleithner, H., and Ludwig, T. (2005) J. Cell. Physiol. 204 767–774 [DOI] [PubMed] [Google Scholar]
- 10.Sato, H., Takino, T., Okada, Y., Cao, J., Shinagawa, A., Yamamoto, E., and Seiki, M. (1994) Nature 370 61–65 [DOI] [PubMed] [Google Scholar]
- 11.Yana, I., and Seiki, M. (2002) Clin. Exp. Metastasis 19 209–215 [DOI] [PubMed] [Google Scholar]
- 12.Golubkov, V. S., Chekanov, A. V., Savinov, A. Y., Rozanov, D. V., Golubkova, N. V., and Strongin, A. Y. (2006) Cancer Res. 66 10460–10465 [DOI] [PubMed] [Google Scholar]
- 13.Golubkov, V. S., Boyd, S., Savinov, A. Y., Chekanov, A. V., Osterman, A. L., Remacle, A., Rozanov, D. V., Doxsey, S. J., and Strongin, A. Y. (2005) J. Biol. Chem. 280 25079–25086 [DOI] [PubMed] [Google Scholar]
- 14.Zhai, Y., Hotary, K. B., Nan, B., Bosch, F. X., Munoz, N., Weiss, S. J., and Cho, K. R. (2005) Cancer Res. 65 6543–6550 [DOI] [PubMed] [Google Scholar]
- 15.Holmbeck, K., Bianco, P., Caterina, J., Yamada, S., Kromer, M., Kuznetsov, S. A., Mankani, M., Robey, P. G., Poole, A. R., Pidoux, I., Ward, J. M., and Birkedal-Hansen, H. (1999) Cell 99 81–92 [DOI] [PubMed] [Google Scholar]
- 16.Itoh, Y., and Seiki, M. (2004) Trends Biochem. Sci. 29 285–289 [DOI] [PubMed] [Google Scholar]
- 17.Wu, Y. I., Munshi, H. G., Sen, R., Snipas, S. J., Salvesen, G. S., Fridman, R., and Stack, M. S. (2004) J. Biol. Chem. 279 8278–8289 [DOI] [PubMed] [Google Scholar]
- 18.Shaw, L. M. (2004) Methods Mol. Biol. 294 97–106 [DOI] [PubMed] [Google Scholar]
- 19.Wang, Z., Juttermann, R., and Soloway, P. D. (2000) J. Biol. Chem. 275 26411–26415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehti, K., Valtanen, H., Wickstrom, S., Lohi, J., and Keski-Oja, J. (2000) J. Biol. Chem. 275 15006–15013 [DOI] [PubMed] [Google Scholar]
- 21.Wang, X., Ma, D., Keski-Oja, J., and Pei, D. (2004) J. Biol. Chem. 279 9331–9336 [DOI] [PubMed] [Google Scholar]
- 22.Jiang, A., Lehti, K., Wang, X., Weiss, S. J., Keski-Oja, J., and Pei, D. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 13693–13698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Julenius, K., Molgaard, A., Gupta, R., and Brunak, S. (2005) Glycobiology 15 153–164 [DOI] [PubMed] [Google Scholar]
- 24.Uekita, T., Itoh, Y., Yana, I., Ohno, H., and Seiki, M. (2001) J. Cell Biol. 155 1345–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roush, D. L., Gottardi, C. J., Naim, H. Y., Roth, M. G., and Caplan, M. J. (1998) J. Biol. Chem. 273 26862–26869 [DOI] [PubMed] [Google Scholar]
- 26.Laughlin, S. T., and Bertozzi, C. R. (2007) Nat. Protoc. 2 2930–2944 [DOI] [PubMed] [Google Scholar]
- 27.Vocadlo, D. J., Hang, H. C., Kim, E. J., Hanover, J. A., and Bertozzi, C. R. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 9116–9121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prescher, J. A., and Bertozzi, C. R. (2006) Cell 126 851–854 [DOI] [PubMed] [Google Scholar]
- 29.Gottardi, C. J., Dunbar, L. A., and Caplan, M. J. (1995) Am. J. Physiol. 268 F285–F295 [DOI] [PubMed] [Google Scholar]
- 30.Caplan, M. J., Forbush, B., III, Palade, G. E., and Jamieson, J. D. (1990) J. Biol. Chem. 265 3528–3534 [PubMed] [Google Scholar]
- 31.Remacle, A. G., Chekanov, A. V., Golubkov, V. S., Savinov, A. Y., Rozanov, D. V., and Strongin, A. Y. (2006) J. Biol. Chem. 281 16897–16905 [DOI] [PubMed] [Google Scholar]
- 32.Hernandez-Barrantes, S., Toth, M., Bernardo, M. M., Yurkova, M., Gervasi, D. C., Raz, Y., Sang, Q. A., and Fridman, R. (2000) J. Biol. Chem. 275 12080–12089 [DOI] [PubMed] [Google Scholar]
- 33.Sato, H., Kinoshita, T., Takino, T., Nakayama, K., and Seiki, M. (1996) FEBS Lett. 393 101–104 [DOI] [PubMed] [Google Scholar]
- 34.Toth, M., Hernandez-Barrantes, S., Osenkowski, P., Bernardo, M. M., Gervasi, D. C., Shimura, Y., Meroueh, O., Kotra, L. P., Galvez, B. G., Arroyo, A. G., Mobashery, S., and Fridman, R. (2002) J. Biol. Chem. 277 26340–26350 [DOI] [PubMed] [Google Scholar]
- 35.Osenkowski, P., Toth, M., and Fridman, R. (2004) J. Cell. Physiol. 200 2–10 [DOI] [PubMed] [Google Scholar]
- 36.Rozanov, D. V., Ghebrehiwet, B., Ratnikov, B., Monosov, E. Z., Deryugina, E. I., and Strongin, A. Y. (2002) FEBS Lett. 527 51–57 [DOI] [PubMed] [Google Scholar]
- 37.Monsigny, M., Roche, A. C., Sene, C., Maget-Dana, R., and Delmotte, F. (1980) Eur. J. Biochem. 104 147–153 [DOI] [PubMed] [Google Scholar]
- 38.Varki, A. (1997) FASEB J. 11 248–255 [DOI] [PubMed] [Google Scholar]
- 39.Lehmann, F., Tiralongo, E., and Tiralongo, J. (2006) Cell Mol. Life Sci. 63 1331–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zanetta, J. P., Pons, A., Iwersen, M., Mariller, C., Leroy, Y., Timmerman, P., and Schauer, R. (2001) Glycobiology 11 663–676 [DOI] [PubMed] [Google Scholar]
- 41.Osenkowski, P., Meroueh, S. O., Pavel, D., Mobashery, S., and Fridman, R. (2005) J. Biol. Chem. 280 26160–26168 [DOI] [PubMed] [Google Scholar]
- 42.Urena, J. M., Merlos-Suarez, A., Baselga, J., and Arribas, J. (1999) J. Cell Sci. 112 773–784 [DOI] [PubMed] [Google Scholar]
- 43.Teuchert, M., Berghofer, S., Klenk, H. D., and Garten, W. (1999) J. Biol. Chem. 274 36781–36789 [DOI] [PubMed] [Google Scholar]
- 44.Ohno, H., Fournier, M. C., Poy, G., and Bonifacino, J. S. (1996) J. Biol. Chem. 271 29009–29015 [DOI] [PubMed] [Google Scholar]
- 45.Ohno, H., Stewart, J., Fournier, M. C., Bosshart, H., Rhee, I., Miyatake, S., Saito, T., Gallusser, A., Kirchhausen, T., and Bonifacino, J. S. (1995) Science 269 1872–1875 [DOI] [PubMed] [Google Scholar]
- 46.Hunziker, W., and Fumey, C. (1994) EMBO J. 13 2963–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loo, T. W., Bartlett, M. C., and Clarke, D. M. (2005) J. Biol. Chem. 280 2522–2528 [DOI] [PubMed] [Google Scholar]
- 48.Schulein, R., Hermosilla, R., Oksche, A., Dehe, M., Wiesner, B., Krause, G., and Rosenthal, W. (1998) Mol. Pharmacol. 54 525–535 [DOI] [PubMed] [Google Scholar]
- 49.Sharma, N., Crane, A., Clement, J. P., Gonzalez, G., Babenko, A. P., Bryan, J., and Guilar-Bryan, L. (1999) J. Biol. Chem. 274 20628–20632 [DOI] [PubMed] [Google Scholar]
- 50.Miranda, K. C., Khromykh, T., Christy, P., Le, T. L., Gottardi, C. J., Yap, A. S., Stow, J. L., and Teasdale, R. D. (2001) J. Biol. Chem. 276 22565–22572 [DOI] [PubMed] [Google Scholar]
- 51.Huet, G., Gouyer, V., Delacour, D., Richet, C., Zanetta, J. P., Delannoy, P., and Degand, P. (2003) Biochimie (Paris) 85 323–330 [DOI] [PubMed] [Google Scholar]
- 52.Yeaman, C., Le Gall, A. H., Baldwin, A. N., Monlauzeur, L., Le, B. A., and Rodriguez-Boulan, E. (1997) J. Cell Biol. 139 929–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rozanov, D. V., Deryugina, E. I., Monosov, E. Z., Marchenko, N. D., and Strongin, A. Y. (2004) Exp. Cell Res. 293 81–95 [DOI] [PubMed] [Google Scholar]
- 54.Brockhausen, I. (1999) Biochim. Biophys. Acta 1473 67–95 [DOI] [PubMed] [Google Scholar]
- 55.Yarema, K. J., and Bertozzi, C. R. (2001) Genome Biol. 2 REVIEWS004. [DOI] [PMC free article] [PubMed] [Google Scholar]