Summary
Klippel-Trenaunay syndrome (KTS) is a severe congenital disorder characterized by capillary malformations, venous malformations or varicose veins, and hypertrophy of the affected tissues. The angiogenic factor gene AGGF1 was previously identified as a candidate susceptibility gene for KTS, but further genetic studies are needed to firmly establish the genetic relationship between AGGF1 and KTS. We analyzed HapMap data and identified two tagSNPs, rs13155212 and rs7704267 that capture information for all common variants in AGGF1. The two SNPs were genotyped in 173 Caucasian KTS patients and 477 Caucasian non-KTS controls, and both significantly associated with susceptibility for KTS (P = 0.004 and 0.013, respectively). Permutation testing also showed a significant empirical P value for the association (empirical P = 0.006 and 0.015, respectively). To control for potential confounding due to population stratification, the population structure for both cases and controls was characterized by genotyping of 38 ancestry-informative markers (AIMs) and the STRUCTURE program. The association between the AGGF1 SNPs and KTS remained significant after multivariate analysis by incorporating the inferred cluster scores as a covariate or after removal of outlier individuals identified by STRUCTURE. These results suggest that common AGGF1 variants confer risk of KTS.
Keywords: AGGF1 (VG5Q), single nucleotide polymorphism (SNP), Klippel-Trenaunay Syndrome (KTS), case-control association study, linkage disequilibrium (LD) block, ancestry-informative markers (AIMs), structured association
Introduction
Klippel-Trenaunay syndrome (KTS, MIM #149000) is a congenital vascular disease characterized by cutaneous capillary malformations, venous malformations or varicose veins and hypertrophy of bone and soft tissues (Berry et al. 1998; Jacob et al. 1998; Kihiczak et al. 2006; Timur et al. 2005). This triad of clinical features affects most KTS patients but the presence of two out of three is sufficient to make a diagnosis (Jacob et al. 1998). The molecular mechanism for the pathogenesis of KTS remains to be fully elucidated. Although KTS is a congenital disorder, most KTS patients develop the disease sporadically and show a mosaic pattern for the affected tissues. To explain these observations, a paradominant hypothesis was proposed, namely, KTS has been hypothesized to be a paradominant disorder where “two hits” in KTS susceptibility genes are required to develop the KTS phenotype (Happle, 1986, 1987, 1993; Tian et al. 2004; Wang, 2005). Due to its sporadic characteristic, KTS may turn out to be a rare complex disease which is caused by multiple genetic factors, environmental factors, and gene-gene and gene-environment interactions.
Previous genetics studies have identified three chromosomal abnormalities in three separate KTS patients: two balanced translocations t(5.11)(q13.3;p15.1) and t(8,14)(q22.3;q13), and an extra supernumerary ring chromosome 18 (Tian et al. 2004; Timur et al. 2004; Wang et al. 2001; Whelan et al. 1995). Molecular characterization of KTS-associated translocation t(5.11)(q13.3;p15.1) advanced far ahead of the other two cytogenetic anomalies. Both the 5q13.3 and 11p15.1 breakpoints were successfully cloned and sequenced (Tian et al. 2004). No gene was identified within 100 kb region at the 11p15.1 breakpoint, but a novel gene named as AGGF1 (previously VG5Q) was at the 5q13.3 breakpoint (Tian et al. 2004). AGGF1 encodes a novel angiogenic factor with G-patch and FHA domains 1 (Tian et al. 2004). Full-length human AGGF1 mRNA is spliced from a transcript of 14 exons spanning 34,807 nucleotides of genomic DNA and encodes a protein consisting of 714 amino acids, which is highly expressed in the blood vessels (Tian et al. 2004). The 5q breakpoint is located in the promoter region of AGGF1. The t(5:11) translocation increased transcription of AGGF1 by three-fold. Purified recombinant AGGF1 promoted angiogenesis as potently as VEGF (Tian et al. 2004), and over-expression of AGGF1 in human umbilical endothelial cells increased vessel formation in a Matrigel angiogenesis assay (Timur et al. 2005; Wang 2005). AGGF1 knockout mice showed markedly reduced vessel density (data not shown). Together these results strongly suggest that AGGF1 is a gene that can increase susceptibility for KTS. However, more genetic studies are needed to firmly establish the relationship between AGGF1 and KTS.
In this study, we employed state-of-the-art genetic tools to study KTS as a complex genetic trait. Recent development of the HapMap database and technological advances for the estimate of linkage disequilibrium (LD) block and identification of tagging single nucleotide polymorphisms (tagSNPs) have revolutionized our strategy to comprehensively assess the genetic association of a candidate gene with the disease under study. In the current study, we have performed a larger scale case-control association analysis of AGGF1 variants using genomic DNA from 173 Caucasian KTS patients and 477 non-KTS Caucasian controls. To minimize the confounding effect of population admixture, we used the STRUCTURE program to determine the population structure of cases and controls, and then carried out the structured association analysis. We found that two tagSNPs that capture the information of all common variants in AGGF1 were significantly associated with KTS in this Caucasian cohort by both traditional association and structured association analyses.
Materials and Methods
Study Subjects and Preparation of Genomic DNA
KTS patients (173 cases) and non-KTS controls (477 controls) were Caucasian. Patients and controls of other ethnic origins were excluded to avoid the confounding effects of population admixture. There were 76 males and 97 females in the KTS case group and 225 males and 252 females in the control group. The cases were recruited in North America for this study. The diagnosis of KTS was based on published reports (Berry et al. 1998; Jacob et al. 1998; Timur et al. 2004, 2005; Wang et al. 2001). For the case-control study, we applied stringent criteria for the diagnosis, and all cases must satisfy all three cardinal features of KTS to be included: capillary malformations, varicose veins or venous malformations, and hypertrophy of affected tissues. The controls were selected from more than 9,800 individuals who underwent coronary angiography in Cardiac Catheterization Laboratories at Cleveland Clinic (Cleveland GeneBank), and only Caucasian individuals without detectable atherosclerotic lesions by angiography were selected (Shen et al. 2007). A total of 560 individuals without coronary artery disease were identified. Then, 83 individuals with peripheral atherosclerotic diseases were excluded. Finally, 477 individuals without any vascular disease were included as controls. The 173 KTS cases did not show any evidence of coronary artery disease or peripheral atherosclerotic diseases either.
This study has been approved by the Cleveland Clinic Foundation and Mayo Clinic Institutional Review Boards on Human Subject Research. Informed consent was obtained from all participants according to the standards established by the local Institutional Review Boards. Genomic DNA was prepared from 10 ml of whole blood using the DNA Isolation Kit for Mammalian Blood (Roche Diagnostics Co., Indianapolis, IN).
SNP Genotyping
We analyzed the SNP genotyping data in the HapMap database (http://www.hapmap.org/) using the Haploview software version 3.0 package (www.broad.mit.edu/mpg/haploview/ download.php) and found that the entire AGGF1 gene is located within one single LD block. The Tagger program (Paul de Bakker, http://www.broad.mit.edu/mpg/tagger) (Ke & Cardon, 2003) was used to select the haplotype-tagged SNP (tagSNP), rs7704267. We also selected SNP rs13155212 for analysis due to its exonic location.
High throughput SNP genotyping was performed using the 5′ nuclease allelic discrimination assay (TaqMan assay) on an ABI PRISM 7900HT Sequence Detection System. SNP probes were purchased through TaqMan Assays-by-Demand or Assays-by-Design from ABI (Applied Biosystems, Foster City, CA, USA). Genotyping was performed in a total 5 μl of PCR reaction volume containing 2.5 μl of TaqMan Universal PCR Master Mix, 0.25 μl of 20X TaqMan MGB Assay Mix, and 25 ng of human genomic DNA. Automatic allele calling was carried out by ABI PRISM 7900HT data collection and analysis software version 2.1.
Statistic Analysis
Statistical analyses of genotyping data were carried out as described previously (Shen et al. 2007, 2008; Shen et al. 2008). Briefly, SNPs were tested for Hardy-Weinberg equilibrium among cases and controls using the Haploview software version 3.0 package (www.broad.mit.edu/mpg/haploview/download.php). Both SNPs were in Hardy-Weinberg equilibrium (P > 0.05, Table 1). Haplotypes were estimated using the PHASE software (www.stat.washington.edu/stephens/software.html). Pairwise linkage disequilibrium values (D′ where D′ = D/Dmax) were obtained from haplotypic data using the Haploview v3.0 program. Association of a SNP or a SNP haplotype with susceptibility to KTS development was assessed using Pearson's 2 × 2 contingency table Chi-square test (SAS Ver 9.00 or Haploview version 3.0). Association was also analyzed for genotypic frequencies of each SNP using logistic regression assuming either an additive, dominant, or recessive model (SAS Ver 9.00). Odds ratios and 95% confidence intervals were estimated using SAS Ver 9.00. An empirical P value was calculated using 10,000 Monte Carlo simulations by the CLUMP program (http://www.mds.qmw.ac.uk/statgen/dcurtis/software.html) or the Haploview software version 3.0 package.
Table 1.
Genotype frequencies of SNPs in the AGGF1 gene
| SNP | Allele | Case (n = 173) | HWE (case) | Control (n = 477) | HWE (control) |
|---|---|---|---|---|---|
| rs7704267 | CC | 10.39% | 0.75 | 19.70% | 0.11 |
| CG | 45.45% | 45.13% | |||
| GG | 44.16% | 35.17% | |||
| rs13155212 | CC | 3.68% | 0.33 | 8.74% | 1.00 |
| CT | 37.42% | 41.58% | |||
| TT | 58.90% | 49.68% |
HWE, P value for Hardy-Weinberg equilibrium.
Selection of Ancestry-Informative Markers (AIM) and Tests of Associations by Adjusting Population Admixture
To analyze the structure of cases or controls, we selected and genotyped 38 ancestry-informative markers (AIMS) that are able to distinguish Europeans, Chinese, and Africans. These AIMs were previously characterized and published (Enoch et al. 2006). The 38 AIMS were genotyped in both KTS cases and controls using the 5′ nuclease allelic discrimination assay (TaqMan assay) as described above. Eight cases were excluded from further analysis because their genotyping failed for >20% of AIMS. The genotyping data for AIMS was analyzed to obtain estimates of the population structure of KTS cases and controls using the program STRUCTURE 2.1 (http://pritch.bsd.uchicago.edu/structure.html) (Pritchard et al. 2000; Falush et al. 2003). STRUCTURE was run with the MCMC scheme assuming K of 3 (number of populations) and a length burn-in period of 50,000 and 50,000 iterations after burn-in. To adjust for potential population admixture in the association analysis, multivariate analysis was performed by incorporating the inferred cluster scores as a covariate using multivariate logistic regression (SAS Ver 9.00). Alternatively, association analysis was repeated after outlier individuals identified by STRUCTURE were removed from the cases and controls.
Results
We applied the Tagger program to HapMap data to select tagSNPs to capture the AGGF1 gene using the criteria of a minor allele frequency = 0.30 and r2 = 0.80. This resulted in the selection of one AGGF1 tagSNP (rs7704267) that is located in intron 11. SNP rs7704267 was genotyped in 173 KTS cases and 477 controls and the data were calculated (Table 1). SNP rs7704267 frequencies did not display a statistically significant deviation from Hardy-Weinberg equilibrium (P > 0.05).
Allelic association was analyzed by contingency table Chi-square tests. The association of intronic SNP rs7704267 with susceptibility for developing KTS was statistically significant (Table 2, odds ratio or OR = 1.48, P = 0.004). Results for allelic association remained significant following permutation testing (Table 2, Pemp = 0.005).
Table 2.
Association analysis of allelic frequencies of SNPs in AGGF1 with KTS
| SNP Name | P-obs | P-emp | P-adj | Odds Ratio (95%CI) |
|---|---|---|---|---|
| rs7704267 | 0.0044 | 0.0051 | 0.0072 | 1.48 (1.13–1.94) |
| rs13155212 | 0.0131 | 0.0145 | 0.0091 | 1.45 (1.08–1.95) |
P-obs, uncorrected P-value; P-emp, permutation P-value calculated using 10,000 Monte Carlo simulations; P-adj, corrected P-value after adjustment for the inferred cluster scores derived from the STRUCTURE analysis; CI, Confidence Intervals.
In order to provide further support to the association of AGGF1 SNP rs7704267 with KTS susceptibility, we studied one exonic SNP located in exon 7 of the AGGF1 gene (rs13155212, I405I). SNP rs13155212 frequencies also did not deviate from Hardy-Weinberg equilibrium (Table 1, P > 0.05). Allelic association of SNP rs13155212 with KTS susceptibility was also statistically significant (P = 0.013) and remained significant after permutation testing (Table 2, Pemp = 0.015).
Association analysis of both SNPs using recessive, additive and dominant models were then carried out. For both SNPs rs7704267 and rs13155212, all three models were statistically significant: for association of SNP rs7704267 with KTS using the dominant model P = 0.009, OR = 2.12; recessive model P = 0.045, OR = 1.46; additive model P = 0.016, OR = 1.45; for association of SNP rs13155212 with KTS using the dominant model P = 0.038, OR = 2.51; recessive model P = 0.043, OR = 1.45; additive model P = 0.036, OR = 1.46 (Table 3).
Table 3.
Association analysis of SNPs in AGGF1 with KTS using various genetic models
| SNP Name | P-obs | P-emp | P-adj | Odds Ratio (95%CI) | |
|---|---|---|---|---|---|
| rs7704267 | Dominant | 0.009 | 0.010 | 0.013 | 2.12 (1.20–3.72) |
| Additive | 0.016 | 0.017 | 0.023 | 1.45 (1.11–1.89) | |
| Recessive | 0.045 | 0.054 | 0.046 | 1.46 (1.01–2.11) | |
| rs13155212 | Dominant | 0.038 | 0.039 | 0.037 | 2.51 (1.04–6.02) |
| Additive | 0.036 | 0.038 | 0.032 | 1.46 (1.08–1.97) | |
| Recessive | 0.043 | 0.046 | 0.030 | 1.45 (1.01–2.08) |
P-obs, uncorrected P-value; P-emp, permutation P-value calculated using 10,000 Monte Carlo simulations; P-adj, corrected P-value after adjustment for the inferred cluster scores derived from the STRUCTURE analysis.
Haplotype analyses indicated that the TG haplotype (Hap A) was associated with risk for developing KTS (Table 4, P = 0.005, OR = 1.47) while the CC haplotype (Hap B) conferred protection from developing KTS (Table 4, P = 0.012, OR = 0.69).
Table 4.
Protective and risk haplotypes associated with KTS in the AGGF1 gene
| Haplotype | Case | Control | P-obs | P-emp | Odds Ratio (95% CI) |
|---|---|---|---|---|---|
| Hap A (TG) | 66.50% | 57.40% | 0.0048 | 0.0145 | 1.467 (1.13–1.91) |
| Hap B (CC) | 22.26% | 29.40% | 0.0117 | 0.0333 | 0.687 (0.51–0.92) |
P-obs, uncorrected P-value; P-emp, permutation P-value calculated using 10,000 Monte Carlo simulations.
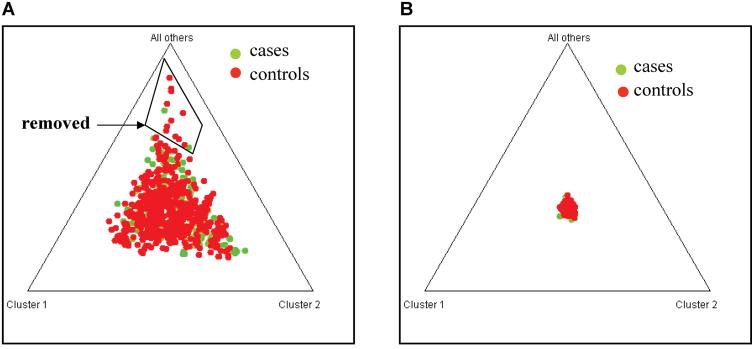
To minimize the confounding effects of population stratification, we analyzed and compared the population structure of both cases and controls. Thirty eight ancestry-informative markers (AIMS) were genotyped in cases and controls and the genotyping data was analyzed using the STRUCTURE program, which has been widely used for analyzing population structure of humans with multi-locus genotype data. The frequencies of the reference alleles of 38 AIMS are shown in Table 5, and demonstrated remarkable similarity to the HapMap frequencies for the Caucasian population, but much disparity from the Chinese population or the African population. Nevertheless, the STRUCTURE program identified 14 individuals, two cases and 12 controls that are outlier individuals (Figure 1). When the 14 outliers were removed, the structure of the cases completely overlapped with that of the controls (Figure 1). Association was re-analyzed by excluding the 14 outlier individuals. Both allelic and genotypic associations between the AGGF1 SNPs and KTS remained significant after the structured association analysis (Tables 6-8). The two most common haplotypes were also significantly associated with KTS after the structured association analysis (Table 9).
Table 5.
Frequencies of reference alleles of 38 AIMs in three ethnic populations and the study populations (KTS cases, Controls)
| SNP ID | CEU Freq of Ref-Allele |
CHB Freq of Ref-Allele |
YRI Freq of Ref-Allele |
KTS Freq of Ref-Allele |
Control Freq of Ref-Allele |
|---|---|---|---|---|---|
| rs10745288 | 0.375 | 0.022 | 0.9 | 0.419 | 0.416 |
| rs10750836 | 0.008 | 0.456 | 0.783 | 0.015 | 0.014 |
| rs10847171 | 0.267 | 0.033 | 0.958 | 0.245 | 0.269 |
| rs1368928 | 0.342 | 0.056 | 0.958 | 0.359 | 0.454 |
| rs1552314 | 0.308 | 0.044 | 0.95 | 0.192 | 0.235 |
| rs1871534 | 1 | 1 | 0.017 | 0.994 | 0.997 |
| rs1885167 | 0.217 | 0.033 | 0.992 | 0.21 | 0.205 |
| rs2625956 | 0.858 | 0.389 | 0.033 | 0.801 | 0.823 |
| rs2730891 | 0.958 | 0.478 | 0.075 | 0.949 | 0.941 |
| rs4721415 | 0.942 | 0.011 | 0.358 | 0.934 | 0.906 |
| rs4968382 | 0.792 | 0.389 | 0.067 | 0.761 | 0.758 |
| rs6593430 | 0.075 | 1 | 1 | 0.077 | 0.067 |
| rs6670693 | 1 | 1 | 0.025 | 0.987 | 1 |
| rs6785846 | 0.208 | 0.067 | 0.958 | 0.167 | 0.15 |
| rs679832 | 1 | 0.011 | 0.183 | 0.664 | 0.668 |
| rs692713 | 0.042 | 0.122 | 0.958 | 0.071 | 0.063 |
| rs9388989 | 0.883 | 0.244 | 0.017 | 0.77 | 0.767 |
| rs12125484 | 0.65 | 0.011 | 0.908 | 0.688 | 0.694 |
| rs733370 | 0.342 | 0.889 | 0.075 | 0.369 | 0.345 |
| rs6595142 | 0.367 | 0.856 | 0.033 | 0.409 | 0.396 |
| rs10488401 | 0.433 | 0.944 | 0.008 | 0.422 | 0.446 |
| rs2927385 | 0.45 | 0.889 | 0.042 | 0.445 | 0.407 |
| rs556399 | 0.525 | 0.078 | 0.875 | 0.546 | 0.553 |
| rs1869237 | 0.625 | 0.011 | 0.833 | 0.574 | 0.572 |
| rs2193595 | 0.483 | 0.933 | 0.092 | 0.481 | 0.496 |
| rs333113 | 0.183 | 1 | 0.075 | 0.255 | 0.232 |
| rs2387137 | 0.167 | 0.833 | 0.042 | 0.191 | 0.162 |
| rs6718709 | 0.55 | 0.022 | 0.825 | 0.513 | 0.555 |
| rs36110 | 0.45 | 0.889 | 0.058 | 0.349 | 0.41 |
| rs1538956 | 0.575 | 0.011 | 0.85 | 0.513 | 0.508 |
| rs6074585 | 0.95 | 0.011 | 0.008 | 0.833 | 0.829 |
| rs16877243 | 0.817 | 0.378 | 0.033 | 0.873 | 0.796 |
| rs326626 | 0.05 | 0.933 | 0.725 | 0.087 | 0.073 |
| rs1507086 | 0.3 | 0.022 | 1 | 0.261 | 0.301 |
| rs2842063 | 0.817 | 0.944 | 0.05 | 0.815 | 0.849 |
| rs10249419 | 0.617 | 1 | 0.083 | 0.692 | 0.686 |
| rs6023367 | 0.717 | 1 | 0.083 | 0.786 | 0.801 |
| rs1894450 | 0.333 | 0.078 | 1 | 0.378 | 0.396 |
CEU, Caucasian ancestry; CHB, Chinese ancestry; YRI, African ancestry. The 38 Ancestry-Informative Markers (AIMS) were selected from 186 AIMS compiled by Enoch et al (Enoch et al. 2006). The reference allele frequencies of the European, Chinese, and African populations were from the HapMap database (http://www.hapmap.org/). Note that the frequencies of reference alleles of all 38 AIMs in both KTS cases and control populations are similar to that in the Caucasian population.
Figure 1.
Identification of population stratification using the STRUCTURE program. (A) Genotyping data for 38 AIMS from cases (green) and controls were analyzed to identify outlier individuals (boxed). The 14 outliers include 2 cases and 12 controls. (B) After removal of 14 outliers, the new cases and controls match each other very well as noted by compact overlapping of cases and controls in the center of the graph.
Table 6.
Genotype frequencies of SNPs in the AGGF1 gene
| SNP | Allele | Case (n = 163) | HWE (case) | Control (n = 465) | HWE (control) |
|---|---|---|---|---|---|
| rs7704267 | CC | 10.53% | 0.78 | 19.09% | 0.19 |
| CG | 45.39% | 45.77% | |||
| GG | 44.08% | 35.14% | |||
| rs13155212 | CC | 3.73% | 0.35 | 8.73% | 0.92 |
| CT | 37.27% | 41.92% | |||
| TT | 59.01% | 49.34% |
HWE, P value for Hardy-Weinberg equilibrium.
Table 8.
Association analysis of SNPs in AGGF1 with KTS using various genetic models
| SNP Name | P-obs | P-emp | Odds Ratio (95%CI) | |
|---|---|---|---|---|
| rs7704267 | Dominant | 0.0147 | 0.0171 | 2.12 (1.20–3.72) |
| Additive | 0.0251 | 0.017 | 1.45 (1.11–1.89) | |
| Recessive | 0.0482 | 0.053 | 1.45 (1.00–2.11) | |
| rs13155212 | Dominant | 0.0372 | 0.0378 | 2.51 (1.04–6.02) |
| Additive | 0.0384 | 0.038 | 1.46 (1.08–1.97) | |
| Recessive | 0.0348 | 0.036 | 1.48 (1.03–2.13) |
P-obs, uncorrected P-value; P-emp, permutation P-value calculated using 10,000 Monte Carlo simulations.
Table 9.
Protective and risk haplotypes associated with KTS in the AGGF1 gene
| Haplotype | Case | Control | P-obs | P-emp | Odds Ratio(95% CI) |
|---|---|---|---|---|---|
| Hap A (TG) | 65.95% | 57.74% | 0.0070 | 0.0071 | 1.35 (1.04–1.76) |
| Hap B (CC) | 22.08% | 29.57% | 0.0100 | 0.0110 | 0.72 (0.53–0.97) |
P-obs, uncorrected P-value; P-emp, permutation P-value calculated using 10,000 Monte Carlo simulations.
Structured association analysis was also carried out using multivariate analysis by including the inferred cl uster scores derived for each case and control from the STRUCTURE program as a covariate. The two SNPs, rs7704267 and rs13155212, showed significant allelic association with KTS with P-adj of 0.0072 and 0.0091, respectively (Table 4). Genotypic association was also significant (Table 3).
Discussion
In this study, we demonstrated that two AGGF1 SNPs located in intron 11 and exon 7 were significantly associated with susceptibility for KTS in a cohort of 173 Caucasian KTS patients and 477 controls. This underscores the complex genetic etiology of KTS. Due to the sporadic nature of most KTS cases and the mosaic pattern of KTS features, KTS development may be explained by a two-hit hypothesis and Happle's concept of paradominant inheritance (Happle, 1986, 1987, 1993; Wang, 2005). Thus, a rare or common germline mutation in a KTS susceptibility gene may increase risk for the disease but alone is not sufficient for disease development. A somatic mutation resulting in a “second hit” in the same KTS susceptibility gene or another is required for the development of clinical KTS symptoms. The study presented here, supports this hypothesis where common alleles in AGGF1 are associated with KTS susceptibility but it is likely that other somatic or germline mutations as well as other genetic and environmental factors are required for the disease to manifest.
An E133K variant in AGGF1 was previously found to be associated with risk of KTS in a small case control study (P = 0.009) (Tian et al. 2004). However, Barker et al. later found that SNP E133K was present at a frequency of 3.3% in 275 healthy, European individuals (Barker et al. 2006), a frequency similar to that in the previous KTS cases (Tian et al. 2004). We analyzed the frequency of SNP E133K in 163 KTS cases and 465 controls characterized in this study. SNP E133K was not in LD with rs7704267 and rs13155212 in both cases and controls (r2 measure of LD equals 0 in both cohorts). The frequency of SNP E133K did not differ significantly between cases and controls (P = 0.59). The new result from this larger case control study suggests that SNP E133K is unlikely to be associated with susceptibility to KTS. The earlier association in Tian et al (2004) may be due to the small sample size and population admixture. On the other hand, the present study clearly demonstrates that two common SNPs in AGGF1, rs7704267 and rs13155212, are associated with risk of KTS even after adjustment for the population structure of both cases and controls. We conclude that AGGF1 remains an interesting candidate gene associated with risk of KTS. This conclusion is strongly supported by the following findings: (i) AGGF1 was the only gene residing within 50 kilobases of the 5p13 breakpoint involved in translocation t(5;11) associated with KTS (Tian et al. 2004). Furthermore, the t(5:11) translocation was found to increase transcription of AGGF1 by three-fold (Tian et al. 2004). These results provide the strongest evidence implicating AGGF1 in the pathogenesis of KTS; (ii) High levels of AGGF1 mRNA and protein expression was detected in blood vessels and cells relevant to KTS; (iii) Purified AGGF1 protein was shown to be a potent angiogenic factor that can promote angiogenesis and knockdown of AGGF1 expression inhibited endothelial vessel formation (Tian et al. 2004); (iv) AGGF1 knockout mice showed markedly reduced vessel density and various vascular phenotypes (data not shown). Despite the above strong evidence that links AGGF1 to KTS, many more future studies are needed to truly establish the true association between AGGF1 and KTS.
Although the polymorphisms in exon 7 and intron 11 did not result in amino acid changes in the AGGF1 protein (I405I), it is still possible that these genetic alterations may function in the regulation of AGGF1 transcription. Many reports suggest that synonymous or intronic SNPs are significantly associated with different diseases such as diabetes, Alzheimer's disease, and schizophrenia by regulating the transcription of the some relevant genes (Law et al. 2007; Moritani et al. 2007; Tokuhiro et al. 2003; Wiener et al. 2007). Recently, a synonymous SNP in exon 26 of the MDR1 gene was shown to exert its functional effect through a novel mechanism of altered timing of co-translational folding and trafficking of P-glycoprotein (Kimchi-Sarfaty et al. 2007; Sauna et al. 2007). It is possible that the synonymous AGGF1 SNP located in exon 7 similarly has a functional effect on the folding and trafficking of the AGGF1 protein. However, further investigations are needed to confirm this hypothesis.
In summary, this study is the largest to date for a case-control association design in studying the genetics of susceptibility to KTS. Our results suggest that KTS is a complex genetic trait for which common variants in the AGGF1 gene may confer a risk.
Table 7.
Association analysis of allelic frequencies of SNPs in AGGF1 with KTS
| SNP Name | P-obs | P-emp | Odds Ratio (95%CI) |
|---|---|---|---|
| rs7704267 | 0.0069 | 0.0072 | 1.45 (1.11–1.91) |
| rs13155212 | 0.0114 | 0.0178 | 1.47 (1.09–1.98) |
P-obs, uncorrected P-value; P-emp, permutation P-value calculated using 10,000 Monte Carlo simulations; CI, Confidence Intervals.
Acknowledgements
We are grateful to all KTS patients for their enthusiastic participation, Judy Vessey at the KT Support Group for her strong support to our genetic research on KTS, and Mary-Anne Enoch at the NIH for her valuable advice and help with the detailed list of AIMs. We acknowledge assistance, advice and discussion from the other members of Wang Laboratory. This study was supported in part by the Scott Hamilton CARES research grant from the Cleveland Clinic Tausig Cancer Center, the NIH grants P50 HL77107, P50 HL81011, and R01 HL66251, an AHA Established Investigator award (0440157N), and the State of Ohio Wright Center of Innovation grant and Biomedical Research and Technology Transfer Partnership Award (BRTT, Ohio's Third Frontier Project).
References
- Barker KT, Foulkes WD, Schwartz CE, Labadie C, Monsell F, Houlston RS, et al. Is the E133K allele of VG5Q associated with Klippel-Trenaunay and other overgrowth syndromes? J Med Genet. 2006;43:613–614. doi: 10.1136/jmg.2006.040790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry SA, Peterson C, Mize W, Bloom K, Zachary C, Blasco P, et al. Klippel-Trenaunay syndrome. Am J Med Genet. 1998;79:319–326. [PubMed] [Google Scholar]
- Enoch MA, Shen PH, Xu K, Hodgkinson C, Goldman D. Using ancestry-informative markers to define populations and detect population stratification. J Psychopharmacol. 2006;20:19–26. doi: 10.1177/1359786806066041. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happle R. Cutaneous manifestation of lethal genes. Hum Genet. 1986;72:280. doi: 10.1007/BF00291899. [DOI] [PubMed] [Google Scholar]
- Happle R. Lethal genes surviving by mosaicism: a possible explanation for sporadic birth defects involving the skin. J Am Acad Dermatol. 1987;16:899–906. doi: 10.1016/s0190-9622(87)80249-9. [DOI] [PubMed] [Google Scholar]
- Happle R. Klippel-Trenaunay syndrome: is it a paradominant trait? Br J Dermatol. 1993;128:465–466. doi: 10.1111/j.1365-2133.1993.tb00214.x. [DOI] [PubMed] [Google Scholar]
- Jacob AG, Driscoll DJ, Shaughnessy WJ, Stanson AW, Clay RP, Gloviczki P. Klippel-Trenaunay syndrome: spectrum and management. Mayo Clin Proc. 1998;73:28–36. doi: 10.1016/S0025-6196(11)63615-X. [DOI] [PubMed] [Google Scholar]
- Ke X, Cardon LR. Efficient selective screening of haplotype tag SNPs. Bioinformatics. 2003;19:287–288. doi: 10.1093/bioinformatics/19.2.287. [DOI] [PubMed] [Google Scholar]
- Kihiczak GG, Meine JG, Schwartz RA, Janniger CK. Klippel-Trenaunay syndrome: a multisystem disorder possibly resulting from a pathogenic gene for vascular and tissue overgrowth. Int J Dermatol. 2006;45:883–890. doi: 10.1111/j.1365-4632.2006.02940.x. [DOI] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Law AJ, Kleinman JE, Weinberger DR, Weickert CS. Disease-associated intronic variants in the ErbB4 gene are related to altered ErbB4 splice-variant expression in the brain in schizophrenia. Hum Mol Genet. 2007;16:129–141. doi: 10.1093/hmg/ddl449. [DOI] [PubMed] [Google Scholar]
- Moritani M, Nomura K, Tanahashi T, Osabe D, Fujita Y, Shinohara S, et al. Genetic association of single nucleotide polymorphisms in endonuclease G-like 1 gene with type 2 diabetes in a Japanese population. Diabetologia. 2007;50:1218–1227. doi: 10.1007/s00125-007-0631-2. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen GQ, Li L, Girelli D, Seidelmann SB, Rao S, Fan C, et al. An LRP8 variant is associated with familial and premature coronary artery disease and myocardial infarction. Am J Hum Genet. 2007;81:780–791. doi: 10.1086/521581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen GQ, Rao S, Martinelli N, Li L, Olivieri O, Corrocher R, et al. Association between four SNPs on chromosome 9p21 and myocardial infarction is replicated in an Italian population. J Hum Genet. 2008;53:144–150. doi: 10.1007/s10038-007-0230-6. [DOI] [PubMed] [Google Scholar]
- Shen GQ, Li L, Rao S, Abdullah KG, Ban JM, Lee BS, et al. Four SNPs on chromosome 9p21 in a South Korean population implicate a genetic locus that confers high cross-race risk for development of coronary artery disease. Arterioscler Thromb Vasc Biol. 2008;28:360–365. doi: 10.1161/ATVBAHA.107.157248. [DOI] [PubMed] [Google Scholar]
- Sauna ZE, Kimchi-Sarfaty C, Ambudkar SV, Gottesman MM. Silent polymorphisms speak: how they affect pharmacogenomics and the treatment of cancer. Cancer Res. 2007;67:9609–9612. doi: 10.1158/0008-5472.CAN-07-2377. [DOI] [PubMed] [Google Scholar]
- Tian XL, Kadaba R, You SA, Liu M, Timur AA, Yang L, et al. Identification of an angiogenic factor that when mutated causes susceptibility to Klippel-Trenaunay syndrome. Nature. 2004;427:640–645. doi: 10.1038/nature02320.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timur AA, Sadgephour A, Graf M, Schwartz S, Libby ED, Driscoll DJ, et al. Identification and molecular characterization of a de novo supernumerary ring chromosome 18 in a patient with Klippel-Trenaunay syndrome. Ann Hum Genet. 2004;68:353–361. doi: 10.1046/j.1529-8817.2004.00095.x. [DOI] [PubMed] [Google Scholar]
- Timur AA, Driscoll DJ, Wang Q. Biomedicine and diseases: the Klippel-Trenaunay syndrome, vascular anomalies and vascular morphogenesis. Cell Mol Life Sci. 2005;62:1434–1447. doi: 10.1007/s00018-005-4523-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuhiro S, Yamada R, Chang X, Suzuki A, Kochi Y, Sawada T, et al. An intronic SNP in a RUNX1 binding site of SLC22A4, encoding an organic cation transporter, is associated with rheumatoid arthritis. Nat Genet. 2003;35:341–348. doi: 10.1038/ng1267. [DOI] [PubMed] [Google Scholar]
- Wang Q, Timur AA, Szafranski P, Sadgephour A, Jurecic V, Cowell J, et al. Identification and molecular characterization of de novo translocation t(8;14)(q22.3;q13) associated with a vascular and tissue overgrowth syndrome. Cytogenet Cell Genet. 2001;95:183–188. doi: 10.1159/000059343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QK. Update on the molecular genetics of vascular anomalies. Lymphat Res Biol. 2005;3:226–233. doi: 10.1089/lrb.2005.3.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan AJ, Watson MS, Porter FD, Steiner RD. Klippel-Trenaunay-Weber syndrome associated with a 5:11 balanced translocation. Am J Med Genet. 1995;59:492–494. doi: 10.1002/ajmg.1320590416. [DOI] [PubMed] [Google Scholar]
- Wiener HW, Perry RT, Chen Z, Harrell LE, Go RC. A polymorphism in SOD2 is associated with development of Alzheimer's disease. Genes Brain Behav. 2007;6:770–775. doi: 10.1111/j.1601-183X.2007.00308.x. [DOI] [PubMed] [Google Scholar]