Summary
The MtrC-MtrD-MtrE efflux pump system confers resistance to macrolide antibiotics and antimicrobial substances of the host innate defense. Clinical isolates with increased resistance to erythromycin and azithromycin frequently harbor mutations in the mtrR structural gene, which encodes a repressor of the mtrCDE operon, or the mtrR promoter region. The MtrC-MtrD-MtrE system is important for gonococcal survival in the murine genital tract, and derepression of the mtrCDE operon via deletion of mtrR confers increased fitness in vivo. Here we compared isogenic strains with naturally occurring mtrR locus mutations for differences in mtrCDE expression and pump-related phenotypes. Mutations upstream of mtrC, including those within the MtrR binding region and a novel mutation that increases mtrC RNA stability conferred the highest levels of derepression as measured by mtrCDE transcription and resistance to antibiotics, progesterone, and antimicrobial peptides. In contrast, mutations within the mtrR coding sequence conferred low to intermediate levels of derepression. In vivo, the mtr mutants were more fit than the wild type strain, the degree to which paralleled in vitro resistance gradients. These studies establish a hierarchy of mtrR locus mutations with regard to regulation of pump efflux, and suggest selection for more derepressed mutants may occur during mixed infections.
Keywords: Neisseria gonorrhoeae, active efflux, MtrC-MtrD-MtrE efflux pump, antibiotic resistance, in vivo fitness, antimicrobial peptides, genital tract
Introduction
Gonorrhea is the second most commonly reported infection in the United States (Jajosky et al., 2006) and it occurs at a high incidence in the developing world with an estimated 62 million annual cases worldwide (Gerbase et al., 1998). Neisseria gonorrhoeae is primarily a mucosal pathogen of the lower urogenital tract. The main site of infection is the cervix and the urethra in men. The female urethra is also often infected and rectal and pharyngeal infections are common in both genders. Lower genital tract infections are usually uncomplicated and often asymptomatic, particularly in women. Ascension to the upper reproductive tract results in more serious disease, including epididymitis, endometritis, salpingitis, and pelvic inflammatory disease (PID). Since no vaccine currently exists for N. gonorrhoeae, antibiotic therapy is a primary measure of infection control. The emergence of antibiotic resistant strains, however, continually challenges the effectiveness of antibiotics as a control strategy as exemplified by the recent removal of fluoroquinolones from the recommended therapy for gonorrhea (CDC, 2004).
A thorough understanding of gonococcal antibiotic resistance mechanisms is critical for the development of new and effective antimicrobial agents. Mechanisms of antibiotic resistance in N. gonorrhoeae include mutation of the antibiotic target, enzymatic breakdown of the antibiotic, decreased permeability of the bacterial membrane, the presence of active efflux systems, and derepression of transmembrane efflux through mutation of pump repressor genes [reviewed in (Alekshun et al., 2007)]. The MtrC-MtrD-MtrE efflux pump system is one of four known efflux pumps of N. gonorrhoeae (Hagman et al., 1995a, Lee et al., 1999, Pan et al., 1994, Rouquette-Loughlin et al., 2003, Rouquette-Loughlin et al., 2005). The MtrC-MtrD-MtrE efflux pump exports macrolide antibiotics and is implicated in high level penicillin resistance (Veal et al., 2002). We previously reported that MtrC-MtrD-MtrE-deficient mutants were highly attenuated in a female mouse model of lower genital tract infection. In contrast, mutants in the FarA-FarB-MtrE efflux pump, which are hypersusceptible to long chain fatty acids were not attenuated in this model (Jerse et al., 2003). These results are consistent with the hypothesis that the MtrC-MtrD-MtrE efflux pump system, which predates the clinical use of antibiotics, protects the gonococcus from antimicrobial substances that may be encountered during genital tract infection. Host substrates that may challenge the gonococcus include bile salts (Delahay et al., 1997, Hagman et al., 1995a), progesterone (Jerse et al., 2003), and LL37, a host-derived antimicrobial peptide found in the human genital tract (Shafer et al., 1998).
The MtrC-MtrD-MtrE pump is a member of the resistance-nodulation-division (RND) family of efflux pumps, and like other homologous pumps, is under the tight control of both a repressor, MtrR (Hagman et al., 1995b) and an activator MtrA (Rouquette et al., 1999). The mtrR gene is located 250-bp upstream and is divergently transcribed from the mtrCDE operon (Pan et al., 1994). The 24.5 kDa MtrR protein is a TetR-type repressor and negatively controls mtrCDE expression by the binding of two MtrR homodimers to pseudo-direct repeats within the mtrCDE promoter (Hoffmann et al., 2005); this DNA-binding action is similar to the QacR repressor that controls efflux pump gene expression in Staphylococcus aureus (Schumacher et al., 2002). A helix-turn-helix (HTH) DNA binding motif exists between residues 32-53 and missense mutations that cause radical amino acid replacements at residue 39 (A39T) or 45 (G45D) can enhance gonococcal resistance to hydrophobic antimicrobials presumably because they abrogate MtrR binding to the target DNA upstream of mtrCDE (Shafer et al., 1995, Hagman et al., 1995b). Other mutations that cause radical amino acid replacements in the center of coding sequence (H105Y) or the C-terminal domain can also impact MtrR function, possibly by altering MtrR multimer formation.
Strains that bear mtr mutations have been recovered in a variety of outbreak investigations. The most common mutation is a single base pair deletion located in the inverted repeat that is between the -10 and -35 hexamers of the mtrR promoter (Cousin et al., 2004, Dewi et al., 2004, Lucas et al., 1995, Ng et al., 2002, Tanaka et al., 2006, Xia et al., 2000, Zarantonelli et al., 1999, Zarantonelli et al., 2001) Other commonly described mutations are located in the structural gene of mtrR, and include a G45D mutation (Shafer et al., 1995, Tanaka et al., 2006, Dewi et al., 2004, Vereshchagin et al., 2004, Zarantonelli et al., 1999) and an A39T mutation (Dewi et al., 2004). While all of these mutations confer increased resistance to hydrophobic antimicrobials, differences exist with respect to the level of resistance afforded by the given mutation (Hagman et al., 1995a, Shafer et al., 1995). Thus, the missense mutations in the mtrR coding sequence typically result in a low to intermediate level of resistance while the promoter mutation affords high levels of resistance. Hagman and Shafer (Hagman et al., 1995b) proposed that the promoter mutation abrogates mtrR transcription. In the promoter mutant mtrCDE transcription is elevated because MtrR is absent and RNA polymerase is better able to interact with the mtrCDE promoter that partially overlaps with the mtrR promoter at the -35 region (Lucas et al., 1997).
While the molecular mechanisms by which the mtrCDE efflux pump-encoding operon is regulated at the level of transcription are well characterized, little information is available regarding the biologic significance of the most common mtrR mutations found among clinical isolates. Additionally, while an mtrR deletion mutant demonstrated increased fitness in the lower genital tract of female mice as predicted by the demonstrated importance of the MtrC-MtrD-MtrE pump in vivo (Warner et al., 2007), the impact of naturally occurring mtr mutations on in vivo fitness has not been tested. Accordingly, here we compared five different mtr mutations found in clinical isolates or vaginal isolates from experimentally infected mice for differences in mtrCDE operon derepression as measured by levels of RNA transcript, protein production, and resistance to erythromycin (Em), azithromycin (Az), and the non-ionic detergent Triton X-100 (TX-100), all of which are substrates for the MtrC-MtrD-MtrE efflux pump. We also evaluated the consequence of each mutation on resistance to progesterone and the murine cathelicidin-related antimicrobial peptide (CRAMP-38), and on gonococcal fitness during experimental murine genital tract infection. Additionally, we report that the frequently used laboratory strain MS11 is a natural mtr mutant that harbors two mtr locus mutations, including a novel mutation that results in increased mtrCDE transcript stability. As with the mutated mtr loci amplified from clinical isolates, we show here that the mtr locus of strain MS11 confers increased resistance to CRAMP-38 and other substrates of the MtrC-MtrD-MtrE efflux pump system, including the human cathelicidin LL-37.
Results
Construction of a series of mtr locus mutations in strain FA19SmR
Investigations of clinical outbreaks have shown that gonococcal isolates with increased resistance to Em, Az, and penicillin frequently harbor naturally occurring mutations in the mtrR gene or its upstream region. Here we created a series of mutants in strain FA19, which represent the majority of natural promoter and structural gene mutants that have been reported in the mtr loci of clinical isolates (Table 1). The FA19 strain used was a spontaneous streptomycin resistant mutant, which is a phenotype that is required for mouse infection studies. A schematic that shows the position of each mutation within the mtr locus is shown in Figure 1. Mutant strain KH15 is a transformant of strain FA19SmR that contains a T deletion on the end of the 13-bp inverted repeat within the mtrR promoter that is adjacent the MtrR-binding region that encompasses the mtrCDE promoter, and was previously described (Lucas et al., 1997). The A39T and G45D mutations, which are in the HTH DNA-binding domain of MtrR (Hoffmann et al., 2005, Lucas et al., 1997, Pan et al., 1994), were amplified from the urogenital isolates LG7 and LG5, respectively (McKnew et al., 2003, Garvin et al., 2008) and transformed into FA19SmR to create strains DW39 and DW45, respectively. We also included mutant DW9 in our survey, which has a one base pair change that results in a glycine residue instead of glutamic acid (E202G) near the C-terminus of the MtrR protein. The E202G mutation was originally identified in a spontaneous EmR mutant that was isolated from experimentally infected mice (Warner et al., 2007). Mutant JF1 carries an internal deletion within mtrR (Folster et al., 2005) and was used for comparisons with mutants that carry naturally occurring mtr mutations.
Table 1. Bacterial strains used in this study.
| Strain1 | Relevant Genotype | Reference |
|---|---|---|
| FA19SmR | Parent strain | (Jerse et al., 2003) |
| KH15 | -T at MtrR binding site (mtr-79) | (Hagman et al., 1995b) |
| DW39 | mtrRA39T (DNA-binding region) | This study |
| DW45 | mtrRG45D (DNA-binding region) | This study |
| DW9 | mtrRE202G | (Warner et al., 2007) |
| JF1 | ΔmtrR | (Folster et al., 2005) |
| MS11 | Wild type strain, mtr120, mtrRA39T | (Swanson et al., 1988) |
| DW120 | FA19SmR mtr120 | This study |
| FA19MS11mtr | FA19SmR mtr120, mtrRA39T | This study |
| JF3 | mtrA∷aphA3 | (Rouquette-Loughlin et al., 2002) |
| DW120A | FA19SmR mtr120, mtrA∷aphA3 | This study |
| JF1 mtr120 | mtr120, ΔmtrR | This study |
| DW3 | mtrE∷cat | (Warner et al., 2007) |
| DW3MS11 | Strain MS11 with mtrE∷cat | This study |
| FA19CmR | cat gene inserted between aspC and lctP loci | (Warner et al., 2007) |
| DW39CmR | mtrRA39T and cat gene inserted between aspC and lctP loci | This study |
| DW45CmR | mtrRG45D and cat gene inserted between aspC and lctP loci | This study |
| DW9CmR | mtrRE202G and cat gene inserted between aspC and lctP loci | This study |
All recombinant strains were constructed strain FA19SmR with the exception of DW3MS11, which was made in strain MS11.
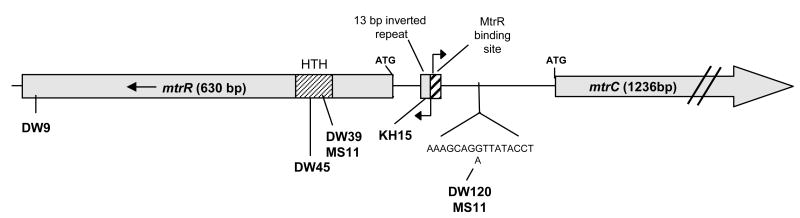
Figure 1. Location of mtr locus mutations used in this study.
A schematic of the N. gonorrhoeae mtr locus is shown. The α-helix-encoded region (HTH) of MtrR used for DNA binding is indicated by the hatched pattern. This region is the location of the mutations found in strains DW39, DW45, and MS11. The E202G mutation harbored in strain DW9 is located at the C-terminal end of the MtrR protein, which is hypothesized to be involved in the dimerization of MtrR to itself. The mtrR and mtrCDE transcriptional start sites are indicated by the arrows and are within the intergenic region that contains the MtR binding site and the 13 bp inverted repeat, which is the region where the mutation in strain KH15 is found (Hagman et al., 1995b). The mtr120 mutation occurs further upstream of the DNA binding region and is present in strains MS11 and DW120. This mutation has not been described previously, and the G to A change is shown in the detailed DNA sequence.
We also included the commonly used laboratory strain MS11 in our analysis. Strain MS11 has been used in numerous pathogenesis studies including experiments with male volunteers (Ramsey et al., 1995, Schmidt et al., 2001, Schneider et al., 1991, Swanson et al., 1987b). Based on the observation in our laboratory that MS11 bacteria exhibit higher levels of in vitro resistance to TX-100, Em, and progesterone than strain FA19, we hypothesized that the MS11 strain may contain one or more mtr locus mutations. Sequence analysis of the MS11 mtr locus revealed the A39T mutation in the MtrR DNA binding motif and an adenine to guanine transition located between the mtrR and mtrC start codons (Fig. 1); this single base pair mutation is in the noncoding region of the mtrC transcript as it is positioned 120-bp upstream of the mtrC start codon and 42-bp downstream of the transcriptional start. This mutation (mtr120) has not been described previously among clinical isolates; however, we have isolated the mtr120 mutation when measuring the rate of spontaneous Em resistance in strain FA19 in vitro (data not shown). Here we moved the mtr120 mutation into strain FA19 to create mutant DW120. The capacity of each of these mutations to modulate levels of mtrCDE expression, resistance to antimicrobial substances and in vivo fitness is described below.
Natural mtr mutations confer differential levels of antibiotic resistance
Hagman and Shafer (1995) previously reported that the mtr mutant strain KH15 displays increased levels of antimicrobial resistance compared to an mtrR deletion mutant (Hagman et al., 1995a, Shafer et al., 1995). As a first step towards further phenotypic characterization of these naturally occurring mutations, we determined the MICs for three substrates of the MtrC-MtrD-MtrE efflux pump (TX-100, Em, and Az) against our collection of mtr locus mutants. Km was used as a non-efflux pump substrate control. Based on the MIC for Em, we observed a stepwise gradient of resistance that could be defined by three classes of resistant strains (Table 2). Compared to drug-susceptible wild type strain FA19, FA19-based mutant strains DW9 (E202G change in MtrR), JF1 (mtrR deletion), and DW45 (G45D change in MtrR) showed only a slight (two-fold), but reproducible increase in Em resistance. A 2-fold increase in the MIC of Az was also exhibited by these strains; resistance to TX-100 was 2-fold (DW9) and 4-fold higher (JF1, DW45) than that of the wild type bacteria. The next level of resistance (intermediate) was defined as a 4-fold increase in the MIC of Em, and was exhibited by mutant DW39 (A39T change in MtrR). The third group of mutants, KH15 and DW120, displayed the highest levels of resistance to Em (16-fold increase). Interestingly, strains KH15 and DW120 both carry mutations outside of the MtrR structural gene. The mutation in strain KH15 is a -T mutation located 79-bp upstream of the mtrR start codon but within the mtrR promoter, and is one of the most commonly described mutations isolated in clinics. The mtr120 mutation in strain DW120, which is carried by laboratory strain MS11, is novel because it is positioned 131-bp downstream from the MtrR-binding site (Lucas et al., 1997), and is predicted to be within the mtrCDE transcript. Mutants KH15 and DW120 also exhibited levels of resistance to Az and TX-100 that were notably higher than the other strains, with increases in MIC of 8-fold and 256-fold, respectively. We tried to construct a mutant that carried both the mtrR-79 mutation in KH15 and the mtr120 mutation by transforming strain DW120 with the appropriate PCR fragment from KH15 and selecting on media with 16 μg/ml Em. No transformants were isolated. This result that suggests each of these mutations confers the highest level of MtrC-MtrD-MtrE-mediated Em resistance that can be achieved.
Table 2. Sensitivity to TX-100 and antibiotic substrates of the MtrC-MtrD-MtrE efflux system.
|
Minimum Inhibitory Concentration MIC (αg/ml) |
|||||
|---|---|---|---|---|---|
| Strain | Genotype | Em | Az | TX-100 | Km |
| FA19 SmR | wild type | 0.5 | 0.125 | 62 | 30 |
| DW9 | FA19 mtrRE202G | 1 | 0.25 | 125 | 30 |
| JF1 | FA19 ΔmtrR | 1 | 0.25 | 250 | 30 |
| DW45 | FA19 mtrRG45D | 1 | 0.25 | 250 | 30 |
| DW39 | FA19 mtrRA39T | 2 | 0.5 | 500 | 30 |
| FA19 MS11mtr | FA19 mtr120, mtrRA39T | 8 | 0.5 | 8000 | 30 |
| DW120 | FA19 mtr120 | 8 | 1 | >16000 | 30 |
| KH15 | FA19 mtrR-79 | 8 | 1 | >16000 | 30 |
| MS11 | wild type; natural mtr120, mtrRA39T mutant | 8 | 1 | >16000 | 30 |
| DW3MS11 | MS11 mtr120, mtrRA39T, mtrE∷Cm | 0.04 | ND | 0.24 | 30 |
| DW3 | FA19 mtrE∷Cm | 0.04 | ND | 0.24 | 30 |
| DW120A | FA19 mtr120, mtrA∷aphA-3 | 8 | 1 | >16000 | 60 |
| JF1mtr120 | FA19 mtr120, mtrRA39T, ΔmtrR | 8 | 1 | >16000 | 30 |
ND, not determined
Analysis of the mtr locus in strain MS11
To further characterize the mtr locus of strain MS11, we moved the entire mutated mtr sequence of strain MS11 into strain FA19SmR. This was accomplished by transformation of a PCR product that was generated using primers that anneal 48-bp after the translational stop site of mtrR and 391-bp within the mtrC gene as described in the Experimental Procedures. The resultant 1355-bp PCR product carried both the A39T and mtr120 mutations. The resistance phenotypes of a representative transformant (FA19MS11mtr) established the genetic linkage between the MS11 mtr locus and resistance to Em, Az, and TX-100 (Table 2). Mutant DW120, which carries the mtr120 mutation in the FA19SmR background as described above, showed increased resistance to Em at a level that was comparable to that exhibited by strain FA19MS11mtr, which carries both mutations and a modest (two-fold) increase in Az and TX-100 resistance. To confirm the role of the MtrC-MtrD-MtrE efflux pump in the increased resistance phenotype exhibited by strain MS11, we introduced an mtrE mutation from plasmid pCR-mtrE (Warner et al., 2007) into strain MS11, and compared the resistance phenotypes of the resultant strain (DW3MS11) and an mtrE mutant of strain FA19 (DW3). Disruption of the mtrE gene in strain MS11, which encodes the outer membrane protein channel of the pump (Delahay et al., 1997) conferred levels of Em and TX-100 resistance that were equal to that of the FA19 mtrE mutant DW3 (Table 2).
Effect of mtr Locus Mutations on mtrCDE Expression and mRNA half-life
To verify that the mtr mutations described above impacted levels of the MtrC-MtrD-MtrE efflux pump, we next determined whether differences exist in the amount of the MtrE protein produced by our test strains. We detected increased levels of MtrE for all mtr mutant strains compared to wild type strain FA19 by immunoblot analysis of outer membrane proteins, and in general, the results reflected the gradient (Fig. 2). DW9 and JF1 gonococci had the lowest increase in band intensity (a 1.8- and 1.94-fold difference relative to the wild type strain), and both strains are at the low end of the gradient as defined by Em MIC. Strains DW45 and DW11 are classified as low and intermediate strains, respectively, and had 2.0-fold increases in MtrE compared to wild type gonococci. Mutants DW39 (intermediate) and DW120 and KH15 (both high) had band intensities that were 2.12- to 2.15-fold greater than the wild type strain. The one strain for which MtrE levels did not correspond well to the Em MIC was FA19MS11mtr, which had a 1.82-fold increase in band intensity, and was thus similar to that of mutants in the low end of the MIC gradient.
Figure 2. Expression of MtrE by wild type and mtr mutant bacteria.
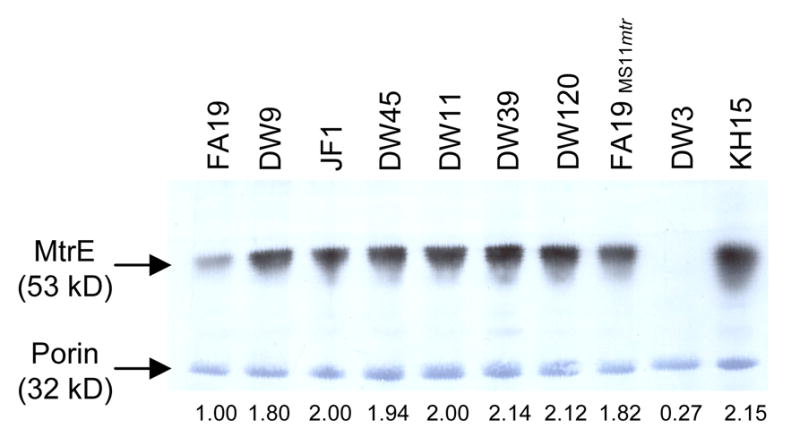
Outer membrane proteins were separated by SDS-PAGE gel electrophoresis and transferred to a PVDF membrane. The 53-kDa MtrE protein was detected by a rabbit MtrE-specific polyclonal antibody described in the Materials and Methods. With the exception of mtrE mutant DW3, protein samples were loaded in ascending order of the levels of antibiotic resistance. The 32-kDa porin protein is constitutively expressed in N. gonorrhoeae and was detected by staining the PVDF membrane with amido black to show equal loading of the samples. The intensity of the MtrE band relative to strain FA19 and after normalization to porin is reported for each strain below the blot. Strain DW11, not described here, carries a frame-shift mutation in the mtrR gene, that is predicted to cause a 52 kD truncated MtrR protein (Warner et al., 2007).
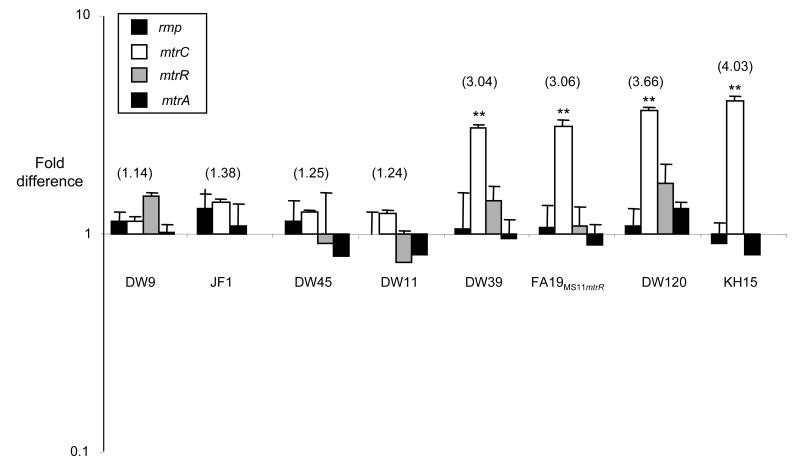
We also used RT-PCR to examine the effect of the mutations on the levels of mtrC, mtrR, and mtrA transcript from each of the mutants (Fig. 3). As reported previously (Hagman et al., 1995b), no mtrR mRNA was detectable in the deletion strain (JF1) or strain KH15 (data not shown). Except for levels of the mtrR transcript produced by strains JF1 and KH15, no significant differences in the expression of the transcriptional activator MtrA or repressor MtrR were detected among mtr mutant and the wild type bacteria. Differences in the level of mtrC transcript, in contrast, were consistent with the observed differences in antibiotic resistance. Mutants DW39, DW120, FA19MS11mtr, and KH15 expressed 3- to 6-fold greater amounts of mtrC mRNA than the wild type bacteria. These strains showed intermediate (DW39) to high (DW120, FA19MS11mtr, and KH15) levels of resistance to Em and Az (Table 2). Mutants DW9, JF1, and DW45 showed 1.1-to 1.4-fold greater amounts of mtrC expression, which is consistent with the lower increases in antibiotic resistance exhibited by these strains. Taken together, these results confirm that the derepressed phenotypes of the mutants are due to increased transcription of the mtrCDE operon and not to changes in the level of mtrA or mtrR transcription. These results also suggest that subtle differences at the mRNA level can alter the functional measures of the phenotype.
Figure 3. Transcriptional analysis of mtrA, mtrR, mtrC, and rmp.
Quantitative RT-PCR was used to assess the fold difference in levels of mRNA compared to the wild type FA19 bacteria. One representative experiment of the three biological replicates tested is shown. There were no significant differences between levels of rmp or mtrA mRNA. mtrR levels were highly down-regulated or non-existent in strains JF1 and KH15 as described previously (Hagman et al., 1995b, Shafer et al., 1995); these values have been omitted to preserve the scale of the figure. Numbers in parentheses denote the fold increase in mtrC levels compared to wild type FA19 bacteria. Levels of mtrC expression were compared using a students t-test to evaluate differences between DW39, DW120, FA19MS11mtr, and KH15 gonococci, which exhibit high levels of mtrC expression, and JF1 bacteria, which demonstrate a low-level increase in MIC and mtrC mRNA. ** Denotes p<0.001. The increase in mtrR transcription shown for strain DW120 was 1.7-fold greater than that of strain FA19, but not statistically significant when the averages from three experiments were compared.
We also found an interesting but subtle difference between the presence of the mtrRA39T and mtrR120 mutations versus the mtrR120 mutation alone. As shown above, FA19MS11mtr gonococci, which carry both mutations, produced less MtrE than expected based on the Em MIC for this strain (Fig. 2). Strain FA19MS11mtr also does not fit squarely into the “high” end of the gradient when Az and TX-100 MICs were evaluated (Table 2). However, the levels of mtrC transcripts were the same for FA19MS11mtr and DW120 bacteria, the latter of which carry only the mtrR120 mutation (Fig. 3). At present we can not readily explain this difference, but we hypothesize there may be regulatory factors that influence transcription or translation of mtrE independently of mtrCD and these are factors are influenced by the presence of one versus two of these mutations.
The location of the mtr120 mutation was of interest because it was positioned downstream of the MtrR binding site (Hoffmann et al., 2005, Lucas et al., 1997). We therefore sought to understand how this mutation increases gonococcal resistance to antimicrobial substances. The transcriptional profile of DW120 showed no difference in the level of mtrR or mtrA compared to the wild type strain, which is consistent with the mutation increasing levels of resistance in an MtrA- and MtrR- independent manner. To further test whether the mtr120 mutation requires MtrR (repressor of mtrCDE) or MtrA (activator of mtrCDE) for its activity, we transformed the mtr120 mutation into strains JF1 and JF3, which are mtrR and mtrA mutants of strain FA19SmR, respectively. The resultant double mutants DW120A (mtrR120, mtrA∷aphA3) and JF1mtr120 (mtr120, ΔmtrR) were tested for sensitivity to Em, Az, and TX-100. DW120A and JF1mtr120 bacteria showed no difference in the MIC of TX-100 or Em compared to DW120 (Table 2). From these results, we conclude that the basis of the increased resistance conveyed by the mtr120 mutation is MtrR- and MtrA-independent. Moreover, the mtrA locus in strain MS11 is identical to that of strain FA19 based on nucleotide sequence analysis (data not shown); this finding further confirms that the resistance phenotype exhibited by wild type MS11 bacteria is not due to differences in the mtrA promoter or structural gene.
Although the mtr120 mutation acted independently of MtrR and MtrA, it does impact mtrCDE transcription (Fig. 3) and causes increased production of MtrE protein (Fig. 2). The mtr120 mutation is located downstream of the MtrR-binding site and the mtrCDE promoter and is within the 5′ untranslated region of the mtrC RNA. The position of this mutation coupled with the detection of high levels of mtrC transcript in this strain in the absence of changes in the expression of known regulators of the mtrCDE operon suggested that the mutation impacts mtrC mRNA transcript stability. Indeed, a similar mutation in the 5′ untranslated region of an efflux pump gene was described for the NorB efflux pump of Staphylococcus aureus, which led to an increase in norA mRNA half-life (Fournier et al., 2001). Therefore, we tested whether there was a difference in the mtrC mRNA half-life between mutant DW120 and the parent strain FA19. We found a rapid decay in the mtrC transcript in wild type bacteria but not in DW120 bacteria compared to that of the constitutively expressed gene rmp (Fig. 4). This result suggests the mtr120 mutation alters the structure of the mtrC transcript or its interaction with either RNA degradation factors or the ribosome to allow for a longer mRNA half-life.
Figure 4. Assessment of RNA decay in strain DW120.
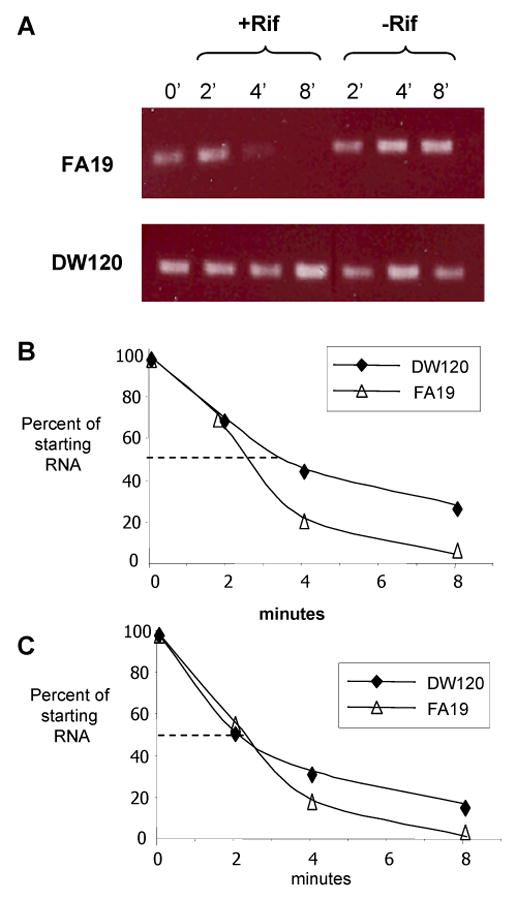
(A) Equal quantities of RNA from strain FA19 (top) and DW120 (bottom) were reverse transcribed and used in a PCR reaction to determine if there was a difference in the degradation rate of mtrC message. Samples were taken pre (T0) and post rifampicin treatment (+Rif); a second set of cultures to which no rifampicin was added (-Rif) were grown in parallel. (B) RNA samples were also reverse-transcribed and used in a qRT-PCR protocol to quantify amounts of mtrC and rmp RNA from strains FA19 and DW120. These values were used to calculate the RNA half life for mtrC and rmp. The experiment was performed twice and the results were similar.
Gradients of resistance to antimicrobial peptides and progesterone
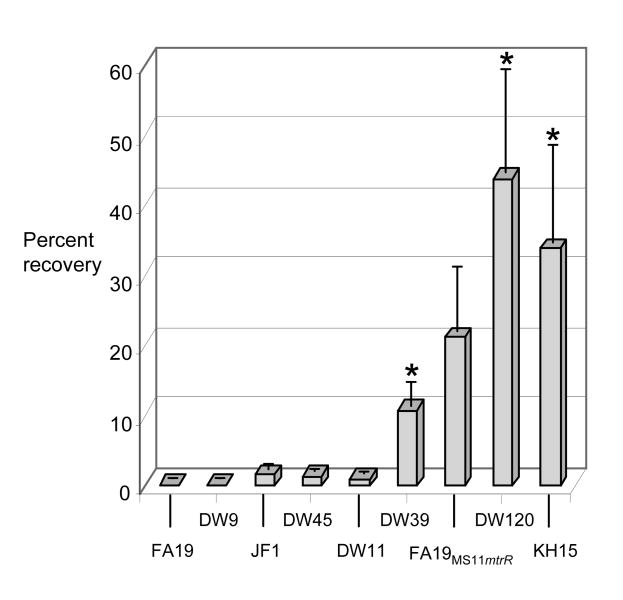
Loss of MtrR production due to an mtrR null mutation is known to increase both levels of the mtrCDE transcript and the in vivo fitness of gonococci in a murine vaginal infection model (Warner et al., 2007). While there are various potential explanations for the in vivo fitness advantage conveyed by increased production of the MtrC-MtrD-MtrE efflux pump, we chose to examine resistance levels to progesterone and CRAMP-38, the murine homologue of the human cathelicidin LL-37 (Nizet et al., 2001) as two host-derived substrates that may challenge N. gonorrhoeae in vivo. We found mutants DW39, DW120, and KH15 to be significantly more resistant to progesterone than the wild type strain, based on the average percentage of gonococci recovered on GC agar with progesterone versus GC agar alone from three independent experiments (Fig. 5). Consistent with the intragenic mutations conferring a higher level of antimicrobial resistance (Table 2), the percent of DW120 and KH15 bacteria recovered on agar with progesterone was consistently higher than that of DW39 bacteria, although not statistically significant, most likely due to variability in the assay. Similarly, a statistically insignificant but reproducibly higher percentage of FA19MS11mtr bacteria was recovered on agar with progesterone compared to wild type FA19 bacteria; this observation is consistent with the mtr locus of MS11 conferring increased resistance to this substrate and with the lower production of MtrE in this strain compared to strain DW120 as shown above.
Figure 5. Progesterone resistance.
Suspensions of wild type and mutant gonococci were quantitatively cultured on GC agar plates supplemented with progesterone (35 μg/ml) or without progesterone and incubated overnight. Results are expressed as 100 × (# CFU on GC agar with progesterone divided by # CFU on GC alone). The average % recovery calculated from three independent experiments is shown; bars represent the standard error. Asterisks indicate a significant difference between DW39 (p < 0.05) or DW120 and KH15 (p < 0.01) (unpaired t test). The p value for FA19MS11mtr versus wild type FA19 was 0.068.
Differences were also found in the sensitivity of the mtr mutants to the cathelicidin CRAMP-38, which is known to possess anti-gonococcal activity in vivo. All mtr mutant bacteria, with the exception of strain DW9, were consistently more resistant to CRAMP-38 compared to wild type FA19SmR gonococci (Fig. 6A). Consistent with the gradient of resistance as defined by Em resistance, the highly Em resistant mutants KH15 and DW120 were more resistant to CRAMP-38 when compared to intermediate and low level Em resistant strains. We also found that wild type strain MS11 was more resistant to both the human cathelicidin LL-37 and CRAMP-38 than strain FA19 (Fig. 6B). Consistent with the mtr locus of strain MS11 conferring increased resistance to these antimicrobial peptides, transformation of the MS11 mtr region into strain FA19 markedly increased the resistance of FA19 to CRAMP- and LL-37, and resistance to the peptides in strain MS11 was negated by the disruption of the MS11 mtrE gene.
Figure 6. Resistance to antimicrobial peptides.
Bacteria were incubated with LL-37, CRAMP, or no peptides and then quantitatively cultured on GC agar. Results are expressed as the percentage of bacteria recovered from wells without peptide, and the graphs shown are representative of one of two or three experiments. Panels A shows the percent recovery of strain FA19 and mtr mutants incubated in CRAMP-38 (4 μg/ml); panel B shows the percent recovery of FA19SmR, FA19MS11mtr, MS11, and DW3MS11 after incubation in LL-37 or CRAMP-38 (16 μg/ml).
In vivo fitness levels correspond to antimicrobial resistance levels
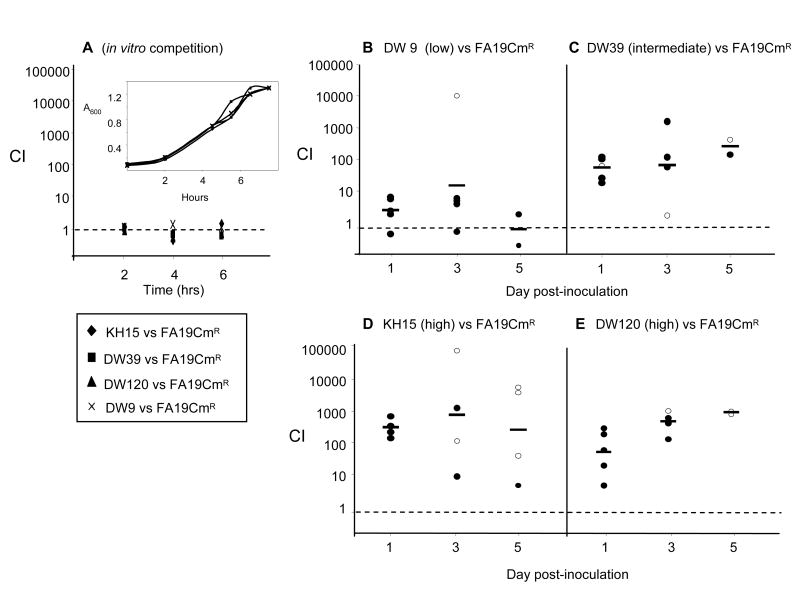
We recently reported that the mtrR deletion mutant JF1 was more fit than the wild type parent bacteria in the lower genital tract of female mice (Warner et al., 2007). JF1 bacteria exhibit a low resistance phenotype in all assays tested here. To better define the resistance gradient as it relates to in vivo fitness, here we tested the fitness of mutants that represent high (KH15 and DW120), intermediate (DW39), and low (DW9) classes of antibiotic resistant strains relative to the wild type parent strain. Competitive infections were performed in which similar numbers of a CmR-marked derivative of wild type strain FA19 and each of the mtr mutant strains were inoculated intravaginally into BALB/c mice. All four mutants demonstrated a fitness advantage over the strain FA19CmR within one day post-inoculation, as defined by CI values greater than 1.0 (Fig. 7B-E). Co-culture of each mutant with strain FA19CmR in GC broth (in vitro competition assays) showed that there were no growth or survival advantages in vitro (Fig. 7A). The mere presence of fitness advantages was not unexpected as this was observed previously with the mtrR deletion mutant JF1 (Warner et al., 2007). These findings are novel, however, because these naturally occurring mutations, which are less disruptive to the mtrR locus, also conferred an advantage over the wild type strain. We also observed a fitness gradient that paralleled the differences in antibiotic resistance. Strain DW9 (E202G change in MtrR), a mutant that showed a 2-fold increase in resistance to Em, TX-100 and Az, displayed mean CIs of 5 and 12 on days 1 and 3, respectively (Fig. 7B). Mutant DW39 (A39T change in MtrR) bacteria, which showed an intermediate level of resistance to Em and Az displayed a stronger advantage over FA19CmR gonococci, with CIs of 100 and 150 on days 1 and 3, respectively (Fig. 7C). Importantly, the intergenic mutations in strains KH15 and DW120, which confer the highest increases in MIC (16-fold for Az and Em, and >250-fold for TX-100), conferred the strongest fitness advantages. Mean CI values for mutant KH15 bacteria were 195 and 509 on days 1 and 3 (Fig. 7D), with 2 of 4 mice clearing FA19CmR bacteria by day 3, and 3 of 4 mice by day 5 (open circles, Fig. 7D). Similarly, strain DW120 displayed mean CI values of 48 and 455 on days 1 and 3 (Fig. 7E), with DW120 bacteria but no FA19CmR gonococci recovered from 1 of 4 mice on day 3, and from 0 of 2 mice on day 5 (open circles, Fig. 7E). In summary, these experiments show that the levels of mtrCDE derepression as measured by antibiotic resistance are mirrored by fitness advantages in vivo.
Figure 7. Differential in vivo fitness of mtr mutants compared to the wild type strain.
Defined ratios of FA19 CmR and mtr mutant bacteria were inoculated into GC broth or estradiol-treated mice to assess the relative fitness of each strain under in vitro and in vivo conditions. The ratio of strains in each inoculum was used in the competitive index (CI) equation as defined in Experimental Procedures. (A) Mixed suspensions were cultured in GC broth (in vitro competition); recovery over the course of growth is expressed as CI. In vivo competition assays between wild type strain FA19CmR and strains (B) DW9, (C) DW39, (D) KH15 and (E) DW120 show different degrees of fitness as measured by CI. Each circle in panels B-E represents the CI from each individual mouse; open circles represent cultures from which no CmR wild type bacteria were recovered. The bars represent the geometric mean of the data, and the dotted line delineates a CI value of 1.0. In cases where a strain was no longer recovered, the limit of detection (4 CFU/100 μl of vaginal wash) was used to calculate the competitive index.
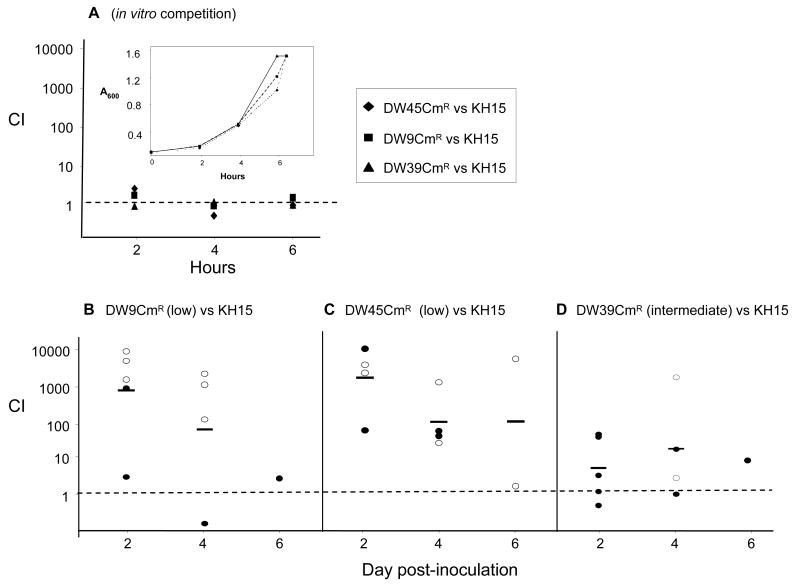
A more sensitive method of detecting a fitness gradient is to perform competitive infections between two mtr mutant strains. Thus, we compared the commonly described and highly resistant promoter mutation found in strain KH15 against the structural gene mutations carried by strains DW39, DW45, and DW9. Strains DW39 and DW45 contain HTH mutations that are common in clinical isolates. Strain DW9, which was isolated as a spontaneous mutant from experimentally infected mice contains a single missense mutation at the 3′-end of the mtrR coding sequence and exhibits a clearly “low” resistance phenotype. To facilitate the testing of these mixtures, we marked strains DW39, DW45, and DW9 with a CmR gene as described in the Experimental Procedures. Mixtures of KH15 and each of the CmR-marked mtrR mutant bacteria were inoculated into mice and the relative recovery of each CmR mutant relative to KH15 bacteria was followed over 6 days. Our hypothesis was that the KH15 mutant would out-compete the other three mutants, but in a step-wise manner with DW9CmR bacteria displaying the most disadvantaged phenotype, followed by DW45CmR and lastly DW39CmR gonococci.
There were no differences in the growth and recovery of each mutant strain versus KH15 bacteria in vitro (Fig. 8A). Mutants DW9CmR and DW45CmR were greatly out-competed by KH15 gonococci, with CI values around 1000 on day 2 (Fig. 8A and 8B). Additionally, high numbers of KH15, but no DW9CmR bacteria were recovered from 3 of 5 mice (day 2) and 3 of 4 mice (day 4) inoculated with a mixture of KH15 and DW9CmR bacteria (open circles, Fig. 8B). Similarly, DW45CmR bacteria were not recovered from 2 of 4 mice on days 2 and 4, and only KH15 was recovered from the two mice that were culture positive on day 6 (open circles, Fig. 8C). Mutant DW39CmR was also less fit than KH15 gonococci; however, as predicted, CI values were markedly lower than that calculated for the other two mutants with a mean value of 12 by day 2 (Fig. 8D). These results confirm our conclusion that the commonly isolated –T promoter mutation carried by mutant KH15 provides a greater in vivo fitness advantage to N. gonorrhoeae than MtrR structural mutations. These data are also consistent with the A39T mutation that is carried by strain DW39 as having an intermediate fitness advantage as predicted from the MIC data.
Figure 8. In vivo competition of mtr mutants.
Defined ratios of mutant KH15 bacteria and CmR-marked mtr mutants were inoculated into GC broth or estradiol-treated mice to compare the relative fitness under in vitro and in vivo conditions. The ratio of strains in the inoculum was used in the competitive index (CI) equation defined below. (A) Mixed suspensions were cultured in GC broth (in vitro competition); recovery over the course of growth is expressed as CI. The inset shows the optical density of the liquid cultures over time. In vivo competition between strains KH15 and (B) DW9CmR (C) DW45CmR and (D) DW39CmR are shown. In panels B-D, each circle represents the CI value for each individual mouse, and open circles signify mice from which only strain KH15 was recovered, and closed circles represent mice from which both strains were recovered. The CI of 1.0 is denoted by a dashed line, while the geometric mean of each distribution is represented by a bar. In cases where a strain was no longer recovered, the limit of detection (4 CFU/100 μl of vaginal wash) was used to calculate the competitive index. The CI was defined as: (KH15/mtrRCmR)output/(KH15/mtrRCmR)input, where mtrRCmR corresponds to strains DW9CmR, DW45CmR, or DW39CmR.
Discussion
Since the discovery of the MtrR repressor by Pan & Spratt (1994) and the realization that it controls mtrCDE expression in N. gonorrhoeae, several studies have examined the impact of mtr mutations on gene expression and antibiotic resistance endowed by the MtrC-MrD-MtrE efflux pump. Presently, investigations of antibiotic resistance outbreaks now include this locus among others in screens for basic resistance determinants. Curiously, in such studies certain mtr mutations are repeatedly isolated. These mutations include missense mutations that cause radical amino acid changes (e.g., A39T or G45D) in the MtrR DNA-binding domain or mutations that impact the C-terminal region of MtrR [e.g., H105Y and the E202G mutation that we isolated from experimentally infected mice (Warner et al., 2007)] (Dewi et al., 2004, Herida et al., 2004, Lundback et al., 2006, Mavroidi et al., 2001, McLean et al., 2004, Shafer et al., 1995, Sutrisna et al., 2006, Tanaka et al., 2006). In general, these coding sequence mutations elevate gonococcal resistance to structurally diverse hydrophobic antimicrobials by 2- to 4-fold. In contrast, promoter mutations are also frequently isolated in the absence of coding sequence mutations, and some enhance antimicrobial resistance by ≥ 10 fold. These promoter mutations consist of a single bp deletion or a dinucleotide insertion in a 13 bp inverted repeat sequence located within the mtrR promoter. These promoter mutations change the spacing between the -10 and -35 hexamers of the mtrR promoter from 17 to 16 or 19 nucleotides and these changes are sufficient to abrogate mtrR transcription (Hagman et al., 1995b, Zarantonelli et al., 1999, Zarantonelli et al., 2001). Moreover, because the divergent mtrR and mtrCDE promoters partially overlap at the -35 regions, the mtrCDE promoter is better recognized by RNA polymerase The +/-T promoter mutations in particular are frequently isolated and known to cause a greater derepression state of the mtrCDE operon than structural mtrR mutants (Hagman et al., 1995b, Shafer et al., 1995).
The goal of the study presented here was to compare the effect of commonly isolated mtrR locus mutations and a novel mutation, mtr120, which we discovered in the laboratory strain MS11 for levels of antibiotic resistance and in vivo fitness. We also tested the relative susceptibility of the mutants to host factors that might challenge the gonococcus during infection. We found that the intergenic mutations conferred the highest level of resistance to antibiotic substrates of the pump and to progesterone and CRAMP-38, and that in general, in vitro resistance phenotypes were predictive of in vivo fitness advantages. Interestingly, mutations within the mtrR structural gene did not fall into one category, but in fact conferred different levels of susceptibility. Among the structural gene mutations, the A39T mutation consistently stood out as having higher in vitro resistances compared to the other three mtrR structural gene mutations. Gonococci that expressed the A39T mutation also competed favorably in vivo with the most fit derepressed mutant, KH15. In contrast, the E202G mutation conferred the lowest levels of in vitro resistance and in vivo fitness. While the crystal structure for MtrR has not yet been determined, it is relevant to note that the A39T mutation is within the first helical domain of the predicted HTH while the G45D mutation is positioned in the second helical domain. It may be that the former mutation has a more significant impact on MtrR-binding to its target site upstream of mtrC. We also confirmed that the mtrR deletion mutant, JF1, is less derepressed for in vitro phenotypes than the promoter mutations as reported by Hagman and Shafer (Hagman et al., 1995b). The mtrR deletion in JF1 was defined in Hagman et al. (Hagman et al., 1995a) and Folster et al. (Folster et al., 2007a), and represents a >90% deletion of the mtrR coding sequence. It is important to note that mutant JF1 has a wild type mtrR promoter that will more effectively compete with the mtrCDE promoter for RNA polymerase binding than the promoter in mutant KH15 to cause a lower level of mtrCDE transcription than that observed for KH15 (Hagman et al., 1995b).
Analysis of mtrC transcript levels provided further evidence of a differential level of mtrCDE expression, and in general the levels of mtrC transcript mimicked that of the MIC values. Our results confirm the previously reported high levels of mtrC transcription and the absence of an mtrR transcript in strain KH15 (Hagman et al., 1995b), however, examination of strains DW39, DW45, DW9, and DW120 provides new insights into the mechanisms of derepression of the mtrCDE operon. Thus, as suggested above, our results predict that the alanine residue at position 39 in the MtrR protein is more important to DNA binding than the glycine residue located six residues away. We also hypothesize that the glutamic acid residue at position 202 plays a minor role in the ability of the repressor to regulate mtrCDE expression, the specifics of which could include the ability of the MtrR proteins to dimerize. Alternatively, this mutation may alter the ability of the MtrR protein to recognize MtrC-MtrD-MtrE substrates at the C-terminal end of the repressor, which has been seen with other TetR repressors (reviewed in (Ramos et al., 2005)), including QacR (Schumacher et al., 2001, Schumacher et al., 2002).
The mtr120 mutation yields one of the highest reported levels of mtr-based resistance. While the mtr120 mutation has not yet been detected among clinical isolates apart from strain MS11, it is important to note that in other efflux pump systems, such as the nor system in Staphylococcus aureus, similar mutations in the 5′ untranslated region yielded derepressed operon transcription (Fournier et al., 2001). Here we present genetic evidence and RT-PCR results to show that the mtr120 mutation is independent of both MtrR and MtrA. Additionally, we showed that the mtr120 mutation conferred an increase in mtrC mRNA half-life, which is a novel mechanism by which increased resistance to MtrC-MtrD-MtrE substrates can occur. The mtr120 mutation suggests a role for RNA stability and the possibility of post-transcriptional regulation in the control of mtrCDE expression. Additional investigation is required to establish why such a mutation has not been described in clinical samples to date.
The impact of the mtr120 mutation on expression of the mtrCDE-encoded efflux pump and levels of resistance and in vivo fitness is of particular interest because it was identified in strain MS11, which has been used extensively in gonococcal research. In particular, MS11 has served as a test strain for understanding mechanisms of pilus variation (Haas et al., 1987, Jonsson et al., 1994, Koomey et al., 1991, Seifert et al., 1988, Swanson et al., 1987a) and transformation (Aas et al., 2002, Chaussee et al., 1998, Hamilton et al., 2005), the kinetics of phase and antigenic variation of surface molecules during experimental urethral infection of male volunteers (Schneider et al., 1991, Swanson et al., 1988, Swanson et al., 1987b), and the host response to experimental urethral infection (Ramsey et al., 1995, Schmidt et al., 2001). Strain MS11 was originally isolated from the cervix of an uncomplicated genital tract infection (Swanson et al., 1988). It now is evident that this strain over-expresses the MtrC-MtrD-MtrE efflux pump system. How derepression of this efflux pump system impacts the biologic activity of strain MS11 is of considerable interest. Interestingly, MS11 bacteria are more infectious than strain FA1090 bacteria based on the ID50 of these strains in the male volunteer urethritis model (Cohen et al., 1994, Schmidt et al., 2001) and in female mice (Jerse, 1999). It is not known whether this increased infectivity is due to the presence of the mtr mutations, since we previously showed that an mtr mutation did not significantly alter the infectious dose of strain FA19 for mice (Warner et al., 2007). However, the results shown here contribute to further understanding and characterization of strain MS11 and emphasize the need to examine how the mtr120 mutation impacts the ability of this strain to cause disease. We note here that repeated attempts to transform the FA19 mtr locus into strain MS11 were unsuccessful, which suggests the mtr120 mutation (and possibly the A39T mutation) may have been selected in this laboratory strain to compensate for a separate mutation that negatively affects its fitness in vitro or in vivo.
An additional objective of this study was the investigation of how these mtr mutations affect fitness. We previously reported that the MtrC-MtrD-MtrE pump is critical for experimental murine genital tract infection (Jerse et al., 2003) and that mutant JF1 (ΔmtrR) exhibits increased in vivo fitness when compared to wild type parent strains (Warner et al., 2007). Results here show that naturally occurring mtr mutations demonstrate a gradient of fitness advantage in vivo. Strain KH15, which bears a common promoter mutation, strongly out-competed all other mtr mutants tested by 1,000-fold except for gonococci that bear the A39T mutation, which showed only a 12-fold reduction in fitness compared to KH15 bacteria. Such a hierarchy in fitness could allow selection for more efficient pathogens in cases of mixed infections with more than one N. gonorrhoeae strain, which occur frequently (Lynn et al., 2005). It seems reasonable to also hypothesize that the increased fitness afforded to mtr mutants could allow for the accumulation of other mutations that compensate for detrimental effects that may occur due to changes in the expression of mtrCDE and other MtrR-regulated genes (Folster et al., 2007b). Modulation of mtrCDE expression appears to be the main reason the mtrR mutant JF1 was more fit in the mouse model based on competitive infection experiments between an mtrR, mtrE double mutant and an mtrE mutant (Warner et al., 2007). However, MtrR can transcriptionally activate and repress over 70 genes, directly or indirectly, and some of these gene products may be of importance (Folster et al, unpublished).
In summary, these findings add to the growing body of literature that support a role for certain RND-type pumps in Gram-negative pathogens beyond the antibiotic resistance phenotype for which they were discovered (Rouquette-Loughlin et al., 2003, Bina et al., 2001, Nishino et al., 2006, Stone et al., 1995). We propose that the MtrC-MtrD-MtrE efflux pump evolved to protect the pathogen from innate immune effectors such as antimicrobial peptides. Increased resistance to CRAMP-38 may explain in part the basis for the observed fitness advantage in mice. The mutants also showed increased resistance to progesterone (Jerse et al., 2003); however, it seems less likely that resistance to progesterone is responsible for our in vivo observations because progesterone concentrations in the lower genital tract are likely to be 100-fold lower than that tested here. It is also unlikely that exposure to high levels of estradiol used in the mouse model gives mtrR locus mutants an advantage, since estradiol does not kill N. gonorrhoeae, even when tested at levels that are greater than 1000-fold higher than normal serum levels and 100-fold higher than serum levels in mice treated with 17-β-estradiol slow-release pellets. Estradiol is also not a substrate of the MtrC-MtrD-MtrE efflux pump system (Jerse et al., 2003). Our demonstration that mtrR locus mutants are more fit in vivo suggests that wild type levels of MtrR-repression are anti-pathogenic and are destined to be selected against with further use of antibiotic therapy. Conversely, it is important to note that this infection model is a surrogate for the lower genital tract infection of females, and other body sites may not confer such an advantage to derepressed mutants. This caveat suggests a role for the MtrR repressor in nature and a reason for the existence of naturally repressed strains. Finally, we note here that the gradient in azithromycin resistance is equally important as the fitness gradient detected in vivo, since azithromycin is increasingly utilized as a therapy for non-complicated urethritis (McNabb et al., 2007).
Experimental Procedures
Bacterial Strains and Culture Conditions
Neisseria gonorrhoeae strain FA19 SmR is a spontaneous streptomycin resistant mutant of strain FA19 (porB1A, serum resistant) and was described previously (Jerse et al., 2003). Strain FA19 CmR and mtrR mutants JF1, DW9, and KH15 were described previously (Warner et al., 2007). A series of additional mutants in the mtrR gene or promoter region were constructed in strain FA19SmR and are described in Table 1. Mutant mtr sequences were PCR-amplified from strains LG5 or LG7 (Garvin et al., 2008), or strain MS11 with primers R-Xho and C-Xho as described (Warner et al., 2007), and used to transform strain FA19SmR following gel purification. Transformants were isolated on GC agar supplemented with Em (0.5 μg/ml). pGCC-5 (provided by H.S. Seifert, Northwestern University) was used to mark strains FA19 SmR, DW9, DW39, and DW45 by insertion of a cat gene into an untranscribed region between the lctP and aspC chromosomal genes as described (Simms et al., 2006). The resultant strains were FA19CmR DW9CmR, DW39CmR, and DW45CmR, respectively. mtrA mutant DW120A was created by PCR amplification of the mutant mtrA∷aphA3 allele from strain JF3 using primers mtrAfor and mtrArev (Warner et al., 2007). The resultant 1900-bp product was transformed into strains DW120, and transformants were selected on GC agar supplemented with kanamycin (Km). To construct mutant JF1-120, which contains both a ΔmtrR and the mtr120 mutation, the PCR-amplified mtr locus from strain JF1 was used to transform strain DW120. All transformations were performed as described (Gunn et al., 1996). The nucleotide sequences of the mtr regions of strains DW9, DW39, DW45, MS11, DW120, and FA19MS11mtr were determined by PCR amplification and subsequent sequencing with primers C-Xho and R-Xho. All sequencing was performed using the Big Dye Terminator V3.1 Cycle Sequencing kit (Applied Biosystems), by the Biomedical Instrumentation Center of the Uniformed Services University of the Health Sciences (USUHS). All bacteria were cultured on supplemented GC agar (Difco) as described (Wu et al., 2006). All antibiotics were from Sigma and were used to supplement selective media at the following concentrations: Cm, 0.6 μg/ml; Em, 0.5 μg/ml; and Km, 50 μg/ml.
Resistance to MtrC-MtrD-MtrE pump substrates
The minimum inhibitory concentration (MIC) of Triton X-100, Em, Az, and Km against wild type parent strain FA19 SmR and the mtr mutants was established using a standard two-fold agar dilution assay (Warner et al., 2007). Sensitivity to a single concentration of water-soluble progesterone (35 μg/ml) (Sigma) was tested using a plating efficiency assay as described (Jerse et al., 2003). Susceptibility of FA19SmR and mtr mutant bacteria in this background to CRAMP-38 was determined by incubating 106 CFU of each strain suspended in PBS alone or with PBS containing CRAMP-38 (4 μg/ml) in a total volume of 100 μl. The mixtures were incubated at 37°C in a CO2 incubator for 45 min before serial dilution and culture on GC agar. For studies with FA19SmR versus wild type MS11 bacteria and the recombinant strains FA19MS11mtr and DW3MS11, 104 CFU were exposed to 16 μg/ml of CRAMP-38 or LL-37 in minimal essential media (total volume 80 μl). GCB (40ul) was added after 55 min incubation at 37°C, and the suspensions were quantitatively cultured on GC agar. The number of CFU recovered from wells with peptide was divided by the number of CFU recovered from wells without peptide and the results were multiplied by 100. All assays were performed in triplicate and repeated at least once to test reproducibility. Peptides LL-37 and CRAMP-38 were synthesized and purified at the Microchemical Facility of Emory University as described (Shafer et al., 1998).
Western Blot Analysis
Outer membrane proteins were fractionated on a 12% SDS/PAGE gel, transferred to polyvinylidene difluoride (PVDF) membranes, and incubated with rabbit polyclonal antibodies against MtrE as described previously (Warner et al., 2007). Detection was performed with anti-rabbit IgG secondary antibody conjugated to horseradish peroxidase (Bethyl) followed by chemiluminescence with the ECL reagent (GE Healthcare). Densitometry was performed using the Image J program.
Measurement of Transcript Levels
We used real time quantitative reverse transcriptase PCR (qRT-PCR) to measure levels of specific transcripts produced by gonococcal strains. Gonococcal strains were grown to early log phase (OD600 = 0.2-0.4) in supplemented GC broth with aeration. RNA-later reagent (Ambion) was added to a 300 μl aliquot and samples were stored at −80° C. Cells were lysed and RNA was prepared as recommended in the Qiagen RNAeasy kit (Qiagen) followed by treatment with Turbo DNAse (Ambion). RNA (100 ng) from each sample were then reverse-transcribed using the Superscript III RT Kit (Invitrogen). Briefly, 10 μl of RNA were mixed with 150 ng of random primers, 10 mM dNTPs, 10 mM dithiothreonol, synthesis buffer, and the Superscript III enzyme. The mixture was incubated at room temperature for 10 min and then 50°C for one hour before deactivation of the enzyme for 10 min at 95°C. Real time PCR was carried out in 96 well plates with a Sybr-Green reaction mix (Applied Biosystems). All primers are described in Table 3. Relative levels of transcript for 16S rRNA, rmp, mtrC, mtrR, and mtrA were determined by the relative method of comparison for each of the mutants to the wild type strain. All transcripts were assayed in triplicate with a no reverse transcriptase control. Dissociation curves were used to confirm the specificity of each reaction and two biological samples were tested to confirm results. The threshold for significance expressed as fold differences was established by examining the fold change between strains with respect to the constitutively expressed gene rmp. All delta Ct values were normalized to 16S rRNA.
Table 3. Oligonucleotide primers used for RT-PCR.
| Primer | Sequence (5′-3′) |
|---|---|
| 16S-for | GCGTGGGTAGCAAACAGGAT |
| 16S-rev | CGCGTTAGCTACGCTACCAAG |
| Rmp-forRT | CAAACAACCTGGTCAGCAAC |
| Rmp-revRT | TTCGGCTTGACAAACTTGAG |
| mtrC-forRT | CCGCTTTAACGCTTGCTTCG |
| mtrC-revRT | CGTTACAAACCGCTGGTTTC |
| mtrA-forRT | GTGGTTTCAATGCTGCAACT |
| mtrA-revRT | AGGATAAGCACCAGCAGGAC |
| mtrR-forRT | AAAATTACCGCCGTTTTGAC |
| mtrR-revRT | CCAACGTCGATTTGATGAAG |
RNA half-life Determination
The half-lives for mtrC and rmp mRNA from strains FA19, JF1, and DW120 were determined by qRT-PCR. Broth cultures were grown to early log phase (OD= 0.2), treated with rifampicin (200 μg/ml), and sampled at times 0, 2, 4, 8, 12, and 20 minutes post-treatment. Samples were suspended in RNA-later reagent (Ambion) and frozen at -80° C before RNA was extracted with the RNAeasy Kit (Qiagen). RNA was quantified by spectophotometric analysis, and 100 ng per sample were converted to cDNA. cDNA was then used with the primers rmp-forRT, rmp-revRT, mtrC-forRT, and mtrC-revRT (Table 3) in a Sybr-Green Real-Time PCR reaction plate as detailed above. Each reaction plate also contained dilutions of FA19 genomic DNA to establish a standard curve for both primer sets. Ct values were converted to actual RNA values using the standard curve, and an RNA degradation curve was established for both genes in all three bacterial strains. These degradation curves were used to calculate the amount of time for a given strain to degrade one half of the starting amount of mRNA. cDNA was also used in a standard PCR reaction using Taq DNA polymerase, followed by separation and visualization on an agarose gel, to visualize the decay of rmp and mtrC message from strains FA19 and DW120. The experiment was performed twice using different biological samples.
Experimental Murine Infection
Female BALB/c mice were treated with 17-β-estradiol and antibiotics to promote long term infection (Jerse, 1999). The competitive infection technique was used to assess fitness in vivo as described (Warner et al., 2007), in which a defined mixture of two strains were co-inoculated into a mouse, with one strain marked with a CmR cassette. Bacterial suspensions were prepared as described (Wu et al., 2006), adjusted to an OD600 of 0.07 (∼1 × 106 CFU/10 μl), and mixed to obtain a 1:1 ratio for each pair of strains. Twenty μl of the mixture were inoculated intravaginally into mice (n = 5 mice per mixture) and vaginal mucus was cultured every other day. The titer of each strain within the inoculum and vaginal mucus was determined by quantitative culture on GC agar with Sm (total CFU) and with Sm and Cm (wild type or mtrR mutant CFU, depending on the experiment). Competitive indices were calculated as described (Unsworth et al., 2000) in which a ratio of (mtrR mutant / wild type)output / (mtrR / wild type)input was calculated. A CI >1.0 was considered a fitness advantage for the mtrR mutant. In cases in which one strain was not recovered, the limit of detection (4 CFU/100 μl) was used to calculate CIs. All strains were tested in in vitro competition assays as described (Warner et al., 2007). For infections in which CmR wild type bacteria were tested, randomly selected colonies were screened on GC media with Em (0.5 μg/ml) to confirm the correct ratio of mtrR mutants to wild type bacteria. Animal experiments were conducted in the laboratory animal facility at USUHS, which is fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care, under a protocol approved by the USUHS Institutional Animal Care and Use Committee.
Acknowledgments
This work was supported by the National Institutes of Health grants RO1-AI42053 and STI-TM-CRC grant U19 AI31496 (A.E.J.), and RO1-AI062755 (W.M.S.). W.M.S. was supported in-part by a Senior Research Career Scientist Award from the VA Medical Research Service. We thank Afrin Begum for technical assistance and Lotisha Garvin for technical assistance and help with preparation of the manuscript.
References
- Aas FE, Wolfgang M, Frye S, Dunham S, Lovold C, Koomey M. Competence for natural transformation in Neisseria gonorrhoeae: components of DNA binding and uptake linked to type IV pilus expression. Mol Microbiol. 2002;46:749–760. doi: 10.1046/j.1365-2958.2002.03193.x. [DOI] [PubMed] [Google Scholar]
- Alekshun MN, Levy SB. Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007;128:1037–1050. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Bina JE, Mekalanos JJ. Vibrio cholerae tolC is required for bile resistance and colonization. Infection and immunity. 2001;69:4681–4685. doi: 10.1128/IAI.69.7.4681-4685.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. Increases in fluoroquinolone-resistant Neisseria gonorrhoeae among men who have sex with men--United States, 2003, and revised recommendations for gonorrhea treatment, 2004. MMWR Morb Mortal Wkly Rep. 2004;53:335–338. [PubMed] [Google Scholar]
- Chaussee MS, Hill SA. Formation of single-stranded DNA during DNA transformation of Neisseria gonorrhoeae. J Bacteriol. 1998;180:5117–5122. doi: 10.1128/jb.180.19.5117-5122.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MS, Cannon JG, Jerse AE, Charniga LM, Isbey SF, Whicker LG. Human experimentation with Neisseria gonorrhoeae: rationale, methods, and implications for the biology of infection and vaccine development. J Infect Dis. 1994;169:532–537. doi: 10.1093/infdis/169.3.532. [DOI] [PubMed] [Google Scholar]
- Cousin S, Jr, Roberts MC, Whittington WL. Insertion of a thymine (+T) in the 13 base pair inverted repeat of the Neisseria gonorrhoeae mtr promoter region and antibiotic susceptibility. Int J Antimicrob Agents. 2004;23:418–419. doi: 10.1016/j.ijantimicag.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Delahay RM, Robertson BD, Balthazar JT, Shafer WM, Ison CA. Involvement of the gonococcal MtrE protein in the resistance of Neisseria gonorrhoeae to toxic hydrophobic agents. Microbiology. 1997;143(Pt 7):2127–2133. doi: 10.1099/00221287-143-7-2127. [DOI] [PubMed] [Google Scholar]
- Dewi BE, Akira S, Hayashi H, Ba-Thein W. High occurrence of simultaneous mutations in target enzymes and MtrRCDE efflux system in quinolone-resistant Neisseria gonorrhoeae. Sex Transm Dis. 2004;31:353–359. doi: 10.1097/00007435-200406000-00007. [DOI] [PubMed] [Google Scholar]
- Folster JP, Dhulipala V, Nicholas RA, Shafer WM. Differential regulation of ponA and pilMNOPQ expression by the MtrR transcriptional regulatory protein in Neisseria gonorrhoeae. J Bacteriol. 2007a;189:4569–4577. doi: 10.1128/JB.00286-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folster JP, Dhulipala V, Nicholas RA, Shafer WM. Differential Regulation of ponA and pilMNOPQ Expression by the MtrR Transcriptional Regulatory Protein in Neisseria gonorrhoeae. J Bacteriol. 2007b doi: 10.1128/JB.00286-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folster JP, Shafer WM. Regulation of mtrF expression in Neisseria gonorrhoeae and its role in high-level antimicrobial resistance. J Bacteriol. 2005;187:3713–3720. doi: 10.1128/JB.187.11.3713-3720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier B, Truong-Bolduc QC, Zhang X, Hooper DC. A mutation in the 5′ untranslated region increases stability of norA mRNA, encoding a multidrug resistance transporter of Staphylococcus aureus. Journal of bacteriology. 2001;183:2367–2371. doi: 10.1128/JB.183.7.2367-2371.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvin LE, Bash MC, Keys C, Warner DM, Ram S, Shafer WM, Jerse AE. Phenotypic and genotypic analyses of Neisseria gonorrhoeae isolates that express frequently recovered PorB PIA variable region types suggest that certain P1a porin sequences confer a selective advantage for urogenital tract infection. Infect Immun. 2008;76:3700–3709. doi: 10.1128/IAI.00265-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbase AC, Rowley JT, Heymann DH, Berkley SF, Piot P. Global prevalence and incidence estimates of selected curable STDs. Sex Transm Infect. 1998;74 1:S12–16. [PubMed] [Google Scholar]
- Gunn JS, Stein DC. Use of a non-selective transformation technique to construct a multiply restriction/modification-deficient mutant of Neisseria gonorrhoeae. Mol Gen Genet. 1996;251:509–517. doi: 10.1007/BF02173639. [DOI] [PubMed] [Google Scholar]
- Haas R, Meyer TF. Molecular principles of antigenic variation in Neisseria gonorrhoeae. Antonie Van Leeuwenhoek. 1987;53:431–434. doi: 10.1007/BF00415498. [DOI] [PubMed] [Google Scholar]
- Hagman KE, Pan W, Spratt BG, Balthazar JT, Judd RC, Shafer WM. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology. 1995a;141(Pt 3):611–622. doi: 10.1099/13500872-141-3-611. [DOI] [PubMed] [Google Scholar]
- Hagman KE, Shafer WM. Transcriptional control of the mtr efflux system of Neisseria gonorrhoeae. J Bacteriol. 1995b;177:4162–4165. doi: 10.1128/jb.177.14.4162-4165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton HL, Dominguez NM, Schwartz KJ, Hackett KT, Dillard JP. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol Microbiol. 2005;55:1704–1721. doi: 10.1111/j.1365-2958.2005.04521.x. [DOI] [PubMed] [Google Scholar]
- Herida M, Sednaoui P, Goulet V. Gonorrhoea surveillance system in France: 1986-2000. Sexually transmitted diseases. 2004;31:209–214. doi: 10.1097/01.olq.0000118426.66742.9e. [DOI] [PubMed] [Google Scholar]
- Hoffmann KM, Williams D, Shafer WM, Brennan RG. Characterization of the multiple transferable resistance repressor, MtrR, from Neisseria gonorrhoeae. J Bacteriol. 2005;187:5008–5012. doi: 10.1128/JB.187.14.5008-5012.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jajosky RA, Hall PA, Adams DA, Dawkins FJ, Sharp P, Anderson WJ, et al. Summary of notifiable diseases--United States, 2004. MMWR Morb Mortal Wkly Rep. 2006;53:1–79. [PubMed] [Google Scholar]
- Jerse AE. Experimental gonococcal genital tract infection and opacity protein expression in estradiol-treated mice. Infect Immun. 1999;67:5699–5708. doi: 10.1128/iai.67.11.5699-5708.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerse AE, Sharma ND, Simms AN, Crow ET, Snyder LA, Shafer WM. A gonococcal efflux pump system enhances bacterial survival in a female mouse model of genital tract infection. Infect Immun. 2003;71:5576–5582. doi: 10.1128/IAI.71.10.5576-5582.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson AB, Ilver D, Falk P, Pepose J, Normark S. Sequence changes in the pilus subunit lead to tropism variation of Neisseria gonorrhoeae to human tissue. Mol Microbiol. 1994;13:403–416. doi: 10.1111/j.1365-2958.1994.tb00435.x. [DOI] [PubMed] [Google Scholar]
- Koomey M, Bergstrom S, Blake M, Swanson J. Pilin expression and processing in pilus mutants of Neisseria gonorrhoeae: critical role of Gly-1 in assembly. Mol Microbiol. 1991;5:279–287. doi: 10.1111/j.1365-2958.1991.tb02108.x. [DOI] [PubMed] [Google Scholar]
- Lee EH, Shafer WM. The farAB-encoded efflux pump mediates resistance of gonococci to long-chained antibacterial fatty acids. Mol Microbiol. 1999;33:839–845. doi: 10.1046/j.1365-2958.1999.01530.x. [DOI] [PubMed] [Google Scholar]
- Lucas CE, Balthazar JT, Hagman KE, Shafer WM. The MtrR repressor binds the DNA sequence between the mtrR and mtrC genes of Neisseria gonorrhoeae. J Bacteriol. 1997;179:4123–4128. doi: 10.1128/jb.179.13.4123-4128.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas CE, Hagman KE, Levin JC, Stein DC, Shafer WM. Importance of lipooligosaccharide structure in determining gonococcal resistance to hydrophobic antimicrobial agents resulting from the mtr efflux system. Mol Microbiol. 1995;16:1001–1009. doi: 10.1111/j.1365-2958.1995.tb02325.x. [DOI] [PubMed] [Google Scholar]
- Lundback D, Fredlund H, Berglund T, Wretlind B, Unemo M. Molecular epidemiology of Neisseria gonorrhoeae- identification of the first presumed Swedish transmission chain of an azithromycin-resistant strain. Apmis. 2006;114:67–71. doi: 10.1111/j.1600-0463.2006.apm_332.x. [DOI] [PubMed] [Google Scholar]
- Lynn F, Hobbs MM, Zenilman JM, Behets FM, Van Damme K, Rasamindrakotroka A, Bash MC. Genetic typing of the porin protein of Neisseria gonorrhoeae from clinical noncultured samples for strain characterization and identification of mixed gonococcal infections. J Clin Microbiol. 2005;43:368–375. doi: 10.1128/JCM.43.1.368-375.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavroidi A, Tzouvelekis LS, Kyriakis KP, Avgerinou H, Daniilidou M, Tzelepi E. Multidrug-resistant strains of Neisseria gonorrhoeae in Greece. Antimicrobial agents and chemotherapy. 2001;45:2651–2654. doi: 10.1128/AAC.45.9.2651-2654.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnew DL, Lynn F, Zenilman JM, Bash MC. Porin variation among clinical isolates of Neisseria gonorrhoeae over a 10-year period, as determined by Por variable region typing. J Infect Dis. 2003;187:1213–1222. doi: 10.1086/374563. [DOI] [PubMed] [Google Scholar]
- McLean CA, Wang SA, Hoff GL, Dennis LY, Trees DL, Knapp JS, et al. The emergence of Neisseria gonorrhoeae with decreased susceptibility to Azithromycin in Kansas City, Missouri, 1999 to 2000. Sex Transm Dis. 2004;31:73–78. doi: 10.1097/01.OLQ.0000109514.91508.FC. [DOI] [PubMed] [Google Scholar]
- McNabb SJ, Jajosky RA, Hall-Baker PA, Adams DA, Sharp P, Anderson WJ, et al. Summary of notifiable diseases --- United States, 2005. MMWR Morb Mortal Wkly Rep. 2007;54:1–92. [PubMed] [Google Scholar]
- Ng LK, Martin I, Liu G, Bryden L. Mutation in 23S rRNA associated with macrolide resistance in Neisseria gonorrhoeae. Antimicrob Agents Chemother. 2002;46:3020–3025. doi: 10.1128/AAC.46.9.3020-3025.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino K, Latifi T, Groisman EA. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol Microbiol. 2006;59:126–141. doi: 10.1111/j.1365-2958.2005.04940.x. [DOI] [PubMed] [Google Scholar]
- Nizet V, Ohtake T, Lauth X, Trowbridge J, Rudisill J, Dorschner RA, et al. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature. 2001;414:454–457. doi: 10.1038/35106587. [DOI] [PubMed] [Google Scholar]
- Pan W, Spratt BG. Regulation of the permeability of the gonococcal cell envelope by the mtr system. Mol Microbiol. 1994;11:769–775. doi: 10.1111/j.1365-2958.1994.tb00354.x. [DOI] [PubMed] [Google Scholar]
- Ramos JL, Martinez-Bueno M, Molina-Henares AJ, Teran W, Watanabe K, Zhang X, et al. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey KH, Schneider H, Cross AS, Boslego JW, Hoover DL, Staley TL, et al. Inflammatory cytokines produced in response to experimental human gonorrhea. J Infect Dis. 1995;172:186–191. doi: 10.1093/infdis/172.1.186. [DOI] [PubMed] [Google Scholar]
- Rouquette-Loughlin C, Dunham SA, Kuhn M, Balthazar JT, Shafer WM. The NorM efflux pump of Neisseria gonorrhoeae and Neisseria meningitidis recognizes antimicrobial cationic compounds. J Bacteriol. 2003;185:1101–1106. doi: 10.1128/JB.185.3.1101-1106.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquette-Loughlin C, Stojiljkovic I, Hrobowski T, Balthazar JT, Shafer WM. Inducible, but not constitutive, resistance of gonococci to hydrophobic agents due to the MtrC-MtrD-MtrE efflux pump requires TonB-ExbB-ExbD proteins. Antimicrobial agents and chemotherapy. 2002;46:561–565. doi: 10.1128/AAC.46.2.561-565.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquette-Loughlin CE, Balthazar JT, Shafer WM. Characterization of the MacA-MacB efflux system in Neisseria gonorrhoeae. J Antimicrob Chemother. 2005;56:856–860. doi: 10.1093/jac/dki333. [DOI] [PubMed] [Google Scholar]
- Rouquette C, Harmon JB, Shafer WM. Induction of the mtrCDE-encoded efflux pump system of Neisseria gonorrhoeae requires MtrA, an AraC-like protein. Mol Microbiol. 1999;33:651–658. doi: 10.1046/j.1365-2958.1999.01517.x. [DOI] [PubMed] [Google Scholar]
- Schmidt KA, Schneider H, Lindstrom JA, Boslego JW, Warren RA, Van de Verg L, et al. Experimental gonococcal urethritis and reinfection with homologous gonococci in male volunteers. Sex Transm Dis. 2001;28:555–564. doi: 10.1097/00007435-200110000-00001. [DOI] [PubMed] [Google Scholar]
- Schneider H, Griffiss JM, Boslego JW, Hitchcock PJ, Zahos KM, Apicella MA. Expression of paragloboside-like lipooligosaccharides may be a necessary component of gonococcal pathogenesis in men. J Exp Med. 1991;174:1601–1605. doi: 10.1084/jem.174.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher MA, Miller MC, Grkovic S, Brown MH, Skurray RA, Brennan RG. Structural mechanisms of QacR induction and multidrug recognition. Science (New York, NY. 2001;294:2158–2163. doi: 10.1126/science.1066020. [DOI] [PubMed] [Google Scholar]
- Schumacher MA, Miller MC, Grkovic S, Brown MH, Skurray RA, Brennan RG. Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR. Embo J. 2002;21:1210–1218. doi: 10.1093/emboj/21.5.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert HS, Ajioka RS, Marchal C, Sparling PF, So M. DNA transformation leads to pilin antigenic variation in Neisseria gonorrhoeae. Nature. 1988;336:392–395. doi: 10.1038/336392a0. [DOI] [PubMed] [Google Scholar]
- Shafer WM, Balthazar JT, Hagman KE, Morse SA. Missense mutations that alter the DNA-binding domain of the MtrR protein occur frequently in rectal isolates of Neisseria gonorrhoeae that are resistant to faecal lipids. Microbiology (Reading, England) 1995;141(Pt 4):907–911. doi: 10.1099/13500872-141-4-907. [DOI] [PubMed] [Google Scholar]
- Shafer WM, Qu X, Waring AJ, Lehrer RI. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc Natl Acad Sci U S A. 1998;95:1829–1833. doi: 10.1073/pnas.95.4.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms AN, Jerse AE. In vivo selection for Neisseria gonorrhoeae opacity protein expression in the absence of human carcinoembryonic antigen cell adhesion molecules. Infect Immun. 2006;74:2965–2974. doi: 10.1128/IAI.74.5.2965-2974.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone BJ, Miller VL. Salmonella enteritidis has a homologue of tolC that is required for virulence in BALB/c mice. Mol Microbiol. 1995;17:701–712. doi: 10.1111/j.1365-2958.1995.mmi_17040701.x. [DOI] [PubMed] [Google Scholar]
- Sutrisna A, Soebjakto O, Wignall FS, Kaul S, Limnios EA, Ray S, et al. Increasing resistance to ciprofloxacin and other antibiotics in Neisseria gonorrhoeae from East Java and Papua, Indonesia, in 2004 - implications for treatment. International journal of STD & AIDS. 2006;17:810–812. doi: 10.1258/095646206779307595. [DOI] [PubMed] [Google Scholar]
- Swanson J, Barrera O, Sola J, Boslego J. Expression of outer membrane protein II by gonococci in experimental gonorrhea. J Exp Med. 1988;168:2121–2129. doi: 10.1084/jem.168.6.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson J, Bergstrom S, Boslego J, Koomey M. Gene conversion accounts for pilin structural changes and for reversible piliation “phase” changes in gonococci. Antonie Van Leeuwenhoek. 1987a;53:441–446. doi: 10.1007/BF00415500. [DOI] [PubMed] [Google Scholar]
- Swanson J, Robbins K, Barrera O, Corwin D, Boslego J, Ciak J, et al. Gonococcal pilin variants in experimental gonorrhea. J Exp Med. 1987b;165:1344–1357. doi: 10.1084/jem.165.5.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Nakayama H, Huruya K, Konomi I, Irie S, Kanayama A, et al. Analysis of mutations within multiple genes associated with resistance in a clinical isolate of Neisseria gonorrhoeae with reduced ceftriaxone susceptibility that shows a multidrug-resistant phenotype. Int J Antimicrob Agents. 2006;27:20–26. doi: 10.1016/j.ijantimicag.2005.08.021. [DOI] [PubMed] [Google Scholar]
- Unsworth KE, Holden DW. Identification and analysis of bacterial virulence genes in vivo. Philos Trans R Soc Lond B Biol Sci. 2000;355:613–622. doi: 10.1098/rstb.2000.0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veal WL, Nicholas RA, Shafer WM. Overexpression of the MtrC-MtrD-MtrE efflux pump due to an mtrR mutation is required for chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. J Bacteriol. 2002;184:5619–5624. doi: 10.1128/JB.184.20.5619-5624.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vereshchagin VA, Ilina EN, Malakhova MV, Zubkov MM, Sidorenko SV, Kubanova AA, Govorun VM. Fluoroquinolone-resistant Neisseria gonorrhoeae isolates from Russia: molecular mechanisms implicated. The Journal of antimicrobial chemotherapy. 2004;53:653–656. doi: 10.1093/jac/dkh145. [DOI] [PubMed] [Google Scholar]
- Warner DM, Folster JP, Shafer WM, Jerse AE. Regulation of the MtrC-MtrD-MtrE efflux-pump system modulates the in vivo fitness of Neisseria gonorrhoeae. J Infect Dis. 2007;196:1804–1812. doi: 10.1086/522964. [DOI] [PubMed] [Google Scholar]
- Wu H, Jerse AE. Alpha-2,3-sialyltransferase enhances Neisseria gonorrhoeae survival during experimental murine genital tract infection. Infect Immun. 2006;74:4094–4103. doi: 10.1128/IAI.00433-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M, Whittington WL, Shafer WM, Holmes KK. Gonorrhea among men who have sex with men: outbreak caused by a single genotype of erythromycin-resistant Neisseria gonorrhoeae with a single-base pair deletion in the mtrR promoter region. J Infect Dis. 2000;181:2080–2082. doi: 10.1086/315510. [DOI] [PubMed] [Google Scholar]
- Zarantonelli L, Borthagaray G, Lee EH, Shafer WM. Decreased azithromycin susceptibility of Neisseria gonorrhoeae due to mtrR mutations. Antimicrob Agents Chemother. 1999;43:2468–2472. doi: 10.1128/aac.43.10.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarantonelli L, Borthagaray G, Lee EH, Veal W, Shafer WM. Decreased susceptibility to azithromycin and erythromycin mediated by a novel mtr(R) promoter mutation in Neisseria gonorrhoeae. J Antimicrob Chemother. 2001;47:651–654. doi: 10.1093/jac/47.5.651. [DOI] [PubMed] [Google Scholar]