Abstract
X11 and X11-like proteins (X11L) are neuronal adaptor proteins whose association to the cytoplasmic domain of amyloid β-protein precursor (APP) suppresses the generation of amyloid β-protein (Aβ) implicated in Alzheimer disease pathogenesis. The amyloidogenic, but not amyloidolytic, metabolism of APP was selectively increased in the brain of mutant mice lacking X11L (Sano, Y., Syuzo-Takabatake, A., Nakaya, T., Saito, Y., Tomita, S., Itohara, S., and Suzuki, T. (2006) J. Biol. Chem. 281, 37853–37860). To reveal the actual role of X11 proteins (X11s) in suppressing amyloidogenic cleavage of APP in vivo, we generated X11 and X11L double knock-out mice and analyzed the metabolism of APP. The mutant mice showed enhanced β-site cleavage of APP along with increased accumulation of Aβ in brain and increased colocalization of APP with β-site APP-cleaving enzyme (BACE). In the brains of mice deficient in both X11 and X11L, the apparent relative subcellular distributions of both mature APP and its β-C-terminal fragment were shifted toward the detergent-resistant membrane (DRM) fraction, an organelle in which BACE is active and both X11s are not nearly found. These results indicate that X11s associate primarily with APP molecules that are outside of DRM, that the dissociation of APP-X11/X11L complexes leads to entry of APP into DRM, and that cleavage of uncomplexed APP by BACE within DRM is enhanced by X11s deficiency. Present results lead to an idea that the dysfunction of X11L in the interaction with APP may recruit more APP into DRM and increase the generation of Aβ even if BACE activity did not increase in brain.
APP3 is a type I membrane protein and is the precursor of amyloid β-protein (Aβ), which is the principal component of senile plaques, a pathological hallmark in the Alzheimer disease (AD) brain. The production, aggregation, and deposition of Aβ are widely believed to be central to the pathogenesis in AD (reviewed in Ref. 1). APP is subjected to two types of cleavage in both a potentially amyloidogenic pathway and an amyloidolytic (or nonamyloiodgenic) pathway. Amyloidogenic cleavage of APP is mediated by β-site APP-cleaving enzyme (β-secretase or BACE), which generates the C-terminal fragment CTFβ, which contains an intact Aβ sequence, whereas amyloidolytic cleavage of APP is mediated by one of several α-site APP-cleaving enzymes (α-secretases, including ADAM10 and ADAM17), thereby generating CTFα, which contains only the C-terminal half of Aβ peptide (reviewed in Refs. 1 and 2). These CTFs are further cleaved within the transmembrane domain by the γ-secretase aspartyl protease complex composed of presenilin (PS), nicastrin, Aph-1, and Pen-2. The γ-secretase cleavage generates Aβ and the APP intracellular domain from CTFβ and, alternatively, γ-secretase generates p3 peptide and APP intracellular domain from CTFα (reviewed in Refs. 3 and 4). However, it is still unclear exact where along the intracellular trafficking itinerary of APP each of these cleavages occurs. The molecular mechanisms regulating secretase reactions are also poorly understood (reviewed in Ref. 5). Recent growing evidence suggests that APP processing by BACE occurs in cholesterol- and sphingolipid-rich detergent-resistant membrane domains (DRM domains) also known as lipid rafts (6–9). DRM are suggested to be the principal compartments containing intracellular monomeric and oligomeric Aβ in brain (10, 11). The γ-secretase complex also associates with DRM and is especially active there (12–15), indicating that Aβ may be largely the product of a series of intra-DRM reactions.
X11 proteins (X11s) comprise a family of three evolutionarily conserved molecules: 1) X11/X11α/Mint1; 2) X11-like (X11L)/X11β/Mint2; and 3) X11-like2 (X11L2)/X11γ/Mint3. X11s are composed of an independent N-terminal half, a conserved central phosphotyrosine interaction domain, and two C-terminal PDZ domains. X11 and X11L are largely brain-specific, whereas X11L2 is expressed ubiquitously (reviewed in Ref. 16). X11s bind to the 681GYENPTY687 motif within APP cytoplasmic region through its phosphotyrosine interaction domain and suppress APP metabolism in cultured cells (17–19). Transgenic mice expressing the APP Swedish mutation and also overexpressing X11 or X11L show a decreased level of cerebral Aβ and a reduction of Aβ plaques in the cerebral cortex and hippocampus in comparison with the mice expressing Swedish mutant APP alone (20, 21). Conversely, the amyloidogenic metabolism of endogenous APP and the generation of Aβ are facilitated in the brains of X11L-deficient mice (22).
These previous observations indicate that X11 and X11L work to suppress the amyloidogenic metabolism of APP selectively, although it is still unclear how X11 and X11L function in the regulation of APP metabolism in the intact brain. To address these issues, we generated X11 and X11L genes homozygous doubly deficient (X11-/-, X11L-/-) mice by crossing X11 gene-deficient (X11-/-) mice with X11L gene-deficient (X11L-/-) mice. We report here that the doubly deficient mice show increased levels of CTFβ and Aβ in hippocampus and cerebral cortex, apparently because of the relatively increased colocalization of vesicles containing both APP and BACE1 in neurites of primary cultured neurons and that of APP and CTFβ in DRM in brain when compared with what occurs in wild-type mice. These results provide evidence that X11 proteins suppress translocation of APP into BACE- and γ-secretase-rich DRM domains, raising the possibility that dysfunction of X11 proteins might play a role in the pathogenesis of AD even if the BACE activity did not increase in AD patients.
EXPERIMENTAL PROCEDURES
Generation of X11/X11L Doubly Deficient Mice—All of the animal studies were conducted in compliance with the guidelines of the Animal Studies Committee of Hokkaido University. The original X11-deficient mice were hybrids of 129/Sv and C57BL/6 (23). This line was further backcrossed to C57BL/6 line over 10 times during 6 years in our laboratory. X11L gene knock-out mice were generated coisogenic to C57BL/6 and backcrossed to same strain over 10 times during 7 years (22). The respective deficient mutant mice were mated to generate the [X11+/-, X11L+/-] heterozygous mutant line. The heterozygous mutant mice were further mated to generate [X11+/-, X11L-/-] and [X11-/-, X11L+/-] mutant mice. To obtain [X11-/-, X11L-/-] doubly mutant mice, we crossed [X11+/-, X11L-/-] and/or [X11-/-, X11L+/-] mutant mice, respectively (supplemental Table S1). The genotype was determined by PCR analysis of the genomic DNA prepared from the tails. PCR product of 250 and 540 bp are detected from wild-type and knock-out mutant alleles of X11, and those of 550 and 310 bp are detected from wild-type and knock-out mutant alleles of X11L (supplemental Fig. S1). We planned mating schedules as shown (supplemental Table S1). Because offspring with the X11/X11L doubly deficient genotype showed unexpected numbers when their genotyping was performed at postnatal day 28, we performed the genotyping of individuals at postnatal day 0 and embryonic days 18.5 and 15.5.
Antibodies—Polyclonal anti-X11 rabbit UT153 antibody was raised against a peptide that is composed of Cys plus the sequence between position 235 and 252 (C+LHHYDERSDGESDSPEKE) of human X11. The specificity was examined by immunoblotting and immunostaining (supplemental Fig. S2). Monoclonal anti-BACE1 antibody BACE-Cat1 (3D5) was described (24). Polyclonal anti-APP cytoplasmic domain G369 antibody was described (25). Monoclonal anti-X11/mint1, anti-X11L/mint2, anti-X11L2/mint3, anti-microtubule-associated protein 2, and anti-flotillin-1 antibodies were purchased from BD Bioscience. Monoclonal anti-APP extracellular domain 22C11 (Chemicon), anti-actin (Chemicon), anti-PS1 C-terminal (Chemicon), anti-α-tubulin (Zymed Laboratories Inc.), anti-transferrin receptor (Zymed Laboratories Inc.), anti-sAPPα 2B3 (IBL), anti-glial fibrillary acidic protein (PROGEN) and anti-synaptophysin (DAKO), and polyclonal anti-APP cytoplasmic domain 8717 (Sigma), anti-sAPPβ (IBL), and anti-calnexin (Stressgen Biotechnologies) antibodies were purchased.
Immunoblot Analysis—The mouse brains were homogenized in 8-volume of radioimmune precipitation assay buffer containing 1% (w/v) SDS, a protease inhibitor mixture (5 μg/ml chymostatin, 5 μg/ml leupeptin, and 5 μg/ml pepstatin), and 1 μm microcystin-LR and then lysed by sonication in ice. The resulted supernatant was used for immunoblot analysis. In a separate study, the brain tissues were homogenized in 8-volume of buffer A (10 mm Hepes-NaOH, pH 7.4, 0.32 m sucrose) containing a protease inhibitor mixture and 1 μm microcystin-LR with 20 strokes of a Dounce homogenizer. After centrifugation twice at 1,000 × g for 10 min of homogenate, the post-nuclear supernatant was further centrifuged at 100,000 × g for 60 min. The precipitated membrane fraction (P100) was solubilized in 8 volumes of radioimmune precipitation assay buffer containing 1% (w/v) SDS by sonication. This P100 was used to detect APP CTFs, and the supernatant (S100) was used to detect sAPP in immunoblot analysis. The immunoreactants were detected using ECL plus detection system (GE Healthcare) and quantified with a Versa Doc model 3000 (Bio-Rad).
Immunohistochemistry—Mouse brain tissue sections were prepared and incubated with primary antibodies as described (22) and further incubated with horse anti-mouse IgG or goat anti-rabbit IgG antibodies conjugated to biotin (Vector Laboratories), followed by the ABC complex (peroxidase activity was revealed using diaminobenzidine as the chromogen) or goat anti-mouse IgG coupled with Alexa Fluor 488 or goat anti-rabbit IgG coupled with Alexa Fluor 546 in PBS containing 5% (v/v) horse and goat serum for 2 h at room temperature. The sections were viewed using Axioplan 2 microscope or the confocal laser scanning microscope LSM510 (Carl Zeiss).
Neuronal Culture and Immunocytochemistry—The primary culture of cortical neurons was described (26). In short, the cortex of mice at embryonic day 15.5 was dissected, and neurons were spread in a buffer containing papain and cultured at 5 × 104 cells/cm2 in Neurobasal medium containing B27, Glutamax, and antibiotics (Invitrogen) on poly-d-lysine-treated glasses. At in vitro day 7, the neurons were fixed with methanol at -20 °C. After thorough washing with PBS containing 0.1% Triton X-100 (PBS-X), neurons were blocked with PBS-X containing 3% (v/v) goat and 3% (v/v) horse serum (blocking solution). Primary antibodies (mouse monoclonal anti-BACE1 antibody BACE-Cat1 (3D5) and rabbit polyclonal anti-APP cytoplasmic domain G369) were incubated for overnight at 4 °C in blocking solution. The neurons were then washed three times with PBS-X and incubated with secondary antibodies (goat anti-mouse IgG coupled with Alexa Fluor 488 or goat anti-rabbit IgG coupled with Alexa Fluor 546) in blocking solution for 2 h at room temperature. The neurons were washed with PBS-X and mounted onto slides with Shandon IMMU-MOUNT (Thermo). The neurons were observed using the BZ-9000 microscope (KEYENCE).
Quantification of Aβ40 and Aβ42—Mouse endogenous Aβ40 and Aβ42 were measured by sandwich enzyme-linked immunosorbent assay system (Wako Pure Chemical Industries Co., Osaka, Japan) described previously (22). Hippocampus (tissues from three mice were combined in each assay) and cerebral cortex (tissues from one mouse) dissected from 12-month-old mice were used.
Isolation of DRM—DRM fractions were prepared according to the established procedure with minor modifications (27). The cortex from each mouse (8–12 weeks old) or its P100 fraction was homogenized in TNE buffer (10 mm Tris-HCl, pH 7.5, 0.15 m NaCl, 5 mm EDTA) containing 1% (v/v) Triton X-100, a protease inhibitor mixture, and 1 μm microcystin-LR. The sucrose concentration of the extract was adjusted to 42.5% (w/v) by the addition of 85% sucrose in TNE buffer; the extract was then placed at the bottom of an ultracentrifuge tube and overlaid with a 5% (1 ml), 10% (1 ml), 20% (3 ml), and 30% (3 ml) discontinuous sucrose gradient in TNE. The gradients were centrifuged at 39,000 rpm for 20 h with SW41 rotor (Beckman Coulter). Fractions (1 ml) were collected from the top of the ultracentrifuge tubes, and proteins in respective fractions were analyzed by immunoblot analysis.
RESULTS
Generation of X11 and X11L Doubly Deficient Mice and Expression of X11 Proteins in Wild-type Mouse Brain—Although the X11 and X11L doubly deficient mice (X11-/-, X11L-/-) genotypes of embryos followed standard Mendelian genetics (supplemental Table S1), ∼50% of the doubly deficient mice were weaker and paler than littermates and died at postnatal day 0 with little or no milk in their stomachs. The doubly deficient (X11-/-, X11L-/-) mice survived but were significantly smaller in size when compared with the wild-type (X11+/+, X11L+/+), X11-deficient (X11-/-, X11L+/+), or X11L-deficient (X11+/+, X11L-/-) mice at all ages examined (supplemental Fig. S1). The same phenotype was reported in the mutant mice produced as an independent line (28). We used these survivors in further analysis of APP metabolism. Prior to biochemical analysis of APP metabolism, we examined brain morphology, and expression and distribution of APP, X11s, and some standard neuronal proteins (supplemental Fig. S3). No remarkable abnormality was observed in overall brain structure and regional structures (supplemental Fig. S3), indicating that no major developmental abnormalities occurred in the X11 and X11L doubly deficient mice. No differences were observed for expression and localization of APP, microtubule-associated protein 2B, glial fibrillary acidic protein, or synaptophysin in the hippocampal CA1 region of wild-type and doubly deficient mouse brain (supplemental Fig. S3), indicating that no gliosis, synaptic loss, or dendritic atrophy was detectable in brains of the X11 and X11L doubly deficient mice.
Because deletion of one X11 family protein did not induce the compensatory expression of other family proteins (supplemental Fig. S3), we first confirmed the expression profile of X11s in brain regions (supplemental Fig. S4). It is well analyzed that APP is ubiquitously expressed in brain tissues including hippocampus and cerebral cortex (29), which are regions suffering from neurodegeneration in human AD. Immunoblot analysis showed that X11s expressed in various brain tissues along with APP, although X11L is relatively abundant in olfactory bulb, cerebral cortex, thalamus/hypothalamus, hippocampus, and cerebellum when compared with other regions (supplemental Fig. S4) (30). Localization of neuron-specific X11 and X11L was examined in hippocampus and cerebral cortex by immunostaining (supplemental Fig. S4). Interestingly, distinct expression between X11 and X11L was observed; X11L was largely expressed in the pyramidal neurons, whereas X11 was expressed in other types of neurons. These results suggest that X11 and X11L function in different neurons but in the same roles for APP metabolism as shown in many studies (17, 19–21, 30); thus biochemical analysis with lysate of the X11 and X11L doubly deficient mice brain allows accurate determination of X11s function in APP metabolism.
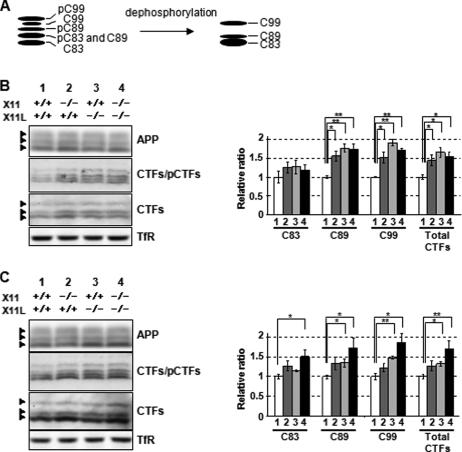
Enhanced β-Cleavage of APP in the Hippocampus and the Cerebral Cortex of the X11 and X11L Doubly Deficient Mice—Amyloidogenic metabolism of APP is facilitated as illustrated by the elevation of CTFβ levels in X11L-deficient mice (22). However, it is unclear whether X11 shows the same proamyloidogenic activity as X11L and whether the X11/X11L doubly deficient mice facilitate the amyloidogenic metabolism of APP beyond that observed with either X11 or X11L singly. To address these issues, we first quantified APP CTFs in hippocampus and cerebral cortex of the X11-deficient (X11-/-, X11L+/+) mice and the X11/X11L doubly deficient (X11-/-, X11L-/-) mouse brains along with those of the wild-type (X11+/+, X11L+/+) mice and the X11L-deficient (X11+/+, X11L-/-) mouse brains (Fig. 1). Quantitative accuracy for the amount of APP-CTFs detected by immunoblot analysis with anti-APP C-terminal antibody (antibody 8171; Sigma) was examined (supplemental Fig. S5). Quantification by this procedure is reliable with a dose-dependent reactivity by the antibody. APP α-secretase produces CTFα, which is composed of C-terminal 83 amino acids (C83), whereas β-secretase or BACE generates two CTFβ, C99 and C89 (31). Because CTFs are phosphorylated at Thr668 residue in brain, six CTF species can be detected as five protein bands by immunoblotting (see schematic picture in Fig. 1A); phosphorylated C99 (pC99), C99, pC89, a mixture of C89 and pC83, and C83 (reviews in Ref. 32, 33). To simplify the quantification of each CTFs (C99, C89, and C83), respective CTFs were subjected to dephosphorylation with λ protein phosphatase as described (34), and the levels of dephosphorylated CTFs in hippocampus (Fig. 1B) and cerebral cortex (Fig. 1C) were determined. The band densities were standardized to the density of transferrin receptor, and ratios of CTF/transferrin receptor were generated (right panels in Fig. 1, B and C). When compared with wild-type mice (lane 1), increased levels of the amyloidogenic fragments C89 and C99, but not the nonamyloidogenic fragment C83, were observed in the hippocampi of: 1) X11-deficient mice (lane 2); 2) X11/X11L doubly deficient mice (lane 4); and 3) X11L-deficient mice (column 3) (Fig. 1B). A similar trend toward elevated CTF levels was observed in the cerebral cortex (Fig. 1C), but the increase of CTFβ species in the X11-deficient mice was not significant. We could also verify the increase of C99 in the cortex of X11L-deficient and X11/X11L doubly deficient mice by using anti-C99 N-terminal G530 antibody (35) (supplemental Fig. S6). G530 antibody specifically recognized the mouse C99 (supplemental Fig. S6), although the titer of G530 was lower than antibody 8717. We quantified C99 in cerebral cortex of the wild-type and X11s-deficient mouse brains by immunoblotting with G530 as did it with antibody 8717 (supplemental Fig. S6). The increased level of C99 quantified with G530 well agreed with that quantified with antibody 8717 in X11s-deficient mice, indicating that quantification of CTF with antibody 8717 is reliable. Therefore, in following experiments, we used antibody 8717 to detect and quantify the brain APP-CTFs. In cerebral cortex, the X11L-deficient mice and the X11/X11L doubly deficient mice showed significantly elevated levels of C89 and C99. The doubly deficient mice showed a less pronounced increase of C83 that was not observed in singly X11 or X11L-deficient mice. These results indicate that, like X11L, X11 also suppresses amyloidogenic metabolism of APP in brain in vivo, although the contribution of X11 to this pathway is apparently less pronounced, maybe by a smaller population of neurons expressing X11 alone, than is the contribution of X11L. X11/X11L doubly deficient mice show an increase in levels of total CTFs in addition to a more specific elevation of CTFβ levels. Therefore, the X11/X11L doubly deficient mice are potentially suitable for studies aimed at elucidating the mechanism by which X11 proteins control Aβ levels.
FIGURE 1.
Quantification of APP CTFs in hippocampus and cerebral cortex of the wild-type, X11-deficient, X11L-deficient, or X11/X11L doubly deficient mice. A, scheme of APP CTFs; C99 and C89 are the CTFβ, and C83 is the CTFα. p indicates species phosphorylated at Thr668 (numbering for APP695 isoform) (32–34). B and C, immunoblot analysis of APP and APP CTFs in the hippocampus (B) and cerebral cortex (C) of wild-type and deficient mice (12 months). Lysates (40 μg of protein), with (third row) or without (second row) λ protein phosphatase treatment, of membrane fractions of hippocampus and cerebral cortex were analyzed by immunoblotting with anti-APP C-terminal (Sigma product) and anti-transferrin receptor (TfR) antibodies. The densities of CTF protein bands were standardized to the density of transferrin receptor and compared with those for wild-type mice, to which a reference value of 1.0 was assigned (the values represent the means ± S.E.). The data were analyzed by a one-way analysis of variance followed by the Tukey's test (n = 4; *, p < 0.05; **, p < 0.01). Lane 1, wild-type mice (X11+/+, X11L+/+); lane 2, X11-deficient mice (X11-/-, X11L+/+); lane 3, X11L-deficent mice (X11+/+, X11L-/-); lane 4, X11 and X11L doubly deficient mice (X11-/-, X11L-/-).
In a previous study of X11L-deficient mouse brains, the levels of sAPP were relatively decreased despite increased levels of CTFβ (22), a result that was attributed to the role of X11s in exocytosis (36). We examined sAPP levels in the brains of X11-deficient mice and the X11/X11L doubly deficient mice (supplemental Fig. S7). Levels of sAPP in the hippocampus (supplemental Fig. S7) and cerebral cortex (supplemental Fig. S7) of X11-deficient mice and the X11/X11L doubly deficient mice were quantified by immunoblot with anti-pan sAPP, anti-sAPPα, and anti-sAPPβ antibodies as were the respective sAPP peptides in the hippocampus and cerebral cortex of X11L-decifient mice. As expected and in contrast to the altered levels of CTFs (Fig. 1), the levels of total sAPP, sAPPα, and sAPPβ were significantly decreased in hippocampus and cerebral cortex of the doubly deficient mice and of the X11L-deficient mice when compared with those of the wild-type mouse brain. However, sAPP levels in the X11-deficient mice brain were not affected significantly. The decrease of sAPP was more remarkable in the X11/X11L doubly deficient mice brain than in singly X11L-deficient mouse brain, suggesting that X11L plays a more important role than X11 in controlling sAPP level. We asked whether the decrease of sAPP levels in the X11s-deficient mice brain is due to the insufficient maturation of APP in the mutant mice. Full-length APP levels in the mutant mice brain were again quantified and were compared with the levels in wild-type mice brain (supplemental Fig. S5). We could not detect a significant difference for both mAPP and imAPP levels between the mutant and the wild-type mice brain, although we know that the ectopic overexpression of X11 and X11L suppresses the maturation of APP in cells in vitro (17, 19). Therefore, the decrease in the sAPP levels in the X11s-deficient mice brain is not caused by the suppression of maturation of APP. These results, as expected from a previous study of the X11L-deficient mouse brain (22), indicate that sAPP levels are relatively insensitive indicators of α- and β-site cleavage of APP in brain in vivo. To reveal the mechanism of how sAPP levels decrease in the X11s-deficient mice brain may help to understand the multiple functions of X11 family proteins in neuron, but it is our next issue to analyze in future.
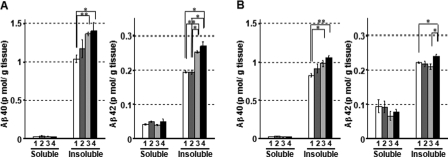
In the X11L-deficient mouse brain, the levels of Aβ were increased, as anticipated by a report that CTFβ levels are increased (22). Therefore, we next examined the levels of Aβ in the X11-deficient mouse brain and in the X11/X11L doubly deficient mouse brain (Fig. 2). Total brain Aβ40 and Aβ42 were largely recovered in the Tris-buffered saline (50 mm Tris-HCl, PH 7.4, 150 mm NaCl)-insoluble fraction as reported (37). Aβ40 and Aβ42 levels in hippocampus (Fig. 2A) and in cerebral cortex (Fig. 2B) were increased in the X11/X11L doubly deficient mice as compared with the corresponding levels in wild-type mice, whereas Aβ40 and Aβ42 levels in the X11-deficient mice were indistinguishable from wild-type levels. These results suggest that Aβ generation was enhanced in brain in the absence of X11 and X11L, which agree with the fact that the β-site cleavage of APP increased in the X11/X11L doubly deficient mouse brain (Fig. 1). Moreover, in hippocampus and cerebral cortex X11L is apparently a more efficient suppressor of Aβ generation than is X11.
FIGURE 2.
Quantification of Aβ40 and Aβ42 in hippocampus and cerebral cortex of wild-type, X11-deficient, X11L-deficient, or X11/X11L doubly deficient mice. Protein extracts from hippocampus (A) and cerebral cortex (B) of wild-type and deficient mice (12 months) were divided into Tris-buffered saline (50 mm Tris-HCl, pH 7.4, 150 mm NaCl)-soluble and insoluble fractions. Aβ40 (left panels) and Aβ42 (right panels) were quantified with sandwich enzyme-linked immunosorbent assay, and their concentrations are normalized to tissue weight. The data were analyzed by a one-way analysis of variance followed by the Tukey's test. Statistical significance is indicated with asterisks (n = 4; *, p < 0.05; **, p < 0.01). The error bars are S.E. Column 1, wild-type mice (X11+/+, X11L+/+); column 2, X11-deficient mice (X11-/-, X11L+/+); column 3, X11L-deficent mice (X11+/+, X11L-/-); column 4, X11/X11L doubly deficient mice (X11-/-, X11L-/-).
X11L Deficiency Enhances APP Translocation into DRM and Generation of APP CTFβ—To reveal the molecular basis for suppression of amyloidogenic cleavage of APP by X11s, we first examined the colocalization of APP and BACE1 within primary cultured neurons. Primary cultured cortical neurons derived from wild-type and X11L-deficient mice brain were fixed and labeled with antibodies specific for APP and BACE1, and localization of these two endogenous proteins was analyzed (Fig. 3). Because the effect of X11L deficiency on β-site cleavage of APP was greater than that of X11 deficiency (Fig. 1), we studied primary cultures from X11L-deficient mice. APP and BACE1 were detectable in neurites, which is where APP vesicles were transported (26); thus, we focused our analysis on the observation of localization of APP and BACE1 in neurites. In wild-type neurons (Fig. 3A, left panels), the majority (over 90%) of APP and BACE were localized at distinct regions of the neurites independently. Colocalized signals (indicated with white and blue arrows in Fig. 3A) to APP signals were 4.6%, and those to BACE signals were 3.6% (Fig. 3B, open columns). In contrast to this, 9.1% of APP signals and 8.0% of BACE signals to respective signals were colocalized in neurites in X11L-deficient neurons (Fig. 3, A, right panels, and B, closed columns). The ratio of APP-containing vesicles to BACE-containing vesicles was not significantly changed between wild-type and X11L-deficient neurons (Fig. 3C), suggesting that neither type of vesicles increased their numbers in X11L-deficient neurons. Moreover, a similar result was observed in neurons expressing APP-green fluorescent protein and BACE1-mRFP (supplemental Fig. S8), and enhanced colocalization of APP and BACE in X11L-deficient mice brain is also suggested from an analysis of biochemical fractionation (22). X11 and X11L can associate with APP (17, 19), but not with BACE.4 These results suggest that X11s regulate subcellular localization of APP but not BACE and that the loss of X11L facilitates the colocalization of APP and BACE within neurites.
FIGURE 3.
Localization of APP and BACE1 in primary cultured cortical neurons of wild-type and X11L-deficient mice. A, localization of endogenous APP and BACE1 in primary cultured cortical neurons. Cortical neurons prepared from brains of wild-type (X11+/+, X11L+/+) and X11L-deficient (X11+/+, X11L-/-) mice (embryonic day 15.5) were cultured for 7 days and immunostained with anti-BACE1 (3D5, green) and anti-APP (G369, red) antibodies. The white and blue arrows indicate colocalized signals. Scale bar,5 μm. B, percentage of colocalized vesicles. Visualized APP-containing vesicles (wild type, 436 vesicles; X11L-deficient, 472 vesicles) and BACE1-containing vesicles (wild type, 554 vesicles; X11L-deficient, 538 vesicles) were counted and merged signals (wild type, 20 vesicles; X11L-deficient, 43 vesicles) to APP or BACE1 signals were shown as percentages (%). C, ratio of APP-containing vesicles to BACE1-containing vesicles in wild-type and X11L-deficient mice neurons. Open column, wild-type (X11+/+, X11L+/+) mice; closed column, X11L-deficient (X11+/+, X11L-/-) mice.
Based on the observation that APP and BACE were extensively colocalized in neurons in the absence of X11L, we analyzed biochemical colocalization of APP and BACE in the X11s-deficient mouse brain, focusing our analysis on the metabolism and localization of APP in the subcellular region. Several lines of evidence indicate that amyloidogenic cleavage of APP occurs in cholesterol- and sphingolipid-enriched DRM, and indeed BACE localizes in DRM together with APP (6, 7). Furthermore, the buoyant, cholesterol-rich DRM has been reported to contain higher γ-secretase specific activity than does the heavy membrane fraction (12–15). These reports led us to the possibility that, in the absence of X11s, more APP might be translocated into DRM and colocalized there with BACE, leading to enhanced amyloidogenic cleavage of APP.
To examine this possibility, we studied the APP holoproteins and CTFs that were localized in DRM isolated from the cortex of the wild-type, X11-deficient, X11L-deficient, and X11 and X11L doubly deficient mouse brains (8–12 weeks old) using immunoblot analysis (Fig. 4). Brain post-nuclear supernatant was fractionated by ultracentrifugation on a sucrose density gradient, and the buoyant DRM fraction (fraction 4) was identified by the recovery of flotillin-1 (DRM marker), whereas non-DRM heavy fractions (fractions 8–10) contained calnexin (non-DRM marker) (Fig. 4A). The content of APP, APP CTFs, BACE, and PS1 C-terminal fragment (PS1-C) were examined in DRM (fraction 4) and non-DRM (fraction 9) by immunoblotting, and the quantities were standardized to the amount of flotillin-1 (fraction 4) or calnexin (fraction 9), respectively (Fig. 4, B and C). Interestingly, higher levels of mature APP (mAPP) and APP CTFβ (C99 + pC99) were recovered in DRM of the X11L-deficient and the X11/X11L doubly deficient mouse brain, but not in DRM of either the wild-type or the X11-deficient mouse brain (Fig. 4B). No obvious genotype-related changes were observed for the levels of BACE and PS1-C. In contrast to the differential localization of proteins in DRM, the levels of APP, APP CTFs, BACE, and PS1-C were identical in the non-DRM, regardless of any deficiency in X11, X11L, or a double deficiency in both X11 and X11L (Fig. 4C).
FIGURE 4.
Localization of APP and APP CTFβ into DRM in the cortex of wild-type, X11-deficient, X11L-deficient, and X11/X11L doubly deficient mice. A, isolation of DRM of cortex. Post-nuclear supernatant from the wild-type and the deficient mouse (8–12 weeks) brain cortex was fractionated by sucrose density gradient centrifugation. Each fraction was subjected to immunoblot analysis with anti-calnexin (non-DRM marker) and anti-flotillin-1 (DRM marker) antibodies. B and C, proteins in DRM and non-DRM fractions. DRM (B; fraction 4 in A) and non-DRM (C; fraction 9 in panel A) samples (40 μg of proteins) were probed with anti-APP (Sigma product), anti-BACE-Cat1, and anti-PS1-C antibodies. The densities of protein bands were standardized to the density of flotillin-1 for DRM (B) and calnexin for non-DRM (C), which was compared with the ratios for wild-type mice, which were assigned a reference value of 1.0 (values represent the means ± S.E.). The data were analyzed by a one-way analysis of variance followed by the Tukey's test (n = 4; *, p < 0.05; **, p < 0.01). mAPP, mature APP species (N- and O-glycosylated APP695 isoforms); imAPP, immature APP (N-glycosylated APP695 isoform).
Although X11s are cytoplasmic adaptor proteins, we observed that X11s were recovered in membrane fractions (P100) along with APP (Fig. 5A), and we also found that these membrane-attached X11s were largely localized in non-DRM (Fig. 5B), suggesting that APP in DRM is liberated from attachment to either X11s. Interestingly, only mAPP was recovered in DRM, which agrees with the observation that Aβ is generated in the late secretory pathway (reviewed in Ref. 5) because mAPP (N- and O-glycosylated APP695 isoform) is concentrated in a late Golgi compartment and in the trans-Golgi network. Furthermore, APP-CTFs in DRM was largely CTFβ (C99 + pC99) (Fig. 5B). Taken together, these results indicate that mature holoAPP associates with X11s in non-DRM, resulting in stabilization of the APP. Thus, association of APP with X11s prevents amyloidogenic cleavage of APP by BACE, which is active in DRM. In the absence of X11s, more APP enters into DRM and is subjected to β-site cleavage by BACE (Fig. 6). These results indicate that X11s function to anchor APP out of DRM and reduce the frequency of APP to encounter with BACE. Present observations suggest that the dysfunction of X11s enhances the translocation of APP into DRM where amyloidogenic metabolism of APP is facilitated.
FIGURE 5.
Membrane-attached X11 proteins in DRM and non-DRM fractions. A, distribution of X11s in membrane fraction of the cortex of wild-type mice (12 weeks) brain. The samples (10 μg protein) of post-nuclear supernatant (PNS), a cytosolic fraction (S100) and a membrane fraction (P100) were immunoblotted with anti-X11 (UT153), anti-X11L (BD Bioscience), anti-X11L2 (BD Bioscience), anti-APP (Sigma), and anti-actin antibodies. B, distribution of membrane anchoring X11 proteins, APP, and APP CTFs in DRM and non-DRM fractions. The samples (10 μg of protein) of DRM prepared from P100 fraction, non-DRM prepared from P100 fraction, and P100 fractions of the cortex of wild-type mice (12 weeks) brain were probed with antibodies described above. mAPP, mature APP species; imAPP, immature APP.
FIGURE 6.
Possible role of X11 proteins in regulating DRM-association and β-site cleavage of APP. X11s associate with APP outside of DRMs and prevent translocation of APP into DRM. When X11L dissociates from APP, the APP translocates into DRMs, and that fraction of APP molecules is cleaved by BACE which is active in DRM (upper panel). In the absence of X11s, APP molecules are not anchored outside of DRMs, and more APP translocates into DRMs, resulting in increased β-site cleavage of APP (lower panel). The arrows indicate translocation direction of APP.
DISCUSSION
The X11 proteins (X11s) were identified as APP-binding proteins, and the association of APP with X11s stabilizes holoAPP and suppresses APP metabolism including Aβ generation in cells (17–19, 30, 38, 39). Transgenic mice that overexpress Swedish mutant APP and X11 or X11L show lower levels of Aβ in their brains as compared with transgenic mice overexpressing Swedish mutant APP alone (20, 21). Moreover, X11L-deficient mice show enhanced amyloidogenic metabolism of endogenous brain APP (22). These observations indicate that X11 family proteins, especially X11 and X11L (both of which are expressed primarily in neurons) play important roles in the regulation of amyloidogenic metabolism of APP. In addition to the generation of X11L-deficient mice (22), X11-deficient mice and X11/X11L/X11L2 triple-deficient mice have been generated (23, 28, 40), but analysis of APP metabolism has not been performed in these lines.
In this study, we focused on the APP metabolism in mice lacking the expression of X11 and X11L. Although an analysis of endogenous APP metabolism in X11L-deficient mouse brain indicates that X11L suppresses β-cleavage of APP in vivo (22), the molecular mechanism was still unclear, and the function of endogenous X11 in APP metabolism in brain has not been analyzed heretofore. To elucidate these issues, we generated X11/X11L doubly deficient mice that did not show obvious morphological alterations in brain structures or in expression and localization of several neural proteins, although the body size of the double knock-outs was smaller than normal, as was also observed in the X11/X11L/X11L2 triply deficient mice (28). Along with the analysis of the X11/X11L doubly deficient mice, we analyzed APP metabolism in X11-deficient mice because X11 is expressed in a population of neurons distinct from those expressing X11L. We confirmed that amyloidogenic metabolism of APP was facilitated in brain of the X11-deficient mice, although the contribution of X11 in the suppression of amyloidogenic metabolism of APP in brain was less than that of X11L, because of relatively smaller population of X11-expressing neurons. In conclusion, enhanced amyloidogenic metabolism of APP was observed in the X11/X11L doubly deficient mice brain, suggesting that X11 and X11L function similarly to suppress the β-site cleavage of APP, but each acts in a different neuronal population.
In the present study, we sought to reveal the mechanism of X11 family protein function in amyloidogenic metabolism of APP. We paid special attention to the increase of CTFβ in the knock-out mouse brain and explored alterations in the relationship between X11L and BACE in the knock-out mouse brain and/or neurons cultured from the brains of those mice. Recent reports indicated that the generation of Aβ is facilitated in DRM and that BACE activity is concentrated in DRM (6, 7). Indeed, we detected a significant increase in the levels of APP and CTFβ in DRM of the X11/X11L doubly deficient mice brain. This finding indicates that X11s associate with APP outside of the DRM, and they suppress the translocation of APP into DRM where BACE activity is concentrated. A schematic diagram of X11 family protein functions is depicted in Fig. 6. In the absence of X11s, more APP molecules are recruited into cholesterol- and sphingolipids-enriched DRM and subjected to amyloidogenic cleavages. This phenomenon may be relevant to our understanding of the cause(s) of common sporadic AD, if, e.g. dysfunction of X11s occurs, leading to a weakening of the interaction between APP and X11s. Although the mechanism regulating the interaction of APP with X11s is yet to be fully elucidated, it is worth noting that X11s contain three protein-interaction domains and interact with other proteins through their phosphotyrosine interaction and/or PDZ domains (Refs. 39 and 41, and reviewed in Ref. 42). Therefore, we cannot rule out the possibility that X11s suppress amyloidogenic metabolism of APP through their interaction with other proteins. These possibilities not withstanding, the current study shows that one means of suppressing β-cleavage of APP is by facilitating the anchoring of APP outside of DRM by X11s. This is the first report of a molecular mechanism underlying how X11s, which bind to cytoplasmic region of APP, regulate amyloidogenic cleavage of APP in brain, thereby revealing a new potential therapeutic opportunity for lowering Aβ levels by modulating the action of X11s.
Supplementary Material
Acknowledgments
We thank the Otsuka GEN Research Institute (Otsuka Pharmaceutical Co. Ltd., Tokushima, Japan) for the supply of X11 gene knock-out mice.
This work was supported, in whole or in part, by National Institutes of Health Grant P01 AG10491 (to S. G.). This work was also supported by Grants-in-aid for Scientific Research 207900049 (to T. N.) and 20390018 (to T. S.) and 20023001 (to T. S.) from MEXT Japan. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S8.
Y. Saito, Y. Sano, T. Nakaya, and T. Suzuki, unpublished observation.
Footnotes
The abbreviations used are: APP, amyloid β-protein precursor; AD, Alzheimer disease; Aβ, amyloid β-protein; mAPP, mature APP; BACE, β-site-cleaving enzyme; PS, presenilin; CTF, C-terminal fragment; DRM, detergent-resistant membrane; X11s, X11 proteins; X11L, X11-like; PBS, phosphate-buffered saline.
References
- 1.Gandy, S. (2005) J. Clin. Investig. 115 1121-1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vassar, R. (2004) J. Mol. Neurosci. 23 105-114 [DOI] [PubMed] [Google Scholar]
- 3.Selkoe, D. J., and Wolfe, M. S. (2007) Cell 131 215-221 [DOI] [PubMed] [Google Scholar]
- 4.Wolfe, M. S. (2008) Curr. Top. Med. Chem. 8 2-818220927 [Google Scholar]
- 5.Small, S. A., and Gandy, S. (2006) Neuron 52 15-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cordy, J. M., Hussain, I., Dingwall, C., Hooper, N. M., and Turner, A. J. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 11735-11740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ehehalt, R., Keller, P., Haass, C., Thiele, C., and Simons, K. (2003) J. Cell Biol. 160 113-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puglielli, L., Ellis, B. C., Saunders, A. J., and Kovacs, D. M. (2003) J. Biol. Chem. 278 19777-19783 [DOI] [PubMed] [Google Scholar]
- 9.Kalvodova, L., Kahya, N., Schwille, P., Ehehalt, R., Verkade, P., Drechsel, D., and Simons, K. (2005) J. Biol. Chem. 280 36815-36823 [DOI] [PubMed] [Google Scholar]
- 10.Lee, S. J., Liyanage, U., Bickel, P. E., Xia, W., Lansbury, P. T., and Kosik, K. S. (1998) Nat. Med. 4 730-734 [DOI] [PubMed] [Google Scholar]
- 11.Kawarabayashi, T., Shoji, M., Younkin, L. H., Wen-Lang, L., Dickson, D. W., Murakami, T., Matsubara, E., Abe, K., Ashe, K. H., and Youkin, S. G. (2004) J. Neurosci. 24 3801-3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wada, S., Morishima-Kawashima, M., Qi, Y., Misono, H., Shimada, Y., Ohno-Iwashita, Y., and Ihara, Y. (2003) Biochemistry 42 13977-13986 [DOI] [PubMed] [Google Scholar]
- 13.Vetrivel, K. S., Cheng, H., Lin, W., Sakurai, T., Li, T., Nukina, N., Wong, P. C., Xu, H., and Thinakaran, G. (2004) J. Biol. Chem. 279 44945-44954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vetrivel, K. S., Cheng, H., Kim, S., Chen, Y., Barnes, N. Y., Parent, A. T., Sisodia, S. S., and Thinakaran, G. (2005) J. Biol. Chem. 280 25892-25900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urano, Y., Hayashi, I., Isoo, N., Reid, P. C., Shibasaki, Y., Noguchi, N., Tomita, T., Iwatsubo, T., Hamakubo, T., and Kodama, T. (2005) J. Lipid Res. 46 904-912 [DOI] [PubMed] [Google Scholar]
- 16.Miller, C. C., McLoughlin, D. M., Lau, K. F., Tennant, M. E., and Rogeli, B. (2006) Trends Neurosci. 29 280-285 [DOI] [PubMed] [Google Scholar]
- 17.Borg, J. P., Yang, Y. N., De Taddeo-Borg, M., Margolis, B., and Turner, R. S. (1998) J. Biol. Chem. 273 14761-14766 [DOI] [PubMed] [Google Scholar]
- 18.Tanahashi, H., and Tabira, T. (1999) Biochem. Biophys. Res. Commun. 255 663-667 [DOI] [PubMed] [Google Scholar]
- 19.Tomita, S., Ozaki, T., Taru, H., Oguchi, S., Takeda, S., Yagi, Y., Sakiyama, S., Kirino, Y., and Suzuki, T. (1999) J. Biol. Chem. 274 2243-2254 [DOI] [PubMed] [Google Scholar]
- 20.Lee, J. H., Lau, K. F., Perkinton, M. S., Standen, C. L., Shemilt, S. J., Mercken, L., Cooper, J. D., McLoughlin, D. M., and Miller, C. C. (2003) J. Biol. Chem. 278 47025-47029 [DOI] [PubMed] [Google Scholar]
- 21.Lee, J. H., Lau, K. F., Perkinton, M. S., Standen, C. L., Rogelj, B., Falinska, A., McLoughlin, D. M., and Miller, C. C. (2004) J. Biol. Chem. 279 49099-49104 [DOI] [PubMed] [Google Scholar]
- 22.Sano, Y., Syuzo-Takabatake, A., Nakaya, T., Saito, Y., Tomita, S., Itohara, S., and Suzuki, T. (2006) J. Biol. Chem. 281 37853-37860 [DOI] [PubMed] [Google Scholar]
- 23.Mori, A., Okuyama, K., Horie, M., Taniguchi, Y., Wadatsu, T., Nishino, N., Shimada, Y., Miyazawa, N., Takeda, S., Niimi, M., Kyushiki, H., Kondo, M., and Mitsumoto, Y. (2002) Neurosci. Res. 43 251-257 [DOI] [PubMed] [Google Scholar]
- 24.Zhao, J., Fu, Y., Yasvoina, M., Shao, P., Hitt, B., O'Connor, T., Logan, S., Maus, E., Citron, M., Berry, R., Binder, L., and Vassar, R. (2007) J. Neurosci. 27 3639-3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oishi, M., Nairn, A. C., Czernik, A. J., Lim, G. S., Isohara, T., Gandy, S. E., Greengard, P., and Suzuki, T. (1997) Mol. Med. 3 111-123 [PMC free article] [PubMed] [Google Scholar]
- 26.Araki, Y., Kawano, T., Taru, H., Saito, Y., Wada, S., Miyamoto, K., Kobayashi, H., Ishikawa, H. O., Ohsugi, Y., Yamamoto, T., Matsuno, K., Kinjyo, M., and Suzuki, T. (2007) EMBO J. 26 1475-1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sawamura, N., Morishima-Kawashima, M., Waki, H., Kobayashi, K., Kuramochi, T., Frosch, M. P., Ding, K., Ito, M., Kim, T. W., Tanzi, R. E., Oyama, F., Tabira, T., Ando, S., and Ihara, Y. (2000) J. Biol. Chem. 275 27901-27908 [DOI] [PubMed] [Google Scholar]
- 28.Ho, A., Morishita, W., Atasoy, D., Liu, X., Tabuchi, K., Hammer, R. E., Malenka, R. C., and Sudhof, T. C. (2006) J. Neurosci. 26 13089-13101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ouimet, C. C., Baerwald, K. D., Gandy, S. E., and Greengard, P. (1994) J. Ciomp. Neurol. 348 244-260 [DOI] [PubMed] [Google Scholar]
- 30.McLoughlin, D. M., Irving, N. G., Brownlees, J., Brion, J.-P., Leroy, K., and Miller, C. C. J. (1999) Eur. J. Neurosci. 11 1988-1994 [DOI] [PubMed] [Google Scholar]
- 31.Liu, K., Doms, R. W., and Lee, V. M. (2002) Biochemistry 41 3128-3136 [DOI] [PubMed] [Google Scholar]
- 32.Suzuki, T., and Nakaya, T. (2008) J. Biol. Chem. 283 29633-29637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sano, Y., Nakaya, T., Pedrini, S., Takeda, S., Iijima-Ando, K., Iijima, K., Mathews, P. M., Itohara, S., Gandy, S., and Suzuki, T. (2006) PLoS ONE 1 e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakaya, T., and Suzuki, T. (2006) Genes Cells 11 633-645 [DOI] [PubMed] [Google Scholar]
- 35.Oishi, M., Nairn, C. A., Czernik, J. A., Lim, S. G., Isohara, T., Gandy, S., Greengard, P., and Suzuki, T. (1997) Mol. Med. 3 111-123 [PMC free article] [PubMed] [Google Scholar]
- 36.Ciufo, L. F., Barclay, J. W., Burgoyne, R. D., and Morgan, A. (2005) Mol. Biol. Cell 16 470-482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iwata, N., Mizukami, H., Shirotani, K., Takaki, Y., Muramatsu, S., Lu, B., Gerard, N. P., Gerard, C., Ozawa, K., and Saido, C. T. (2004) J. Neurosci. 24 991-998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mueller, H. T., Borg, J. P., Margolis, B., and Turner, R. S. (2000) J. Biol. Chem. 275 39302-39306 [DOI] [PubMed] [Google Scholar]
- 39.Araki, Y., Tomita, S., Yamaguchi, H., Miyagi, N., Sumioka, A., Kirino, Y., and Suzuki, T. (2003) J. Biol. Chem. 278 49448-49458 [DOI] [PubMed] [Google Scholar]
- 40.Ho, A., Morishita, W., Hammer, R. E., Malenka, R. C., and Sudhof, T. C. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 1409-1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Araki, Y., Miyagi, N., Kato, N., Yoshida, T., Wada, S., Nishimura, M., Komano, H., Yamamoto, T., De Strooper, B., Yamamoto, K., and Suzuki, T. (2004) J. Biol. Chem. 279 24343-24354 [DOI] [PubMed] [Google Scholar]
- 42.Rogeli, B., Mitchell, J. C., Miller, C. C., and McLoughlin, D. M. (2006) Brain Res. Rev. 52 305-315 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.