Abstract
Autophagy is a lysosome-dependent degradative pathway that regulates the turnover of intracellular organelles, parasites, and long-lived proteins. Deregulation of autophagy results in a variety of pathological conditions, but little is known regarding the mechanisms that link normal cellular and pathological signals to the regulation of distinct stages in the autophagy pathway. Here we uncover a novel role for the Abl family kinases in the regulation of the late stages of autophagy. Inhibition, depletion, or knockout of of the Abl family kinases, Abl and Arg, resulted in a dramatic reduction in the intracellular activities of the lysosomal glycosidases α-galactosidase, α-mannosidase and neuraminidase. Inhibition of Abl kinases also reduced the processing of the precursor forms of cathepsin D and cathepsin L to their mature, lysosomal forms, which coincided with the impaired turnover of long-lived cytosolic proteins and accumulation of autophagosomes. Furthermore, defective lysosomal degradation of long-lived proteins in the absence of Abl kinase signaling was accompanied by a perinuclear redistribution of lysosomes and increased glycosylation and stability of lysosome-associated membrane proteins, which are known to be substrates for lysosomal enzymes and play a role in regulating lysosome mobility. Our findings reveal a role for Abl kinases in the regulation of late-stage autophagy and have important implications for therapies that employ pharmacological inhibitors of the Abl kinases.
Macroautophagy (hereafter referred to as autophagy) is a catabolic process by which long-lived cytoplasmic proteins, protein complexes, and entire organelles are degraded through a lysosome-dependent pathway. Autophagy is essential to maintain homeostatic processes such as organelle and protein turnover, but it is also critical in the response to stress conditions such as nutrient deprivation, oxidative stress, pathogen infection, and hypoxia (1). Deregulation of autophagy has been implicated in a wide range of pathologies, including cancer, myopathies, and neurodegenerative diseases (1). Autophagy involves the sequestration of cytoplasmic components and intracellular organelles within a double-membrane vesicle, the autophagosome. The outer membrane of the autophagosome fuses with the lysosome, and sequestered components are thereby delivered to the lysosome for degradation by lysosomal enzymes (2).
The low basal level of autophagy in cells is up-regulated under stress conditions. A number of genes that regulate autophagy have been identified, and the majority of these autophagy-related genes appear to function at the initial steps of autophagosome formation (1, 2). The target of rapamycin (TOR)2 kinase is a major inhibitory signal that shuts off autophagy in the presence of growth factors and nutrients. The binding of growth factors to cell surface receptors activates class I phosphoinositide 3-kinase, which in turn activates the Akt1 kinase and its target the mammalian target of rapamycin (mTor) (3), leading to negative regulation of autophagosome formation. The effectors of mTOR signaling critical for the regulation of mammalian autophagy remain to be identified but are likely to be involved in autophagy induction (1, 4, 5). However, increasing evidence supports the existence of mTOR-independent pathways downstream of growth factor signaling involved in regulating distinct stages of autophagy (1).
The Abelson family of cytoplasmic non-receptor tyrosine kinases, Abl (Abl1) and Arg (Abl2), have been implicated in the regulation of cytoskeletal processes important for cell adhesion and migration, as well as cell proliferation and survival (6, 7). Deregulation of Abl kinase activity is implicated in the pathogenesis of chronic myelogenous leukemia as a result of a chromosomal translocation event that produces the BCR-ABL fusion protein with constitutive Abl tyrosine kinase activity (8, 9). Early-stage chronic myelogenous leukemia can be effectively treated with signal transduction inhibitor 571 (STI571), also known as Gleevec or imatinib mesylate, which inhibits Abl kinase activity by binding to the ATP-binding pocket (10).
Recent studies have highlighted important roles for Abl kinase signaling in cellular and pathological processes. These include the regulation of cell-cell adhesion (11), as well as cell proliferation, survival, anchorage-independent growth, and invasion of cancer cells (6, 12). Abl kinases are activated downstream of ligand-activated growth factor receptors for platelet-derived growth factor (13, 14), epidermal growth factor (15, 16), and insulin-like growth factor-1 (12) and elevated levels of Abl kinase activity have been detected in non-small cell lung cancer and breast cancer cell lines (12, 16, 17). Abl kinase signaling has also been implicated in microbial pathogenesis. Abl kinases are catalytically activated upon Shigella flexneri infection and mediate actin comet tail formation, intracellular motility, and cell-to-cell spread of the bacteria (18, 19).
It was recently reported that treatment of cells with the Abl kinase inhibitor, STI571, resulted in the accumulation of autophagosomes, an early-stage marker of autophagy, which led to the conclusion that Abl kinases negatively regulate autophagy (20). However, it is becoming increasingly clear that the accumulation of autophagosomes alone cannot be used as an indicator of increased autophagy. Additional assays, such as measurement of long-lived protein degradation rates and lysosomal enzyme activities, are essential to assess the function of the lysosomal system and flux through the entire autophagy pathway. A blockage in the lysosomal degradation of autophagic proteins may also increase the accumulation of autophagosomes. Thus, the accumulation of autophagosomes alone is not necessarily an indicator of increased autophagy (21, 22).
Here we uncover an essential role for Abl kinases as positive regulators of autophagy. Through the use of pharmacological inhibition as well as knockdown and knockout of Abl family kinases, we show that Abl kinases modulate the later stages of autophagy by regulating the localization and activity of acidic glycosidases and cathepsins and the subcellular trafficking of lysosomes. Inhibition or depletion of Abl kinases results in the accumulation of autophagosomes due to lysosomal dysfunction leading to decreased degradation of long-lived proteins. These findings highlight the importance of assaying the degradation of long-lived proteins to measure autophagy and not relying solely on detection of autophagosome levels, which may be indicative of impaired lysosomal functions or other defects (21). Our findings reveal a role for Abl kinases in the regulation of autophagy through modulation of lysosomal processes and have implications for therapies that target Abl kinase signaling.
EXPERIMENTAL PROCEDURES
Antibodies, Inhibitors, and Reagents—Antibodies used were as follows: rabbit polyclonal anti-phospho-CrkL (Tyr-207, Cell Signaling Technology); mouse monoclonal anti-CrkL (Cell Signaling Technology, 32H4); mouse monoclonal anti-ABL (BD Pharmingen, 8E9), rabbit polyclonal anti-LAMP-1 (Sigma, L1418); mouse monoclonal anti-LAMP-1 (Santa Cruz Biotechnology, H4A3); mouse monoclonal anti-LAMP-2 (Santa Cruz Biotechnology, H4B4); mouse anti-cathepsin L (BD Bioscience); and goat anti-cathepsin D (Santa Cruz Biotechnology, C20). Horseradish peroxidase-conjugated secondary antibodies were from Santa Cruz Biotechnology. STI571 was a kind gift from Novartis Pharmaceuticals. Rapamycin, vinblastine, and peptide N-glycosidase F were purchased from Sigma-Aldrich.
Cell Lines—The retroviral construct pBabe-GFP-microtubule-associated protein 1 light chain 3 (LC3) (21) and the A549 alveolar carcinoma cell line were obtained from Dr. Tso-Pang Yao (Dept. of Pharmacology & Cancer Biology, Duke University Medical Center). The A549 alveolar carcinoma cell line was maintained in Dulbecco's modified eagles medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS). Retrovirus production in 293T cells was performed as previously described (14). A549 cells were transduced with GFP-LC3 virus-containing supernatant in the presence of 8 μg/ml Polybrene and selected with 3 μg/ml puromycin. GFP+ cells were sorted by flow cytometry. Mouse embryonic fibroblasts (MEFs) were generated and maintained in this laboratory from Abl-Arg double knockout mice and littermate controls as previously described (13).
Drug Treatment—A549-GFP-LC3 cells were incubated with vehicle (DMSO), 10 μm STI571, or 2 μg/ml rapamycin for the indicated times. The culture medium was removed every 24 h and replaced with fresh medium containing STI571 or rapamycin.
RNA Interference Using siRNA—A549-GFP-LC3 cells were transfected with a non-targeted control small interfering RNA (siRNA) or on-target specific siRNAs directed to Abl and Arg sequences (Dharmacon RNA Technologies). Briefly, 10 nm Abl(7) siRNA (GAAGGAAAUCAGUGACAUAUU) was combined with 10 nm Arg(5) siRNA (GAAAUGGAGCGAACAGAUAUU) in a 1:1 ratio or 20 nm of a non-targeted control siRNA was transfected into A549-GFP-LC3 cells using Oligofectamine (Invitrogen). After 24 h, cells were retransfected with the indicated siRNAs for an additional 24 h.
RNA Interference Using miRNA—Because long-lived degradation assays are routinely performed over a 4- to 5-day period, it was necessary to down-regulate Abl and Arg expression for longer times than those obtained by transient transfection with siRNAs. Therefore, lentiviral-mediated delivery of microRNA (miRNA) sequences toward Abl and Arg transcripts was employed in A549 cells (23). The BLOCK-iT polymerase II miR RNAi expression vector kit from Invitrogen was used as described in a previous study (23) with the following modifications. A construct encoding both Abl6 miRNA (GGTGTATGAGCTGCTAGAGAA) and Arg9 miRNA (CCTTATCTCACCCACTCTGAA) was cloned into the lentiviral vector FCW attR1-attR2 (23). The resulting construct (Fc EmGFPW-Abl6-Arg9) was then transfected into 293T cells along with pCMV-VSVG, pRSV-Rev, and pMPL plasmids (23). Lentivirus-containing supernatant was collected 24 and 48 h later and placed on A549 cells in the presence of 8 μg/ml Polybrene. After 48 h, transduced cells were assayed for long-lived protein degradation rates as described below.
Autophagosome Detection—For detection of GFP-LC3+ autophagosomes by immunofluorescence, sub-confluent A549-GFP-LC3 cells growing on glass coverslips were incubated with vehicle (DMSO), STI571 or rapamycin for up to 48 h. Alternatively, cells were transfected with control and Abl/Arg siRNAs as described above. Cells were fixed and analyzed by fluorescence microscopy (see below). To biochemically analyze the conversion of GFP-LC3-I to GFP-LC3-II, total cell lysates were subjected to immunoblotting with an anti-GFP mouse antibody (Roche Applied Sciences).
Long-lived Protein Degradation Assays—Long-lived protein degradation assays were performed essentially as described (24). Briefly, cells were seeded in 12-well plates for 16 h at 70% sub-confluency. Cells were incubated in l-leucine-free DMEM containing 10% (v/v) dialyzed FBS for 1 h to deplete endogenous pools of l-leucine. Cells were then metabolically labeled with 5 μCi/ml 3H-labeled l-leucine (PerkinElmer) for 24 h and then chased for an additional 24 h in DMEM containing 10% (v/v) FBS (complete medium) and an excess of l-leucine to permit the degradation of labeled short-lived cytosolic proteins. For autophagy assays under basal conditions the 24-h chase medium was removed and replaced with complete medium containing DMSO (vehicle), 10 μm STI571, or 2 μg/ml rapamycin. For starvation-induced autophagy assays the 24-h chase medium was removed and replaced with Earle's balanced salt solution containing DMSO (vehicle), 10 μm STI571, or 50 μm vinblastine for up to 6 h. After addition of the various compounds for the indicated time points, the amount of radioactivity in the medium and that retained in the cells following cell lysis, was determined using a liquid scintillation counter. The rates of degradation were obtained by dividing the cpm in the medium by the total cpm (medium cpm plus lysate cpm) (24).
Lysosomal Enzyme Assays—Lysosomal enzyme activities were measured with the appropriate 4-methylumbelliferone-conjugated substrates (Sigma-Aldrich). Briefly, cells were seeded in triplicate in 6-well plates. Prior to performing the assay, cells were incubated in medium containing 10% (v/v) heat inactivated FBS for 24-48 h to deplete any lysosomal enzymes derived from bovine calf serum origin. Cells were washed with ice-cold PBS and harvested with a cell lifter using 200 μl of ice-cold sterile water. Aliquots of each whole cell lysate were incubated with 50 μl of 10 mm 4-methylumbelliferone-conjugated substrates at 37 °C for up to 1 h in acetate buffer, pH 4.8. Reactions were stopped by the addition of 200 μl of 0.5 m NaHCO3, pH 10.7, and the relative fluorescence was measured with a Labsystems Fluoroskan fluorometer set at 355 nm excitation and 460 nm emission. A standard curve comprising varying concentrations of free 4-methylumbelliferone (Sigma-Aldrich, 0, 200, 400, 600, and 800 pm, respectively) was also included in each group of lysosomal enzyme assays and used to calculate enzyme activities. An aliquot of each cell lysate was also assayed for total protein using the Bio-Rad DC protein assay reagents A, B, and S as described by the manufacturer (Bio-Rad). Enzyme activities were expressed as nanomoles/min/mg of total protein.
Western Blotting—Cells were washed with ice-cold PBS and harvested using a cell scraper in the presence of mammalian protein extraction reagent (M-PER, Pierce Biotechnology) containing freshly added protease and phosphatase inhibitors. Cell lysates were clarified by microcentrifugation and assayed for total protein as described above. Lysates were immediately diluted to 1 μg/μlin1× Laemmli sample buffer (1% (w/v) SDS, 4 mm urea, 80 mm Tris-HCl, pH 6.8, 0.1% (w/v) Bromphenol Blue), boiled for 5 min and analyzed by SDS-PAGE and Western blotting. Nitrocellulose membranes were blocked in Tris-buffered saline Tween 20 (TBST) containing 5% (w/v) milk powder (blocking solution) for 1 h at room temperature and then incubated with fresh blocking solution containing primary antibody overnight at 4 °C. Blots were washed three times with TBST and then incubated with the appropriate secondary horseradish peroxidase-conjugated antibody in blocking solution for 1 h at room temperature. Blots were then washed and developed using ECL Western blotting detection reagent as recommended by the supplier (Amersham Biosciences GE Healthcare). Quantification of signal intensities on Western blots was done using ImageJ software.3
Pulse-chase Studies and Immunoprecipitation—A549 cells were grown to 70% confluence in 10-cm dishes and metabolically labeled with 100 μCi/ml EXPRESS[35S] protein labeling mix (PerkinElmer Life Sciences) for 24 h in cysteine and methionine-free DMEM (Invitrogen) supplemented with 10% (v/v) dialyzed FBS. Cells were then rinsed with serum-free medium and chased in the presence of DMEM containing 10% FBS and an excess of cysteine and methionine for 48 h. During the chase time 35S-labeled cells were incubated with varying concentrations of STI571. Cells were then harvested with a cell scraper using 1 ml of Brij buffer (50 mm Tris-HCl (pH 7.6), 150 mm NaCl, 5 mm EDTA, 1 mm Na3VO4, 1% v/v Brij97, 1% v/v N-octyl-β-d-glucoside) and clarified by microcentrifugation. Lysates (1 mg) were then immunoprecipitated using 3 μg of a rabbit polyclonal antibody toward LAMP-1 (Sigma-Aldrich) or 3 μg of control rabbit IgG and 40 μl of protein A/G PLUS-agarose (Santa Cruz Biotechnology) as recommended by the supplier. Immunoprecipitated complexes were analyzed by SDS-PAGE and autoradiography.
Fluorescence Microscopy—Cells exponentially growing on glass coverslips were fixed with freshly prepared PBS containing 4% (w/v) paraformaldehyde for 15 min at 4 °C, permeabilized with PBS containing 3% (w/v) bovine serum albumin and 0.1% (v/v) Triton X-100 at room temperature for 15 min, and then blocked overnight at 4 °C with PBS containing 3% (w/v) bovine serum albumin (blocking buffer). Fixed cells were then incubated with primary antibodies diluted in blocking buffer for 1 h followed by a 1-h incubation with Alexa Fluor 488-conjugated or Alexa Fluor 568-conjugated secondary antibodies (Santa Cruz Biotechnology). Samples were washed with PBS and counterstained with Hoescht to visualize nuclei and analyzed using a Zeiss Axio Imager wide field fluorescence microscope (Carl Zeiss MicroImaging). Consecutive images were acquired using the oil objective with the red, green, and blue channels, pseudocolored, and overlaid using MetaMorph software (MDS Analytical Technologies). The “Linescan” application of the MetaMorph software program was used to determine the distance of LAMP+ lysosomes from the nucleus.
Live Cell Imaging—To track the movement of lysosomes in the absence of Abl signaling, A549 cells were treated with DMSO or 10 μm STI571 for 48 h and then labeled with Lyso-Tracker Red DND-99 (Molecular Probes, Invitrogen L-5728) for 30 min. The distribution and mobility of acidic organelles were assessed by live cell imaging using a Zeiss Axio observer live cell station (Carl Zeiss MicroImaging). Twenty-four images were acquired over a 2-min acquisition period and used to generate movies with MetaMorph software (played at six frames per second). The relative mobility of individual lysosomes was analyzed with MetaMorph software and depicted by rainbow analysis as described in the legend of Fig. 3C. The velocity of individual lysosomes was determined using the MetaMorph software “Track Objects” application. Values were plotted in Excel and used to generate graphs of peripheral and perinuclear lysosome movement.
FIGURE 3.
Lysosome localization and motility is altered in the absence of Abl signaling. A, A549-GFP-LC3 cells were treated with DMSO (control) or 10 μm STI571 for 48 h, fixed and analyzed for LAMP-1 immunostaining. LAMP-1+ lysosomes (red) were dispersed widely throughout the cytosol in control cells, whereas lysosomes from STI571-treated cells aggregated in the perinuclear region. The size and intensity of LAMP-1+ lysosomes increased in STI571-treated cells. B, the distribution of individual lysosomes relative to the nucleus was measured in control and STI571-treated cells using MetaMorph software. The distribution of LAMP-1+ lysosomes, expressed as a fraction of the total, was plotted as a function of distance (pixels) from the nucleus. The graph in panel B represents the LAMP-1 distribution data for the control and STI571-treated cell shown in panel A. C, the lysosomes of control or STI571-treated A549 cells (48 h) were labeled with LysoTracker DND-99 and analyzed by live cell imaging over a 2-min acquisition period. The relative mobility of individual lysosomes was assessed by rainbow analysis. The first acquired image at time 0, the image acquired at 1 min, and the final images acquired at 2 min are pseudo-colored green, blue, and red, respectively (small panels) and then overlaid (large panels). Non-motile lysosomes are represented in white, whereas lysosomes that moved over the 2-min acquisition period are pseudo-colored. The results are representative of three independent experiments. D, the average velocity of 20 peripheral lysosomes and 20 perinuclear lysosomes in individual cells from control and STI571-treated cells (n = 5 cells per group) was quantified using Meta Morph software. The data are represented as the average velocity ± S.D. Scale bar = 20 μm.**, p<0.05 compared with the velocity of peripheral and perinuclear lysosomes in DMSO-treated control cells.
Statistical Analysis—Unpaired Student t-tests were performed using GraphPad software (www.graphpad.com) to determine whether the observed differences in autophagosome formation, autophagy rates, lysosome movement, and lysosomal enzyme activity levels were statistically significant.
RESULTS
Inhibition of Abl Kinases Promotes the Accumulation of Autophagosomes—To determine if Abl kinases play a role in the regulation of autophagy, we first analyzed the effect of Abl kinase inhibition with STI571 (Gleevec, Imatinib) in A549 alveolar carcinoma cells. To monitor autophagosome levels we employed a fusion of GFP to LC3, the mammalian ortholog of yeast ATG8. Upon induction of autophagy, LC3 undergoes conjugation with phosphatidylethanolamine and conversion to a non-soluble form (LC3-II) (25), which specifically marks the autophagosome membrane. In agreement with a previous report (20), treatment of A549-GFP-LC3 cells with STI571 altered the pattern of GFP-LC3 staining in the cytosol from diffuse to a punctate pattern, characteristic of increased levels of autophagosomes (Fig. 1, A and B). Treatment with STI571 induced a marked reduction in Abl kinase activity in a dose-dependent manner as determined by assessing site-specific phosphorylation of the Abl substrate CrkL (Fig. 1E, middle panel). A direct correlation was observed between the inhibition of Abl kinase activity with STI571 and the conversion of LC3-I to LC3-II (Fig. 1E and data not shown) and accompanying accumulation of GFP-LC3-positive autophagosomes (Fig. 1, A and B).
FIGURE 1.
Inhibition of Abl kinases increases the accumulation of autophagosomes. A and C, A549 human lung carcinoma cells stably expressing GFP-LC3 (A549-GFP-LC3) were treated with 10 μm STI571 for 24 h (A, right) or transfected with Abl and Arg siRNAs (C, right), and were then analyzed by immunofluorescence. The pattern of GFP-LC3 staining in the cytosol changed from diffuse to predominantly punctate/vesicular. Scale bar = 20 μm. The number of cells with vesicular GFP-LC3 staining in control (DMSO) and STI571-treated cells (B), and in non-targeted control siRNA and Abl/Arg siRNA transfected cells (D), was scored in three individual fields at 20× magnification and expressed as a percentage of the total number of cells present. The results are represented as the mean (n = 3)±S.D. Both pharmacological inhibition and siRNA-mediated depletion of Abl kinases significantly increased autophagosome formation. **, p < 0.05 compared with the percentage of cells in control groups. E, subconfluent A549-GFP-LC3 cells were incubated with 10 μm or 30 μm STI571 for 24 h and analyzed by Western blotting with an anti-GFP antibody to monitor the relative amounts of GFP-LC3-I and GFP-LC3-II, or with phospho-CrkL and CrkL-specific antibodies as indicated. The results are representative of three independent experiments. The-fold changes in LC3-II levels are indicated below the blot. F, A549-GFP-LC3 cells were transfected with control siRNA or Abl/Arg siRNA oligonucleotides, and analyzed 48 h later by Western blotting as in panel E. The lysates were also analyzed for Abl and Arg protein levels using a pan Abl antibody.
It is known that STI571 not only inhibits Abl kinases but also other kinases such as the platelet-derived growth factor receptor and c-Kit, as well as non-kinase targets such as the NADPH:quinone oxireductase QR2 (26). Thus, to directly determine whether Abl kinase inhibition mediates the effects of STI571 on autophagosome accumulation, we silenced the expression of Abl and Arg with specific small interfering RNAs (siRNAs). Western blotting with a pan-Abl antibody demonstrated a marked reduction in the levels of Abl and Arg proteins, which coincided with an increase in LC3-II protein levels (Fig. 1F). Immunofluorescence staining of Abl/Arg siRNA-transfected cells demonstrated significantly increased numbers of cells with punctate GFP-LC3 staining compared with cells transfected with control siRNA (Fig. 1, C and D). Collectively, these results suggest that down-regulation of Abl kinase signaling promotes the accumulation of autophagosomes.
Abl Kinase Inhibition Impairs the Degradation of Long-lived Proteins—The accumulation of GFP-LC3+ autophagosomes in the absence of Abl kinase signaling may be due to either up-regulation of the initial stages of autophagy or to reduced degradation of autophagic material by blockage of downstream events in the autophagy process. Thus, analysis of GFP-LC3 levels alone does not provide a measurement of flux through the autophagy pathway (24). To determine if long-lived cytoplasmic proteins were in fact being delivered to and degraded within lysosomes in the absence of Abl kinase signaling, we performed long-lived protein degradation assays (21, 24). A549-GFP-LC3 cells were metabolically labeled with [l-3H]leucine under normal growth conditions and chased in the presence of “cold” medium containing excess l-leucine for varying times in the presence of STI571 or a known pharmacological stimulator of autophagy. Inhibition of the mTOR kinase with rapamycin activates autophagy and was used as a positive control (25). Inactivation of mTOR signaling with rapamycin completely abolished phosphorylation of the mTOR substrate p70S6K as expected, but had no effect on Abl kinase activity as shown by unchanged phospho-CrkL protein levels (Fig. 2A). Similarly, inhibition of Abl kinases with STI571 did not affect mTOR activity (Fig. 2A). Treatment of A549-GFP-LC3 cells with STI571 or rapamycin both induced the conversion of LC3-1 to LC3-II (Fig. 2B). Conversion of LC3-1 to LC3-II increased persistently over a 48-h period with STI571 treatment, but with rapamycin treatment LC3-II levels peaked at 24 h and decreased slightly by 48 h (Fig. 2B). In addition to inducing the conversion of LC3-I to LC3-II, rapamycin also induced a marked increase in the basal rate of [l-3H]leucine-labeled protein turnover (Fig. 2C), thereby enhancing the flux through the autophagy pathway as reported (25). In contrast, inhibition of Abl kinase activity with STI571 dramatically reduced the basal rate of long-lived protein turnover when compared with control cells (Fig. 2C).
FIGURE 2.
Abl kinase inhibition impairs the degradation of long-lived proteins. A, sub-confluent A549-GFP-LC3 cells were treated with 10 μm STI571 or 2 μg/ml rapamycin for 24 h or 48 h and analyzed for activation markers of Abl kinase signaling (phospho-CrkL) and mTOR signaling (phospho-p70S6K). The results are representative of three independent experiments. B, the same lysates were also analyzed for GFP-LC3-I and GFP-LC3-II levels by Western blotting with an anti-GFP antibody. The -fold changes in LC3-II levels are indicated below the blot. C and D, analysis of basal and starvation-induced long-lived protein degradation in drug-treated A549-GFP-LC3 cells. A549-GFP-LC3 cells were metabolically labeled with [l-3H]leucine. The cells were then chased in complete medium for 6, 24, or 48 h (C) or in Earle's balanced salt solution media for 6 h (D). The media was supplemented with DMSO (control), 2 μg/ml rapamycin, 10 μm STI571, or 50 μg/ml vinblastine as indicated. At each time point the radioactivity released into the medium was measured by scintillation counting. [l-3H]Leucine turnover rates were determined by dividing the cpm in the medium by the total cpm (medium cpm + lysate cpm). E, A549 cells expressing control or Abl and Arg miRNAs were analyzed for starvation-induced long-lived protein degradation as in D except no drugs were added to the medium. F, Western blots of the corresponding cell lysates in E blotted with phospho-CrkL, CrkL and Abl-specific antibodies as indicated. G, analysis of starvation-induced long-lived protein degradation in Abl/Arg double knockout and Abl/Arg heterozygous MEFs. H, Western blots of the corresponding cell lysates in G blotted with phospho-CrkL, CrkL and Abl-specific antibodies as indicated. n = 3 ± S.D. **, p < 0.05 compared with the control (C and D), scramble (E), or Abl/Arg heterozygous (G) groups. The results for each autophagy assay are representative of three independent experiments.
Autophagy can be induced when nutrients are limited, as in the case of growth factor deprivation (27). To examine whether Abl kinases play a role in autophagy stimulated in response to nutrient deprivation, autophagy was induced in 3H-leucine-labeled cells by withdrawal of growth factors and amino acids. Withdrawal of nutrients and growth factors for 4 h induced a 35% turnover of [l-3H]leucine-labeled protein in control cells (Fig. 2D). As expected, addition of vinblastine, a microtubule depolymerizing agent that inhibits autophagy by preventing the fusion between the autophagosomal and lysosomal membranes, significantly inhibited starvation-induced autophagy (Fig. 2D). Notably, inhibition of Abl kinase signaling with STI571 also significantly reduced the rate of starvation-induced autophagy in [3H]leucine-labeled cells (Fig. 2D).
A similar reduction in starvation-induced autophagy as measured by long-lived protein degradation assays was observed in A549 cells depleted of Abl kinases following transduction with Abl and Arg miRNAs (Fig. 2, E and F), which resulted in marked knockdown of Abl proteins (Fig. 2F, top panel) when compared with control cells, and a 95% reduction in Abl kinase signaling as measured by pCrkL protein levels (Fig. 2F, middle panel). Moreover, we also observed significantly reduced levels of starvation-induced long-lived protein degradation rates in MEFs derived from Abl/Arg-double knockout mice compared with those exhibited by Abl/Arg heterozygous MEFs (Fig. 2, G and H). Thus, inhibition of Abl kinases promotes the accumulation of autophagosomes by inhibiting the subsequent degradation of sequestered long-lived proteins.
Abl Kinase Inhibition Alters the Size and Subcellular Distribution of LAMP+ Lysosomes—To further explore the mechanism whereby Abl kinases regulate long-lived protein degradation, we examined the effect of Abl kinase inhibition on the later stages of autophagy, namely the fusion of autophagosomes with lysosomes and the subsequent degradation of sequestered cytosolic proteins. The lysosomal system was analyzed by immunofluorescence staining with antibodies against lysosome-associated membrane proteins 1 and 2 (LAMP-1 and LAMP-2). Both proteins contain a large, heavily glycosylated luminal domain and a short cytosolic tail, and are required for fusion of lysosomes with phagosomes (28). In untreated cells, LAMP-1+ lysosomes (red) were dispersed widely throughout the cell (Fig. 3A, top panel), whereas lysosomes in STI571-treated cells were larger and aggregated in the perinuclear region (Fig. 3A, bottom panel), frequently co-localizing with GFP-LC3+ autophagosomes (Fig. 4A, middle panels). To quantify these differences, the relative distance of lysosomes from the nucleus was measured in individual cells and plotted against the total number of lysosomes. The majority of LAMP-1+ lysosomes in STI571-treated cells were localized in the immediate vicinity of the nucleus, whereas in control cells LAMP-1+ lysosomes were evenly distributed throughout the cytoplasm (Fig. 3B). Aggregation of lysosomes adjacent to the nucleus was also observed by staining with a LAMP-2 antibody following pharmacological inactivation of Abl kinases with STI571 (supplemental Fig. S1A). Similar aggregation of lysosomes in the vicinity of the nucleus was observed in Abl/Arg-depleted A549 cells (supplemental Fig. S1B) and Abl/Arg-double knockout MEFs (supplemental Fig. S2).
FIGURE 4.
STI571 and rapamycin both induce autophagosome accumulation, but only STI571 affects lysosome localization and LAMP-1 protein levels and mobility. A, A549-GFP-LC3 cells were treated with 10 μm STI571 or 2 μg/ml rapamycin for 48 h and analyzed by immunofluorescence. In both STI571-treated cells and rapamycin-treated cells the pattern of GFP-LC3 staining in the cytosol changed from a diffuse to a predominantly punctate/vesicular appearance indicating increased levels of autophagosomes. LAMP-1+ lysosomes (red) were dispersed widely throughout untreated and rapamycin-treated cells, whereas lysosomes from STI571-treated cells clustered adjacent to the perinuclear region and often co-localized with GFP-LC3. These results are representative of three independent experiments. B, quantification of GFP-LC3 and LAMP-1 co-localization. The yellow and red images for individual stained cells described in the A (n = 3) were quantified using ImageJ software and expressed as a percentage to determine the proportion of LAMP-1+ lysosomes (red), which co-localized with GFP-LC3 (yellow in the merged images). **, p < 0.05 compared with the percentage of LAMP-1+/GFP-LC3+-positive lysosomes in control cells. Results are representative of three independent experiments. C, A549-GFP-LC3 cells were treated with 10 μm STI571 or 2 μg/ml rapamycin for up to 48 h as indicated and analyzed by Western blotting for LAMP-1 and β-tubulin levels. The data are representative of three independent experiments.
Abl Kinase Inactivation Induces Perinuclear Aggregation of LAMP+ Lysosomes and Reduces Their Velocity—Analysis of LAMP-1-/- LAMP-2-/- double-deficient cells showed that these proteins are required for the mobility of lysosomes and their subsequent fusion with phagosomes (28). It was reported that cells deficient in both LAMP-1 and LAMP-2 displayed a more peripheral and dispersed distribution of lysosomes, which exhibited reduced overall motility along microtubules and decreased movement toward the microtubule-organizing center (28). The aggregation and altered subcellular localization of LAMP proteins in the absence of Abl kinase signaling prompted us to compare the mobility of peripheral and perinuclear lysosomes in control and STI571-treated cells by time-lapse imaging of lysosomes labeled with LysoTracker Red (Fig. 3C). STI571 treatment significantly reduced the velocity of peripheral lysosomes with that of control cells (Fig. 3D and supplemental Movies S1 and S2). As observed with LAMP-1-stained cells (Fig. 3A), STI571 treatment promoted the perinuclear aggregation of LysoTracker-labeled acidic organelles in live cells (Fig. 3C and supplemental Movies S1 and S2). The mobility of these large, perinuclear lysosomes was also significantly reduced when compared with that of control cells (Fig. 3D). Collectively, these results suggest that inactivation of Abl kinase signaling increases the aggregation of LAMP-containing lysosomes, which in turn, impacts on their subcellular distribution and decreases their mobility.
Abl Kinase Inactivation Increases LAMP Glycosylation and Stability—Inhibition of Abl kinase signaling induced marked changes in the size and subcellular distribution of LAMP-1+ and LAMP-2+ lysosomes (Figs. 3A, 4A, and 4B and supplemental Fig. S1). Analysis of LAMP proteins by Western blotting revealed that down-regulation of Abl kinase activity with STI571 or Abl family protein depletion with Abl/Arg-specific siRNAs resulted in marked increases in the levels of LAMP-1 and LAMP-2 proteins (Figs. 4C and 5). Furthermore, the mobility of both LAMP proteins was affected by loss of Abl signaling, which resulted in the appearance of higher molecular weight forms of LAMP-1 and LAMP-2 (Figs. 4C, 5A, and 5B). As expected, pharmacological inhibition of Abl kinases with STI571 or siRNA-mediated depletion of Abl proteins resulted in over a 80% decrease in Abl kinase signaling as determined by measuring the levels of phosphorylation of the Abl substrate CrkL (Fig. 5, A and B). In contrast, no changes in the mobility of LAMP-1 were observed in A549-GFP-LC3 cells treated with the mTOR inhibitor, rapamycin (Fig. 4C). Moreover, in contrast to STI571-treated cells, cells treated with rapamycin did not exhibit altered subcellular localization of LAMP-1+ lysosomes near the perinuclear region (Fig. 4, A and B). Thus, although rapamycin and STI571 both induce the accumulation of LC3-GFP+ autophagosomes and the conversion of LC3-I to LC3-II, only treatment with STI571 promotes aggregation and perinuclear localization of LAMP+ lysosomes. Notably, the STI571-induced effects on the mobility of LAMP-1 were shown to correlate with the duration of Abl kinase inactivation and were reversible (Fig. 5C).
FIGURE 5.
Abl kinase inactivation increases the stability and glycosylation of LAMP-1. A, Western blot analysis was employed to determine the relative amounts of LAMP-1, LAMP-2, phospho-CrkL, and total CrkL protein levels in A549 cells treated without (-) or with (+) 10 μm STI571 for 24 h. The levels of LAMP-1 and LAMP-2 proteins from STI571-treated cells were increased and migrated with a higher molecular weight on SDS-PAGE gels when compared with LAMP proteins from control cells (top two panels). B, A549-GFP-LC3 cells were transfected with a non-targeted control siRNA or Abl/Arg siRNA oligonucleotides and analyzed 48 h later by Western blotting as in A. The lysates were also analyzed for Abl and Arg protein levels using a pan-Abl antibody. C, the STI571-induced increase in LAMP-1 size is reversible. A549 cells were incubated with STI571 for up to 48 h and then analyzed for LAMP-1 levels by Western blotting. Where indicated, STI571 was removed after 48 h, and the medium was replaced with fresh medium for an additional 24 h prior to analysis (last lane). The absence of STI571 during the 24-h recovery period induced a shift in the mobility of LAMP-1 to a lower molecular weight. D, STI571 induces increased glycosylation of LAMP-1. A549 cells were treated with or without 10 μm STI571 for 24 h. Cell lysates were incubated in deglycosylation buffer with or without peptide N-glycosidase F as indicated for 16 h at 37 °C. The deglycosylation reactions were analyzed by immunoblotting with an anti-LAMP-1 antibody (Sigma). Incubation of A549 cells with STI571 resulted in an increase in the size of LAMP-1. Following deglycosylation of cell lysates with peptide N-glycosidase F (PNGaseF), the size of LAMP-1 was identical in both control and STI571-treated cell lysates. E, pharmacological inhibition of Abl kinase with STI571 increases the stability of LAMP-1. A549 cells were metabolically labeled with EXPRE35S35S-labeled amino acids for 24 h and then chased for 48 h in the presence of increasing concentrations of STI571 as indicated. Cell lysates were immunoprecipitated with anti-LAMP-1 (Sigma) or a control rabbit IgG and analyzed by SDS-PAGE and autoradiography. Duplicate unlabeled cells treated with the indicated concentrations of STI571 were analyzed by Western blotting for phospho-CrkL and total CrkL as indicated to confirm that Abl signaling was reduced in a dose-dependent manner.
The altered mobility of LAMP proteins was suggestive of enhanced post-translational modification upon inactivation of Abl kinases. We found no evidence of increased phosphorylation or acetylation of LAMP-1 and LAMP-2 in STI571-treated cells (data not shown). The higher molecular weight of LAMP-1 in the absence of Abl kinase activity appeared to be due to differential glycosylation of the protein, because LAMP-1 mobility in the presence of STI571 was similar to that in control lysates in the presence of the endoglycosidase peptide N-glycosidase F (Fig. 5D). Similar results were observed for LAMP-2 (data not shown).
To examine whether the turnover of LAMP-1 protein was affected by inhibition of Abl signaling, A549 cells were metabolically pulse-labeled with [35S]methionine and then chased with “cold” methionine for 48 h in the presence of increasing concentrations of STI571. As shown in Fig. 5E, increasing amounts of STI571 resulted in a markedly reduced turnover rate of LAMP-1 protein. Collectively, these results suggest that loss of Abl kinase signaling promotes hyperglycosylation of LAMP-1, which reduces protein turnover. These changes are concomitant with the aggregation and altered intracellular localization of LAMP+ lysosomes in cells with impaired Abl signaling.
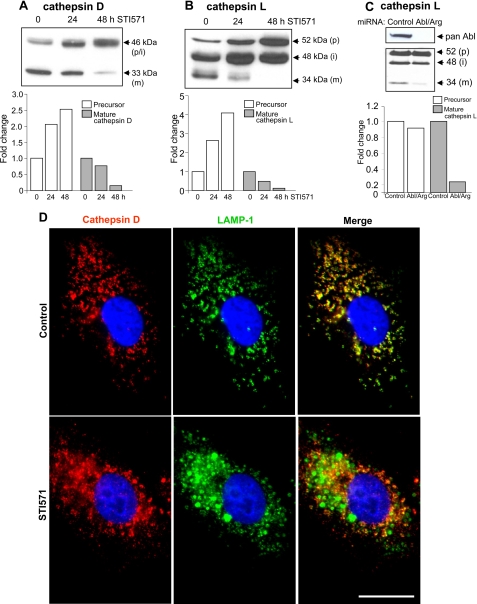
Abl Kinase Inhibition Impairs the Function of Lysosomal Hydrolases—The frequent co-localization of GFP-LC3+ autophagosomes with LAMP+ lysosomes in STI571-treated cells suggested that these organelles have undergone normal fusion in the absence of Abl kinase activity (Fig. 4, A and B). Previous studies have demonstrated that the turnover of the long-lived proteins LAMP-1 and LAMP-2 is regulated by hydrolases present in the acidified lumen of the lysosome (29, 30). These findings together with the altered mobility and turnover of LAMP proteins in the absence of Abl signaling suggested that optimal degradation of sequestered cytosolic proteins, as well as autophagosomal (LC3) and lysosomal (LAMPs) membrane constituents by lysosomal hydrolases may require Abl kinase activity. To test this hypothesis we analyzed the levels of a cohort of lysosomal cathepsins and glycosidases in control and STI571-treated A549-GFP-LC3 cells. Lysosomal cathepsins are synthesized as inactive preproenzymes and undergo proteolytic processing to their active, mature forms as they traffic from the trans-Golgi network to the progressively acidified environments of late endosomes and lysosomes (31). We observed a significant decrease in the processing of the precursor forms of cathepsin D and cathepsin L to their mature, lower molecular weight, lysosomal forms in STI571-treated cells (Fig. 6, A and B). Quantification of these blots demonstrated that the levels of precursor cathepsin D and cathepsin L progressively increased over the 48-h incubation period with STI571, whereas the levels of the mature, active enzymes decreased (Fig. 6, A and B, bottom panels). Moreover, A549 cells depleted of Abl kinases following expression of Abl/Arg miRNAs also displayed markedly reduced levels of the mature form of cathepsin L when compared with A549 cells expressing a control scrambled siRNA (Fig. 6C). Furthermore, whereas in control cells, cathepsin D frequently co-localized with LAMP-1, in the absence of Abl kinase signaling cathepsin D failed to co-localize with LAMP-1 (Fig. 6D). These results suggest that Abl signaling is required for the correct lysosomal targeting and activation of lysosomal cathepsins, which regulate late-stage autophagy by degrading long-lived proteins in the lysosome.
FIGURE 6.
Abl kinase inhibition impairs the processing and lysosomal localization of the cysteine proteases, cathepsin D and cathepsin L. A and B, A549-GFP-LC3 cells were incubated with or without 10 μm STI571 for 24 h or 48 h as indicated. Cell lysates were analyzed by Western blotting with anti-cathepsin D (A) and anti-cathepsin L (B) antibodies. Inhibition of Abl kinases with STI571 resulted in a time-dependent decrease in the processing of the precursor forms of cathepsin D and cathepsin L to their mature (activated), lower molecular weight forms, which characteristically accumulate in the lysosome. p = precursor, i = intermediate, and m = mature. The signal intensities of the precursor and mature cathepsin bands were determined using ImageJ software and demonstrated a progressive loss of the mature forms of cathepsin D and L over the 48-h incubation period with STI571 (graphs below each blot). C, A549 cells expressing a control scrambled miRNA or Abl6/Arg-9 miRNAs were analyzed by Western blotting with anti-cathepsin L antibody. D, the subcellular distribution of cathepsin D and LAMP-1 was analyzed in A549-GFP-LC3 cells by immunofluorescence. Cathepsin D displayed a punctate/vesicular staining pattern in control cells and frequently co-localized with LAMP-1, whereas cathepsin D in STI571-treated cells (48 h) did not co-localize with LAMP-1. Scale bar = 20 μm. The results are representative of three independent experiments.
We also tested the activities of a series of lysosomal glycosidases, which are essential for the stepwise degradation of glycoproteins targeted to the lysosome for degradation. Notably, the enzyme activity levels of lysosomal α-galactosidase, α-mannosidase, and neuraminidase were dramatically reduced following Abl kinase inactivation with STI571 (Fig. 7, A-C). Moreover, Abl/Arg siRNA-treated cells also exhibited significant decreases in the activities of α-galactosidase, α-mannosidase, and neuraminidase when compared with control siRNA-transfected control cells (Fig. 7, E-G). Similar decreases in enzymatic activity were observed for α-galactosidase, α-mannosidase, and neuraminidase in Abl/Arg double knockout MEFs when compared with Abl/Arg heterozygous MEFs (Fig. 7, I-K). The majority of lysosomal hydrolases, including the glycosidases and cathepsins shown here to be affected by loss of Abl signaling, are targeted to the lysosomal compartment by the mannose-6-phosphate receptor (M6PR) pathway (32). In contrast, acid phosphatase is targeted to the lysosome independently of the M6PR pathway and did not appear to be significantly altered following inactivation of Abl kinase signaling with STI571, or by siRNA-mediated depletion of Abl and Arg proteins (Fig. 7, D and H). Also, complete and persistent loss of Abl signaling in Abl/Arg double knockout MEFs did not significantly affect acid phosphatase activity levels when compared with Abl/Arg heterozygous MEFs (Fig. 7L). These findings suggest that Abl kinase inactivation preferentially affects the M6PR-mediated transport of lysosomal glycosidases and cathepsins. Collectively, these results reveal a role for Abl kinase signaling in the regulation of acidic hydrolase function, possibly by affecting their trafficking to the lysosomal compartment.
FIGURE 7.
Abl kinase inhibition impairs the activities of specific lysosomal hydrolases. A-D, the activities of the lysosomal hydrolases α-galactosidase (A), α-mannosidase (B), neuraminidase (C), and acid phosphatase (D) were analyzed in A549-GFP-LC3 cells treated with DMSO (control) or STI571 (10 μm) for up to 72 h using 4-methylumbelliferone-conjugated substrates (Sigma). E-H, A549-GFP-LC3 cells were transfected with a non-targeted control siRNA or Abl and Arg siRNA oligonucleotides, and analyzed 48 h later for α-galactosidase (E), α-mannosidase (F), neuraminidase (G), or acid phosphatase (H). Abl/Arg-double knockout MEFs and Abl/Arg-heterozygous MEFs were assayed for α-galactosidase (I), α-mannosidase (J), neuraminidase (K), or acid phosphatase (L). **, p < 0.05 compared with the DMSO-treated control (time 0, A-C), control siRNA (E-G), or Abl/Arg heterozygous (I-K) groups. No significant differences were observed for acid phosphatase activities (D, H, and L; p > 0.05). The results are representative of three independent experiments. M, schematic representation of the lysosome-related events that arise from inhibition of Abl kinase signaling.
DISCUSSION
Here we report that Abl kinase signaling positively regulates the trafficking and function of lysosomal components. Inhibition of Abl signaling has a number of downstream consequences on processes, which are dependent on lysosomal function. As summarized in Fig. 7M, Abl deficiency resulted in decreased enzymatic activity of a subset of lysosomal enzymes leading to impaired degradation of longlived proteins and the concomitant accumulation of autophagosomes and LAMP+ lysosomes. Loss of Abl kinase does not appear to impair the fusion of autophagosomes with lysosomes, because Abl-deficient cells frequently showed co-localization of GFP-LC3+ autophagosomes with LAMP-1+ lysosomes. The accumulation of autophagosomes in the absence of Abl kinase signaling is likely to occur as a consequence of lysosomal dysfunction.
We demonstrate that Abl kinases are positive regulators of late-stage autophagy and are required for maximal flux through this pathway. It is becoming increasingly clear that, whereas GFP-LC3 and other early autophagy markers are useful for monitoring autophagosome formation, additional assays are required to monitor the flux of longlived proteins through the autophagy pathway (24). Accumulation of autophagosomes in STI571-treated cells as measured by enhanced levels of GFP-LC3-II has been observed in several cell types, which led one group to conclude that STI571 treatment promotes autophagy, and that by inference, Abl kinases negatively regulate autophagy (20). However, these investigators did not assess long-lived protein degradation (20), which is essential for monitoring the later stages in the autophagy pathway (22). It has been shown that a large number of compounds that increase the levels of GFP-LC3-II fail to increase the degradation of proteins and organelles by autophagy (24). Increased levels of GFP-LC3-II may result from increased cell death or from lysosomal defects that block autophagic degradation (24). We did not observe increased cell death following treatment with Abl kinase inhibitors or in cells depleted of Abl kinases (data not shown). However, we found profound defects in lysosomal enzyme function that led to the accumulation of autophagosomes and to a dramatic reduction in the turnover of long-lived proteins, including LAMP proteins, in the absence of Abl signaling. Thus, the accumulation of autophagosomes in cells lacking Abl signaling is due to impaired turnover of long-lived proteins that continue to accumulate in the absence of Abl kinases, and is not due to increased biogenesis of autophagosomes during the early stages of autophagy (20). However, it has been reported that STI571 treatment increased the clearance of pathological prion protein (PRPsc) through a lysosomal pathway (33). It is possible that inhibition of Abl signaling and subsequent down-regulation of lysosomal enzyme function might shift the flux of specific proteins under pathological conditions into alternative degradative pathways.
Inhibition or depletion of Abl kinases elicited marked reductions in the enzyme activities of several lysosomal glycosidases. Furthermore, Abl kinase inhibition significantly reduced the intracellular amounts of the catalytically active, mature forms of cathepsin D and cathepsin L. The altered processing and subcellular distribution of the cathepsins combined with inhibition of lysosomal hydrolases in Abl-deficient cells suggested that Abl kinases may play a role in the regulation of lysosomal enzyme trafficking. Because acid phosphatase activity levels were not significantly altered in the absence of Abl signaling, these findings suggest that the M6PR-mediated trafficking of lysosomal enzymes may be specifically regulated downstream of the Abl kinases.
Abl kinase signaling may affect lysosomal processes through one or more pathways. We have shown that Abl kinase inactivation increased the stability of LAMP proteins, which may be due to altered glycosylation, because N-glycosylation is important for the stability of LAMP-1 and LAMP-2 in the lysosomal membrane (34). Notably, LAMP-2 deficiency results in accumulation of autophagic vacuoles and decreased basal rates of long-lived protein degradation (35). Moreover, LAMP-2-deficient hepatocytes display improper processing of cathepsin D to the mature, active form and have impaired trafficking of some lysosomal enzymes to lysosomes (36). In this study we observe that increased amounts and altered mobility of LAMP proteins in the absence of Abl signaling coincided with reduced lysosomal cathepsin activities and reduced autophagy. It is likely that persistent and excessive glycosylation of LAMP proteins may alter their biological functions. Thus, defective autophagy may occur by altered processing and stability of LAMPs in the absence of Abl kinase function (Fig. 7M).
A recent proteomic study has demonstrated that protein degradation during starvation-induced autophagy occurs in an ordered fashion, where cytosolic proteins are degraded initially followed by organelle-associated proteins (37). In a similar fashion inhibition of Abl kinase signaling may initially up-regulate the cellular pools of cytosolic proteins followed later by organelle-associated proteins.
An alternative and not mutually exclusive mechanism for the impaired autophagic degradation in the absence of Abl signaling may be decreased organelle mobility. Inhibition of Abl kinases results in perinuclear aggregation and decreased mobility of lysosomes. In this regard, LAMP proteins have been shown to regulate phagosome mobility (28), and altered LAMP processing and stability may affect organelle mobility in cells lacking Abl signaling. Abl kinases may regulate autophagy in part through modulation of the microtubule cytoskeleton, which may be required for intracellular mobility of organelles. Microtubules have been shown to facilitate autophagosome formation and fusion with lysosomes and are also required for autophagosome motility (38). Notably, Abl kinases have been shown to regulate microtubule dynamics (39, 40). Alternatively, Abl kinases may affect the mobility of organelles and macromolecular cargoes through regulation of microtubule motors (41).
The Abl-dependent effects on the morphology and localization of lysosomes have also been observed in cells other than cancer cells. We showed that primary mouse embryonic fibroblasts from Abl/Arg double knockout mice exhibit profound changes in the subcellular distribution of LAMP-1+ and LAMP-2+ lysosomes. Disruption of the lysosomal system may contribute to some of the observed effects of imatinib on antigen processing and presentation by dendritic cells (42) and hematopoiesis in the normal bone niche (43). In the context of cancer, a reduction in autophagy in some carcinomas is believed to confer growth advantages (44). However, a low level of autophagy may also be required for cancer progression (2, 45, 46). Autophagy may be up-regulated as an adaptive mechanism to allow cancer cells to overcome a lack of nutrient supply (47). In this regard, we have observed that inhibition of Abl signaling decreased the degradation of long-lived proteins in response to nutrient deprivation. Thus, inhibition of autophagy in STI571-treated cells may contribute to the synergistic cell killing effect reported in cells treated with both STI571 and cisplatin (48).
Our findings suggest that Abl kinases may play a role in processes dependent on integral lysosomal functions, which may extend beyond Abl-dependent regulation of autophagy. Further, the effects of Abl signaling on late-stage autophagy should be of consideration when employing therapeutic strategies that involve inhibition of the Abl kinases.
Supplementary Material
Acknowledgments
We thank Drs. Pang Yao, Yasheng Gao, and Colleen Ring, Elizabeth Chislock, and Ran Li (Duke University Medical Center) for valuable discussions and critical reading of the manuscript. We are grateful to Dr. Sam Johnson from the Duke University Medical Center light microscopy core facility for excellent technical advice and training.
This work was supported, in whole or in part, by National Institutes of Health Grant CA70940 (to A. M. P.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Movies S1 and S2.
Footnotes
The abbreviations used are: TOR, target of rapamycin; mTOR, mammalian target of rapamycin; Abl, Abelson; STI571, signal transduction inhibitor 571; LC3, microtubule-associated protein 1 light chain 3; LAMP-1 and LAMP-2, lysosome-associated membrane proteins 1 and 2; M6PR, mannose-6-phosphate receptor; GFP, green fluorescent protein; DMEM, Dulbecco's modified Eagle's medium; FBS, fetal bovine serum; MEF, mouse embryonic fibroblast; siRNA, small interference RNA; miRNA, microRNA; PBS, phosphate-buffered saline.
W. S. Rasband (1997-2008) ImageJ, U. S. National Institutes of Health, Bethesda, MD, rsb.info.nih.gov/ij/.
References
- 1.Rubinsztein, D. C., Gestwicki, J. E., Murphy, L. O., and Klionsky, D. J. (2007) Nat. Rev. Drug Discov. 6 304-312 [DOI] [PubMed] [Google Scholar]
- 2.Meijer, A. J., and Codogno, P. (2004) Int. J. Biochem. Cell Biol. 36 2445-2462 [DOI] [PubMed] [Google Scholar]
- 3.Sarbassov, D. D., Guertin, D. A., Ali, S. M., and Sabatini, D. M. (2005) Science 307 1098-1101 [DOI] [PubMed] [Google Scholar]
- 4.Maiuri, M. C., Zalckvar, E., Kimchi, A., and Kroemer, G. (2007) Nat. Rev. Mol. Cell. Biol. 8 741-752 [DOI] [PubMed] [Google Scholar]
- 5.Guertin, D. A., and Sabatini, D. M. (2007) Cancer Cell 12 9-22 [DOI] [PubMed] [Google Scholar]
- 6.Pendergast, A. M. (2002) Adv. Cancer Res. 85 51-100 [DOI] [PubMed] [Google Scholar]
- 7.Hernandez, S. E., Krishaswami, M., Miller, A. L., and Koleske, A. J. (2004) Trends Cell Biol. 14 36-44 [DOI] [PubMed] [Google Scholar]
- 8.Heisterkamp, N., Stam, K., Groffen, J., de Klein, A., and Grosveld, G. (1985) Nature 315 758-761 [DOI] [PubMed] [Google Scholar]
- 9.Pendergast, A. M., Muller, A. J., Havlik, M. H., Maru, Y., and Witte, O. N. (1991) Cell 66 161-171 [DOI] [PubMed] [Google Scholar]
- 10.Druker, B. J., Talpaz, M., Resta, D. J., Peng, B., Buchdunger, E., Ford, J. M., Lydon, N. B., Kantarjian, H., Capdeville, R., Ohno-Jones, S., and Sawyers, C. L. (2001) N. Engl. J. Med. 344 1031-1037 [DOI] [PubMed] [Google Scholar]
- 11.Zandy, N. L., Playford, M., and Pendergast, A. M. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 17686-17691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srinivasan, D., Sims, J. T., and Plattner, R. (2008) Oncogene 27 1095-1105 [DOI] [PubMed] [Google Scholar]
- 13.Plattner, R., Ervin, B. J., Guo, S., Blackburn, K., Kazlauskas, A., Abraham, R. T., York, J. D., and Pendergast, A. M. (2003) Nat. Cell Biol. 5 309-319 [DOI] [PubMed] [Google Scholar]
- 14.Plattner, R., Kadlec, L., DeMali, K. A., Kazlauskas, A., and Pendergast, A. M. (1999) Genes Dev. 13 2400-2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanos, B., and Pendergast, A. M. (2006) J. Biol. Chem. 281 32714-32723 [DOI] [PubMed] [Google Scholar]
- 16.Srinivasan, D., and Plattner, R. (2006) Cancer Res. 66 5648-5655 [DOI] [PubMed] [Google Scholar]
- 17.Lin, J., and Arlinghaus, R. (2008) Oncogene 27 4385-4391 [DOI] [PubMed] [Google Scholar]
- 18.Burton, E. B., Plattner, R., and Pendergast, A. M. (2003) EMBO J. 22 5471-5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burton, E. A., Oliver, T. N., and Pendergast, A. M. (2005) Mol. Cell. Biol. 25 8834-8843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ertmer, A., Huber, V., Gilch, S., Yoshimori, T., Erfle, V., Duyster, J., Elasser, H.-P., and Schatzl, H. M. (2007) Leukemia 21 936-942 [DOI] [PubMed] [Google Scholar]
- 21.Klionsky, D. J., Cuervo, A. M., and Seglen, P. O. (2007) Autophagy 3 181-206 [DOI] [PubMed] [Google Scholar]
- 22.Klionsky, D. J., Abeliovich, H., Agostinis, P., Agrawal, D. K., Aliev, G., Askew, D. S., Baba, M., Baehrecke, E. H., Bahr, B. A., Ballabio, A., Bamber, B. A., Bassham, D. C., Bergamini, E., Bi, X., Biard-Piechaczyk, M., Blum, J. S., Bredesen, D. E., Brodsky, J. L., Brumell, J. H., and Brunk, U. T., et al. (2008) Autophagy 4 151-17518188003 [Google Scholar]
- 23.McLaughlin, J., Cheng, D., Singer, O., Lukacs, R. U., Radu, C. G., Verma, I. M., and Witte, O. N. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 20501-20506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang, L., Yu, J., Pan, H., Hu, P., Hao, Y., Cai, W., Zhu, H., Yu, A. D., Xie, X., Ma, D., and Yuan, J. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 19023-19028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levine, B., and Kroemer, G. (2008) Cell 132 27-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rix, U., Hantschel, O., Durnberger, G., Remsing Rix, L. L., Planyavsky, M., Fernbach, N. V., Kaupe, I., Bennett, K. L., Valent, P., Colinge, J., Kocher, T., and Superti-Furga, G. (2007) Blood 110 4055-4063 [DOI] [PubMed] [Google Scholar]
- 27.Takahashi, Y., Coppola, D., Matsushita, N., Cualing, H. D., Sun, M., Sato, Y., Liang, C., Jung, J. U., Cheng, J. Q., Mul, J. J., Pledger, W. J., and Wang, H. G. (2007) Nat. Cell Biol. 9 1142-1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huynh, K. K., Eskelinen, E. L., Scott, C. C., Malevanets, A., Saftig, P., and Grinstein, S. (2007) EMBO J. 26 313-324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuervo, A. M., Mann, L., Bonten, E. J., d'Azzo, A., and Dice, J. F. (2003) EMBO 22 47-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yogalingam, G., Bonten, E. J., van de Vlekkert, D., Hu, H., Moshiach, S., Connell, S. A., and d'Azzo, A. (2008) Dev. Cell 15 74-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kominami, E., Tsukahara, T., Hara, K., and Katunuma, N. (1988) FEBS Lett. 231 225-228 [DOI] [PubMed] [Google Scholar]
- 32.Ghosh, P., Dahms, N. M., and Kornfeld, S. (2003) Nat. Rev. Mol. Cell. Biol. 4 202-212 [DOI] [PubMed] [Google Scholar]
- 33.Ertmer, A., Gilch, S., Yun, S., Flechsig, E., Klebl, B., Stein-Gerlach, M., Klein, M. A., and Schatzl, H. M. (2004) J. Biol. Chem. 279 41918-41927 [DOI] [PubMed] [Google Scholar]
- 34.Eskelinen, E. L. (2006) Mol. Aspects Med. 27 495-502 [DOI] [PubMed] [Google Scholar]
- 35.Tanaka, Y., Guhde, G., Suter, A., Eskelinen, E. L., Hartmann, D., Lullmann-Rauch, R., Janssen, P. M., Blanz, J., von Figura, K., and Saftig, P. (2000) Nature 406 902-906 [DOI] [PubMed] [Google Scholar]
- 36.Eskelinen, E. L., Illert, A. L., Tanaka, Y., Schwarzmann, G., Blanz, J., Von Figura, K., and Saftig, P. (2002) Mol. Biol. Cell 13 3355-3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kristensen, A. R., Schandorff, S., Hoyer-Hansen, M., Nielsen, O., Jaattela, M., Dengjel, J., and Andersen, J. S. (2008) Mol. Cell. Proteomics 10.1074/mcp.M800184-MCP200, in press [DOI] [PubMed]
- 38.Kochl, R., Hu, X. W., Chan, E. Y., and Tooze, S. A. (2006) Traffic 7 129-145 [DOI] [PubMed] [Google Scholar]
- 39.Wang, Y., Miller, A. L., Mooseker, M. S., and Koleske, A. J. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 14856-14870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, A. L., Wang, Y., Mooseker, M. S., and Koleske, A. J. (2004) J. Cell Biol. 165 407-419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin, M., Ahern-Djamali, S. M., Hoffmann, F. M., and Saxton, W. M. (2005) Mol. Biol. Cell 16 4225-4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Appel, S., Rupf, A., Weck, M. M., Schoor, O., Brummendorf, T. H., Weinschenk, T., Grunebach, F., and Brossart, P. (2005) Clin. Cancer Res. 11 1928-1940 [DOI] [PubMed] [Google Scholar]
- 43.Van Glabbeke, M., Verweij, J., Casali, P. G., Simes, J., Le Cesne, A., Reichardt, P., Judson, J. R., van Oosterom, A. T., and Blay, J. Y. (2006) Eur. J. Cancer 42 2277-2285 [DOI] [PubMed] [Google Scholar]
- 44.Mathew, R., Karantza-Wadsworth, V., and White, K. (2007) Nat. Rev. Cancer 7 961-967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amaravadi, R. K., Yu, D., Lum, J. J., Bui, T., Christophorou, M. A. Evan, G. I., Thomas-Tikhonenko, A., and Thompson, C. B. (2007) J. Clin. Invest. 117 326-336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Easton, J. B., and Houghton, P. J. (2006) Oncogene 25 6436-6446 [DOI] [PubMed] [Google Scholar]
- 47.Klionsky, D. J. (2007) Nat. Rev. Mol. Cell. Biol. 8 931-937 [DOI] [PubMed] [Google Scholar]
- 48.Zhang, P., Gao, W. Y., Turner, S., and Ducatman, B. S. (2003) Mol. Cancer 2 1-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.