Abstract
Gravin (AKAP12, SSeCKS) is a scaffolding protein that acts as a potent inhibitor of tumor metastasis in vivo and in vitro, and regulates morphogenesis during vertebrate gastrulation. Despite being implicated in many cellular processes, surprisingly little is known about the mechanism by which Gravin elicits cell shape changes. In this work we use in vitro cell spreading assays to demonstrate that the Gravin N-terminus containing the three MARCKS-like basic regions (BRs) is necessary and sufficient to regulate cell shape in vitro. We show that the conserved phosphorylation sites in the BRs are essential for their function in these assays. We further demonstrate that the Gravin BRs are necessary for in vivo function during gastrulation in zebrafish. Together, these results provide an important step forward in understanding the mechanism of Gravin function in cell shape regulation and provide valuable insight into how Gravin acts as a cytoskeletal regulator.
Keywords: AKAP, Gravin, SSeCKS, Cell spreading, zebrafish
Introduction
Gravin is a member of the AKAP (A kinase anchoring protein) family of multivalent scaffolding proteins that mediate precise spatiotemporal control of Protein Kinase A activity in cells [1-3]. Gravin interacts with many important signaling molecules in addition to PKA, including Protein Kinase C [1], the β2 Adrenergic receptor [4], Src Tyrosine Kinase [5], Calmodulin [6], 1,4 β-galactosidase [7] and others (reviewed in [8, 9]). Gravin is also a potent tumor suppressor down-regulated by several oncogenes in vitro and multiple human tumors in vivo, and a mouse lacking the gravin gene displays increased prostate hyperplasia [10-12]. Ectopic expression of Gravin in v-Src transformed fibroblasts inhibits a number of metastatic cellular behavioral changes [11, 12]. Overexpression of Gravin elicits several cell shape changes including increased cell spreading and loss of stress fibers [11-14]. Gravin also plays a critical role in promoting a switch from migratory to intercalative behaviors of mesodermal cells during the gastrulation stage of early zebrafish development, which is essential for the normal morphogenesis of the embryo [15]. Interestingly, overexpression or loss of Gravin has no affect on cell migration in 2D scratch assays [12], but Gravin is a potent inhibitor of 3D cell invasion through matrigel [13]. This indicates that Gravin is not simply a permissive factor in cell migration but a critical regulator of complex cell behaviors. Although Gravin genetically interacts with the Src and RhoA pathways [11, 13, 15], the mechanism by which Gravin mediates cell shape changes is mysterious. One important feature of Gravin is the presence of three N-terminal Basic Regions (BRs). These BRs have been implicated in mediating reversible plasma membrane interaction, similar to the membrane effector domain of MARCKS or AKAP79 [16-18]. In fact, these BRs are critical for proper Gravin localization and regulation of the β2-adrengeric receptor [6, 17], but their role in controlling cell shape and behavior is unknown.
In this work, we set out to elucidate the functional domains in the Gravin protein that are required for regulation of cell shape and behavior. We used COS7 cell spreading as a straightforward assay for Gravin function. We created a series of deletion and point mutations in zebrafish Gravin, which we previously showed functions the same as human Gravin in this assay [15]. These deletions mutants demonstrated the essential role of the BRs for Gravin's cell shape changes as well as Gravin localization in cultured mammalian cells. Moreover, we found that the conserved phosphorylation sites within the BRs are necessary for Gravin's function in this assay. Finally, we confirmed our cell-based experiments in vivo by demonstration that BRs are required for Gravin's function during early zebrafish development.
Materials and Methods
Zebrafish and Mammalian Cell Culture
Zebrafish (Danio rerio) embryos were obtained, injected and staged, and COS7 cells were maintained and transfected as described [15]. Flattening assays were performed as described [15], modified to allow cells to flatten for the indicated time (1-24 hrs) by trypsinizing, and plating on poly-d lysine coated coverslips.
Cloning of zebrafish gravin Plasmids and mRNA
Zebrafish GFP Gravin, T7-Gravin and the morpholino antisense oligonucleotide were described [15]. Deletions and point mutants were derived from GFP-Gravin (details available on request). Equal expression of all mutants was confirmed by equal fluorescence intensity of the analyzed cells. The conserved serines or threonines in the three basic repeats were mutated to aspartic acid. A T7-Gravin ΔBR 1,2,3 was made by ligating a Xho1-Nco1 fragment from GFP ΔBR 1,2,3 into T7-Gravin. mRNA was produced using the mMessage mMachine T7 ultra kit (Ambion).
Microscopy
Fixed cells and live zebrafish embryos were imaged with a Zeiss Axiovert 200M microscope using AxioVision 4 software. Confocal images were taken on an Olympus FV-1000 Confocal Microscope.
Data Analysis
Data were collected using AxioVision software cell area tool and statistical calculations were performed using Microsoft Excel. Error bars represent standard deviation, and P values were determined using student t-tests, with which each sample compared to GFP transfected cells.
Results and Discussion
One of the characteristic features of Gravin is its ability to promote cultured cells to greatly increase their surface area by spreading across a substrate, a property conserved from fish to mammals [14, 15, 19]. We sought to establish cell spreading as an cell-based assay to study Gravin structure-function. GFP-Gravin transfected cells were fixed after 1, 2, 3, 4, 8, and 24 hours of spreading (Figure 1A). Importantly, Gravin produces a statistically significant increase in the maximum extent of cell spreading (p <.05), but it does not increase the rate of cell flattening during the early stages of cell spreading (Figure 1B). Interestingly, Gravin's subcellular localization changes as cells spread in agreement with reports in endothelial cells (Figure 1A) [14]. Initially, Gravin is localized exclusively to perinuclear puncta, but as cells continue to flatten, Gravin localized to the cytosol (8-24hr) and finally peripheral membrane puncta (24hr). This indicates that Gravin not only promotes cell spreading but its subcellular localization is dynamically regulated during cell spreading. We observed similar localization and increased cell spreading in NIH 3T3 and HeLa cells (data not shown) but COS7 cells are used in this study because they exhibited the strongest effect. This spreading assay gives us a straightforward way to determine which Gravin domains are required for cell shape regulation.
Figure 1. Gravin subcellular localization is dynamically regulated during cell spreading.
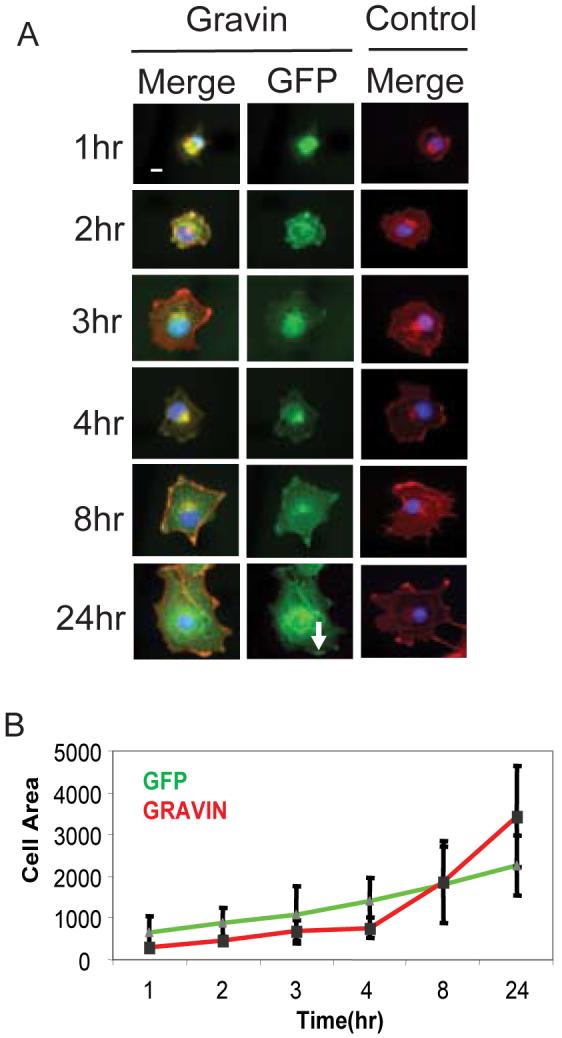
A. COS7 cells expressing either GFP-Gravin or GFP alone were allowed to spread for 1, 2, 3, 4, 8, or 24 hours, then fixed, stained and analyzed for actin (red), DNA (blue) and GFP-fluorescence (green). Merge lanes show DNA, actin and Gravin localization, while the GFP lane shows only Gravin localization. The control cell shows only actin and DNA. White arrow points to peripheral membrane puncta. B. Quantification of the time course of cell flattening. Y-axis is average cell area (μM2) and the x-axis is time in hours. White bar in the first panel indicates 10 μm.
Characterization of Domains Required for Gravin-Induced Cell Spreading
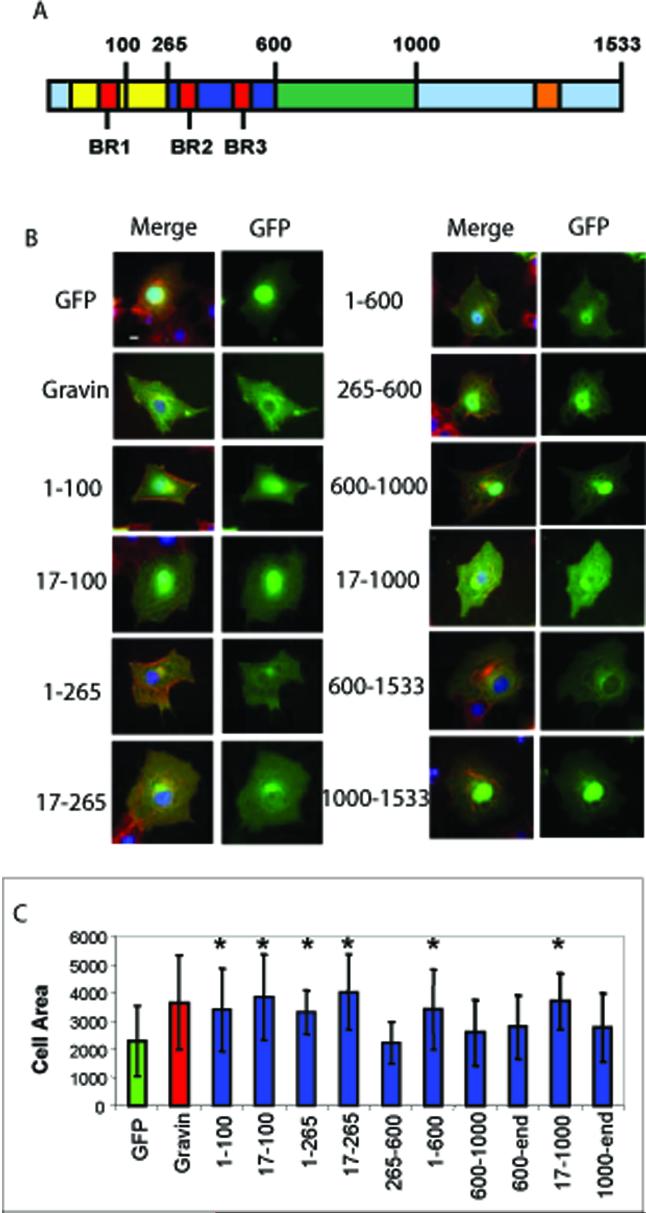
We generated C- and N-terminal Gravin deletion constructs and expressed them as GFP fusions. Specifically, the following truncations were generated (named by amino acid positions in zebrafish Gravin):1-100, 17-100, 1-265, 17-265, 265-600, 1-600, 600-1000, 600-1533, 17-1000, and 1000-1533 (Figure 2A). Expression of the Gravin C-terminus, 1000-1533 or the central domain, 600-1000 or 600-1533, had no effect on cell spreading (Figure 2B, C). However, expression of N-terminal domains of Gravin including 1-100, 17-100, 1-265, 17-265, and 1-600 resulted in cell spreading similar to the extent of full-length Gravin (Figure 2C). One N-terminal fragment, 265-600, did not increase cell flattening (Figure 2C). Thus, we demonstrated that the first 100 amino acids of Gravin, which include BR1, are necessary and sufficient to induce cell flattening.
Figure 2. Truncation mutants of zebrafish Gravin reveal a critical role for the basic regions in cell flattening.
A. A schematic of the proposed functional domains in Gravin and the sites of truncation. Gravin contains a PKC binding domain (blue), a MARCKS-like domain (Yellow), a central domain (green), an unconserved C-terminal region (light blue) and a PKA binding helix (orange). The N-terminus contains 3 basic regions (BRs in red). B. Expression of Gravin truncation constructs in COS7 cells. A merged image showing GFP, Actin and DNA, and also as a GFP alone image are shown. C. Quantification of cell spreading for each deletion construct. The y-axis is cell area (μM2) and x-axis shows the expressed construct. * indicates increased cell flattening relative to the GFP control p<0.05.
The structure-function of mammalian Gravin's subcellular localization has been described [17]. In agreement with previous reports we observed with zebrafish Gravin that the C-terminus was necessary and sufficient for nuclear exclusion (600-1533) and that the all three BRs were necessary for peripheral membrane targeting (1-600 but not 1-100, 1-265 or 265-600; Figure 2A). We also made the unique observation that two domains mediate perinuclear accumulation independently (1-600 and 600-1533). Therefore, multiple domains of Gravin are required for proper subcellular localization, a trait conserved from fish to mammals [17]. Since the short N-terminal fragments containing the BRs are sufficient to promote cell spreading, we next sought to directly test the role of each of the BRs in cell spreading and subcellular localization.
N-terminal BRs are Required for Cell Spreading
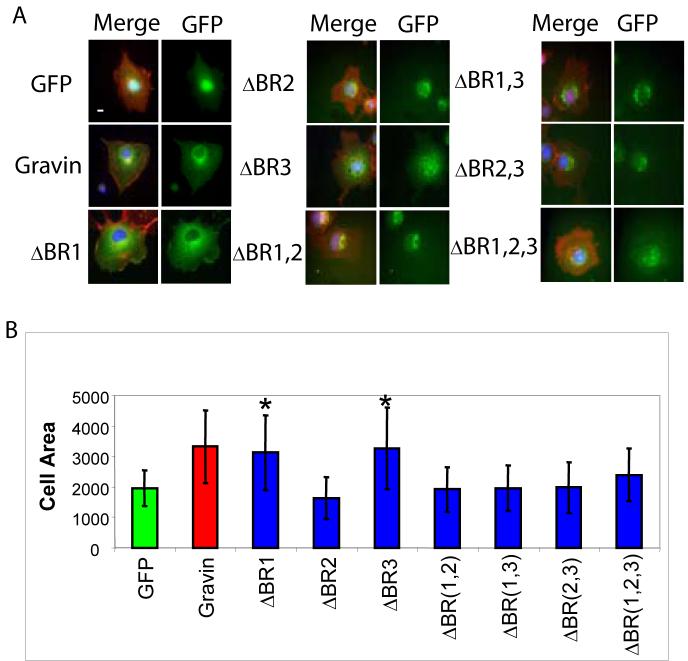
We generated a series of point mutants in each of the BRs in the full-length Gravin (Figure 3A). Each BR contains between one and three conserved serine or threonine residues that may be phosphorylated in mammalian cells and which mediate reversible membrane interaction [17]. We substituted aspartic acid for each of the conserved serine and threonines to mimic phosphorylation, which is proposed to abrogate the BR's electrostatic interaction with the membrane [6, 16, 17]. We generated mutants in BR1 (T82D, T91D), BR2 (S222D) and BR3 (T378D, S386D, S387D) singly; BR1 and BR2, BR2 and BR3, BR1 and BR3 in combination; and all three in combination. Mutation of BR1 or BR3 alone did not reduce the extent of cell spreading relative to full-length Gravin (Figure 3B). However, mutation of BR2 reduced cell flattening to the same extent as the negative control (Figure 3B). Mutation of any two BRs (1 and 2, 2 and 3, or 1 and 3) or all three resulted in no increase in cell flattening compared to the negative control (Figure 3B).
Figure 3. The BRs are required to promote cell spreading.
A. Expression of BR mutants in COS7 cells. A merged images showing GFP (green), Actin (red) and DNA (blue) and as GFP alone are shown. B. Quantification of cell spreading. The y-axis is cell area (μM2) and x-axis shows the expressed construct. * indicates increased cell flattening relative to the GFP control p<0.05.
In addition to our analysis of cell spreading we also confirmed the role of the BRs in subcellular localization. We saw that mutation of any BR increased perinuclear accumulation at the expense of cytoplasmic or peripheral membrane localization (Figure 3A). All three double mutants displayed exclusively perinuclear localization (Figure 3A). The triple mutant lacking function in all three BRs localized primarily to the nucleus and to perinuclear puncta (Figure 3A). This observation is in direct support of previous observations that increasing Gravin phophorylation promotes redistribution from the peripheral membrane to the perinucleus [17, 20].
Thus, in the context of the full-length Gravin, at least two BRs were required to induce cell flattening. However, N-terminal fragments such as 1-100 containing only a single BR could elicit WT levels of cell spreading (Figure 2). This indicates that C-terminal domains may have a negative effect on cell flattening. Thus, these more C-terminal domains can inhibit Gravin-induced cell flattening in the context of full-length Gravin, but in their absence, the extreme N-terminus can elicit cell flattening containing only a single BR. Also, expression of 265-600 of GFP Gravin produced no increase in cell spreading (Figure 2C) even though it contained BR2 and BR3. This is likely due to the localization of 265-600, almost exclusively localized in the nucleus where it is unable to regulate the actin cytoskeleton (Figure 2B).
Therefore, the BRs are necessary for proper Gravin subcellular localization and we provide the first evidence that the N-terminal region and BRs are both necessary and sufficient for Gravin to promote cell spreading. We next sought to determine if the BRs are also essential for Gravin function in vivo.
BRs are Required to Rescue Gravin Loss-of-Function In Vivo
We showed previously that Gravin plays a critical role in regulating a transition in behavior of gastrula stage mesodermal cells from highly migratory to intercalative [17]. Without Gravin function, zebrafish embryos display serious defects in convergence-extension movements of gastrulation and fail to elongate the body axis [15]. These results were obtained using a morpholino antisense oligonucleotide that blocks gravin mRNA splicing [15]. Importantly, normal gastrulation could be rescued by co-injecting gravin mRNA [15]. This provides a very convenient assay for assaying Gravin mutants in vivo. To determine if Gravin requires the BRs in vivo, we injected mRNA encoding Gravin with point mutations in all three BRs [ΔBR(1,2,3)] into morphant embryos and compared the rescue to that caused by wild type gravin mRNA. Whereas wild type gravin mRNA rescued the phenotype in approximately 50% of the embryos, gravin mRNA without the functional BRs failed to rescue the gastrulation phenotype (Figure 4D), even though the constructs express equally well (data not shown). Thus, we have established the first in vivo structure-function assay for Gravin and demonstrated that the BRs are required in vivo.
Figure 4. The Gravin BRs are required to regulate mesodermal cell behavior during zebrafish gastrulation.
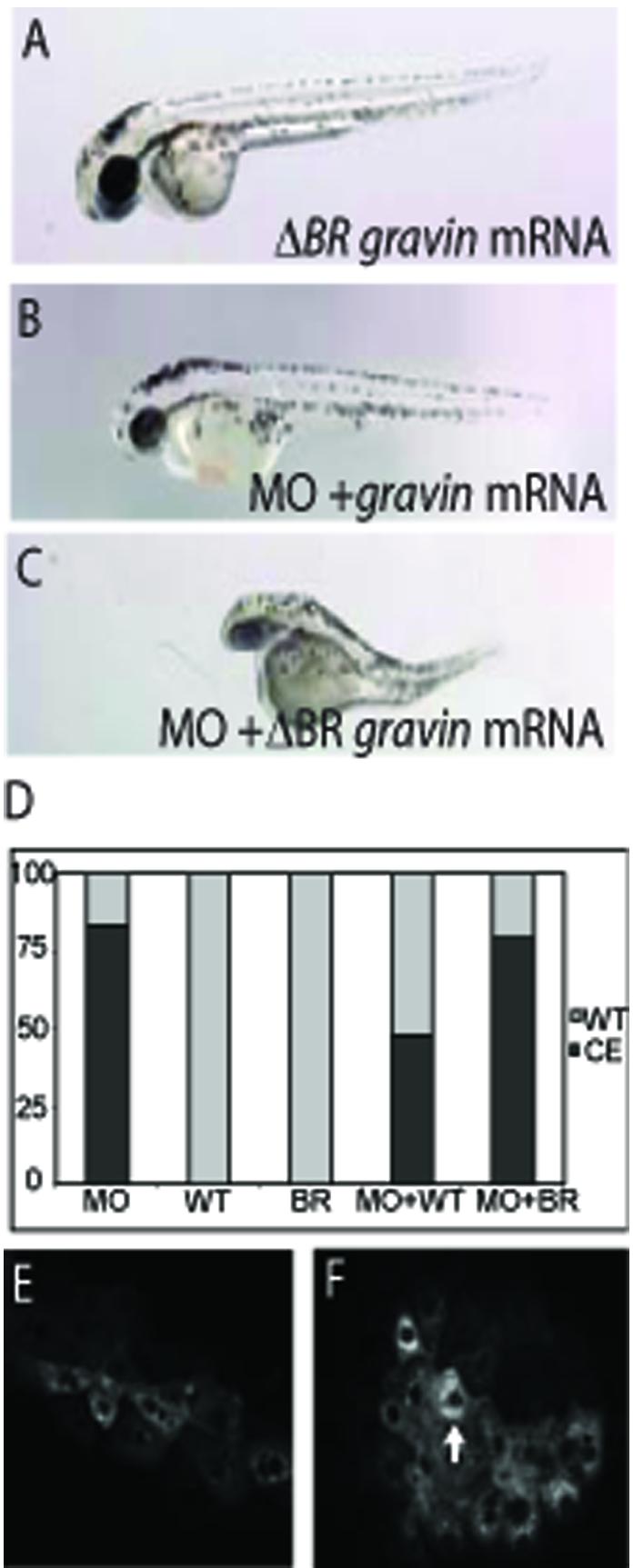
(A) A gravin ΔBR(1,2,3) mRNA injected embryo with a wild type phenotype. (B) An embryo co-injected with the Gravin morpholino (MO) and wild type gravin mRNA with a rescued phenotype. (C) An embryo co-injected with the Gravin morpholino and gravin ΔBR(1,2,3) mRNA displaying a convergent extension (CE) phenotype. (D) Quantification of CE phenotype at 48 hours. Y-axis shows percent of embryos displaying a WT/Rescued phenotype in gray or a CE phenotype in black. The x-axis indicates injected reagent: WT = wild type gravin mRNA, MO = morpholino, BR = gravin ΔBR(1,2,3) mRNA. Expression of GFP-Gravin (E) or ΔBR(1,2,3) Gravin (F) in zebrafish mesodermal cells at the end of gastrulation. Images are at 20X, white arrow indicates increased perinuclear localization.
We also sought to determine the subcellular localization of Gravin ΔBR(1,2,3) in vivo in intact zebrafish embryos. Confocal images of GFP-Gravin and GFP-ΔBR(1,2,3) Gravin in live embryonic mesodermal cells are shown in Figures 4E and F. GFP-Gravin shows a strikingly similar localization in mesodermal cells as it does in COS7 cells with largely diffuse cytoplasmic localization and perinuclear enrichment, with a few cells showing expression at the peripheral membrane. In contrast, ΔBR(1,2,3) GFP-Gravin showed greatly increased localization to the perinucleus, but did not show the nuclear accumulation seen in COS7 cells (Figure 4E). Therefore, the BRs mediate subcellular localization not only in cultured cells but in zebrafish mesodermal cells in vivo.
Conclusion
We demonstrated here that Gravin has a highly dynamic subcellular localization during cell spreading and that this localization is mediated by multiple domains within Gravin. We also provide the first evidence that the N-terminal BRs of Gravin are required for Gravin-induced cell shape changes both in cell spreading assays and in zebrafish mesodermal cells in vivo. Many questions still remain about how Gravin mediates cell behavior changes, including which binding partners are critical, what downstream targets and upstream regulators are involved and the role of possible post-translation modification. Our experimental approach combining in vitro cell flattening assays and in vivo zebrafish experiments will provide a useful tool to elucidate the mechanisms underlying Gravin's role in controlling cell migration during development and cancer cell invasion.
Acknowledgments
We thank Dr. Steve Hauschka for the kind use of his microscope and tissue culture facility, and Hank Farr for technical assistance. This work was supported by NIH grant RO1HD27262 to DK and an NRSA fellowship F32HD053189 to DCW.
Abbreviations
- AKAP
A Kinase Anchoring Protein
- MARCKS
Myristoylated alanine-rich C kinase substrate
- CE
convergent extension
- MO
morpholino antisense oligonucleotide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Nauert JB, Klauck TM, Langeberg LK, Scott JD. Gravin, an autoantigen recognized by serum from myasthenia gravis patients, is a kinase scaffold protein. Curr Biol. 1997;7:52–62. doi: 10.1016/s0960-9822(06)00027-3. [DOI] [PubMed] [Google Scholar]
- [2].Wong W, Scott JD. AKAP signalling complexes: focal points in space and time. Nat Rev Mol Cell Biol. 2004;5:959–970. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- [3].Diviani D, Scott JD. AKAP signaling complexes at the cytoskeleton. J Cell Sci. 2001;114:1431–1437. doi: 10.1242/jcs.114.8.1431. [DOI] [PubMed] [Google Scholar]
- [4].Shih M, Lin F, Scott JD, Wang HY, Malbon CC. Dynamic complexes of beta2-adrenergic receptors with protein kinases and phosphatases and the role of gravin. J Biol Chem. 1999;274:1588–1595. doi: 10.1074/jbc.274.3.1588. [DOI] [PubMed] [Google Scholar]
- [5].Tao J, Wang HY, Malbon CC. Src docks to A-kinase anchoring protein gravin, regulating beta2-adrenergic receptor resensitization and recycling. J Biol Chem. 2007;282:6597–6608. doi: 10.1074/jbc.M608927200. [DOI] [PubMed] [Google Scholar]
- [6].Tao J, Shumay E, McLaughlin S, Wang HY, Malbon CC. Regulation of AKAP-membrane interactions by calcium. J Biol Chem. 2006;281:23932–23944. doi: 10.1074/jbc.M601813200. [DOI] [PubMed] [Google Scholar]
- [7].Wassler MJ, Foote CI, Gelman IH, Shur BD. Functional interaction between the SSeCKS scaffolding protein and the cytoplasmic domain of beta1,4-galactosyltransferase. J Cell Sci. 2001;114:2291–2300. doi: 10.1242/jcs.114.12.2291. [DOI] [PubMed] [Google Scholar]
- [8].Wang HY, Tao J, Shumay E, Malbon CC. G-Protein-coupled receptor-associated A-kinase anchoring proteins: AKAP79 and AKAP250 (gravin) Eur J Cell Biol. 2006;85:643–650. doi: 10.1016/j.ejcb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- [9].Gelman IH. The role of SSeCKS/gravin/AKAP12 scaffolding proteins in the spaciotemporal control of signaling pathways in oncogenesis and development. Front Biosci. 2002;7:d1782–1797. doi: 10.2741/A879. [DOI] [PubMed] [Google Scholar]
- [10].Akakura S, Huang C, Nelson PJ, Foster B, Gelman IH. Loss of the SSeCKS/Gravin/AKAP12 gene results in prostatic hyperplasia. Cancer Res. 2008;68:5096–5103. doi: 10.1158/0008-5472.CAN-07-5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lin X, Gelman IH. Reexpression of the major protein kinase C substrate, SSeCKS, suppresses v-src-induced morphological transformation and tumorigenesis. Cancer Res. 1997;57:2304–2312. [PubMed] [Google Scholar]
- [12].Xia W, Unger P, Miller L, Nelson J, Gelman IH. The Src-suppressed C kinase substrate, SSeCKS, is a potential metastasis inhibitor in prostate cancer. Cancer Res. 2001;61:5644–5651. [PubMed] [Google Scholar]
- [13].Gelman IH, Gao L. SSeCKS/Gravin/AKAP12 metastasis suppressor inhibits podosome formation via RhoA- and Cdc42-dependent pathways. Mol Cancer Res. 2006;4:151–158. doi: 10.1158/1541-7786.MCR-05-0252. [DOI] [PubMed] [Google Scholar]
- [14].Cheng C, Liu H, Ge H, Qian J, Qin J, Sun L, Shen A. Essential role of Src suppressed C kinase substrates in endothelial cell adhesion and spreading. Biochem Biophys Res Commun. 2007;358:342–348. doi: 10.1016/j.bbrc.2007.04.147. [DOI] [PubMed] [Google Scholar]
- [15].Weiser DC, Pyati UJ, Kimelman D. Gravin regulates mesodermal cell behavior changes required for axis elongation during zebrafish gastrulation. Genes Dev. 2007;21:1559–1571. doi: 10.1101/gad.1535007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Disatnik MH, Boutet SC, Pacio W, Chan AY, Ross LB, Lee CH, Rando TA. The bi-directional translocation of MARCKS between membrane and cytosol regulates integrin-mediated muscle cell spreading. J Cell Sci. 2004;117:4469–4479. doi: 10.1242/jcs.01309. [DOI] [PubMed] [Google Scholar]
- [17].Streb JW, Miano JM. Cross-species sequence analysis reveals multiple charged residue-rich domains that regulate nuclear/cytoplasmic partitioning and membrane localization of AKAP 12 (SSeCKS/Gravin) J Biol Chem. 2005;280:28007–28014. doi: 10.1074/jbc.M414017200. [DOI] [PubMed] [Google Scholar]
- [18].Dell'Acqua ML, Faux MC, Thorburn J, Thorburn A, Scott JD. Membrane-targeting sequences on AKAP79 bind phosphatidylinositol-4, 5-bisphosphate. Embo J. 1998;17:2246–2260. doi: 10.1093/emboj/17.8.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gelman IH, Lee K, Tombler E, Gordon R, Lin X. Control of cytoskeletal architecture by the src-suppressed C kinase substrate, SSeCKS. Cell Motil Cytoskeleton. 1998;41:1–17. doi: 10.1002/(SICI)1097-0169(1998)41:1<1::AID-CM1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- [20].Lin X, Tombler E, Nelson PJ, Ross M, Gelman IH. A novel src- and ras-suppressed protein kinase C substrate associated with cytoskeletal architecture. J Biol Chem. 1996;271:28430–28438. doi: 10.1074/jbc.271.45.28430. [DOI] [PubMed] [Google Scholar]