Abstract
Protein phosphatase 2A (PP2A) is a heterotrimer comprising catalytic, scaffold, and regulatory (B) subunits. There are at least 21 B subunit family members. Thus PP2A is actually a family of enzymes defined by which B subunit is used. The B56 family member B56α is a phosphoprotein that regulates dephosphorylation of BCL2. The stress kinase PKR has been shown to phosphorylate B56α at serine 28 in vitro, but it has been unclear how PKR might regulate the BCL2 phosphatase. In the present study, PKR regulation of B56α in REH cells was examined, because these cells exhibit robust BCL2 phosphatase activity. PKR was found to be basally active in REH cells as would be predicted if the kinase supports B56α-mediated dephosphorylation of BCL2. Suppression of PKR promoted BCL2 phosphorylation with concomitant loss of B56α phosphorylation at serine 28 and inhibition of mitochondrial PP2A activity. PKR supports stress signaling in REH cells, as suppression of PKR promoted chemoresistance to etoposide. Suppression of PKR promoted B56α proteolysis, which could be blocked by a proteasome inhibitor. However, the mechanism by which PKR supports B56α protein does not involve PKR-mediated phosphorylation of the B subunit at serine 28 but may involve eIF2α activation of AKT. Phosphorylation of serine 28 by PKR promotes mitochondrial localization of B56α, because wild-type but not mutant S28A B56α promoted mitochondrial PP2A activity. Cells expressing wild-type B56α but not S28A B56α were sensitized to etoposide. These results suggest that PKR regulates B56α-mediated PP2A signaling in REH cells.
Protein phosphatase 2A (PP2A)2 is an important regulator of apoptosis and may be a tumor suppressor (1–4). How PP2A might function as a tumor suppressor is not clear, as the enzyme on the one hand is required for cell survival but on the other is active in processes responsible for cell death (2–7). An explanation for this paradox is that PP2A is not a single enzyme but rather a family of protein phosphatase isoforms (3, 4, 8, 9). PP2A is a heterotrimer composed of a catalytic (C) subunit, a scaffold (A) subunit, and a regulatory (B) subunit. The C and A subunits form a catalytic complex that interacts with one of at least 21 diverse B subunit members from three major families (i.e. B55, B56, and PR72/130; see Refs. 2, 3, and 9). It is becoming evident that the function of PP2A relies on the B subunit because the various B subunit proteins determine substrate specificity and PP2A complex subcellular localization (2, 3, 7, 10–13). The recent crystal structure of PP2A supports this premise (4, 13–15). Thus the PP2A family of protein phosphatase isoforms can best be defined by its regulatory B subunit.
PP2A function is regulated both negatively and positively by post-translational modification (4, 16–18). Phosphorylation of B56γ by ERK at serine 337 inhibits PP2A function (13, 18). Interestingly, this regulatory ERK site is conserved in all of the known B56 family members except B56α. Although ERK may not regulate B56α function negatively, phosphorylation of this particular B56 isoform by the double-stranded RNA-activated protein kinase (PKR) has been suggested to affect PP2A function positively (19). PKR activation is associated with growth inhibition and cell death (20–23). Although PKR is best known as an eIF2α kinase, other targets also have been identified (20–23). PKR phosphorylates B56α in vitro (19).
The PP2A isoform that is responsible for BCL2 dephosphorylation contains B56α (12). Human acute lymphoblastic leukemia (ALL)-derived REH cells express high levels of BCL2, but there is little if any phosphorylation of the protein under basal conditions (24). In the present study, we examined whether PKR is a physiologic B56α kinase in REH cells and how the kinase might regulate B56α-mediated PP2A function in these cells. It was found that PKR was indeed basally active in REH cells as well as in three other ALL-derived cell lines (i.e. CCRF-CEM, RS (4, 11), and MOLT4). Loss of PKR rendered REH cells resistant to etoposide. PKR supports B56α-mediated mitochondrial and BCL2 phosphatase activity. We found that PKR was responsible for B56α phosphorylation at serine 28, which promoted mitochondrial localization of the B subunit resulting in enhanced mitochondrial PP2A activity and dephosphorylation of BCL2. PKR protects B56α from degradation by the proteasome, although this mechanism does not involve the role of PKR as a B56α kinase. Recently, the Koromilas laboratory identified a role for eIF2α phosphorylation in proteasomal regulation (25) and in activation of the PI3K/AKT signaling cascade (26). Preliminary evidence suggests that PKR protection of B56α from the proteasome may involve phosphorylation of eIF2α and activation of the PI3K/AKT/mTor signaling cascade. Consistent with a mechanism whereby phosphorylated eIF2α promotes B56α expression, three ALL cell lines (REH, CCRF-CEM, and MOLT4) displayed phosphorylated eIF2α and expressed B56α, whereas RS(4;11) cells did not display any phosphorylated eIF2α (due to lack of the eIF2α protein) and had little if any B56α. In addition, mitochondrial PP2A activity in the ALL cell lines correlated with B56α expression. In conclusion, the findings presented here suggest that PKR regulates a B56α-mediated apoptotic process that likely includes inactivation of BCL2 by dephosphorylation.
EXPERIMENTAL PROCEDURES
Cell Lines—Phoenix™ Ampho retroviral packaging cells were obtained from Orbigen (San Diego, CA) and maintained in Dulbecco's modified Eagle's medium supplemented with 5% newborn calf serum and 5% fetal bovine serum at 37 °C in 5% CO2. REH, CCRF-CEM, and MOLT4 cells were obtained from the ATCC (Manassas, VA) and maintained in RPMI 1640 plus 5% newborn calf serum plus 5% fetal bovine serum at 37 °C in 5% CO2. RS(4;11) cells were obtained from the ATCC and maintained in RPMI 1640 plus 10% fetal bovine serum at 37 °C in 5% CO2. The REH/B56α transfectant cell lines were obtained from batch transfections using either an N-terminal HA-tagged plasmid encoding the human PPP2R5A gene (B56α) under the cytomegalovirus promoter (CMV; GeneCopoeia, German-town, MD) or its serine 28-mutated counterpart. As a control, a plasmid from GeneCopoeia containing HA-tagged GFP under the cytomegalovirus promoter was used also. To obtain the latter we used a QuikChange® XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) as directed by the manufacturer. A complementary primer pair was used to create the S28A mutation (sense primer, cttcacccggaaagctgtccgcaaggcgca). A second complementary primer pair was used to create the S28E mutation (sense primer, cttcacccggaaagaggtccgcaaggcgca). Primer pairs were annealed to the complementary region of a dammethylated parental plasmid and extended using Pfu polymerase to generate a nicked, nonmethylated daughter plasmid containing the mutation. Following digestion of the parental DNA by DpnI, the nicked daughter plasmid was used to transform competent bacterial cells. Mutant constructs were confirmed by DNA sequence analysis.
The plasmids were introduced into REH cells by electroporation (200 V, 975-microfarad capacitance) using a GenePulser Xcell electroporator (Bio-Rad). Transfectants were selected and maintained in the above medium plus 0.5 mg/ml G418 (Mediatech, Manassas, VA). Where appropriate, PKR inhibitor (Calbiochem), LY294002 (Calbiochem), LiCl (Sigma), salubrinal (Tocris, Evansville, MO), and/or MG-132 (Sigma) was added to cells.
Analysis of Cell Death and Apoptosis by Annexin V Staining or Sub-G0 Content—Cells were treated with varying doses of etoposide (Sigma), MG-132 (Sigma), PKR inhibitor (Calbiochem), AKT inhibitor VIII (Calbiochem), or rapamycin (LC Laboratories, Woburn, MA) for 24 h. Cell viability was measured by trypan blue dye exclusion, and apoptosis was analyzed either by annexin V staining or measurement of sub-G0 content. Apoptosis was evaluated using the Annexin V-PE apoptosis detection kit I (BD Biosciences). The cells were washed twice with phosphate-buffered saline and resuspended in 1× annexin binding buffer. 100 μl was transferred to a 5-ml tube, and 5 μl of annexin V-PE and 5 μl of 7-AAD were added to each tube. The cells were mixed gently and incubated for 15 min in the dark at room temperature. Cells were analyzed on a FACSCalibur flow cytometer (BD Biosciences), placing the PE signal in FL2 and the 7-AAD signal in FL3. Intact cells were gated to exclude small debris. Cells in the lower right quadrant of the FL2/FL3 dot plot (labeled with annexin V-PE only) were considered to be in early apoptosis and cells in the upper right quadrant (labeled with annexin V-PE and 7-AAD) in late apoptosis/necrosis.
To determine apoptosis by sub-G0 content, a variation of a conventional FACS method to detect sub-2N DNA content in nonviable cells was used. Cells were treated with drug, fixed in methanol, incubated with 0.1 mg/ml RNase A, and stained with 0.1 mg/ml propidium iodide (Sigma) prior to analysis using the FACSCalibur flow cytometer.
Retroviral Transduction of shRNA—HuSH shRNA (29-mer) targeting PKR and a negative control plasmid targeting GFP were purchased from OriGene (Rockville, MD). These were transiently transfected into Phoenix™ Ampho cells using Lipofectamine™ 2000 (Invitrogen) as directed by the manufacturer. Retroviral supernatants were harvested at 48 h post-transfection first by centrifugation for 10 min at room temperature at 800 × g and then by filtration through 0.45 μm surfactant-free cellulose acetate to assure complete removal of producing cells. Polybrene (Chemicon, Temecula, CA) was then added to a concentration of 8 μg/ml, and the resulting virus stock was used at once to spinoculate REH cells in a variation of the method of O'Doherty et al. (27). Briefly, REH cells were resuspended at a concentration of 0.5 million cells/ml of virus stock, transferred to the wells of a 12-well tissue culture cluster, and centrifuged for 30 min at 30 °C at 1300 × g. After the addition of 1 volume of fresh virus stock, the cells were subject to a second round of centrifugation followed by incubation at 32 °C in 5% CO2 for 30 min. The infected cells were then washed twice with growth medium to remove the Polybrene and allowed to grow for 24 h after which time they were subject to selection with 0.5 μg/ml puromycin (Calbiochem). Puromycin-resistant pools of infected cells were assessed for PKR knockdown by Western analysis.
Metabolic Labeling, Immunoprecipitation, and Immunoblotting Analysis—Cells were labeled with [32P]orthophosphoric acid, and the phosphorylation status of BCL2 was determined by an immunoprecipitation method that we had used previously (24). We used a similar method to determine the phosphorylation status of B56α. Because an antibody that could immunoprecipitate B56α was not available, REH cells that exogenously express HA-tagged B56α protein or mutant S28A B56α protein were used. HA-tagged B56α protein from radiolabeled cells was isolated using HA antibody conjugated to agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA). The isolated protein was electrophoresed using 10% SDS-PAGE and then transferred to a nitrocellulose filter. The identity of the labeled band as HA-tagged B56α protein was verified by Western analysis using B56α antisera (Imgenex, San Diego, CA). For other Western analysis, 20 μg of total protein was subjected to SDS-PAGE using antibodies specific for the analyzed proteins. Antibodies used were p-PKR (Cell Signaling Technology, Danvers, MA, and Upstate Biotechnology, Lake Placid, NY), PKR (Cell Signaling and Upstate), p-eIF2α (Cell Signaling), eIF2α (Cell Signaling), AKT (Cell Signaling), p-AKT (Cell Signaling), HA (Cell Signaling), lamin A/C (Santa Cruz Biotechnology), BCL2 (Dako, Carpinteria, CA), β-actin (Sigma), GAPDH (Santa Cruz), and tubulin (Sigma). The B56α antibody was produced at Alpha Diagnostics International (San Antonio, TX) using a peptide (DGFTRKSVRKAQRQKR) that represents amino acids 22–37 of the PP2A/B56α protein sequence. Molecular weights are presented using an Invitrogen Benchmark™ prestained protein ladder or Bio-Rad Precision Plus Protein™ standards.
Where appropriate, densitometric analysis of protein bands was performed using Image J software (version 1.38x available in the public domain from the National Institutes of Health Web site, Bethesda, MD).
Isolation of Nuclei and Mitochondria—Subcellular fractionation of cells was performed as described previously (12). Cells (2 × 107 cells) were swelled in ice-cold hypotonic buffer (10 mm Hepes, pH 7.4, 5 mm MgCl2, 40 mm KCl, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 10 μg/ml leupeptin) for 30 min, aspirated repeatedly through a 25-gauge needle (25 strokes), and centrifuged at 200 × g to pellet nuclei. The resulting supernatant was centrifuged at 10,000 × g to pellet the mitochondria.
Protein Phosphatase Assay—Protein phosphatase activity of isolated mitochondria and nuclei were determined as described previously (12). Generation of free phosphate from the phosphopeptide RRA(pT)VA was measured using the molybdate-malachite green-phosphate complex assay as described by the manufacturer (Promega, Madison, WI). The phosphatase assay was performed in a PP2A-specific reaction buffer (50 mm imidazole, pH 7.2, 0.2 mm EGTA, 0.02% 2-mercaptoethanol, 0.1 mg/ml bovine serum albumin) using 100 μm phosphopeptide substrate and 2 μg of protein mitochondria or nuclei isolated as described above. After a 30-min room temperature incubation, molybdate dye was added, and free phosphate was measured by optical density at 590 nm. A standard curve with free phosphate was used to determine the amount of free phosphate generated. Phosphatase activity was defined as pmol of free phosphate generated/μg of protein/min.
Statistics—Statistical analysis was performed using standard t test analysis with SigmaStat computer software (SSPS, Chicago, IL). Results are expressed as means ± S.E. of three separate replicate experiments. Values of p < 0.05 were considered significant.
RESULTS
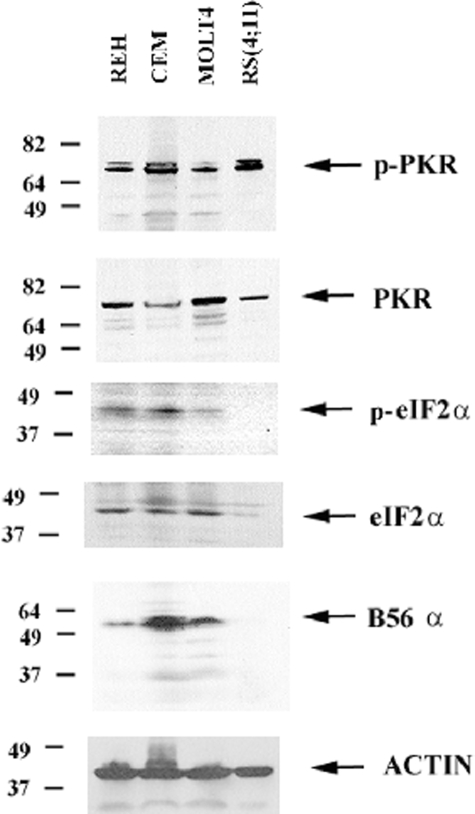
PKR Is Basally Active in a Number of ALL Cell Lines Including REH—PKR is thought to be normally dormant in cells and is expected to be activated only in response to viral infection or stress challenges (22). Thus activation of PKR under survival conditions is generally not expected. PKR expression and activation status was determined in four ALL cell lines: REH, CCRF-CEM, MOLT4, and RS(4;11). As shown in Fig. 1, PKR was expressed and phosphorylated in all four cell lines. The PKR antibody does not detect the presumed phosphorylated (i.e. slower migrating) form of the kinase (Fig. 1). This would suggest that the level of phosphorylated kinase is below the level of detection of the PKR antibody and thus that only a fraction of the PKR kinase is activated in the ALL cell lines. Nevertheless, a constitutively active nonphosphorylated PKR has yet to be reported (28), and the presence of any active PKR in the ALL cell lines suggests that the kinase may be important for cellular homeostasis in these cells.
FIGURE 1.
PKR is basally active in ALL cell lines including REH. Western blot analysis was performed to determine PKR expression and phosphorylation in the ALL cell lines REH, CCRF-CEM, MOLT4, and RS(4;11) using total cell lysates (1 × 106 cell equivalents) and antibodies against PKR, p-PKR, eIF2α, p-eIF2α, B56α, and actin.
The best characterized substrate of PKR is eIF2α (20–23). Phosphorylation of eIF2α was examined to determine whether PKR phosphorylation levels correlated with phosphorylation of the primary substrate of the kinase. Consistent with the presence of active PKR, phosphorylated eIF2α was detected in all of the cell lines except RS(4;11) (Fig. 1). The lack of observed phosphorylated eIF2α in the RS(4;11) cells, however, likely reflects the low abundance of the eIF2α protein. As shown in Fig. 1, there is little eIF2α protein present in the RS(4;11) cells compared with the other ALL cell lines.
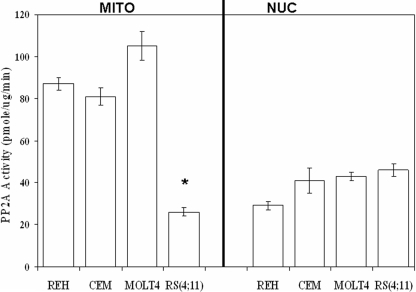
B56α Expression Correlates with Mitochondrial PP2A Activity in ALL Cell Lines—Mitochondrial and nuclear PP2A activity was assessed in the four ALL cell lines using a molybdate dye assay that we routinely use to measure PP2A activity (12). As shown in Fig. 2, REH, CCRF-CEM, and MOLT4 cells exhibited higher mitochondrial PP2A activity (87, 81, and 105 pmol/μg/min, respectively) compared with nuclear PP2A activity in these cells (26, 41, and 43 pmol/μg/min, respectively). As shown in Fig. 2, the RS(4;11) cells exhibited low mitochondrial PP2A activity (26 pmol/μg/min) compared with nuclear PP2A activity in this cell line (46 pmol/μg/min). Although there was no difference by standard t test in the nuclear PP2A activity among the ALL cell lines, there was a significant difference in the mitochondrial PP2A activity in the RS(4:11) cells compared with the other three ALL cell lines (p < 0.001). The lack of mitochondrial PP2A activity in RS(4;11) cells may be because of a lack of B56α in these cells. RS(4;11) cells displayed little if any B56α protein, whereas the other three ALL cell lines expressed the B subunit (Fig. 1). These findings suggest that B56α may be a positive regulator of mitochondrial PP2A activity in ALL cells.
FIGURE 2.
ALL cells display robust mitochondrial PP2A activity with the exception of RS(4;11) cells. Mitochondrial (MITO) and nuclear (NUC) PP2A activity was determined using a molybdate dye assay for REH, CCRF-CEM (CEM), MOLT4, and RS(4;11) cells. Error bars represent the mean ± S.D. from three separate experiments. *, statistically significant differences from REH PP2A activity (standard t test; p < 0.05).
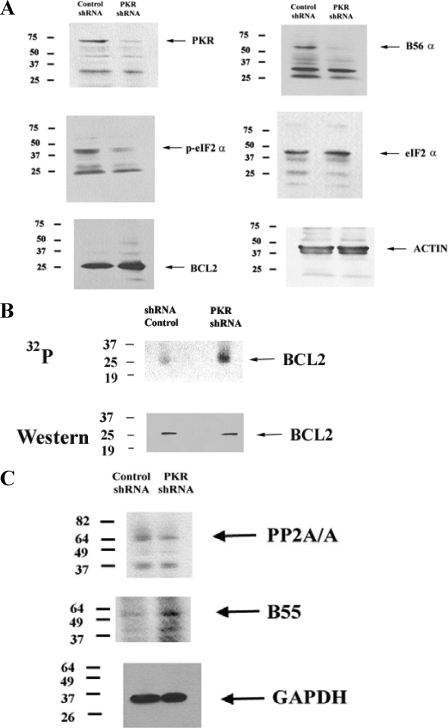
Suppression of PKR in REH Cells by shRNA Promotes BCL2 Phosphorylation, Inhibits Mitochondrial PP2A Activity, and Promotes Chemoresistance—To determine whether PKR supports BCL2 phosphatase, PKR gene expression was suppressed in REH cells using shRNA. A shRNA plasmid targeting PKR (designated TI379192) is commercially available from OriGene. A negative control shRNA plasmid that targets GFP (designated TR30003) is also available from OriGene. The shRNA plasmids were introduced into REH cells by retroviral transduction. As shown in Fig. 3A, shRNA targeting PKR resulted in significant loss of PKR protein when expression was compared with the cells transduced with the negative control. Consistent with a loss of PKR kinase activity, REH cells expressing shRNA against PKR displayed reduced phosphorylation of eIF2α (Fig. 3A), although they did not show any effects on cell viability, cell proliferation, or cell cycle profile (data not shown).
FIGURE 3.
Suppression of PKR expression by shRNA suppresses B56α expression and promotes BCL2 phosphorylation. A, Western blot analysis was performed using antibodies against PKR, B56α, p-eIF2 α, eIF2 α, BCL2, and actin on total lysate (1 × 106 cell equivalents) from REH cells transfected with control shRNA or PKR shRNA. B, REH cells transfected with control shRNA or REH cells transfected with PKR shRNA were labeled with [32P]orthophosphoric acid (32P). BCL2 was immunoprecipitated using polyclonal rabbit antisera (Santa Cruz Biotechnology), electrophoresed using 10% SDS-PAGE, and transferred to a nitrocellulose filter, and phosphorylated bands were detected by autoradiography. The identity of the BCL2 32P-labeled band was established by using a mouse monoclonal antibody against BCL2 (Dako) on the same filter. C, Western blot analysis was performed using antibodies against B55, PP2A/A, and GAPDH on total lysate (1 × 106 cell equivalents) from REH cells transfected with control shRNA or PKR shRNA.
Loss of PKR had no effect on overall BCL2 protein expression (Fig. 3A), but an effect on the phosphorylation status of the anti-apoptotic protein was unknown. To determine the effect of loss of PKR on BCL2 phosphorylation, a metabolic radiolabeling experiment was performed. REH cells transduced with control shRNA plasmid or with PKR shRNA were radiolabeled with 32P, and BCL2 protein was immunoprecipitated using rabbit polyclonal antisera (from Santa Cruz Biotechnology). Immunoprecipitated protein was electrophoresed and transferred to a nitrocellulose filter, and phosphorylated bands on the filter were detected by autoradiography. Western analysis using a mouse monoclonal antibody against BCL2 (from Dako) was used to determine that the band corresponding to 32P-labeled protein was indeed BCL2. As shown in Fig. 3B, loss of PKR potently promoted BCL2 phosphorylation in the REH cells. Such a consequence would be expected if PKR supports B56α-mediated PP2A activity targeting BCL2.
As shown in Fig. 3A, B56α expression is suppressed in REH cells transfected with PKR shRNA. The loss of BCL2 phosphatase function in REH cells with PKR shRNA may reflect loss of this vital component of the BCL2 phosphatase. As noted earlier, PP2A is an obligate heterotrimer, and there is a stoichiometric competition between B subunits for available PP2A catalytic core (i.e. A and C subunit) complexes. Loss of B56α might promote expression of other B subunits to replace B56α as a partner for available catalytic core complexes. To determine whether such a phenomenon occurred in the PKR shRNA REH cells, we examined compared expression of B55, a ubiquitously expressed B subunit, in the PKR shRNA REH cells with REH cells transfected with control shRNA. As shown in Fig. 3C, B55 expression levels were increased in REH cells with PKR shRNA. Densitometric analysis of the B55 and GAPDH bands (see Fig. 3C) using Image J software revealed that B55 (relative to GAPDH expression) was increased 2.9-fold in REH cells with PKR shRNA compared with REH cells with control shRNA. Interestingly, REH cells with PKR shRNA displayed reduced expression of the A scaffold subunit of PP2A (i.e. PP2A/A) compared with cells with control shRNA. Densitometric analysis of the PP2A/A and GAPDH bands (see Fig. 3C) revealed that PP2A/A (relative to GAPDH expression) was less than half (i.e. 43%) in REH cells with PKR shRNA compared with REH cells with control shRNA. These findings suggest that there is a stoichiometric change in expression of the scaffold A subunit and at least one B subunit with loss of B56α. Such changes would be expected to influence the PP2A activity profile in REH cells.
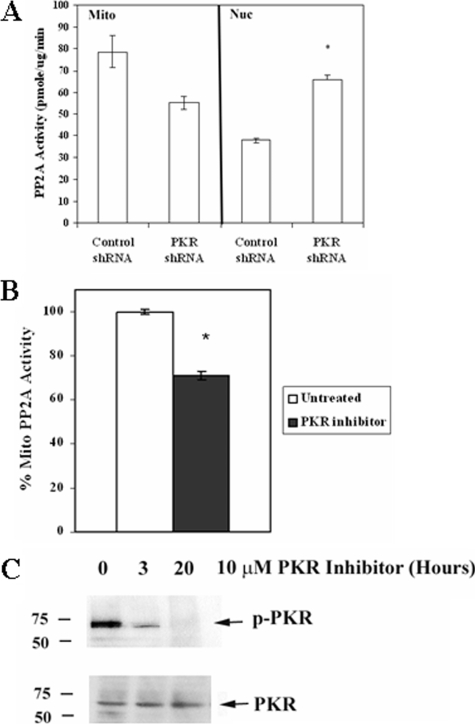
To examine the effect of loss of PKR on PP2A function, PP2A activity from mitochondrial and nuclear fractions were measured in REH cells transduced either with control shRNA or with PKR shRNA. As shown in Fig. 4A, cells with PKR shRNA demonstrated reduced (i.e. ∼25% less) mitochondrial PP2A activity compared with control cells, although the reduction was not statically significant (p = 0.099). As shown in Fig. 4B, this level of reduction is similar to the level of mitochondrial PP2A suppression in parental REH cells treated with 10 μm Calbiochem PKR inhibitor for 3 h (i.e. 29%; p = 0.029). Treatment of REH cells with 10 μm Calbiochem PKR inhibitor for 3 or 20 h potently inhibits the kinase (Fig. 4C). Densitometric analysis of the bands shown in Fig. 4C using Image J software revealed that PKR was inhibited 78% after 3 h and 98% after 20 h when comparing p-PKR/PKR in untreated and treated cells.
FIGURE 4.
Suppression of PKR expression by shRNA suppresses mitochondrial PP2A activity and promotes chemoresistance. A, mitochondrial (Mito) and nuclear (Nuc) PP2A activity was determined using a molybdate dye assay for REH cells transduced with control shRNA or with shRNA against PKR. Error bars represent the mean ± S.D. from three separate experiments. *, statistically significant differences from PP2A activity from REH cells transduced with control shRNA (p < 0.05, standard t test). B, REH parental cells were treated with 10 μm PKR inhibitor for 3 h, and mitochondrial PP2A activity was determined using the molybdate dye assay. Error bars represent the mean ± S.D. from three separate experiments. *, statistically significant differences from PP2A activity in untreated REH cells (p < 0.05, standard t test). C, Western blot analysis was performed to determine PKR activity in untreated REH cells and cells treated with 10 μm Calbiochem PKR inhibitor for 3 or 20 h using antibody against p-PKR. PKR antibody was used to measure total kinase.
Inhibition of PKR by pharmacologic agent or by genetic suppression of expression of the kinase via shRNA blocks mitochondrial PP2A activity. As shown in Fig. 4A, REH cells expressing PKR shRNA exhibited a statistically significant increase in nuclear PP2A activity compared with control shRNA REH cells (p < 0.001). These results demonstrate that PKR has broad effects on PP2A activity in REH cells.
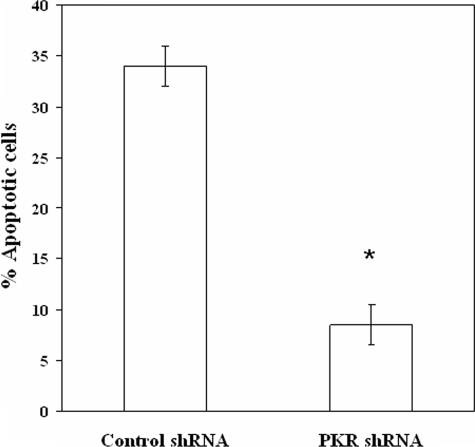
If PKR regulates a mitochondrial PP2A that participates in apoptosis, REH cells expressing PKR shRNA would be expected to be resistant to cell death with stress challenges such as etoposide. Furthermore, phosphorylation of BCL2 has been demonstrated to promote cell survival in REH cells (24), and REH cells expressing PKR shRNA do exhibit elevated phosphorylation of the anti-apoptotic protein compared with control cells (Fig. 3B). Control shRNA REH cells and REH cells expressing PKR shRNA were treated with 1 μm etoposide for 24 h, after which apoptosis was measured by FACScan analysis of sub-G0 content using propidium iodide staining. As shown in Fig. 5, REH cells expressing PKR shRNA were more resistant to the chemotherapeutic drug compared with control shRNA cells (i.e. >30% versus ∼8% cell death, respectively). These results suggest that PKR supports mitochondrial PP2A-mediated death pathways that may involve BCL2.
FIGURE 5.
Suppression of PKR expression by shRNA promotes chemoresistance. Apoptosis of shRNA-transduced REH cells treated with 1 μm etoposide for 24 h was examined using FACScan analysis of sub-G0 content of propidium iodide-stained cells. Error bars represent the mean ± S.D. from three separate experiments. *, statistically significant differences from cell viability in untreated REH cells (p < 0.05, standard t test).
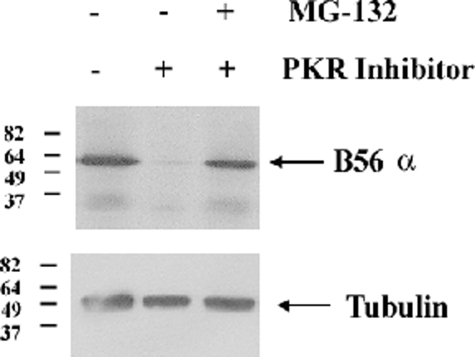
PKR Supports B56α Protein Expression by Blocking Proteasome-mediated Degradation of the B Subunit—The loss of BCL2 phosphatase activity and mitochondrial PP2A activity in REH cells with suppressed PKR expression appears to involve loss of B56α protein. An examination of B56α expression in REH cells transduced with PKR shRNA revealed that reduction of PKR affects B56α protein expression (Fig. 3A). Although B56α was expressed robustly in REH cells transduced with control shRNA, expression of the protein was severely reduced in REH cells transduced with PKR shRNA. This finding raises the possibility that PKR supports B56α-mediated PP2A activity by promoting expression of the B subunit. The possibility that PKR supports B56α expression by a transcriptional mechanism is unlikely because B56α mRNA levels are equivalent in REH cells transduced with control shRNA and REH cells transduced with PKR shRNA as determined by real-time PCR (data not shown). To determine whether a post-translational mechanism may be involved, we examined whether we could promote B56α proteolysis in REH cells using the Calbiochem PKR inhibitor. Cells were treated with 10 μm PKR inhibitor for 20 h and cotreated with the proteasome inhibitor MG-132 (10 μm) where appropriate. As shown in Fig. 6, the PKR inhibitor promoted loss of B56α, but this loss of the B subunit was blocked in cells that were treated with the proteasome inhibitor. This finding suggests that PKR regulation of B56α protein expression involves a post-translational mechanism. As PKR can phosphorylate B56α, it is possible that PKR supports B56α protein stability by phosphorylating the B subunit.
FIGURE 6.
PKR suppresses proteasomal degradation of B56α expression. Western blot analysis was performed using antibody for B56α and tubulin antibody on total lysates (1 × 106 cell equivalents) from untreated REH cells or cells treated with 10 μm PKR inhibitor and/or 10 μm MG-132 for 20 h.
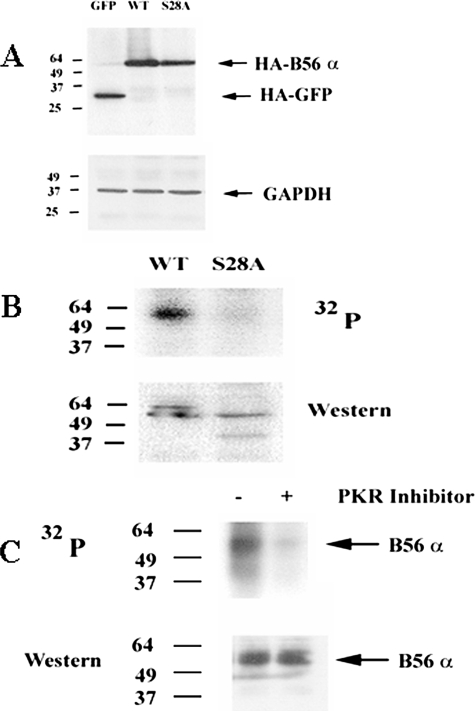
B56α Is Phosphorylated Basally by PKR, Primarily at Serine 28 in REH Cells—The B56 regulatory subunit family members are phosphoproteins (17), but it is not clear how post-translational modification of these proteins may affect their function. Phosphorylation of B56α at serine 28 has been shown to promote in vitro PP2A heterotrimer activity (19). REH cells express the B56α PP2A subunit (Fig. 1A), but it is not known if B56α is phosphorylated in REH cells. At present an antibody that efficiently immunoprecipitates B56α is unavailable; therefore exogenous HA-tagged B56α was introduced into REH cells (REH/B56α cells). In addition, a serine to alanine mutant of HA-tagged B56α at position 28 was created and was introduced into REH cells (REH/S28A cells). As a control, HA-tagged GFP was introduced into REH cells (REH/GFP cells). Expression of exogenous protein in the various REH transfectants is shown in Fig. 7A. If phosphorylation of B56α by PKR at serine 28 is required for protein stability, we would expect not to observe exogenous S28A B56α protein under conditions in which the proteasome is active. However, exogenous S28A B56α protein was detected in transfectant REH cells (Fig. 7A). In fact, exogenous B56α protein levels in the REH transfectants are similar, because cells expressing S28A B56α expressed only slightly less exogenous protein than cells transfected with wild-type (WT) B56α. The level of mutant protein in REH/S28A cells was 0.9-fold that of WT protein in REH/B56α cells when normalized to GAPDH expression. Still, it is not known whether PKR phosphorylates B56α at serine 28 in the REH cells.
FIGURE 7.
B56α is phosphorylated primarily at serine 28 in REH cells. A, Western blot analysis was performed using antibody for HA to detect exogenous control protein (HA-GFP) or exogenous B56α (HA-B56α) on total lysate from REH cells transfected with control plasmid (GFP), WT B56α (WT), or S28A B56α (S28A). Western blot analysis was also performed using antibody against GAPDH. B, REH/B56α cells (WT) and REH/S28A B56α cells (S28A) were labeled with [32P]orthophosphoric acid (32P). B56α was immunoprecipitated using HA antisera, electrophoresed using 10% SDS-PAGE, and transferred to a nitrocellulose filter, and phosphorylated bands were detected by autoradiography. The identity of the B56α 32P-labeled band was established by using an antibody against B56α on the same filter. C, REH/B56α cells were treated with 10 μm MG-132 and 10 μm PKR inhibitor for 20 h where appropriate. Cells were labeled with [32P]orthophosphoric acid, and B56α phosphorylation status (32P) was analyzed by immunoprecipitation as described in B, except that HA tag antibody was used for Western blot analysis.
Xu and Williams (19) demonstrated in an in vitro assay that phosphorylation of B56α promoted PP2A activity; the cell lines (i.e. T98G glioblastoma and 2fTGH fibroblast cells) used in that study did not express basally active PKR and were found to express low levels of B56α. PKR is generally dormant in those cells and is activated in response to stress challenge or viral infection (23). To determine whether PKR phosphorylates B56α at serine 28 in the REH cells, the phosphorylation status of the B regulatory subunit was examined using metabolic radiolabeling. REH/B56α cells (WT) and REH/S28A B56α cells (S28A) were labeled with 32P, and exogenous protein was immunoprecipitated using HA antisera, electrophoresed, and then transferred to a nitrocellulose filter. Phosphorylated bands on the filter were detected by autoradiography. Western analysis using an anti-B56α antibody was used to determine that the 32P-labeled protein immunoprecipitated by the HA antisera was indeed B56α. As shown in Fig. 7B, B56α was basally phosphorylated in the cells expressing the WT protein. Furthermore, cells expressing S28A B56α displayed little phosphorylation, suggesting that the serine 28 site is the major phosphorylation site in REH cells (Fig. 7B).
To determine whether PKR is a physiologic B56α kinase in REH cells, the phosphorylation status of B56α was examined in REH/B56α cells using a pharmacologic inhibitor of PKR. REH/B56α transfectant cells treated with 10 μm PKR inhibitor (Calbiochem) displayed nearly complete inhibition of PKR activity after 20 h (Fig. 4C). The phosphorylation status of B56α was examined in REH/B56α cells that were either untreated or pretreated with 10 μm PKR inhibitor for 20 h prior to the addition of 32P. Cells treated with the PKR inhibitor did not show phosphorylated B56α because the protein was not immunoprecipitated in treated cells (data not shown). This result was anticipated, as B56α protein was lost in REH cells with suppressed PKR expression (Fig. 3A). The metabolic radiolabeling experiment with the PKR inhibitor was repeated, but REH/B56α transfectant cells were also pretreated with MG-132 to prevent proteolysis of B56α. MG-132 did not suppress B56α phosphorylation, as REH/B56α transfectant cells still displayed potent phosphorylation of the B regulatory subunit in the presence of the proteasome inhibitor. As shown in Fig. 7C, MG-132 prevented proteolysis of B56α in the presence of the PKR inhibitor, and the PKR inhibitor potently suppressed B56α phosphorylation. Although PKR is clearly a physiologic B56α kinase in REH cells, it is not clear how phosphorylation of B56α at serine 28 affects its function.
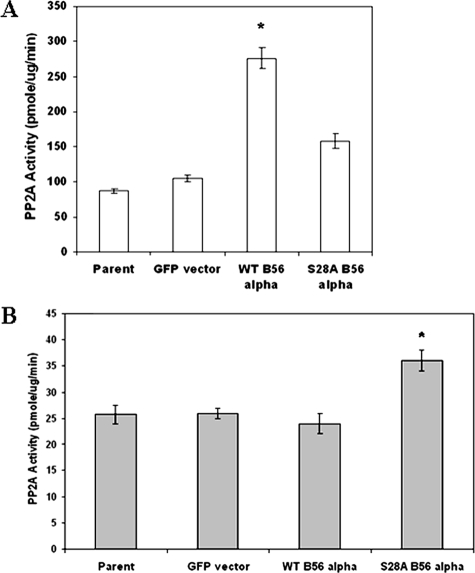
Mutation of Serine 28 Inhibits Mitochondrial PP2A Activity and Impedes B56α Mitochondrial Localization—It was not known how phosphorylation of B56α at serine 28 might affect PP2A activity in REH cells. To address this question, PP2A activity was assessed in the mitochondria and nuclei of REH cells expressing exogenous WT B56α and cells expressing nonphosphorylatable mutant S28A B56α. Mitochondrial protein and nuclear protein lysates were isolated from REH parental cells, REH/GFP cells, REH/B56α cells, and REH/S28A cells. Isolated protein was used in an in vitro PP2A assay. As shown in Fig. 8A, mitochondrial PP2A activity is nearly equivalent in REH parental cells and vector control cells (REH/GFP). Introduction of WT B56α into REH cells resulted in a statistically significant increase in mitochondrial PP2A activity (>3-fold; p < 0.001) compared with the activity observed in REH parental cells (Fig. 8A). On the other hand, introduction of mutant S28A B56α into REH cells resulted in only a modest increase in mitochondrial PP2A activity (∼1.8-fold increase) compared with REH parental cells, which was not statistically significant (p = 0.10). Introduction of B56α into REH cells had no effect on nuclear PP2A activity. There was no significant difference in nuclear PP2A activity in REH/B56α cells and parental cells (Fig. 8B). However, as shown in Fig. 8B, there was a slight but statistically significant increase (∼1.4-fold; p = 0.004) in nuclear PP2A activity in cells expressing mutant S28A B56α compared with parental cells. These results suggest that phosphorylation of serine 28 promotes B56α-mediated PP2A activity in the mitochondria of REH cells.
FIGURE 8.
Exogenous WT B56α promotes mitochondrial but not nuclear PP2A activity. Mitochondria (A) and nuclei (B) were isolated, and PP2A activity was determined using 2 μg of isolated protein with the Promega assay kit, as described under “Experimental Procedures” for parental REH cells and REH cells transfected with control plasmid (GFP), wild-type (WT) B56α, and mutant (S28A) B56α. Error bars represent the mean ± S.D. from three separate experiments. *, statistically significant differences from PP2A activity in REH parental cells (p < 0.05, standard t test).
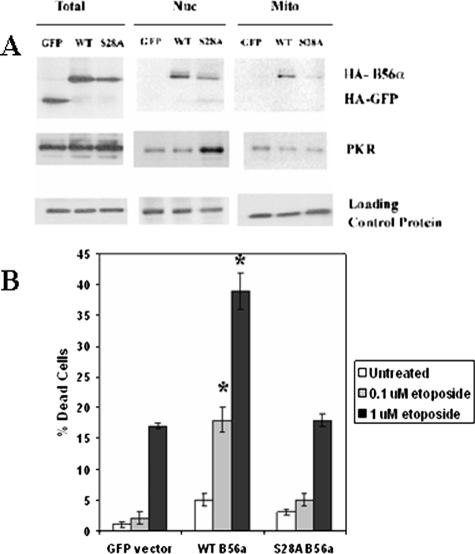
A requirement for an intact serine 28 for efficient localization of B56α to the mitochondria would explain the reduced activation of mitochondrial PP2A by the mutant S28A B56α compared with WT protein. To test this possibility, nuclear and mitochondrial localization of exogenous WT and S28A B56α was examined. As a control, localization of HA-tagged GFP protein was also determined. Nuclei and mitochondria from REH/GFP, REH/WT B56α, and REH/S28A cells were isolated. Total lysate was also obtained to monitor expression of exogenous B subunit or GFP protein. As loading controls, tubulin was used for total lysates, lamin A/C for nuclear fractions, and COX IV for mitochondrial fractions. As shown in Fig. 9A, HA-tagged GFP protein was expressed in total lysates but was not detected by Western analysis in either nuclear or mitochondrial fractions. Both WT and S28A B56α protein were detected in the nuclei. WT B56α was localized to the mitochondria, whereas little S28A B56α protein was detected there (Fig. 9A). The results presented here support a model where phosphorylation of B56α supports mitochondrial translocation of the PP2A subunit.
FIGURE 9.
Exogenous WT B56α localizes to the nuclei and mitochondria and promotes sensitivity to etoposide. A, subcellular fractionation studies were performed using 4 × 107 cells to isolate nuclei and mitochondria from REH cells transfected with control plasmid (GFP), WT B56α (WT), or S28A B56α (S28A). Western blot analysis using 40 μg of protein from total protein, nuclear protein, and mitochondrial protein was performed using antibodies against HA, PKR, and appropriate loading control (tubulin for total protein, lamin for nuclear protein, and COX IV for mitochondrial protein). B, 0.5 × 106 cells were treated with 0.1 or 1 μm etoposide for 24 h. Viability was determined by trypan blue exclusion. Error bars represent the mean ± S.D. from three separate experiments. *, statistically significant differences from cell viability in untreated REH cells transfected with (p < 0.05, standard t test).
As shown in Fig. 9A, both PKR and wild-type B56α are localized in both the mitochondria and nucleus. It should be noted that the isolated mitochondrial fractions contain roughly 10% endoplasmic reticulum, as indicated by the presence of S6 ribosomal protein (data not shown). This contamination raises the possibility that the PKR detected in the mitochondrial fraction may reflect PKR that is present in the endoplasmic reticulum. Thus it remains to be established where B56α phosphorylation by PKR might occur in the cell. Interestingly, the nonphosphorylatable S28A B56α mutant appears to sequester PKR in the nucleus (Fig. 9A). A plausible explanation for this phenomenon is that PKR associates with the mutant B subunit but the kinase remains bound or is slower to disassociate from the target protein upon failing to perform its catalytic function.
As observed in REH cells expressing PKR shRNA, loss of mitochondrial PP2A activity (Fig. 4A) correlated with increased chemoresistance (Fig. 5). If chemosensitivity associated with B56α is dependent on mitochondrial PP2A activity, we would predict that exogenous WT, but not S28A B56α, would promote sensitivity to chemotherapeutic drugs in REH cells. As shown in Fig. 9B, REH/B56α cells were sensitized to etoposide compared with REH/GFP control cells, whereas the sensitivity of REH/S28A cells to etoposide was similar to that of REH/GFP cells. Cells expressing WT B56α displayed a statistically significant increase in IC50 at 24 h compared with GFP-transfected REH cells when cells were treated with 0.1 μm etoposide (>9-fold increase; p < 0.001) or 1 μm etoposide (>2-fold increase; p < 0.001). This finding supports a model of chemosensitivity based on mitochondrial B56α-mediated PP2A activity.
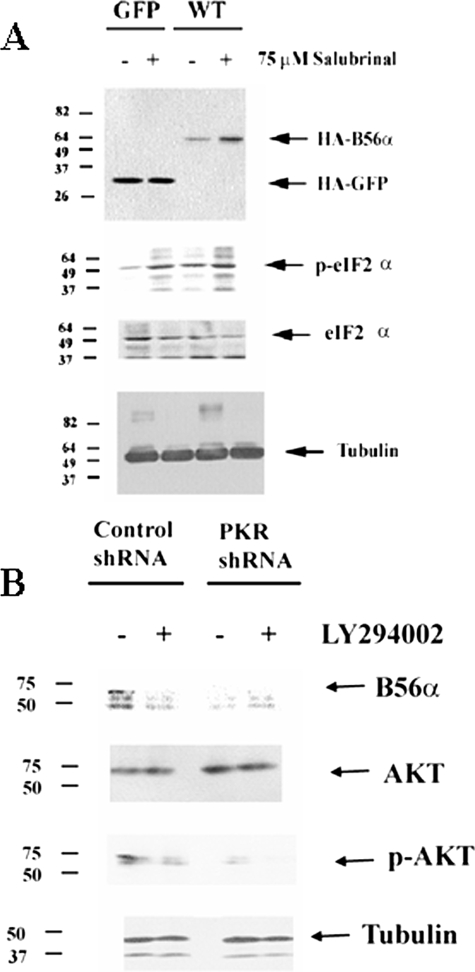
PKR Regulation of B56α Protein Expression Likely Involves eIF2α Activation of AKT and mTor—It appears that PKR does not regulate B56α protein stability by directly phosphorylating the protein, because the S28A mutant was not degraded (Fig. 7A). REH cells with PKR shRNA exhibited reduced B56α expression but also demonstrated loss of eIF2α phosphorylation (Fig. 3A). There is a correlation between B56α protein expression and eIF2α phosphorylation status in the ALL cell lines. As shown in Fig. 1, RS(4;11) cells do not express B56α and display little if any phosphorylated eIF2α, whereas the other three cell lines have phosphorylated eIF2α and express the B subunit. This result suggests that phosphorylated eIF2α may promote expression of B56α. To test this hypothesis, we treated REH cells transfected with either HA-WT B56α or HA-GFP with salubrinal, a drug that has been shown to promote eIF2α phosphorylation by preventing the dephosphorylation of the molecule (29). Densitometric analysis of the bands (in Fig. 10A) using Image J software reveals that salubrinal promotes phosphorylation of eIF2α (i.e. a 16.2-fold increase in p-eIF2 α/eIF2 α compared with untreated) but has no effect on expression of GFP protein (i.e. a 1.03-fold difference when normalized to tubulin) in REH/GFP cells treated with 75 μm salubrinal for 3 h. On the other hand, REH/WT B56α cells treated with 75 μm salubrinal for 3 h display increased phosphorylation of eIF2α (i.e. 6.8-fold increase in p-eIF2 α/eIF2 α compared with untreated) and a 4.4-fold increase in expression of B56α protein (Fig. 10A). This result suggests that PKR may regulate the PP2A subunit via a mechanism involving eIF2α.
FIGURE 10.
eIF2α and AKT promote B56α protein expression. A, Western blot analysis was performed to determine the effect of salubrinal on B56α expression. HA antibody was used to detect HA-GFP protein in REH/GFP cells and exogenous HA-B56α protein in REH/WT B56α cells on total lysates (1 × 106 cell equivalents) from untreated cells or cells treated with 75 μm salubrinal for 24 h. Antibodies against eIF2α, p-eIF2α, and tubulin were used as controls. B, Western blot analysis was performed to determine the effect of LY294002 on B56α expression in REH cells transfected with control shRNA and REH cells transfected with PKR shRNA. Rabbit polyclonal antibody against B56α protein was used on total lysates (1 × 106 cell equivalents) from untreated cells or cells treated with 1 μm LY294002 for 24 h. Antibodies against p-AKT, AKT, and tubulin were used as controls.
Recent studies from Koromilas and colleagues (26) have demonstrated a role for eIF2α phosphorylation in activation of the PI3K/AKT signaling cascade. GSK3β has been demonstrated to have a role in proteolysis of a number of proteins including MYC (30) and MCL-1 (31). AKT can inactivate GSK3β by phosphorylating serine 9 of the kinase (32), and this mechanism has been implicated in the regulation of MCL-1 degradation (31). We postulated that phosphorylation of eIF2α by PKR might support B56α expression by activating AKT to inhibit GSK3β. It appears that the mechanism involves the PI3K/AKT cascade because inhibition of this signaling pathway promotes B56α expression (Fig. 10B). REH cells with control shRNA and REH cells with PKR shRNA were treated with 1 μm LY294002 for 24 h. Densitometric analysis of the bands (in Fig. 10B) using Image J software reveals that LY294002 blocks AKT phosphorylation (i.e. 81% by using p-AKT/AKT ratio) in REH cells expressing control shRNA. A role for AKT in regulation of B56α expression seems likely because LY294002 blocks expression of the B subunit in REH cells with control shRNA (Fig. 10B). Consistent with this mechanism, REH cells expressing PKR shRNA displayed only 5% phosphorylated AKT compared with the REH cells with control shRNA (as determined by Image J software measurement of bands; Fig. 10B), and these cells do not express B56α. However, a role for GSK3β in this process does not seem likely, as we could not restore B56α expression in REH PKR shRNA cells or augment expression of the B subunit in the REH cells with control shRNA (data not shown). If phosphorylation of B56α by GSK3β was involved in degradation of the B subunit, we would have observed phosphorylation of B56α under conditions in which PKR was inhibited and the proteasome was blocked. Inhibition of PKR by a pharmacologic inhibitor in the presence of the MG-132 proteasome inhibitor did not promote phosphorylation but rather resulted in near complete suppression of B56α phosphorylation in the REH cells (Fig. 7C). This finding further suggests that GSK3β is not involved in loss of B56α expression.
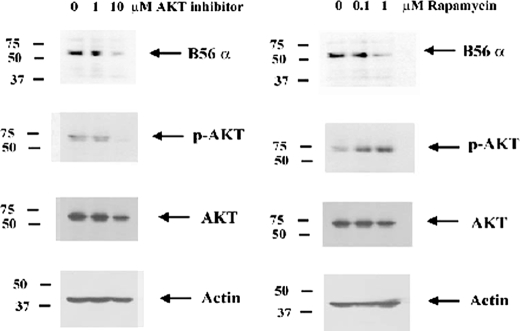
To determine whether inhibition of AKT suppressed B56α expression, REH cells were treated with 1 or 10 μm AKT inhibitor (AKT inhibitor VIII, Calbiochem) for 24 h. The drug had little if any effect on cell viability (data not shown). As shown in Fig. 11, AKT inhibitor VIII at a concentration of 10 μm inhibited AKT phosphorylation status by 94% as determined by densitometric analysis of p-AKT and AKT bands using Image J software. B56α expression was significantly suppressed in REH cells treated with the AKT inhibitor (Fig. 11). Densitometric analysis of B56α and actin bands in Fig. 11 reveals that AKT inhibitor VIII blocks B56α expression by 74% (relative to actin) in REH cells. This finding suggests that AKT is necessary to maintain B56α expression in REH cells.
FIGURE 11.
Inhibition of AKT or mTor suppresses B56α protein expression. Western blot analysis was performed to determine the effect of AKT inhibitor VIII and rapamycin on B56α expression in REH cells. Rabbit polyclonal antibody against B56α protein was used on total lysates (1 × 106 cell equivalents) from untreated cells or cells treated with AKT inhibitor VIII (1 or 10 μm) or rapamycin (0.1 or 1 μm) for 24 h. Antibodies against p-AKT, AKT, and actin were used as controls.
The mTor complex is regulated by AKT and is a critical regulator of protein expression (33, 34). It is possible that B56α expression might be regulated via mTor. To address this question, we treated REH cells with rapamycin, an inhibitor of mTor complex 1 (mTORC1; 33, 34). Cells were treated with 0.1 or 1 μm rapamycin for 24 h. REH cells displayed little toxicity to the drug at these concentrations. As shown in Fig. 11, rapamycin resulted in potent suppression of B56α expression. Densitometric analysis of B56α and actin bands in Fig. 11 reveals that rapamycin blocks B56α expression by 77% (relative to actin) in REH cells. As shown in Fig. 11, REH cells treated with rapamycin demonstrated increased AKT phosphorylation (∼3-fold increase as determined by densitometry). This result likely reflects a shift from rapamycin-sensitive mTORC1 to rapamycin-insensitive mTORC2, which would promote AKT phosphorylation, because the mTORC2 complex phosphorylates AKT on Ser-473 (33, 34). Further studies are under way to determine the mechanism by which eIF2α and AKT support B56α expression.
DISCUSSION
The findings in this study suggest that PKR-mediated phosphorylation of B56α is important for the regulation of PP2A function where this B subunit is involved. Previous studies have suggested that B56α may have a role either in survival or in cell death depending on the cell type involved (6, 7, 12). A plausible explanation for these diverse functions could involve a mechanism whereby the subcellular localization of the B subunit defines whether it supports pro-survival or pro-apoptotic signaling. In the case of REH cells, PKR may promote B56α subunit translocation to the mitochondria where pro-survival molecules such as BCL2 are dephosphorylated and thus inactivated. Although the results presented here demonstrate that PKR may direct localization of B56α, the mechanism of how this is achieved is not clear.
Movement of B56α from one subcellular compartment to another would be expected to affect signal transduction pathways, which would influence cell survival. B56α translocation from the nucleus to the mitochondria could result in PP2A-mediated pro-stress signaling. Localization of B56α to the mitochondria in itself is not toxic, as REH cells that over-expressed WT B56α showed increased mitochondrial PP2A activity but were still viable. Nevertheless, these cells are more sensitive to etoposide-induced cell death, and thus B56α-mediated PP2A supports apoptotic pathways in response to stress challenge.
An interesting finding was that PKR appears to support B56α protein expression by suppressing proteasome-mediated proteolysis. Strack et al. (7) found that C subunit and B55, but not B56, family member degradation was mediated by the proteasome in response to suppression of Aα expression. In the present study, degradation of B56α appears to be mediated by the proteasome because MG-132 blocked loss of the protein in response to the PKR inhibitor. The differences between our study and the Strack study may reflect differences in how B regulatory subunits are processed in neuronal cells and hematopoietic cells. Alternatively, there may be different mechanisms utilized for proteolysis of B56α in response to loss of PKR versus loss of binding to the catalytic core (which would presumably occur with suppression of Aα expression). In the latter case, loss of Aα would promote B56α protein to be in a monomeric state, so perhaps there are differences in the proteolytic mechanisms depending on whether the B subunit is in a complex or is monomeric.
PKR regulation of B56α protein stability could affect B subunit stoichiometry in cells and thus influence PP2A signaling. The loss of B56α that was observed with a pharmacologic inhibitor or suppression of PKR by shRNA could promote expression of other B subunits that might positively affect survival. Interestingly, REH cells transfected with PKR shRNA displayed an increase in B55 expression. B55 has been shown to be a critical component in nuclear PP2A activity (35). The nuclear PP2A isoform containing B55 may have pro-survival functions, because ionizing radiation was found to promote highly specific dissociation of B55 from the PP2A heterotrimeric complex in the nucleus (35). Thus, loss of B56α could promote expression of B55, which could favor nuclear PP2A activity. Of course, such a model is hypothetical. The possibility that suppression of B56α expression promotes B55-mediated PP2A activity is currently being pursued in our laboratory.
At present, the mechanism by which the proteasome degrades B56α is not clear, and how PKR suppresses this process is not known, but it may involve eIF2α. The Koromilas laboratory (26) has identified a novel role for eIF2 α phosphorylation in activation of the PI3K/AKT signaling cascade. The data presented here suggest that PKR phosphorylation of eIF2α promotes activation of AKT and supports expression of B56α. A mechanism whereby AKT might suppress GSK3β-mediated proteolysis of B56α seems possible, as such a mechanism has been discovered for MCL-1 (31). However, a role for GSK3β in regulating B56α proteolysis seems unlikely (at least in REH cells). The mTor kinase appears to be involved in this process because the mTor inhibitor, rapamycin, blocks B56α expression. Although mTor regulates translational and transcriptional pathways (34), it would not be expected that mTor would regulate B56α directly by such pathways because expression of the B subunit can be restored with a proteasome inhibitor. Perhaps mTor supports B56α expression indirectly by promoting expression of an unidentified molecule that acts to directly protect the B subunit from proteasome-mediated proteolysis. At present, although an eIF2α/IP3K/AKT/mTor pathway appears to regulate B56α protein expression, the mechanism of how this pathway supports expression of the B subunit is not clear.
A role for PKR as a tumor suppressor has been suggested in leukemia and other hematopoietic malignancies (36–39). However, a number of years ago, Basu et al. (40) examined PKR expression in various leukemia cells and found that PKR gene expression is actually higher in cells from patients with acute leukemia, suggesting that PKR may not be a tumor suppressor (40). Whether PKR acts as a tumor suppressor or supports malignant growth likely depends on which substrates are involved. Perhaps PKR can act as a tumor suppressor in cell types where PKR regulates PP2A function. Recently Neviani et al. (41) demonstrated that PP2A is suppressed in the blast crisis phase of chronic myeloid leukemia but not during the chronic phase of the disease. BCR/ABL was shown to inactivate PP2A by enhancing expression of the potent PP2A inhibitor protein SET (41, 42). Activation of PP2A by a novel drug (i.e. FTY720) is currently being investigated in BCR-ABLE-positive ALL and chronic myeloid leukemia (42, 43). It will be interesting to determine whether FTY720 activation of PP2A involves PKR. The findings in the present study suggest that PKR plays an important role in determining chemoresistance in ALL-derived REH cells by regulating B56α-mediated PP2A activity and the BCL2 phosphatase. Understanding how PKR functions to regulate apoptosis in acute lymphoblastic leukemia will be important for developing strategies for the treatment of this disease.
This work was supported, in whole or in part, by National Institutes of Health Grants CA30715 (to P. P. R.). This work was also supported by the Hormel Foundation and the Office of the Dean of the Graduate School of the University of Minnesota (to P. P. R.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PP2A, protein phosphatase 2A; ALL, acute lymphoblastic leukemia; PKR, double-stranded RNA-activated protein kinase; ERK, extracellular-signal regulated kinase; HA, hemagglutinin; PE, phycoerythrin; 7-AAD, 7-amino-actinomycin; mTor, mammalian target of rapamycin; PI3K, phosphatidylinositol 3-kinase; shRNA, short hairpin RNA; GFP, green fluorescent protein; FACS, fluorescence-activated cell sorter; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; WT, wild type.
References
- 1.Fujiki, H., and Suganuma, M. (1993) Adv. Cancer Res. 61 143–194 [DOI] [PubMed] [Google Scholar]
- 2.Van Hoof, C., and Goris, J. (2003) Biochim. Biophys. Acta 1640 97–104 [DOI] [PubMed] [Google Scholar]
- 3.Janssens, V., Goris, J., and Van Hoof, C. (2005) Curr. Opin. Genet. Dev. 15 34–41 [DOI] [PubMed] [Google Scholar]
- 4.Mumby, M. (2007) ACS Chem. Biol. 2 99–103 [DOI] [PubMed] [Google Scholar]
- 5.Mumby, M. (1995) Semin. Cancer Biol. 6 229–237 [DOI] [PubMed] [Google Scholar]
- 6.Silverstein, A. M., Barrow, C. A., Davis, A. J., and Mumby, M. C. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 4221–4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strack, S., Cribbs, J. T., and Gomez, L. (2004) J. Biol. Chem. 279 47732–47739 [DOI] [PubMed] [Google Scholar]
- 8.Mumby, M. C., and Walter, G. (1993) Physiol. Rev. 73 673–699 [DOI] [PubMed] [Google Scholar]
- 9.McCright, B., and Virshup, D. M. (1995) J. Biol. Chem. 270 26123–26128 [DOI] [PubMed] [Google Scholar]
- 10.Ruediger, R., Van Wart Hood, J. E., Mumby, M., and Walter, G. (1991) Mol. Cell. Biol. 12 4282–4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seeling, J. M., Miller, J. R., Gil, R., Moon, R. T., White, R., and Virshup, D. M. (1999) Science 283 2089–2091 [DOI] [PubMed] [Google Scholar]
- 12.Ruvolo, P. P., Clark, W., Mumby, M., Gao, F., and May, W. S. (2002) J. Biol. Chem. 277 22847–22852 [DOI] [PubMed] [Google Scholar]
- 13.Cho, U. S., and Xu, W. (2007) Nature 445 53–57 [DOI] [PubMed] [Google Scholar]
- 14.Chen, Y., Xu, Y., Bao, Q., Xing, Y., Li, Z., Lin, Z., Stock, J. B., Jeffrey, P. D., and Shi, Y. (2007) Nat. Struct. Mol. Biol. 14 527–534 [DOI] [PubMed] [Google Scholar]
- 15.Xu, Y., Xing, Y., Chen, Y., Chao, Y., Lin, Z., Fan, E., Yu, J. W., Strack, S., Jeffrey, P. D., and Shi, Y. (2006) Cell 127 1239–1251 [DOI] [PubMed] [Google Scholar]
- 16.Chen, J., Martin, B. L., and Brautigan, D. L. (1992) Science 257 1261–1264 [DOI] [PubMed] [Google Scholar]
- 17.McCright, B., Rivers, A. M., Audlin, S., and Virshup, D. M. (1996) J. Biol. Chem. 271 22081–22089 [DOI] [PubMed] [Google Scholar]
- 18.Letourneux, C., Rocher, G., and Porteu, F. (2006) EMBO J. 25 727–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu, Z., and Williams, B. R. (2000) Mol. Cell. Biol. 20 5285–5299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams, B. R. (1999) Oncogene 18 6112–6120 [DOI] [PubMed] [Google Scholar]
- 21.Williams, B. R. (2001) Sci. STKE 2001, RE2. [DOI] [PubMed]
- 22.Garcia, M. A., Gil, J., Ventoso, I., Guerra, S., Domingo, E., Rivas, C., and Esteban, M. (2006) Microbiol. Mol. Biol. Rev. 70 1032–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.García, M. A., Meurs, E. F., and Esteban, M. (2007) Biochimie (Paris) 89 799–811 [DOI] [PubMed] [Google Scholar]
- 24.Ruvolo, P. P., Deng, X., Carr, B. K., and May, W. S. (1998) J. Biol. Chem. 273 25436–25442 [DOI] [PubMed] [Google Scholar]
- 25.Raven, J. F., Baltzis, D., Wang, S., Mounir, Z., Papadakis, A. I., Gao, H. Q., and Koromilas, A. E. (2008) J. Biol. Chem. 283 3097–3108 [DOI] [PubMed] [Google Scholar]
- 26.Kazemi, S., Mounir, Z., Baltzis, D., Raven, J. F., Wang, S., Krishnamoorthy, J. L., Pluquet, O., Pelletier, J., and Koromilas, A. E. (2007) Mol. Biol. Cell 18 3635–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Doherty, U., Swiggard, W. J., and Malim, M. H. (2000) J. Virol. 74 10074–10080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cole, J. L. (2007) Trends Biochem. Sci. 32 57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyce, M., Bryant, K. F., Jousse, C., Long, K., Harding, H. P., Scheuner, D., Kaufman, R. J., Ma, D., Coen, D. M., Ron, D., and Yuan, J. (2005) Science 307 935–939 [DOI] [PubMed] [Google Scholar]
- 30.Sears, R., Nuckolls, F., Haura, E., Taya, Y., Tamai, K., and Nevins, J. R. (2000) Genes Dev. 14 2501–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maurer, U., Charvet, C., Wagman, A. S., Dejardin, E., and Green, D. R. (2006) Mol. Cell 21 749–760 [DOI] [PubMed] [Google Scholar]
- 32.Cross, D. A., Alessi, D. R., Cohen, P., Andjelkovich, M., and Hemmings, B. A. (1995) Nature 378 785–789 [DOI] [PubMed] [Google Scholar]
- 33.Steelman, L. S., Stadelman, K. M., Chappell, W. H., Horn, S., Bäsecke, J., Cervello, M., Nicoletti, F., Libra, M., Stivala, F., Martelli, A. M., and McCubrey, J. A. (2008) Expert Opin. Ther. Targets 12 1139–1165 [DOI] [PubMed] [Google Scholar]
- 34.Wullschleger, S., Loewith, R., and Hall, M. N. (2006) Cell 124 471–484 [DOI] [PubMed] [Google Scholar]
- 35.Guo, C. Y., Brautigan, D. L., and Larner, J. M. (2002) J. Biol. Chem. 277 4839–4844 [DOI] [PubMed] [Google Scholar]
- 36.Abraham, N., Jaramillo, M. L., Duncan, P. I., Methot, N., Icely, P. L., Stojdl, D. F., Barber, G. N., and Bell, J. C. (1998) Exp. Cell Res. 244 394–404 [DOI] [PubMed] [Google Scholar]
- 37.Beretta, L., Gabbay, M., Berger, R., Hanash, S. M., and Sonenberg, N. (1996) Oncogene 12 1593–1596 [PubMed] [Google Scholar]
- 38.Hii, S. I., Hardy, L., Crough, T., Payne, E. J., Grimmett, K., Gill, D., and McMillan, N. A. (2004) Int. J. Cancer 109 329–335 [DOI] [PubMed] [Google Scholar]
- 39.Murad, J. M., Tone, L. G., de Souza, L. R., and De Lucca, F. L. (2005) Blood Cells Mol. Dis. 34 1–5 [DOI] [PubMed] [Google Scholar]
- 40.Basu, S., Panayiotidis, P., Hart, S. M., He, L. Z., Man, A., Hoffbrand, A. V., and Ganeshaguru, K. (1997) Cancer Res. 57 943–947 [PubMed] [Google Scholar]
- 41.Neviani, P., Santhanam, R., Trotta, R., Notari, M., Blaser, B. W., Liu, S., Mao, H., Chang, J. S., Galietta, A., Uttam, A., Roy, D. C., Valtieri, M., Bruner-Klisovic, R., Caligiuri, M. A., Bloomfield, C. D., Marcucci, G., and Perrotti, D. (2005) Cancer Cell 8 355–368 [DOI] [PubMed] [Google Scholar]
- 42.Perrotti, D., and Neviani, P. (2006) Br. J. Cancer 95 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu, Q., Zhao, X., Frissora, F., Ma, Y., Santhanam, R., Jarjoura, D., Lehman, A., Perrotti, D., Chen, C. S., Dalton, J. T., Muthusamy, N., and Byrd, J. C. (2008) Blood 111 275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]