Abstract
We show that the Saccharomyces cerevisiae ribosomal protein Rpl42ab (the identical product of the RPL42A and RPL42B genes) is monomethylated at Lys-40 and Lys-55. The methylation of Lys-40 is dependent upon the Ybr030w gene product; the methylation of Lys-55 is dependent upon the Set7 gene product. Ybr030w and SET7 genes both encode SET domain containing proteins homologous to known protein lysine methyltransferases, suggesting that their products are the specific enzymes responsible for the monomethylation of the two sites in Rpl42ab. We thus designate Ybr030w as Rkm3 and Set7 as Rkm4. Yeast strains with deletions in both the Ybr030w and SET7 genes produce unmethylated Rpl42ab. A slow growth phenotype was seen for the SET7 deletion strain and the double knock-out when grown in low concentrations of the eukaryotic protein synthesis inhibitor, cycloheximide. These results suggest that modification of Rpl42ab at Lys-55 can fine-tune its structure to avoid inhibition. An intact mass fragmentation approach (“top down mass spectrometry”) was used to quantitate the extent of methylation of Rpl42ab. In wild-type strains, it was found that 78% was monomethylated at both Lys-40 and Lys-55 and that 22% was a mixture of species with either Lys-40 or Lys-55 monomethylated. The top down approach was also used to reevaluate the methylation sites of Rpl12ab. We found that the yeast Rpl12ab protein is dimethylated at the N-terminal proline residue, trimethylated at Lys-3 by Rkm2, and monomethylated at Arg-66.
The emergence of histone protein lysine methyltransferases that regulate the epigenetic state of chromatin highlights the biological importance of this modification (1, 2). Protein lysine methylation has also been found to occur in over a hundred proteins ranging from flagellin in Salmonella typhimurium to RUBISCO in plants to transcription initiation factor TAFT10 in Homo sapiens (3). Physiological functions of lysine methylation of non-histone proteins remain unclear, and the enzymes responsible for the modifications have only recently begun to be characterized.
With the exceptions of the yeast DOT1 and the bacterial PrmA enzymes, known protein lysine methyltransferases all belong to the family of SET domain-containing proteins (4–6). Structurally the SET domain enzymes are distinct from the more common seven β-strand class of methyltransferases. All SET domain methyltransferases studied to date appear to only catalyze protein lysine methylation reactions (3–6). These enzymes are capable of adding up to three methyl groups to the side chain amino group of lysine residues to form mono-, di-, and trimethylated derivatives. Interestingly, this modification can be reversed by members of a large family of demethylases (7).
We have been interested in identifying protein lysine methyltransferases that are involved in the formation of methylation marks on ribosomal proteins. Multiple proteins in both the small and large ribosomal subunits have been shown to be modified at lysine residues in both prokaryotes and eukaryotes (8). Our work has focused on the ribosomal large subunit of the yeast Saccharomyces cerevisiae. Evidence has been presented that six proteins are methylated, Rpl1ab, Rpl3, Rpl12ab, Rpl23ab, Rpl42ab, and Rpl43ab (9). Our laboratory has recently determined the sites and the SET methyltransferases responsible for the methylation of Rpl23ab and Rpl12ab (10–12). We identified the SET domain protein Rkm1 as the enzyme responsible for the dimethylation of Lys-105 and Lys-109 in Rpl23ab (10, 12) and Rkm2 as the enzyme responsible for the trimethylation of a lysine residue of Rpl12ab (11).
Sequence analysis has shown that the S. cerevisiae genome encodes twelve SET domain proteins that can be separated into two subfamilies each containing six members (11). The known Set1 and Set2 histone lysine methyltransferases are members of one family, and Rkm1, Rkm2, and the enzyme responsible for cytochrome c methylation are members of the second family. However, the methyltransferase function of seven of these proteins remains to be identified.
In this work we focused on the modification of the large ribosomal protein encoded by the RPL42A and the RPL42B genes of S. cerevisiae. The protein produced from each of these genes is identical in sequence and we designate it Rpl42ab (9). This protein is conserved in nature and corresponds to the product of the human RPL36A gene (13). This protein is located in the ribosomal tRNA exit site with contacts to both the large subunit rRNA and the tRNA (14). This species has also been associated with cycloheximide sensitivity (15). Previous work has demonstrated that Rpl42ab is monomethylated at two lysine residues (16), but the enzyme or enzymes responsible for the modification remains unidentified. Using a combination of reverse phase liquid chromatography coupled to mass spectrometry, we analyzed Rpl42ab from yeast deletion mutants lacking SET domain proteins. Using a top down mass spectrometric approach, we quantitated modification of Rpl42ab and found that Ybr030w is required for the methylation of Lys-40 and that Set7 is required for the methylation of Lys-55. Finally, we found that monomethylation of Lys-55 imparted resistance to cycloheximide.
EXPERIMENTAL PROCEDURES
Purification of Yeast Ribosomal Large Subunit Proteins from Wild Type and Mutant Strains—Strains used in this study are listed in Table 1. The ΔYbr030w ΔSET7 double knock-out strain was created by crossing the MATα strain of ΔSET7 with the MATa strain of ΔYbr030w and selecting for cells on lysine- and methionine-deficient plates. The resulting diploid strain was induced to sporulate and screening on kanamycin plates for the non-parental ditype identified haploid spores containing both gene deletions. The deletion of both the Ybr030w and SET7 genes was confirmed by PCR analysis using flanking TAG1 and TAG2 primers.
TABLE 1.
Saccharomyces cerevisiae strains used
| Strain | Genotype | Source |
|---|---|---|
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | SGDPa |
| ΔYbr030w (BY4742) | BY4742, Δ Ybr030w::Kanr | SGDPa |
| ΔSET7 (BY4742) | BY4742, Δ SET7::Kanr | SGDPa |
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | SGDPa |
| ΔYbr030w (BY4741) | BY4741, Δ Ybr030w::Kanr | SGDPa |
| ΔSET7 (BY4741) | BY4741, Δ SET7::Kanr | SGDPa |
| ΔYbr030w ΔSET7 | MATα ΔYbr030w::Kanr Δ SET7::Kanr LYS+MET+ | This study |
Strains prepared by the Saccharomyces Genome Deletion Project (SGDP) and purchased from Open Biosystems (Huntsville, AL)
Large ribosomal subunits were isolated from wild-type, ΔYbr030w, ΔSET7, and the double knock-out ΔYbr030w ΔSET7 strains as described previously (11), with the exception that the final supernatant was not lyophilized. The resulting purified large ribosomal subunits were concentrated to a volume of ∼100 μl by vacuum centrifugation and promptly subjected to analysis.
Liquid Chromatography with Electrospray Ionization Mass Spectrometry of Intact Ribosomal Proteins (LC-MS+)3—Ribosomal proteins were fractionated using reverse-phase liquid chromatography. The resulting effluent was split between a fraction collector, collecting 1-min fractions, and an electrospray ionization mass spectrometer as described previously (11, 17). For HPLC fractionation a PLRP-S polymeric column was used that had a pore size of 300 Å, bead size of 5 μm, and a dimensions of 150 × 1 mm (Polymer Laboratories, Amherst, MA). The column was maintained at 50 °C and initially equilibrated in solvent A (0.05% trifluoroacetic acid in water) at 95% and solvent B (0.05% trifluoroacetic acid in acetonitrile) at 5%. The ribosomal proteins were eluted at a flow rate of 60 μl/min using a program of 5 min at 5% B, followed by a 25-min gradient from 5 to 30% B and ending with a 100-min gradient from 30 to 100% B, for a total gradient time of 130 min. The API III+ mass spectrometer (PE Sciex) used was tuned and calibrated as described previously (18) to yield a mass accuracy of 0.01% (±1.0 Da at 10 kDa). The 30-μl fractions collected during chromatography were stored at -20 °C. The data obtained was analyzed using the BioMultiview 1.3.1 software (Applied Biosystems).
Identification of Methylation Sites by Top Down Mass Spectrometry Using Collisionally Activated Dissociation and Electron Capture Dissociation of Intact Proteins—Fractions containing isolated ribosomal protein from LC-MS+ were diluted with 30 μl water/acetonitrile/formic acid (500/500/1, v/v/v) and analyzed by direct nanospray injection. LC fractions were individually loaded into a 2-μm internal diameter externally coated nanospray emitter (New Objective Inc., Woburn, MA) and desorbed using a spray voltage between 1.2 kV and 1.4 kV versus the inlet of the mass spectrometer. These conditions produced a flow rate of 20–50 nl/min. A hybrid linear ion-trap/FTICR mass spectrometer was used for the analysis (LTQ FT Ultra, Thermo Fisher, Bremen, Germany). The m/z resolving power of the FTICR mass analyzer was set at 100,000 (defined by m/Δm50% at m/z 400). Individual charge states of multiply protonated molecular ions were selected for isolation and collisional activation in the linear ion trap followed by detection of the resulting fragments in the FTICR cell. For the collisionally activated dissociation studies, the precursor ions were activated using 35% normalized collision energy at the default activation q-value of 0.25. Additional studies were conducted in which the precursor ions were guided to the FTICR cell and fragmented using electron capture dissociation. The fragmentation efficiency was optimized to maximize fragment ion signal. All FTICR MSMS spectra were processed using ProSightPC software (Thermo Fisher, San Jose, CA) in single protein mode with a 10 ppm mass accuracy threshold. The root mean square deviations for assigned fragments were less than 5 ppm. Corresponding ribosomal protein sequences were taken from Swiss-Prot and used as the reference sequences in ProSightPC. Intact mass measurements were processed using the manual Xtract program, version 1.516 (Thermo Fisher, San Jose, CA). All top down experimental data obtained for this study are available online at proteome commons.
Quantitation of Protein Species from FTICR Mass Spectrometry—In spectra where multiple species of the same protein differing in mass are present, the PIRR method of Pasavento et al. (19, 20) was used to measure the relative amounts of protein species present. We integrated the intensity of the four most abundant isotope peaks (13C5, 13C6, 13C7, 13C8) for the 14+ and 15+ charge states of the wild-type Rpl42ab protein. The values were combined into a weighted average and the percent abundances of each species were calculated. This was repeated for the Rpl42ab protein in the ΔSET7 strain with the exception of only the 15+ charge state used in the calculation due to it being the only charge state with a signal to noise value >10, thus allowing for accurate quantitation (19).
Growth of Yeast Strains in Cycloheximide—Single colonies were seeded into 5 ml of YPD broth (Difco, 1% yeast extract, 2% peptone, and 2% dextrose) and incubated 18 h at 30 °C with shaking (250 rpm/min). Appropriate volumes were taken to inoculate 49 ml of YPD in 125 ml Erlenmeyer flasks to an optical density (600 nm) of 0.05. Cells were grown at 30 °C with shaking as above. When an optical density of 0.1 was reached, 50 μl of a stock solution of cycloheximide in water (1 mg/ml) was added to give a final concentration of 1 μg/ml and growth was monitored by optical density over the next 40 h.
RESULTS AND DISCUSSION
Identification of Two Rpl42ab Lysine Methyltransferases—Ribosomes from wild-type and SET domain knock-out yeast strains were isolated by differential centrifugation, and subunits were separated using high salt sucrose gradients (11). The resulting large subunit proteins were fractionated by reverse-phase HPLC and their intact masses determined by electrospray mass spectrometry as described under “Experimental Procedures.” We focused on the large ribosomal protein Rpl42ab since evidence has been presented for monomethyllysine formation at two sites (16). The presence of two methyl groups on Rpl42ab was supported by the mass spectrometric determination of the intact protein. It was 28 Da larger than expected from the encoded open reading frame (9). Because the loss of a SET domain protein lysine methyltransferase in a given knock-out strain would be expected to result in the loss of one or more methyl groups on a ribosomal protein, we examined the mass of Rpl42ab for differences in wild-type and mutant strains. The accuracy of the triple quadrupole mass spectrometer used (100 ppm) permitted us to detect differences as low as a few Da in mass.
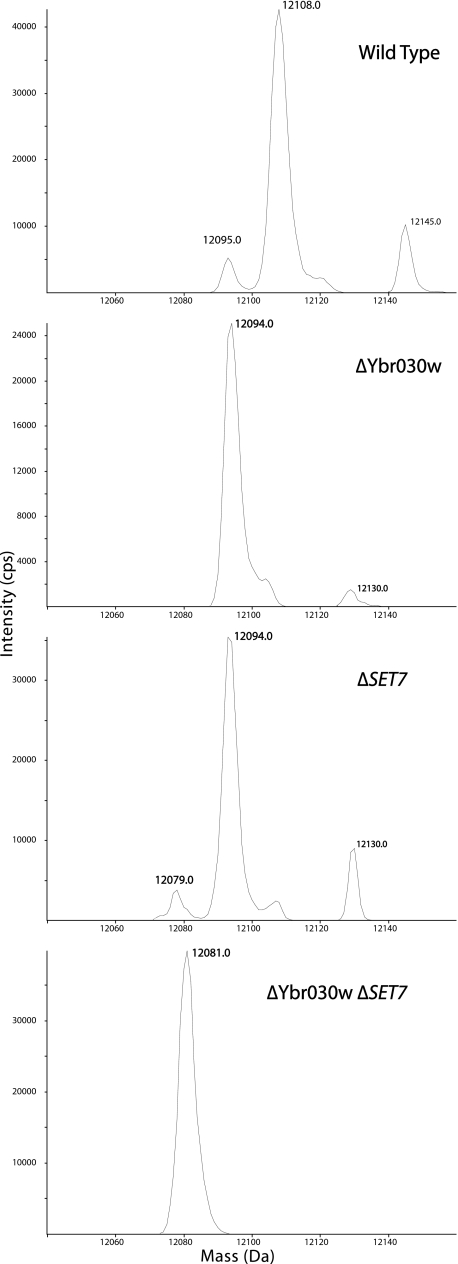
This approach led to the identification of two SET domain-containing proteins, Set7 and Ybr030w, that are required for Rpl42ab modification. In the wild-type BY4742 strain, an intact average mass of 12,108 Da was seen for a species eluting at 27 min on reverse-phase HPLC (Fig. 1). This mass corresponds to the mass of the gene product of Rpl42ab with the loss of the initiator methionine and the presence of two methyl groups (9, 16). This wild-type 12,108 species was also observed when the analysis was repeated for ribosomal proteins from a number of SET domain mutant strains, including RKM2, SET2, SET3, SET4, SET5, SET6, and Yhl039w.
FIGURE 1.
Intact mass reconstruct of Rpl42ab derived from wild type (BY4742), ΔYbr030w (BY4742), ΔSET7 (BY4742), and the double knock-out ΔYbr030w ΔSET7 strains. Ribosomal large subunit proteins were isolated and separated by reverse phase HPLC as described under “Experimental Procedures.” The resulting effluent was split, with half directed to the source of an electrospray mass spectrometer (PE Sciex III) and the other portion collected in a fraction collector for later analysis. The resulting deconvoluted MS spectra of Rpl42ab are displayed with the average masses of the significant peaks shown.
However, no species with the mass 12,108 Da was detected in the Rpl42ab fraction from the SET7 and Ybr030w mutant strains. Here, we detected a new species in each strain at 12,094 Da, or 14 Da lower than that of the wild-type species (Fig. 1). Similar results were found for the SET7 and Ybr030w mutants in the independent BY4741 strain background (Table 1 and data not shown). The loss of 14 Da corresponds to the loss of a single methyl group in the protein; the loss in both mutant strains suggests that the SET7 and Ybr030w genes each encode a methyltransferase that modifies a distinct site on Rpl42ab. To confirm this hypothesis, we constructed a double knock-out strain with deletions in both the SET7 and Ybr030w genes. Ribosomal proteins from this strain were subjected to the same analysis. Here, we found the Rpl42ab fraction contained a species of 12,081 Da (Fig. 1). This mass is within the instrument error of the calculated mass of unmodified Rpl42ab at 12,080 Da. This loss of 28 Da from the wild type protein along with the previous data showing the presence of two monomethylated lysine residues (9, 16) provides additional evidence for Set7 and Ybr030w as lysine methyltransferases, each adding one methyl group to Rpl42ab. The modification of Rpl42ab in wild-type cells may not be complete because we do find a small peak corresponding to the singly methylated species of mass 12,094 Da (Fig. 1).
Determination and Quantitation of the Methylation Sites of Rpl42ab by Top Down MSMS—To identify the sites of methylation in Rpl42ab, we first performed intact mass top down analysis. Rpl42ab fractions isolated from the LC-MS+ method were analyzed on a hybrid linear ion trap Fourier transform ion cyclotron resonance mass spectrometer (Thermo Fisher LTQ FT Ultra) as described under “Experimental Procedures.” This instrument resolves the 13C isotopomers allowing for the accurate determination of the monoisotopic masses of the all-12C species. The deconvoluted spectrum of wild type Rpl42 shows two molecular ions differing by the mass of one methyl group. The most abundant monoisotoopic signal was at 12,100.71 Da corresponding to the doubly methylated protein (Table 2 and supplemental Fig. S1). A second minor signal was seen at 12086.70 Da corresponding to a singly methylated form. We confirmed that the Rpl42ab protein from the deletion strains ΔYbr030w and ΔSET7 contains the monomethylated form at 12,086.73 Da and 12,086.75 Da, respectively, as expected. The ΔYbr030w strain has no detectable amount of unmodified Rpl42ab; however, the ΔSET7 strain shows the presence of a small amount of the unmodified species (Table 2 and supplemental Fig. S1). Rpl42ab from the ΔYbr030w ΔSET7 double mutant displays only the unmodified species (Table 2 and supplemental Fig. S1). These observations confirm the average mass results obtained with triple-quadrupole instrument described above and shown in Fig. 1.
TABLE 2.
Fourier transform ion cyclotron resonance of intact Rpl42ab
| Rpl42ab purified from strain | Observed monoisotopic mass | Calculated monoisotopic mass | Error | Modifications observed | % Abundance of methylation statea | |
|---|---|---|---|---|---|---|
| Da | Da | Δmass Da | ppm | % | ||
| Wild type | 12100.71 | 12100.70 | 0.01 | 0.84 | Two methyl groups | 78 |
| 12086.70 | 12086.69 | 0.01 | 0.71 | One methyl group | 22 | |
| ΔYbr030w | 12086.73 | 12086.69 | 0.04 | 3.57 | One methyl group | 100 |
| ΔSET7 | 12086.75 | 12086.69 | 0.07 | 5.38 | One methyl group | 88 |
| 12072.72 | 12072.67 | 0.05 | 4.34 | None | 12 | |
| ΔYbr030w ΔSET7 | 12072.68 | 12072.67 | 0.01 | 0.93 | None | 100 |
Calculated as described under “Experimental Procedures”
Initially the mass spectral data were deconvoluted and the individual intensities of each peak were used to determine the relative levels of each modified protein species. However, we found a bias for the most abundant ions when the signal to noise level was less than 10. We then followed the approach used by Pesavento et al. (19, 20) to quantitate human histone H4 variants to determine the relative amounts of the fully and partially methylated Rpl42ab species in yeast cells. We applied the protein ion relative ratio calculation described in “Experimental Procedures” to determine that 78% of Rpl42ab is dimethylated in wild-type cells and 22% is monomethylated. For the ΔSET7 strain the unmodified version accounted for 12% and the monomethylated accounted for 88% of the total Rpl42ab present. It is worthwhile to note that the FTICR instrumentation used in this study is a commercially available hybrid linear ion-trap/FTICR while Pesavento et al. (19, 20) used a custom quadrupole-FTICR hybrid.
Single charge states of intact species were then individually isolated and fragmented to localize the sites of methylation on the Rpl42ab protein. Two fragmentation methods were used, collisionally activated dissociation producing b and y ions, and electron capture fragmentation producing c and z ions. Multiple MSMS spectra representing different charge states were collected of Rpl42ab for each yeast strain, mating type, and methylation state. These data were combined to create the fragmentation map shown in Fig. 2. The MSMS data for the wild-type fully methylated Rpl42ab confirms the sites of monomethylation to Lys-40 and Lys-55. Chemical sequence analysis of Rpl42ab (16) had previously identified a monomethyllysine residue in a Gly-Arg-Lys-Arg sequence; it now appears that this sequence should be Gly-Lys-Arg-Arg corresponding to the genomic and mass spectral determined sequence of residues 39–42 (Fig. 2). These results, however, confirm the previous identification of the Gln-Thr-Lys-Pro sequence as a possible second site of lysine monomethylation at residues 53–56 (16).
FIGURE 2.
Fragmentation pattern of intact ribosomal protein Rpl42ab from wild type (BY4742), ΔYbr030w, ΔSET7, and the double knock-out ΔYbr030w ΔSET7 strains. Fractions containing Rpl42ab isolated from the LC-MS+ chromatography from wild type and knock-out strains were subjected to top down analysis where the intact protein was subjected to fragmentation as described under “Experimental Procedures.” Fragmentation is shown for the portion of the intact protein from residues 36–48; monomethylation sites on lysine residues are indicated. b and y ions from collisionally activated dissociation are indicated by sloping bars; c and z ions from electron capture dissociation are indicated by horizontal bars.
Interestingly, we find that the less abundant monomethylated form of Rpl42ab from the wild-type strain is a mixture of two modification states. One form contains a single methyl group at Lys-40 and the other contains a single methyl group at Lys-55 (Fig. 2). We estimated the amount of each monomethylated species by the signal intensity of the informative c43 fragment ions (residues 1–43; supplemental Fig. S2). We found that the singly methylated wild-type form at Lys-40 is present in three times the amount as the form methylated at Lys-55. The unmodified c43 ion intensity was weak, with a signal to noise below 10 rendering accurate quantitation difficult (19).
Rpl42ab isolated from the ΔYbr030w strain lacks methylation at Lys-40, indicating the gene product of Ybr030w is required for monomethylation of this site (Fig. 2). The ΔSET7 mutant has a loss of methylation at Lys-55, demonstrating the protein encoded by the SET7 gene is required for monomethylation of Lys-55 (Fig. 2). In agreement with the intact mass data, the ΔYbr030w ΔSET7 strain is unmodified at both Lys-40 and Lys-55 (Fig. 2 and Table 2).
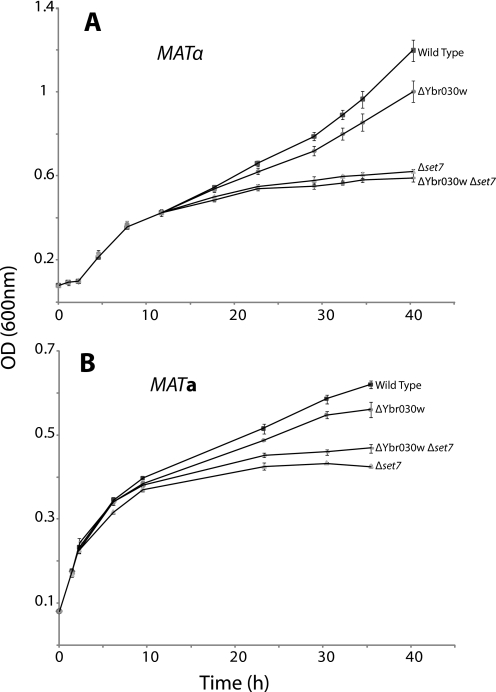
Growth Defect of SET7-deficient Yeast in the Presence of Cycloheximide—Mutation of the Rpl42ab Pro-56 residue to a glutamine residue confers resistance to cycloheximide, an inhibitor of eukaryotic protein synthesis produced by the soil bacterium Streptomyces griseus (15). The close proximity Lys-55 and Pro-56 in Rpl42ab raises the question of whether methylation may affect the sensitivity of yeast to cycloheximide. We thus monitored the growth of yeast strains containing or lacking the genes for Rpl42ab methylation in the presence of a low concentration of cycloheximide (Fig. 3). SET7 mutants in two strain backgrounds (Table 1) demonstrated decreased resistance to cycloheximide, while the Ybr030w mutants grew similarly to the wild-type strains. The double mutant also displayed sensitivity to cycloheximide. Control experiments performed in the absence of cycloheximide displayed no growth differences under these conditions (data not shown). Thus, methylation of Lys-55 by SET7 may have evolved to provide a protective effect against cycloheximide in the environment; a similar protection may be obtained in organisms such as Candida maltosa that naturally contain a glutamine residue at position 56.
FIGURE 3.
Differential effect of low cycloheximide levels (1 μg/ml) on the growth of wild type, ΔYbr030w, ΔSET7, and the double knock-out ΔYbr030w ΔSET7 strains. Cells were grown in YPD media and translational elongation inhibitor cycloheximide was added at 0.1 OD as described under “Experimental Procedures.” Panel A shows results from triplicate experiments with strains in the MATα background; panel B shows results from triplicate experiments of strains in the MATa background. The standard deviation values are indicated by bars.
Utilization of Top Down Mass Spectrometry to Determine the Sites of Methylation of Rpl12ab—Previously we reported the SET domain-containing protein Rkm2 trimethylated the ribosomal protein Rpl12ab at lysine 10 and an unknown enzyme dimethylated the protein at lysine 3 (11). Since that time, the methylation sites for two orthologs of Rpl12ab have been reported for Arabidopsis thaliana and the fission yeast Schizosaccharomyces pombe. In Arabidopsis, evidence was presented for the dimethylation of the N-terminal proline residue and the trimethylation of lysine 3 (21); a similar modification pattern was seen in S. pombe (22). Sadaie et al. (22) also identified the methyltransferase responsible for the trimethylation of lysine 3, SET11 in S. pombe. In light of sequence identity of the Rpl12ab protein from all of these species at the N terminus, we isolated wild-type S. cerevisiae Rpl12ab protein and subjected it to intact mass top down MSMS analysis as described above for Rpl42ab (Fig. 4). Unlike Rpl42ab, we found a single modified species for Rpl12ab containing six methyl groups. MSMS fragmentation revealed two methyl groups on Pro-1, three on Lys-3, and one on Arg-66. Close evaluation of previously published data on the MSMS of an N-terminal ArgC fragment of Rpl12ab from a ΔRKM2 deletion strain shows the loss of three methyl groups at the third lysine and the presence of the N-terminal dimethyl proline (Fig. 4). These results suggest that Rkm2 modifies Rpl12a largely at Lys-3, although modification at Lys-10 cannot be ruled out, especially when considering the b3, b8, b9, b10, and y9 ions in the original mass spectrum (11). Fragmentation evidence for the presence of methylation at Lys-10 was not seen in the intact mass top down analysis, possibly due to the low abundance of this form of modification. In any case, it now appears that the major modifications of Rpl12 at the N terminus are similar in S. cerevisiae, S. pombe, and Arabidopsis.
FIGURE 4.
Fragmentation pattern of intact wild-type ribosomal protein Rpl12ab and a peptide from the ΔRKM2 mutant protein. Fractions containing Rpl12ab isolated from the LC-MS+ chromatography were subjected to either top down analysis using electron capture dissociation and collisionally activated dissociation of the wild-type protein (panel A) or collisionally activated dissociation of the tryptic N-terminal peptide from the ΔRKM2 protein (panel B, Ref. 11) as described in the legend of Fig. 2 and the “Experimental Procedures” section.
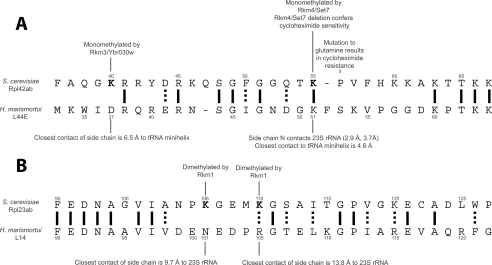
Structural Significance of Lysine Methylation in the Yeast Large Ribosomal Subunit—Cryo-EM structures are available for the free yeast 80S ribosome at a resolution of ∼15 Å (23) and for the 80S subunit bound to eEF2 at a resolution of 11.7 Å (24). In both structures, the 25S, 5.8S, and 5S rRNA, and the 43 yeast ribosomal proteins were computationally docked onto the cryo-EM structure using the previously solved crystal structures of the 30S subunit of T. thermophilus and the 50S subunit of H. marismortui as a guide (25, 26). Both structures show Rpl42ab positioned close to the tRNA exit site. The docked structure showed the positions of the monomethylated lysine residues in close proximity to rRNA. The ε-N of Lys-40 is within 3.6 Å and 3.9 Å of the 25S rRNA and ε-N of Lys-55 overlaps the 25S rRNA. This region of overlap appears to be an error in the docking and shows the limitations of the composite structures.
To overcome this limitation, the position of the corresponding sites of yeast Rpl42ab Lys-40 and Lys-55 in the homologous L44E protein of the 60S H. marismortui ribosomal subunit was determined from its x-ray crystal structure in complex with a deacylated tRNA minihelix (14). In this structure, L44E localizes to the tRNA exit site and has direct contacts to the bound tRNA minihelix (14). The aspartate at position 37 (corresponding to monomethyl Lys-40 of Rpl42ab) is within 6.5 Å of the tRNA minihelix, presenting the possibility that methyl lysine plays a role in binding tRNA in yeast (Fig. 5, panel A). The ε-N of Lys-51 (corresponding to the monomethyl Lys-55 residue of Rpl42ab) is 2.9 Å and 3.7 Å from the 23S rRNA and is within 4.5 Å of the bound tRNA minihelix (Fig. 5, panel A). These structural data suggest that the monomethyl lysine residues in Rpl42ab may contact rRNA and tRNA, and may not be exposed at the surface of the ribosome. If this is the case, the methyl marks in Rpl42ab may not be “read” by protein species including the tudor, MBT, and chromo domains that specifically recognize methylated lysine residues in histone tails (27). We suggest instead that lysine methylation in Rpl42ab may modulate hydrogen bonding interactions to RNA.
FIGURE 5.
Summary of methylation sites and interactions of large subunit ribosomal proteins Rpl42ab and Rpl23ab. Methylated sites are indicated in bold.In panel A, the alignment of yeast Rpl42ab to the archaebacterium H. marismortui L44E protein is shown along with the contacts of the residues of H. marismortui corresponding to the methylated yeast residues as determined in the x-ray crystal structure (PDB file 1QVF, Ref. 14). In panel B, the alignment of yeast Rpl23ab to the H. marismortui L14 protein is shown along with the closest RNA contacts for the L14 protein residues corresponding to the methylated lysine residues in Rpl23ab. ClustalW was used to align sequences.
To determine whether the possible interaction of ribosomal protein methylated lysine residues with rRNA is a general phenomenon, we examined the location of the dimethylated lysine residues of yeast Rpl23ab at Lys-106 and Lys-110 (11). The position of the dimethylated lysine residues in the yeast composite cryo-EM structure of the 80S eEF2 complex show the closest contact of the ε-N of dimethyl Lys-106 to be 7.6 Å and 8.1 Å from the 25S rRNA, and the ε-N of dimethyl Lys-110 closest contact to be 8.6 Å to the 25S rRNA. In the 60S H. marismortui crystal structure, Rpl23ab is homologous to the L14 protein with dimethyl Lys-106 corresponding to Asn-101. The closest contact of this asparagine is the 23S rRNA at 9.7 Å. Rpl23ab dimethyl Lys-110 aligns with Arg-105 and is 13.8 Å from its closest contact with the 23S rRNA. Thus, it appears that the methylated lysine residues in Rpl23ab do not contact the 23S rRNA. The Rpl23ab protein is on the binding face of the large ribosomal subunit and is involved in multiple contacts with the 18S small ribosomal rRNA (24). This localization opens the possibilities that the methylated lysine residues may interact with the 18S rRNA on the small subunit. Finally, we were not able to position the methylated Lys-3 residue in Rpl12ab to these structures because the N terminus is disordered (23, 24). Taken together, these results suggest that methylated lysine residues in ribosomal proteins may play structural roles distinct from the recognition marking roles seen in histones (27).
Supplementary Material
Acknowledgments
We thank Dr. Joseph Loo for his support and helpful advice on mass spectrometry.
This work was supported, in whole or in part, by National Institutes of Health Grant GM026020 and Grant S10 RR023045 (purchase of the LTQ FT Ultra mass spectrometer). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: LC-MS+, liquid chromatography with electrospray-ionization mass spectrometry and fraction collection; FTICR, Fourier-transform ion cyclotron resonance; MSMS, tandem mass spectrometry.
References
- 1.Couture, J. F., and Trievel, R. C. (2006) Curr. Opin. Struct. Biol. 16 753-760 [DOI] [PubMed] [Google Scholar]
- 2.Xiao, B., Wilson, J. R., and Gamblin S. J. (2003) Curr. Opin. Struct. Biol. 13 699-705 [DOI] [PubMed] [Google Scholar]
- 3.Dirk, L. M. A., Trievel, R. C., and Houtz, L., R. (2006) The Enzymes,3rd Ed., 24 179-228 [DOI] [PubMed] [Google Scholar]
- 4.Marmostein, R. (2003) Trends Biochem. Sci. 28 59-62 [DOI] [PubMed] [Google Scholar]
- 5.Qian, C., and Zhou, M. M. (2006) Cell Mol. Life. Sci. 63 2755-2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng, D. W-K., Wang, T., Chandrasekharan, M. B., Aramayo, R., Kertbundit, S., and Hall, T. C. (2007) Biochim. Biophys. Acta 1769 316-329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Culhane, J. C., and Cole, P. A. (2007) Curr. Opin. Chem. Biol. 11 561-568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polevoda, B., and Sherman, F. (2007) Mol. Microbiol. 65 590-606 [DOI] [PubMed] [Google Scholar]
- 9.Lee, S. W., Berger, S. J., Martinovic, S., Pasa-Tolic, L., Anderson, G. A., Shen, Y., Zhao, R., and Smith, R. D. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 5942-5947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porras-Yakushi, T. R., Whitelegge, J. P., Miranda, T. B., and Clarke, S. (2005) J. Biol. Chem. 280 34590-34598 [DOI] [PubMed] [Google Scholar]
- 11.Porras-Yakushi, T. R., Whitelegge, J. P., and Clarke, S. (2006) J. Biol. Chem. 281 35835-35845 [DOI] [PubMed] [Google Scholar]
- 12.Porras-Yakushi, T. R., Whitelegge, J. P., and Clarke, S. (2007) J. Biol. Chem. 282 12368-12376 [DOI] [PubMed] [Google Scholar]
- 13.Mager, W. H., Planta, R. J., Ballesta, J.-P. G., Lee, J. C., Mizuta, K., Suzuki, K., Warner, J. R., and Woolford, J. (1997) Nucleic Acids Res. 25 4872-4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmeing, T. M., Moore, P. B., and Steitz, T. A. (2003) RNA 9 1345-1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawai, S., Murao, S., Mochizuka, M., Shibuya, I., Yano, K., and Takagi, M. (1992) J. Bacteriol. 174 254-262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itoh, T., and Wittmann-Liebold, B. (1978) FEBS Lett. 96 399-402 [DOI] [PubMed] [Google Scholar]
- 17.Whitelegge, J. P. (2004) Methods Mol. Biol. 251 323-340 [DOI] [PubMed] [Google Scholar]
- 18.Whitelegge, J. P., Gundersen, C. B., and Faull, K. F. (1998) Protein Sci. 7 1423-1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pesavento, J. J., Mizzen, C. A., and Kelleher, N. L. (2006) Anal. Chem. 78 4271-4280 [DOI] [PubMed] [Google Scholar]
- 20.Pesavento, J. J., Bullock, C. R., LeDuc, R. D., Mizzen, C. A., and Kelleher, N. L. (2008) J. Biol. Chem. 283 14927-14937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carroll, A. J., Heazlewood, J. L., Ito, J., and Millar, A. H. (2008) Mol. Cell. Proteomics 7 347-369 [DOI] [PubMed] [Google Scholar]
- 22.Sadaie, M., Shinmyozu, K., and Nakayama, J. I. (2008) J. Biol. Chem. 283 7185-7195 [DOI] [PubMed] [Google Scholar]
- 23.Spahn, C. M. T., Beckmann, R., Eswar, N., Penczek, P. A., Sali, A., Blobel, G., and Frank, J. (2001) Cell 107 373-386 [DOI] [PubMed] [Google Scholar]
- 24.Spahn, C. M. T., Gomez-Lorenzo, M. G., Grassucci, R. A., Jorgensen, R., Andersen, G. R., Beckmann, R., Penczek, P. A., Ballesta, J. P. G., and Frank, J. (2004) EMBO J. 23 1008-1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wimberly, B.T., Brodersen, D. E., Clemons, W. M. Jr., Morgan-Warren, R. J., Carter, A. P., Vonrhein, C., Hartsch, T., and Ramakrishnan, V. (2000) Nature 407 327-339 [DOI] [PubMed] [Google Scholar]
- 26.Ban, N., Nissen, P., Hansen, J., Moore, P. B., and Steitz, T. A. (2000) Science 289 905-920 [DOI] [PubMed] [Google Scholar]
- 27.Kim, J., Daniel, J., Espejo, A., Lake, A., Krishna, M., Xia, L., Zhang, Y., and Bedford, M. T. (2006) EMBO Rep. 7 397-403 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.