Abstract
The Saccharomyces cerevisiae Mec1-Ddc2 protein kinase (human ATR-ATRIP) initiates a signal transduction pathway in response to DNA damage and replication stress to mediate cell cycle arrest. The yeast DNA damage checkpoint clamp Ddc1-Mec3-Rad17 (human Rad9-Hus1-Rad1: 9-1-1) is loaded around effector DNA and thereby activates Mec1 kinase. Dpb11 (Schizosaccharomyces pombe Cut5/Rad4 or human TopBP1) is an essential protein required for the initiation of DNA replication and has a role in checkpoint activation. In this study, we demonstrate that Dpb11 directly activates the Mec1 kinase in phosphorylating the downstream effector kinase Rad53 (human Chk1/2) and DNA bound RPA. However, DNA was not required for Dpb11 to function as an activator. Dpb11 and yeast 9-1-1 independently activate Mec1, but substantial synergism in activation was observed when both activators were present. Our studies suggest that Dpb11 and 9-1-1 may partially compensate for each other during yeast checkpoint function.
Phosphatidylinositol kinase-like protein kinases function as sensors in response to genotoxic stress to initiate a series of phosphorylation events that lead to a slowing down of cell cycle progression and promoting DNA repair. Ataxia telangiectasia mutated and ATR2 are the major protein kinases that initiate DNA damage checkpoint in mammalian cells, although this function is carried out by Mec1 and Tel1 kinases in yeast Saccharomyces cerevisiae and Rad3 and Tel1 in Schizosaccharomyces pombe (reviewed in Refs. 1-3). The Ddc2 (human ATRIP) subunit of the heterodimeric Mec1-Ddc2 complex protein is required for association of Mec1 with single-stranded DNA coated with the single-stranded DNA-binding protein RPA (4, 5).
DNA damage is also recognized by another group of sensor proteins called the checkpoint clamp-clamp loader complex (reviewed in Refs. 6, 7). The yeast 9-1-1 checkpoint clamp is a heterotrimer of the Ddc1, Rad17, and Mec3 proteins and is structurally similar to the replication clamp proliferating cell nuclear antigen. The yeast clamp loader for 9-1-1 is Rad24-RFC, and it consists of the four small subunits of replication factor C (Rfc2-5) together with the Rad24 subunit, the ortholog of Rad17 in S. pombe and human. The checkpoint clamp and clamp loader are required for the DNA damage checkpoint. During the G1 phase of the cell cycle, RPA-coated ssDNA gaps that are generated during repair of DNA damage, e.g. by the nucleotide excision repair machinery, serve as a loading site for both the 9-1-1 clamp and Mec1 (8). We have recently reconstituted this checkpoint machinery in vitro with purified proteins (9). A partial duplex DNA with a 5′-primer-template junction and with the ssDNA region coated with RPA forms an efficient substrate for the loading of 9-1-1 by Rad24-RFC in an ATP-dependent reaction (10). This complex specifically activates Mec1 to phosphorylate its downstream targets. Among these targets are the Rpa1 and Rpa2 subunits of RPA, the Rad24 subunit of the clamp loader, and the Ddc1 and Mec3 subunits of the clamp. The effector kinase Rad53 is also efficiently phosphorylated by the activated form of Mec1.
Our in vitro studies are supported by cellular studies of the phosphorylation cascades put into effect by DNA damage and 9-1-1-promoted activation of Mec1 (9, 11). The Rad53 and Mec1 kinases also function to stabilize stalled replication forks in response to hydroxyurea-mediated nucleotide depletion (12). However, based upon the analysis of yeast cells arrested with hydroxyurea, the participation of 9-1-1 in the DNA replication checkpoint appears to be of minor importance (13).
S. cerevisiae Dpb11 is an essential protein with a critical role in replisome assembly. The BRCT repeats in Dpb11 interact with the Sld2 and Sld3 initiator proteins that have been phosphorylated by the S phase CDK-cyclin complex, and this interaction is a crucial step in the initiation of DNA replication (14, 15). Dpb11 was first identified as a multicopy suppressor of conditional lethal mutations in DNA polymerase ε (16). The association of Dpb11 and DNA polymerase ε with the pre-replicative complex shows a mutual dependence (17). Proteins involved in DNA replication can also function in the DNA replication checkpoint (13). Among those are Dpb11, DNA polymerase ε, the Sgs1 helicase, Tof1, and Mrc1 (16, 18-21). The Dpb11 homolog in S. pombe is Cut5/Rad4, and in human and Xenopus it is TopBP1, and these essential proteins also function in DNA replication and the replication checkpoint (22-25).
S. cerevisiae Dpb11 interacts with the Ddc1 (human Rad9) subunit of the 9-1-1 clamp, and this interaction is conserved in S. pombe, human, and Xenopus. Phosphorylation of a Ser/Thr residue in the C-terminal tail of Ddc1 is critical for this interaction (26-30). In S. pombe and higher eukaryotes, one function of 9-1-1 appears to be the recruitment of Cut5/TopBP1 to stalled replication forks, with subsequent activation of ATR by TopBP1. In vitro studies with purified Xenopus TopBP1 and immunoprecipitated ATR show that TopBP1 can directly activate ATR without the necessity of DNA or RPA (31). However, another report notes that the presence of DNA further stimulates the activation of human ATR by TopBP1 (32). Activation ATR by human TopBP1 is mediated by interactions with both ATR and ATRIP, the regulatory subunit of ATR (33).
In S. cerevisiae, the 9-1-1 clamp loaded onto DNA directly activates the Mec1 kinase, without a requirement for Dpb11, an activity not yet observed in other model systems (9). To understand whether checkpoint initiation in S. cerevisiae is fundamentally different from that in other organisms, we asked whether yeast Dpb11 is capable of activating the Mec1 kinase in vitro, and if so, whether it functions together with the 9-1-1 clamp. Here we show that Dpb11 is able to activate Mec1 independent of 9-1-1, and full activation of Mec1 by Dpb11 is comparable with that by the 9-1-1 clamp. However, at subsaturating concentrations of Dpb11 and 9-1-1, we observe a significant synergism between Dpb11 and 9-1-1 indicating that these factors may act together to accomplish a more robust activation of Mec1. Activation is abolished by mutations in the C-terminal tail of Dpb11.
EXPERIMENTAL PROCEDURES
Plasmids and DNA Substrates—The DPB11 gene was cloned in a multicopy yeast shuttle vector pRS426-GALGST (2 μm origin, URA3, GAL1-10) and named pBL512. The glutathione S-transferase gene (GST) is separated from the N terminus of Dpb11 by a rhinoviral 3C protease cleavage site. A stop codon was generated by site-directed mutagenesis in pBL512 at position 572 to generate pBL512-571 (1-571 amino acids). Plasmids and their sequences are available upon request. Deca-primed ssDNA was made by annealing single-stranded bluescript SKII+ DNA with 10 28-mer primers spaced approximately equally around the ssDNA.
Proteins—Mec1-Ddc2, Rad53-kd, RPA, Rad24-RFC, and Rad17-Mec3-Ddc1 (yeast 9-1-1) were purified as described earlier (9, 34). To make GST-Dpb11, pBL512 was transformed in yeast strain BJ2168 (MATa: ura3-52, trp1-298, leu2-3, 112, prb1-1122, prc1-407, pep4-3). Cells were grown and induced as described (35). Yeast cells (50 g) were blended in dry ice, followed by nucleic acid precipitation and ammonium sulfate fractionation, and purified over 1 ml of glutathione-Sepharose as described (35). The GST-Dpb11 preparation was treated with 20 units of 3C protease overnight at 4 °C. Dpb11 was further purified over a 1-ml heparin-agarose column (GE Healthcare) in buffer A100 (buffer A0 = 50 mm Hepes-NaOH, pH 7.5, 10% glycerol, 1 mm dithiothreitol, 0.01% Nonidet P-40, 1 mm EDTA, 1 mm EGTA, 1 mm phenylmethylsulfonyl fluoride, 2 μm pepstatin A, 2 μm leupeptin, 10 mm NaHSO3, 1 mm benzamidine; the subscript refers to mm KCl in the buffer). GST and 3C protease do not bind to heparin-agarose. The column was washed with 10 ml of buffer A100, 10 ml of buffer A180, and Dpb11 was eluted with buffer A500. The truncated protein was purified in a similar way.
Protein Phosphorylation Assays—Standard 20-μl phosphorylation assays contained 25 mm Hepes-NaOH, pH 7.8, 8 mm MgCl2, 1 mm dithiothreitol, 100 μg/ml bovine serum albumin, 125 mm NaCl, 100 μm unlabeled ATP, and 0.5 μCi of [γ-32P]ATP. Other components when added were 1 nm Deca-primed ssDNA, 150 nm RPA, 100 nm Rad53-kd. Concentrations of Dpb11 and Rad17/3/1, clamp loader when added are indicated in legends. Reactions were preheated for 1 min at 30 °C and initiated by the addition of 5 nm Mec1/Ddc2 kinase. After 10 min at 30 °C, or the indicated times, the assays were stopped with 5 μl of 5 × SDS-polyacrylamide gel loading dye. The samples were run on 10 or 12% SDS-PAGE, dried, and exposed to a phosphor screen (GE Healthcare). Protein phosphorylation was quantified using ImageQuant software.
RESULTS AND DISCUSSION
Dpb11 Directly Activates the Mec1/Ddc2 Kinase—Dpb11 was overproduced in yeast from a multicopy plasmid with the DPB11 gene placed under control of the galactose-inducible GAL1-10 promoter. A cleavable GST tag was added to aid in purification (see under “Experimental Procedures”). Following the glutathione-affinity column, the GST tag was proteolytically cleaved, and Dpb11 was further purified by heparin-agarose chromatography to more than 98% purity (Fig. 1A). Dpb11 was free of (contaminating) nuclease or protein kinase activity.
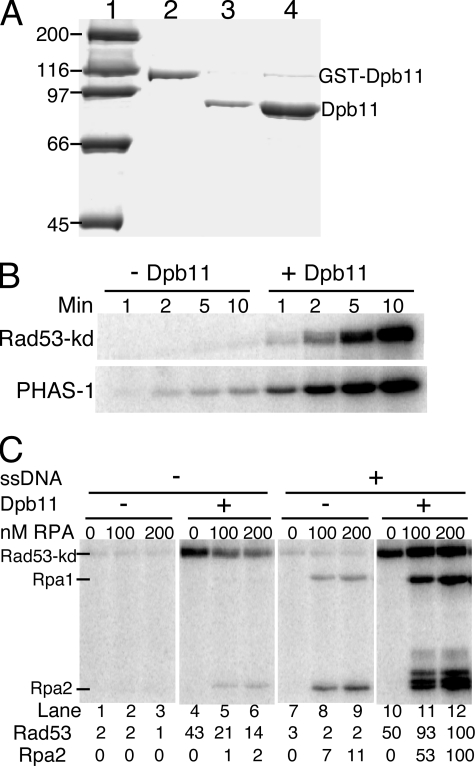
FIGURE 1.
Dpb11 activates Mec1 kinase activity. A, purification of Dpb11 from S. cerevisiae. Coomassie-stained 10% SDS-polyacrylamide gel showing GST-Dpb11 after glutathione-Sepharose column purification (lane 2), after cutting of the purified preparation with 3C protease (lane 3), and after heparin-agarose column purification (lane 4). Lane 1, molecular weight markers. B, standard phosphorylation assays contained 100 nm Rad53kd (upper panel) or 2.5 μm PHAS-I (lower panel) in assay buffer with or without 20 nm Dpb11. The reaction was initiated with 5 nm Mec1, and aliquots were taken after 1, 2, 5, and 10 min and analyzed by 10 and 12% SDS-PAGE, respectively. C, standard phosphorylation assays contained 5 nm Mec1, 100 nm Rad53-kDa, the indicated concentration of RPA, and where indicated, 1 nm deca-primed ssSK2 DNA and 20 nm Dpb11. Phosphorylation of Rad53-kd and Rpa2 was quantified with that in lane 12 set at 100.
We studied the effect of Dpb11 on Mec1 kinase activity. The phosphorylation of Rad53 by Mec1 is critical in initiating the checkpoint response. Rad53 is also a downstream effector kinase that can undergo autophosphorylation to activate its functions (36, 37). To eliminate the Rad53 autophosphorylation activity that might complicate the analysis, we used its kinase-defective form Rad53-kd (Rad53K227A) as a phosphorylation substrate (9). The presence of Dpb11 caused a dramatic enhancement of the Mec1 kinase activity, and phosphorylated Rad53-kd was visible from as early as 1 min from the start of the reaction (Fig. 1B, upper panel). We also studied Mec1-catalyzed phosphorylation of human PHAS-I, a commonly used substrate for PIK-like kinases (38), in the absence and presence of Dpb11. Dpb11 caused an approximate 20-fold activation of Mec1 in phosphorylating PHAS-I (Fig. 1B, lower panel). Thus, Dpb11 appears to function as a general activator of Mec1, analogous to our previous analysis of Mec1 activation by the S. cerevisiae 9-1-1 clamp, and to ATR activation by vertebrate TopBP1 (9, 31).
We also studied the phosphorylation of the single-stranded DNA-binding protein RPA. In yeast, both the Rpa1 and Rpa2 subunits of RPA are phosphorylated by Mec1 in response to DNA damage (39). In vitro, these two RPA subunits are phosphorylated by Mec1; however, phosphorylation is greatly stimulated when the RPA is bound to ssDNA (40, 41). Addition of the 9-1-1 clamp loaded onto DNA stimulates the Mec1-mediated phosphorylation of DNA-bound RPA even further (9). In this study, a rather complex interaction between RPA and Mec1 or its Dpb11-activated form was delineated.
First, we determined the rate of Rad53-kd phosphorylation by Mec1. In the absence of DNA, addition of increasing RPA caused a substantial inhibition of basal or Dpb11-activated Mec1 activity on Rad53-kd (Fig. 1C, lanes 1-6). This inhibition by RPA disappeared when DNA was also added to the assay. In fact, a small stimulation by RPA-coated ssDNA in comparison with the absence of both factors was consistently detected (Fig. 1C, compare lane 4 with 11 and 12). A similar phosphorylation pattern was observed with PHAS-I as target for Mec1 kinase (data not shown). A more detailed analysis of the role of DNA in Mec1 activation is presented below. Next, we determined the role of Dpb11 and DNA in RPA phosphorylation. In agreement with previous studies (9, 41), only DNA-bound RPA is an efficient target for phosphorylation by Mec1 (Fig. 1C). In the absence of DNA and the presence of Dpb11, a weak phosphorylation of RPA was observed (Fig. 1C, lanes 5 and 6). Both the basal and Dpb11-activated phosphorylation of Rpa1 and Rpa2 were greatly enhanced when ssDNA was added to the assay (Fig. 1C, compare lanes 8 and 9 with 2 and 3 and lanes 11 and 12 with 5 and 6).
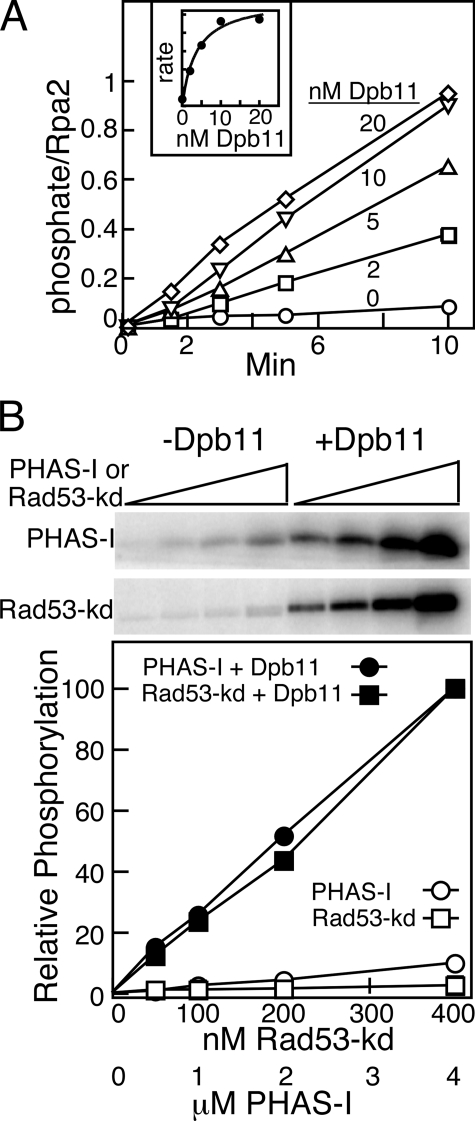
Affinity of Mec1 for Dpb11 and for Phosphorylation Targets—Two additional parameters of the Dpb11-mediated stimulation of Mec1 were investigated. First, we used the activation assay to determine the strength of interaction between Dpb11 and Mec1. RPA-coated DNA and Rad53-kd were incubated with Mec1 and with increasing levels of Dpb11, from 0 to 20 nm, and phosphoproteins were quantified as a function of time (Fig. 2A). The rate curves allowed us to determine a Km value of ∼5 nm for Dpb11 (Fig. 2A, inset). Within error, the same Km value was determined regardless whether phosphorylation rates of Rpa1 or Rpa2 or of Rad53-kd were used for the Km calculation. Second, we titrated target proteins PHAS-I and Rad53-kd into the Dpb11-stimulated Mec1 phosphorylation assay to obtain an estimate of the affinity of these target proteins for Mec1. However, even at 4 μm PHAS-I or at 400 nm Rad53-kd, the highest levels of these proteins that were practically attainable, phosphate incorporation still increased linearly with target concentration making an estimate of the relative affinity of these proteins for activated Mec1 impossible (Fig. 2B). Thus, although the activator Dpb11 has a high affinity for Mec1, target proteins such as Rad53-kd and PHAS-I have a relatively low affinity. The same low affinity of activated Mec1 for its phosphorylation targets was previously obtained with the checkpoint clamp 9-1-1 as activator of Mec1 (9). In this study, Rad53-kd was routinely used at 100 nm.
FIGURE 2.
Affinity of Mec1 for Dpb11 and targets. A, standard phosphorylation assays contained 1 nm deca-primed ssSK2 DNA, 150 nm RPA, and 5 nm Mec1 with the indicated concentrations of Dpb11. Aliquots were taken after the indicated times and analyzed by 10% SDS-PAGE, and phosphorylation of Rpa2 was quantified and plotted. The inset shows the phosphorylation rate as a function of Dpb11 concentration. B, standard phosphorylation assays contained 5 nm Mec1 with or without 20 nm Dpb11 and increasing Rad53-kDa or PHAS-I. Assays were analyzed by 10% SDS-PAGE for Rad53-kd (top panel) and 12% SDS-PAGE for PHAS-I (bottom panel), quantified, and plotted.
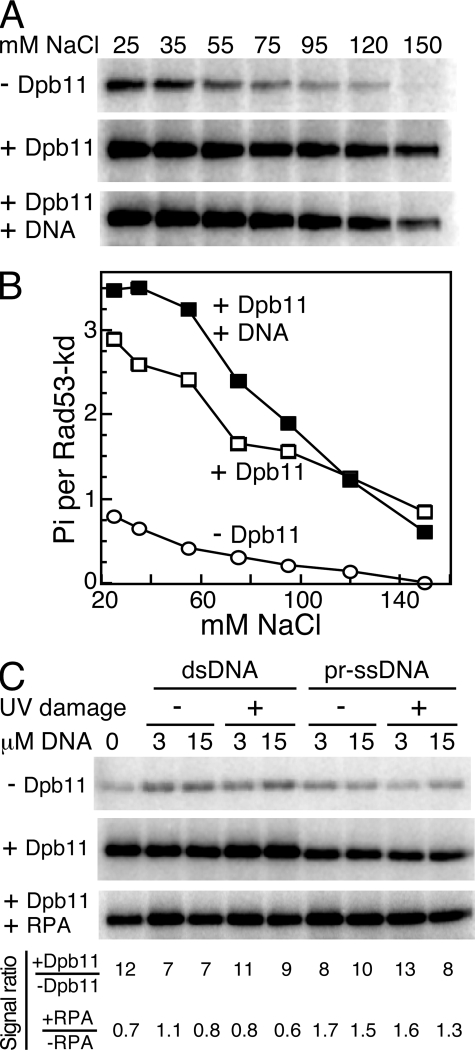
Role of DNA in Dpb11 Activation of Mec1—Under physiological salt conditions, i.e. ∼125 mm NaCl, the yeast 9-1-1 clamp only stimulates Mec1 when the clamp is loaded onto DNA by its clamp loader Rad24-RFC (9). However, at low salt (∼40 mm NaCl), this specificity for a loaded clamp is relaxed, and some stimulation by the clamp alone can be observed. Although this type of stimulation without DNA at low salt has been useful for mechanistic studies, we think it has no physiological relevance (9). Interestingly, recent studies of human TopBP1 indicate a similar type of stimulation pattern (32). At low salt, TopBP1 stimulated ATR without DNA present, whereas at higher salt, the presence of DNA aided in the TopBP1 stimulation of ATR. An even greater stimulation was observed when the DNA contained damage (32).
We studied if the presence of DNA affected Dpb11 in its ability to activate Mec1, and the effect of salt on the potential for DNA stimulation. Initially, we used a 3-kb deca-primed single-stranded phagemid circle as effector DNA. Rad53-kd phosphorylation by Mec1 was monitored at increasing NaCl levels in the absence of Dpb11, in the presence of Dpb11, or Dpb11 with deca-primed single-stranded circular DNA. In the absence of Dpb11, weak Rad53-kd phosphorylation was observed at low salt conditions, and this diminished with increasing NaCl levels. Without DNA present, Dpb11 was able to efficiently activate Mec1 phosphorylation of Rad53kd at all salt levels (Fig. 3, A and B). In fact, at 150 mm NaCl, Mec1 kinase activity was completely dependent on Dpb11. The addition of DNA showed only a slight increase in Mec1 activation by Dpb11 at low NaCl concentrations, but this effect disappeared with increasing salt (Fig. 3B). We then tested if the presence of damaged DNA had an effect on Dpb11 activation of Mec1. We used two levels (1 and 5 μm nucleotide) of double-stranded supercoiled DNA or deca-primed ssDNA with or without prior irradiation with 1500 J/m2 of UV254. Both basal and Dpb11-stimulated Mec1 activity on Rad53 was largely unaffected by the presence of either single-stranded or double-stranded DNA with or without UV damage. The fold stimulation (±Dpb11) varied from 7- to 13-fold. The additional presence of RPA caused an approximate 2-fold inhibition in the absence of DNA, as shown before (Fig. 3C). However, in the presence of ssDNA, a low but significant ∼1.5-fold stimulation of Rad53-kd phosphorylation was observed. We conclude from these studies that RPA-coated ssDNA but not double-stranded DNA causes a minor stimulation of Dpb11-activated Mec1 activity regardless of damage. Whether this low stimulation occurs through binding of Mec1 to RPA-coated ssDNA (5) or through the binding of other factors remains to be established.
FIGURE 3.
Dpb11 activation of Mec1 is largely independent of DNA. A, Rad53-kd phosphorylation by Mec1 (5 nm) was determined at the indicated NaCl levels in the absence of Dpb11 (1st panel), with 5 nm Dpb11 (2nd panel), or with 5 nm Dpb11 and 15 μm (nucleotide) deca-primed single-stranded Bluescript DNA (3rd panel). B, Rad53-kd phosphorylation from A was quantified and plotted as a function of mm NaCl. C, Rad53-kd phosphorylation by Mec1 (5 nm) was determined at 80 mm NaCl in the absence of Dpb11 (1st panel), or with 5 nm Dpb11 (2nd panel), or with 5 nm Dpb11 and 150 nm RPA (3rd panel). Double-stranded or deca-primed single-stranded Bluescript DNA, with or without prior UV (1500 J/m2) treatment, was added as indicated. The phosphorylation signal ratios (2nd panel/1st panel) and (3rd panel/2nd panel) are shown.
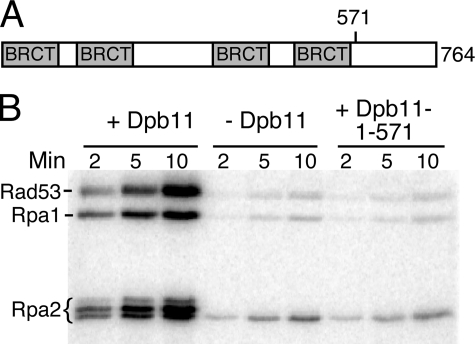
Dpb11 C-terminal Tail Is Required for the Activation of Mec1 Kinase—The ATR activating domain lies between the sixth and seventh BRCT repeat in the C-terminal domain of vertebrate TopBP1 (31). There is no obvious similarity to this domain in S. cerevisiae Dpb11 or S. pombe Cut5. Dpb11 is a protein of 764 amino acids and has four BRCT repeats. A dpb11-1 mutant strain with a premature stop codon at amino acid 583 is temperature-sensitive for growth and checkpoint defective at the permissive temperature (16). Therefore, in a preliminary analysis, we retained all the BRCT repeats and deleted the C-terminal end of the protein to generate Dpb11-(1-571). This truncated form of Dpb11 had lost its Mec1 activation ability. Either 20 nm wild-type or 20 nm Dpb11-(1-571) was added to the standard Mec1 stimulation assay that included Rad53-kd and RPA-coated deca-primed ssDNA (Fig. 4). Phosphorylation by Mec1 of the three target proteins in this assay, Rpa1, Rpa2, and Rad53-kd, was not stimulated by Dpb11-(1-571) compared with the control without Dpb11, indicating that this region of Dpb11 is essential for Mec1 activation.
FIGURE 4.
The C-terminal tail of Dpb11 is required for stimulation. A, domain structure of Dpb11. B, assays contained 1 nm deca-primed ssDNA, 150 nm RPA, and 100 nm Rad53-kd in assay buffer with 20 nm full-length Dpb11 (1st to 3rd lanes) or 20 nm Dpb11 (1-571 amino acids) in 7th to 9th lanes. No activator was present in 4 to 6 lanes. Aliquots were analyzed by 12% SDS-PAGE.
Dpb11 and the 9-1-1 Clamp Show Synergism for Mec1 Activation—In S. pombe and higher eukaryotes, the 9-1-1 clamp is a critical factor in the S phase checkpoint. The phosphorylated Rad9 subunit of the clamp interacts with Cut5/TopBP1, and Cut5/TopBP1 activates the ATR kinase (27-29, 31). So far, no direct activation of Mec1/ATR by the clamp has been reported in organisms other than budding yeast, where we have previously shown that the 9-1-1 clamp itself mediates a robust activation of the Mec1 kinase (9). The Rad24-RFC checkpoint-specific clamp loader loads 9-1-1 in an ATP-dependent reaction onto partially double-stranded DNA. The loader shows a high preference for a 5′-junction when the ssDNA is coated with RPA (10, 42). The loaded clamp is an efficient activator of Mec1. At physiological salt concentrations, no activation of Mec1 was observed by the unloaded form of 9-1-1. Therefore, efficient Mec1 activation by 9-1-1 requires appropriate effector DNA, RPA, and the Rad24-RFC loader. Now that we show here that Dpb11 can also independently activate the Mec1 kinase, it is of interest to determine whether these two activators function independently, synergistically, or compete for the same site on Mec1. Phosphorylation assays were carried out under experimental conditions that allow for both 9-1-1 and Dpb11 to activate Mec1.
Phosphorylation of Rpa1 and Rpa2 bound to deca-primed circular ssDNA was monitored at increasing levels of Dpb11 (Fig. 5A). Mec1 kinase activity increased with Dpb11 concentration, in agreement with the data in Fig. 2A. Similarly, Mec1 kinase activity increased with increasing 9-1-1 concentrations, provided the checkpoint clamp loader Rad24-RFC was also present. However, when both activators were included, a large hyperstimulation of Mec1 was observed, particularly in the low concentration range, at ∼1 nm of Dpb11 and 9-1-1. At that concentration, Mec1 activity was 3-fold that expected by adding the separate contributions made by Dpb11 and 9-1-1 (Fig. 5B). We observed a similar synergy of Mec1 activity at low activator concentrations when phosphorylation of Rpa1 was measured (Fig. 5B). In four independent experiments, the synergy measured at 0.5-1 nm of the activators varied between 2.5- and 3.5-fold. At higher concentrations of the activators, the degree of synergy gradually decreased, and at 10 nm Dpb11 and 9-1-1 synergy was no longer observed.
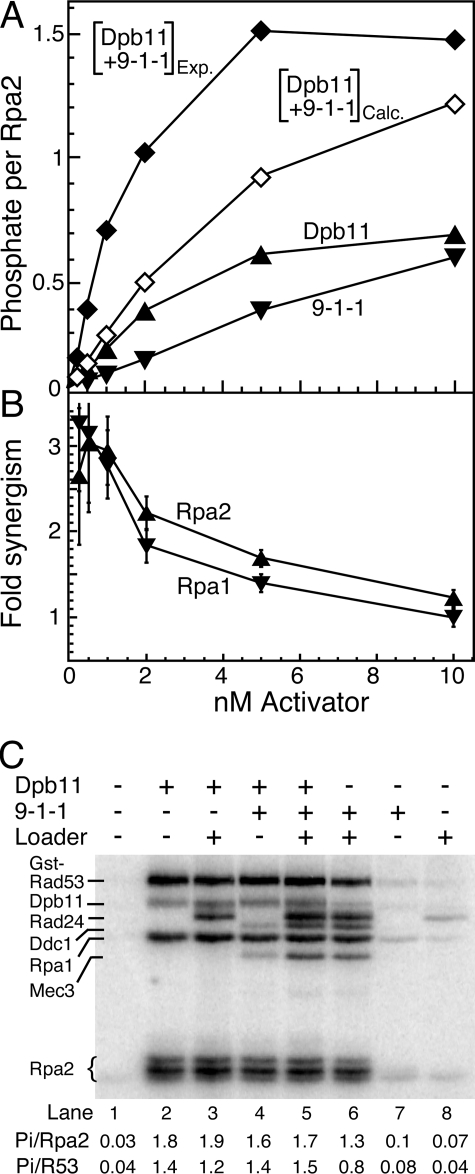
FIGURE 5.
Synergism between Dpb11 and 9-1-1. A, standard phosphorylation assays contained 1 nm deca-primed ssSK2 DNA, 150 nm RPA, 10 nm Rad24-RFC, and 5 nm Mec1, with the indicated concentrations of activator, either Dpb11, 9-1-1, or Dpb11 plus 9-1-1, at equimolar concentration. Assays were analyzed by 10% SDS-PAGE, and phosphorylation of Rpa2 was quantified and plotted. Open diamonds, calculated activity obtained by adding together the Dpb11 and 9-1-1-stimulated activities; closed diamonds, actual measured activity from the equimolar mixture of Dpb11 and 9-1-1. B, synergism, defined as the ratio of Mec1 stimulation activity by Dpb11 + 9-1-1 measured over that calculated from the individual assays (Dpb11 + 9-1-1)Exp/(Dpb11 + 9-1-1)Calc for Rpa2 phosphorylation from the data in A, and analogously for Rpa1 phosphorylation. C, assays as in A with 30 nm Dpb11, 30 nm 9-1-1, and 30 nm Rad24-RFC, as indicated. Quantification of Rpa2 and GST-Rad53-kd (R53) is given below the figure. In this assay GST-Rad53-kd was used rather than Rad53-kd to obtain a clear separation from other phosphorylated species. The GST tag does not alter the rate of phosphorylation (9).
At much higher near-saturating concentrations of the activators, the opposite was actually observed (Fig. 5C). We measured phosphorylation of Rpa2 and of Rad53-kd in the presence of either 30 nm Dpb11, 30 nm 9-1-1 (and 30 nm Rad24-RFC), or 30 nm of both. Dpb11 alone caused a large stimulation of Mec1 as judged by Rpa2 phosphorylation (Fig. 5C, lane 2). The 9-1-1 clamp, with loader, also gave a large stimulation (Fig. 5C, lane 6). However, when both were present, no further increase in stimulation was observed (Fig. 5C, lane 5). Control experiments showed the following: (i) that the presence of the Rad24-RFC loader did not affect activation by Dpb11 (Fig. 5C, lane 3); (ii) that 9-1-1 without loader did affect activation by Dpb11 (Fig. 5C, lane 4); and (iii) that in the absence of Dpb11, no activation was observed when either Rad24-RFC or 9-1-1 was present, but not both (Fig. 5C, compare lanes 7 and 8 with lane 6). The same pattern of Mec1 activation was observed when we quantified phosphorylation of Rad53-kd (Fig. 5C). These data indicate that high levels of either activator suffice for complete activation of Mec1.
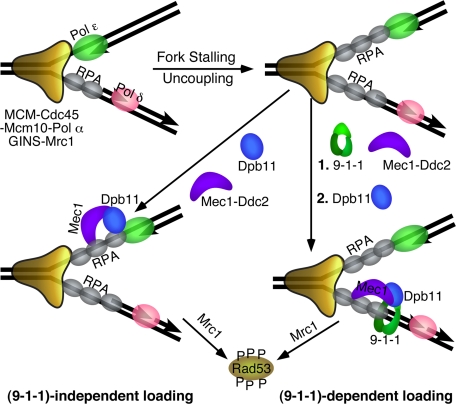
Dpb11 and 9-1-1 Define Both Independent and Synergistic Checkpoint Pathways for Mec1 Activation—Our studies suggest that 9-1-1 and Dpb11 may represent two independent and overlapping pathways that function to activate Mec1. In the G1 phase of the cell cycle, the 9-1-1 clamp is absolutely required for Mec1 activation in response to damage that is repaired by nucleotide excision repair (8). Whether Dpb11 also contributes to the G1 checkpoint is difficult to determine because it is an essential gene, and DPB11 mutants show pleiotropic phenotypes (30). However, during S phase, Dpb11 fulfills an essential checkpoint function. In S. cerevisiae, 9-1-1 is stimulatory for the S phase checkpoint, whereas in S. pombe the clamp plays a more essential role (43). The current model in this organism and in metazoans is that the phosphorylated Rad9 subunit of 9-1-1 recruits Cut5/TopBP1, which in turn activates ATR. However, in S. cerevisiae, Dpb11 can be recruited to stalled forks via interactions with pol ε (Fig. 6), and certain POL2 mutants defective for this interaction show S phase checkpoint defects. Interestingly, in S. pombe, pol ε does not function in the replication checkpoint (44), and therefore, recruitment of Cut5 via 9-1-1 may be the only pathway in these cells (Fig. 6). That Dpb11 needs to be recruited specifically to a stalled fork follows from studies indicating that, despite its function in replisome assembly, it actually is not part of the moving replication machinery (17).
FIGURE 6.
Model for the DNA replication checkpoint. Stalling of the fork results in uncoupling of the leading and lagging strands, and in the formation of extended RPA-coated ssDNA regions that can recruit Mec1 through RPA-Ddc2 interactions. In S. cerevisiae, S. pombe, and in human recruitment of Dpb11/Cut5/TopBP1 is facilitated by loading of the 9-1-1 clamp (26-30). The clamp is shown loaded onto the 5′-junction of an Okazaki fragment because of its demonstrated loading specificity (10, 42), but it might also load onto the leading strand after fork restart by Polα-primase (45). Interactions are mediated through the C-terminal tail of Ddc1/Rad9. In S. cerevisiae, both 9-1-1-dependent and 9-1-1-independent recruitment of Dpb11 may occur, the latter likely through interactions with pol ε (16). Dpb11 activates Mec1 to phosphorylate Rad53 and additional factors. Mrc1 is the proposed downstream chromatin-associated mediator that binds Rad53 and promotes its further transphosphorylation resulting in its activation as an effector kinase (46).
Our current studies indicate a model where Dpb11 can independently activate Mec1 at stalled replication forks. However, activation of Mec1 through interaction with 9-1-1 is more efficient in vitro as well as in vivo. Our studies do not provide a rational explanation for the puzzling observation that 9-1-1 alone does not efficiently activate the replication checkpoint in the absence of Dpb11 function, e.g. in the dpb11-1 mutant. Because such DPB11 mutants are pleiotropic, the checkpoint defect may in part be indirectly caused by the associated replication defects. Progress in this area will require the generation of DPB11 mutants that clearly separate its replication function from its checkpoint function.
Acknowledgments
We thank John Majors for critical discussions during the course of this work and Jurek Majka for advice during the initial stages. Carrie Stith provided expert technical support.
This work was supported, in whole or in part, by National Institutes of Health Grant GM32431. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ATR, ATM and Rad3-related protein; ssDNA, single-stranded; RPA, replication protein A; Rad24-RFC, complex of Rad24 and Rfc2-5 subunits; 9-1-1, complex of Ddc1, Rad17, Mec3 (human Rad9, Rad1, Hus1); GST, glutathione S-transferase; pol, polymerase.
References
- 1.Abraham, R. T. (2001) Genes Dev. 15 2177-2196 [DOI] [PubMed] [Google Scholar]
- 2.Bakkenist, C. J., and Kastan, M. B. (2004) Cell 118 9-17 [DOI] [PubMed] [Google Scholar]
- 3.Lambert, S., and Carr, A. M. (2005) Biochimie (Paris) 87 591-602 [DOI] [PubMed] [Google Scholar]
- 4.Rouse, J., and Jackson, S. P. (2002) Mol. Cell 9 857-869 [DOI] [PubMed] [Google Scholar]
- 5.Zou, L., and Elledge, S. J. (2003) Science 300 1542-1548 [DOI] [PubMed] [Google Scholar]
- 6.Parrilla-Castellar, E. R., Arlander, S. J., and Karnitz, L. (2004) DNA Repair 3 1009-1014 [DOI] [PubMed] [Google Scholar]
- 7.Majka, J., and Burgers, P. M. (2004) Prog. Nucleic Acids Res. Mol. Biol. 78 227-260 [DOI] [PubMed] [Google Scholar]
- 8.Giannattasio, M., Lazzaro, F., Longhese, M. P., Plevani, P., and MuziFalconi, M. (2004) EMBO J. 23 429-438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majka, J., Niedziela-Majka, A., and Burgers, P. M. (2006) Mol. Cell 24 891-901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Majka, J., Binz, S. K., Wold, M. S., and Burgers, P. M. (2006) J. Biol. Chem. 281 27855-27861 [DOI] [PubMed] [Google Scholar]
- 11.Majka, J., and Burgers, P. M. (2007) Cell Cycle 6 1157-1160 [DOI] [PubMed] [Google Scholar]
- 12.Tercero, J. A., Longhese, M. P., and Diffley, J. F. (2003) Mol. Cell 11 1323-1336 [DOI] [PubMed] [Google Scholar]
- 13.Longhese, M. P., Clerici, M., and Lucchini, G. (2003) Mutat. Res. 532 41-58 [DOI] [PubMed] [Google Scholar]
- 14.Zegerman, P., and Diffley, J. F. (2007) Nature 445 281-285 [DOI] [PubMed] [Google Scholar]
- 15.Tanaka, S., Umemori, T., Hirai, K., Muramatsu, S., Kamimura, Y., and Araki, H. (2007) Nature 445 328-332 [DOI] [PubMed] [Google Scholar]
- 16.Araki, H., Leem, S. H., Phongdara, A., and Sugino, A. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 11791-11795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masumoto, H., Sugino, A., and Araki, H. (2000) Mol. Cell. Biol. 20 2809-2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navas, T. A., Zhou, Z., and Elledge, S. J. (1995) Cell 80 29-39 [DOI] [PubMed] [Google Scholar]
- 19.Dua, R., Levy, D. L., and Campbell, J. L. (1999) J. Biol. Chem. 274 22283-22288 [DOI] [PubMed] [Google Scholar]
- 20.Frei, C., and Gasser, S. M. (2000) Genes Dev. 14 81-96 [PMC free article] [PubMed] [Google Scholar]
- 21.Katou, Y., Kanoh, Y., Bando, M., Noguchi, H., Tanaka, H., Ashikari, T., Sugimoto, K., and Shirahige, K. (2003) Nature 424 1078-1083 [DOI] [PubMed] [Google Scholar]
- 22.Saka, Y., Fantes, P., Sutani, T., McInerny, C., Creanor, J., and Yanagida, M. (1994) EMBO J. 13 5319-5329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makiniemi, M., Hillukkala, T., Tuusa, J., Reini, K., Vaara, M., Huang, D., Pospiech, H., Majuri, I., Westerling, T., Makela, T. P., and Syvaoja, J. E. (2001) J. Biol. Chem. 276 30399-30406 [DOI] [PubMed] [Google Scholar]
- 24.Yamane, K., Wu, X., and Chen, J. (2002) Mol. Cell. Biol. 22 555-566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parrilla-Castellar, E. R., and Karnitz, L. M. (2003) J. Biol. Chem. 278 45507-45511 [DOI] [PubMed] [Google Scholar]
- 26.Wang, H., and Elledge, S. J. (2002) Genetics 160 1295-1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furuya, K., Poitelea, M., Guo, L., Caspari, T., and Carr, A. M. (2004) Genes Dev. 18 1154-1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delacroix, S., Wagner, J. M., Kobayashi, M., Yamamoto, K., and Karnitz, L. M. (2007) Genes Dev. 21 1472-1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, J., Kumagai, A., and Dunphy, W. G. (2007) J. Biol. Chem. 282 28036-28044 [DOI] [PubMed] [Google Scholar]
- 30.Puddu, F., Granata, M., Di Nola, L., Balestrini, A., Piergiovanni, G., Lazzaro, F., Giannattasio, M., Plevani, P., and Muzi-Falconi, M. (2008) Mol. Cell. Biol. 28 4782-4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumagai, A., Lee, J., Yoo, H. Y., and Dunphy, W. G. (2006) Cell 124 943-955 [DOI] [PubMed] [Google Scholar]
- 32.Choi, J. H., Lindsey-Boltz, L. A., and Sancar, A. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 13301-13306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mordes, D. A., Glick, G. G., Zhao, R., and Cortez, D. (2008) Genes Dev. 22 1478-1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Majka, J., and Burgers, P. M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 2249-2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bylund, G. O., Majka, J., and Burgers, P. M. (2006) Methods Enzymol. 409 1-11 [DOI] [PubMed] [Google Scholar]
- 36.Gilbert, C. S., Green, C. M., and Lowndes, N. F. (2001) Mol. Cell 8 129-136 [DOI] [PubMed] [Google Scholar]
- 37.Sweeney, F. D., Yang, F., Chi, A., Shabanowitz, J., Hunt, D. F., and Durocher, D. (2005) Curr. Biol. 15 1364-1375 [DOI] [PubMed] [Google Scholar]
- 38.Lawrence, J. C., Lin, T. A., McMahon, L. P., and Choi, K. M. (2004) Curr. Top. Microbiol. Immunol. 279 199-213 [DOI] [PubMed] [Google Scholar]
- 39.Brush, G. S., and Kelly, T. J. (2000) Nucleic Acids Res. 28 3725-3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim, H. S., and Brill, S. J. (2003) DNA Repair 2 1321-1335 [DOI] [PubMed] [Google Scholar]
- 41.Bartrand, A. J., Iyasu, D., and Brush, G. S. (2004) J. Biol. Chem. 279 26762-26767 [DOI] [PubMed] [Google Scholar]
- 42.Ellison, V., and Stillman, B. (2003) PLoS Biol. 1 231-243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marchetti, M. A., Kumar, S., Hartsuiker, E., Maftahi, M., Carr, A. M., Freyer, G. A., Burhans, W. C., and Huberman, J. A. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 7472-7477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Durso, G., and Nurse, P. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 12491-12496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.You, Z., Kong, L., and Newport, J. (2002) J. Biol. Chem. 277 27088-27093 [DOI] [PubMed] [Google Scholar]
- 46.Alcasabas, A. A., Osborn, A. J., Bachant, J., Hu, F., Werler, P. J., Bousset, K., Furuya, K., Diffley, J. F., Carr, A. M., and Elledge, S. J. (2001) Nat. Cell Biol. 3 958-965 [DOI] [PubMed] [Google Scholar]