Abstract
Innervation is important for normal metabolism in skeletal muscle, including insulin-sensitive glucose uptake. However, the transcription factors that transduce signals from the neuromuscular junction to the nucleus and affect changes in metabolic gene expression are not well defined. We demonstrate here that the orphan nuclear receptor Nur77 is a regulator of gene expression linked to glucose utilization in muscle. In vivo, Nur77 is preferentially expressed in glycolytic compared to oxidative muscle and is responsive to β-adrenergic stimulation. Denervation of rat muscle compromises expression of Nur77 in parallel with that of numerous genes linked to glucose metabolism, including GLUT4 and genes involved in glycolysis, glycogenolysis, and the glycerophosphate shuttle. Ectopic expression of Nur77, either in rat muscle or in C2C12 muscle cells, induces expression of a highly overlapping set of genes, including GLUT4, muscle phosphofructokinase, and glycogen phosphorylase. Furthermore, selective knockdown of Nur77 in rat muscle by shRNA or genetic deletion of Nur77 in mice reduces the expression of a battery of genes involved in skeletal muscle glucose utilization in vivo. Finally, we show that Nur77 binds the promoter regions of multiple innervation-dependent genes in muscle. These results identify Nur77 as a potential mediator of neuromuscular signaling in the control of metabolic gene expression.
Keywords: Nur77, glucose metabolism, skeletal muscle
Introduction
Skeletal muscle is the largest glucose-utilizing organ in the human body. In response to exercise or stress, skeletal muscle metabolizes glucose substrates prior to mobilizing lipid or protein for energy production. This well-orchestrated process involves glucose transport, glycolysis, and glycogenolysis. Perturbation of each of these processes can disrupt glucose utilization and cause untoward physiologic effects. For instance, mice with muscle specific deletions of the insulin-sensitive glucose transporter GLUT4 are insulin resistant (1). Patients with type 2 diabetes mellitus have impaired glucose uptake in skeletal muscle (2). Conversely, glucose uptake is enhanced with exercise, and is mirrored by an increase in GLUT4 protein level as well as transporter translocation (2-4). It has been hypothesized that increasing Glut4 expression in muscle may be a possible route to type 2 diabetes therapy, a notion supported by studies showing that increasing Glut4 expression in db/db transgenic mice improve glucose disposal (5).
Several members of the nuclear receptor superfamily have been shown to regulate glucose metabolism in vivo, including the glucocorticoid receptor (GR), hepatocyte nuclear factor 4 (HNF4), peroxisome proliferator-activated receptors (PPARs), liver X receptors (LXRs), and the NR4A orphan nuclear receptors (6-11). The NR4A subfamily consists of 3 well-conserved members – NR4A1, NR4A2, and NR4A3, otherwise known in mouse as Nur77, Nurr1, and NOR1, respectively. Unlike PPARs and LXR, the NR4As do not possess ligand-binding cavities (12, 13). Instead, NR4As are immediate early genes whose expression is induced rapidly by various stimulus including cAMP, growth factors, inflammatory signals, and parathyroid hormone (14). Regulation of the NR4A receptors is thought to occur predominantly at the level of protein expression and post-transcriptional regulation. Previous work has established pleotropic and cell-type dependent functions for this widely expressed group of receptors (15-17). The potential role of NR4A receptors in metabolism has not been well explored. We have shown that NR4A receptors contribute to the hormonal response of gluconeogenic genes in the liver, pointing to an unexpected function for this receptor family in glucose metabolism (10). Maxwell and colleagues have reported that Nur77 is induced in response to adrenergic stimulation in the C2C12 murine skeletal muscle cell line and that stable expression of an siRNA construct targeting Nur77 suppresses lipolysis (18). The function of NR4A receptors in skeletal muscle metabolism in vivo is currently unknown.
In order to identify potential transcriptional mediators of neural control of muscle gene expression, we performed expression profiling on denervated rat muscle. We discovered that expression of Nur77 was profoundly downregulated in response to denervation. Interestingly, loss of Nur77 expression correlated not only with decreased expression of GLUT4, but also with decreased expression of a battery of genes involved in glucose and glycogen metabolism. Ectopic expression of Nur77 in differentiated C2C12 cells and denervated EDL muscle induced expression of this same set of genes, whereas genetic deletion of Nur77 in mice compromised their expression. These results establish Nur77 as a potential mediator of neuromuscular signaling in the control of glucose metabolic gene expression.
Results
To identify innervation-dependent gene expression, we performed expression profiling on rat extensor digitorum longus (EDL) muscle. Total RNA was extracted from the EDL muscle of sham-operated and denervated legs three days later, and subjected to microarray profiling. Expression data for selected genes from denervated animals is shown in Table 1. Glut4 expression was markedly reduced by denervation, as reported previously (19). We also observed a marked decrease in expression of a battery of genes involved in glucose utilization in denervated muscle. These genes included those involved in glycolysis (muscle phosphofructokinase – Pfkm, phosphoglycerate mutase 2 – Pgam2, 2,3-bisphosphoglycerate mutase – Bpgm), glycerophosphate shuttle (glycerol-3-phosphate dehydrogenase 1 – GPD1), and glycogenolysis (phosphorylase kinase gamma 1 – Phkg1, and muscle glycogen phosphorylase – Pygm). We also endeavored to identify transcriptional regulators whose expression was altered by denervation. Strikingly, Nur77 expression was reduced in denervated muscle to a similar extent as Glut4 (Table 1), suggesting a link between Nur77 and glucose metabolic gene expression in this tissue.
Table 1.
Denervation downregulates glucose metabolism gene expression
| Metabolic Pathway | Accession Number | Gene Name | Symbol | Fold reduction (control/denervation) | Fold induction* (Ad-Nur77/Ad-GFP) |
|---|---|---|---|---|---|
| Glucose uptake | NM_009204 NM_012751 | insulin sensitive glucose transporter | Slc2a4 (Glut4) | 4.6 | 8.2 |
| Glycolysis | NM_021514 NM_031715 | phosphofructokinase, muscle | Pfkm | 3.8 | 2.0 |
| NM_018870 NM_017328 | phosphoglycerate mutase 2 | Pgam2 | 3.4 | 3.2 | |
| NM_007563 NM_199382 | 2,3-bisphosphoglycerate mutase | Bpgm | 3.1 | 2.0 | |
| Glycerophosphate shuttle | NM_010271 NM_022215 | glycerol-3-phosphate dehydrogenase 1 (soluble) | Gpd1 | 3.6 | 3.9 |
| Glycogenolysis | J03293 NM_031573 | phosphorylase kinase gamma 1 | Phkg1 | 5.1 | 40.4 |
| NM_011224 XM_001076049 | muscle glycogen phosphorylase | Pygm | 1.5 | 4.1 | |
| Miscellaneous | NM_007994 NM_053716 | fructose bisphosphatase 2 | Fbp2 | 20.0 | 171 |
| NM_007498.2 NM_012912 | activating transcription factor 3 | Atf3 | 0.5 | 0.14 | |
| NM_010444 NM_024388 | nuclear receptor subfamily 4, group A, member 1 | Nr4a1 (Nur77) | 4.8 | 17.1 |
For each gene, the top accession number represents mouse and the bottom number represents rat.
Expression profiling result of C2C12 myotubes ectopically expressing Nur77 vs control (GFP)
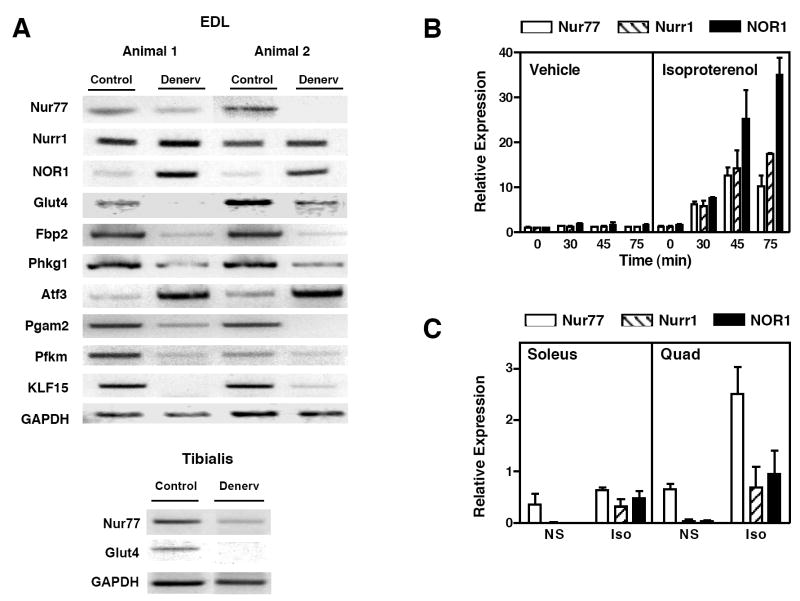
Microarray results were verified by RT-PCR (Figure 1a). Expression of Glut4, Nur77, Phkg1, Pgam2, and Pfkm was significantly reduced in denervated muscles, in complete agreement with the data of Table 1. We also examined the expression of the other two NR4A receptors – Nurr1 and NOR1. While Nurr1 expression appeared unchanged, NOR1 mRNA was upregulated in response to denervation.. As controls, we also confirmed expected changes in the expression of genes reported to increase upon denervation, including myosin heavy chain, the acetylcholine receptor, myogenin and MyoD (data not shown) (20-22). Atf3 expression was also upregulated, consistent with previous reports of increased Atf3 expression following peripheral nerve injury (23). We also observed a profound loss of Fbp2 expression upon denervation. Fbp2 shows sequence homology to the liver fructose bisphosphatase (Fbp1), a key regulator of gluconeogenesis, but its function in skeletal muscle is presently unknown.
Figure 1.
Innervation-dependent expression of Nur77 and glucose metabolism genes. (a) One leg each of two rats was denervated (“denerv”), and the other leg was sham-operated (“control”). Total RNA of EDL muscle was extracted three days after surgery and RT-PCR was performed. (b) β-adrenergic receptor activation of NR4A expression in vitro. Differentiated C2C12 cells were treated with isoproterenol 100 nM. Cells were harvested for RNA isolation at time points shown. (c) Isoproterenol activation of NR4A expression in vivo. Fed-wildtype C57BL/6 mice (n=2, male) were injected with isoproterenol 10 mg/kg (“Iso”) or normal saline (“NS”). Tissues were collected for RNA isolation 90 minutes after injection.
Previous studies have shown that two NR4A family members – Nur77 and NOR1, are induced by β-adrenergic stimulation (18, 24), thereby raising the possibility that denervation-induced loss of Nur77 may be attributable to interruption of sympathetic innervation. We confirmed that all three NR4A receptors are rapidly induced by isoproterenol in differentiated C2C12 muscle cells (Figure 1b). To determine if NR4A receptors were similarly upregulated in vivo, we injected C57BL/6 mice with vehicle or 10 mg/kg isoproterenol. Expression of all three receptors was increased in response to β-adrenergic stimulation (Figure 1c). Quantitation by real-time PCR demonstrated that Nur77 was the most abundant receptor in skeletal muscle, suggesting that it may be the most physiologically important NR4A isoform in this tissue.
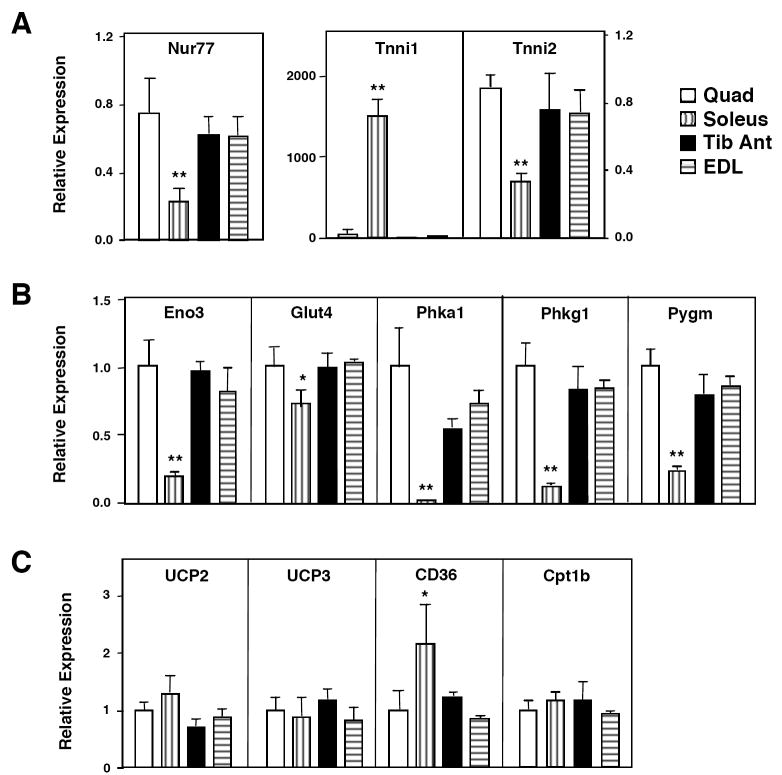
To investigate whether Nur77 might play a direct role in control of glucose metabolism in skeletal muscle, we determined the relative expression of Nur77 and denervation-sensitive glucose metabolic genes in various skeletal muscles. Gene expression in quadriceps, soleus, EDL, and tibialis anterior muscles of C57bl/6 mice was analyzed by real-time PCR. Interestingly, basal expression of Nur77 was substantially higher in quadriceps, EDL, and tibialis anterior muscles (predominantly type 2, fast-twitch, glycolytic fibers) compared to soleus muscle (predominantly type 1, slow-twitch, oxidative fibers; Figure 2a). As expected, Troponin I 1(Tnni1) was specifically expressed in oxidative muscle, whereas Troponin I 2 (Tnni2) was more highly expressed in glycolytic muscle. This observation suggested that Nur77 might play a particularly important role in control of gene expression in muscles that primarily utilize glucose as an energy source. Remarkably, the muscle type-selective expression of a number of glucose-utilization genes mirrored the tissue distribution of Nur77 (Figure 2b). Specifically, enolase 3 (Eno3), Glut4, phosphorylase kinase alpha 1 (Phka1), Phkg1, and Pygm were all expressed at higher levels in fast-twitch muscles (quadriceps, EDL, and tibialis anterior) compared to the slow-twitch soleus muscle. By contrast, the fatty acid transporter CD36 was more highly expressed in slow-twitch muscle. Genes encoding other factors involved in oxidative metabolism such as UCP2, UCP3, and CPT-1b were similarly expressed in all muscle groups. The common fiber type-selective expression of Nur77 and glucose-utilization genes is consistent with a regulatory role for Nur77 in glucose metabolism in muscle.
Figure 2.
Expression of Nur77 and multiple glucose metabolism genes in vivo. Quadriceps, soleus, extensor digitorum longus, and tibialis anterior muscles were isolated from fed-wildtype C57BL/6 mice (n=5, male). RNA was isolated and real-time PCR was performed. (a) Nur77 was predominantly expressed in fast-twitch glycolytic muscles (Quad, EDL, Tib Anterior) compared to slow-twitch oxidative muscle (Soleus). Tnni1 and Tnni2 expression are shown as control for muscle groups. (b) Expression of glucose-utilization genes in different muscle groups mirrors that of Nur77. (c) Expression of lipid oxidation genes. * p<0.05, **p<0.01.
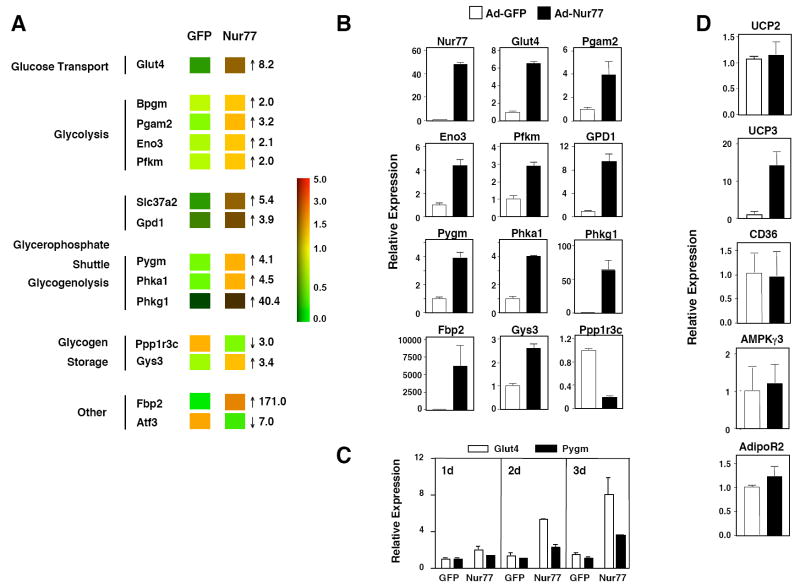
To examine the effect of Nur77 on skeletal muscle metabolism in vitro, we expressed Nur77 in C2C12 myotubes using an adenoviral vector. Control cells were infected with adenovirus expressing green fluorescent protein (GFP) in parallel. Expression profiling was performed on RNA samples harvested three days after infection using Affymetrx Arrays. Remarkably, pathways involved in glucose and glycogen metabolism were prominently affected by Nur77 expression. Not only did genes linked to glucose metabolism represent one of the largest group of genes induced by Nur77 (Supplemental Table 3), this group also contained the genes most highly induced by Nur77. Moreover, we observed a striking concordance between genes induced by Nur77 expression and those downregulated in denervated muscle (Figure 3a and Table 1). The metabolic pathways affected by Nur77 expression, namely glucose uptake, glycolysis, glycerophosphate shuttle, and glycogenoloysis, collectively would be expected to increase glucose utilization and ATP production.
Figure 3.
Adenovirus-Nur77 induces glucose metabolism genes in differentiated C2C12 muscle cells. (a) Expression profiling of C2C12 myotubes infected with Ad-GFP or Ad-Nur77. RNA was harvested three days after infection, and cRNA samples were hybridized to Affymetrix Mouse 430A v2.0 arrays in replicates. Numbers represent fold change of Nur77 relative to GFP. (b) Real-time PCR of genes shown in panel (a). (c) RNA was harvested 1, 2, or 3 days after adenovirus infection. (d) Expression of lipid metabolism and energy balance genes. Results represent the average of two experiments. White bars – Ad-GFP, black bars – Ad-Nur77. p<0.01 for all comparisons in (b), and for UCP3 in (d).
Real-time PCR confirmed increased expression of Glut4, Pgam2, Eno3, Pfkm, GPD1, Pygm, phosphorylase kinase alpha 1 (Phka1), and Phkg1 in Ad-Nur77-transduced cells compared to controls (Figure 3b). Of note, protein phosphatase 1, regulatory subunit 3C (Ppp1r3c) was markedly down-regulated in response to Nur77. Ppp1r3c is a scaffolding protein that binds glycogen and protein phosphatase-1 to enhance glycogen synthesis. Reduction of Ppp1r3c expression would be expected to limit glycogen accumulation, concordant with the upregulation of glycogenolytic genes (25, 26). Glycogen synthase expression was modestly increased, which may reflect a compensatory response to glycogenolysis. These changes in gene expression were observed as early as 24 hours after adenoviral infection, suggesting that Nur77 may directly regulate many of these glucose-utilization genes (Figure 3c). Furthermore, Nur77-induced changes in gene expression were not secondary to effects on myotube differentiation, as Tnni1 and Tnni2 expression was unchanged between Ad-GFP and Ad-Nur77 infected samples (data not shown). With the exception of UCP3, we did not observe changes in expression of several other genes previously suggested to be Nur77-responsive in C2C12 cells (Figure 3d; UCP2, CD36, AMPKγ3, and AdipoR2) (18).
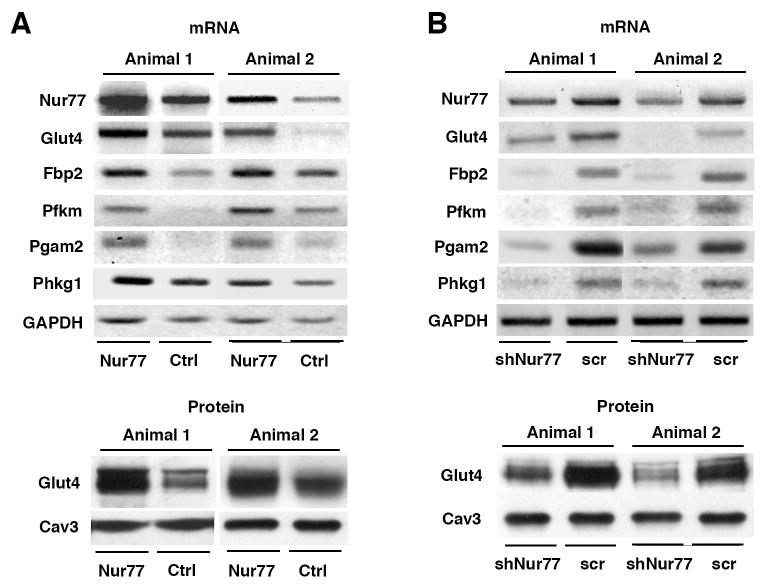
We proceeded to test the effect of Nur77 on muscle gene expression in vivo. We performed “rescue” experiments in denervated rat muscle by re-introducing wild type Nur77 by electroporation. We compared Glut4 expression levels between the leg that received the control vector and one that received the Nur77 expression vector. Glut4 expression was enhanced in Nur77-electroporated denervated EDL muscle at both the mRNA and protein level (Figure 4a). Nur77 expression also rescued expression of Fbp2, Pfkm, Pgam2, and Phkg1. The demonstration that ectopic Nur77 expression restores the expression of glucose metabolic genes in denervated muscle strongly suggests that loss of Nur77 expression contributes to the downregulation of these genes in response to denervation.
Figure 4.
Nur77-dependent expression of multiple glucose utilization genes. (a) Electroporation of Nur77-cDNA into denervated rat hind limbs rescues glucose metabolism gene expression. Both hind legs of each rat were denervated as described. 60 μg of Nur77 cDNA was injected and electroporated into EDL muscle in one leg, and pcDNA 3.1 empty vector (Ctrl) was injected as control in the other leg of the same animal. Animals were sacrificed after three days. We observed Nur77-dependent rescue in 2 of 4 animals. (b) Nur77 knockdown reduces the expression of glucose utilization genes. 100 μg of pSUPER vector encoding shRNA of Nur77 (shNur77) was electroporated into rat EDL muscle in one leg and scramble sequence (scr) was used in the other leg as control. Rats were sacrificed after 6 days. For both (a) and (b), total RNA was extracted for RT-PCR, and total crude membrane was extracted for western blot. GAPDH and Cav3 served as controls for each case.
In order to establish the physiologic relevance of Nur77 for expression of this metabolic gene program we utilized two loss-of-function models. First, we knocked down Nur77 in vivo using small hairpin (sh) RNAs. Candidate shRNA sequences directed against Nur77 were selected from the literature and commercial design engines and cloned into pSuper vector. The efficiency of knockdown was tested in 293T cells by transfecting the vector-driven shRNA and Nur77 cDNA. A sequence was chosen (see Methods) that attenuated Nur77 expression >70% by Western blotting (data not shown). This shRNA was injected, then electroporated into EDL muscle; a scrambled sequence was injected in the other leg of the same animal. Muscles were isolated 6 days after surgery. We observed a 60% loss of Glut4 mRNA expression in the Nur77 shRNA-treated leg as comparing to the leg treated with a scrambled shRNA construct (Figure 4b). Western blotting also revealed diminished Glut4 protein expression. Similarly, Nur77 shRNA reduced the mRNA expression of multiple other glucose-utilization genes, including Fbp2, Phkg1, Pgam2, and Pfkm. Collectively, the data of Figure 4 suggest that endogenous GLUT4, Fbp2, Phkg1, Pgam2, and Pfkm expression in rat EDL is dependent on Nur77 activity.
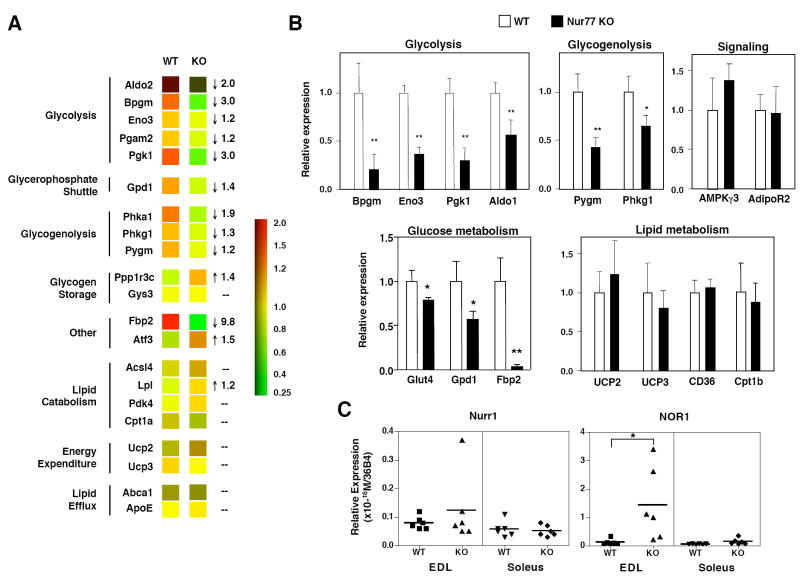
We next analyzed the effect of Nur77 deletion in mice on muscle gene expression in vivo. We isolated RNA from quadriceps muscle of fasted wildtype and Nur77-null mice and performed expression profiling using Affymetrix arrays. Figure 5a shows that in the absence of Nur77, expression of several genes involved in glycolysis and glycogenolysis genes were attenuated, including phosphoglycerokinase 1 (Pgk1), Bpgm, and Fbp2. Although changes in other glucose utilization genes were not predicted by these microarray studies, real-time PCR analysis revealed statistically significant reductions in aldolase 1(Aldo1), Eno3, Pygm, Phkg1, Glut4, and Gpd1 (Figure 5b). Consistent with the results of both the denervation (Table 1) and Nur77-shRNA knockdown studies (Figure 4b), Fbp2 was dramatically reduced in Nur77 null muscle in vivo. Furthermore, in general agreement with our C2C12 over-expression system, we did not observe any change in the expression of a number of genes previously suggested to be responsive to Nur77, including UCP2, UCP3, CD36, AMPKγ3, and AdipoR2. These results definitively establish that Nur77 is required for physiologic expression of glucose metabolic genes in skeletal muscle.
Figure 5.
Expression of a subset of glucose-metabolism genes is reduced in fast-twitch muscle of Nur77-null mice. (a) Expression profiling of quadriceps muscle of wildtype (WT) or Nur77-null (KO) mice (n=4 to 5, fasted, male). RNA was prepared from quadriceps muscle, and cRNA samples were hybridized to Affymetrix Mouse 430A v2.0 arrays in replicates. – no change. Numbers represent fold-change of KO relative to WT. (b) Real-time PCR validated expression changes seen with microarray. (c) Expression of Nurr1 and NOR1 in EDL and soleus of WT or KO mice (n=6, fasted, male). * p<0.05, ** p<0.01.
To determine if Nur77 deletion was accompanied by changes in expression of other NR4A family members, we examined the expression of Nurr1 and NOR1. As shown in Figure 5c, Nurr1 expression was not significantly altered in Nur77 null mice, whereas expression of NOR1 was upregulated. This effect was observed in EDL (fast-twitch glycolytic) muscle, but not in the soleus (slow-twitch oxidative) muscle. It is likely that the persistent expression of these 2 other NR4A family members partially compensates for loss of Nur77 expression.
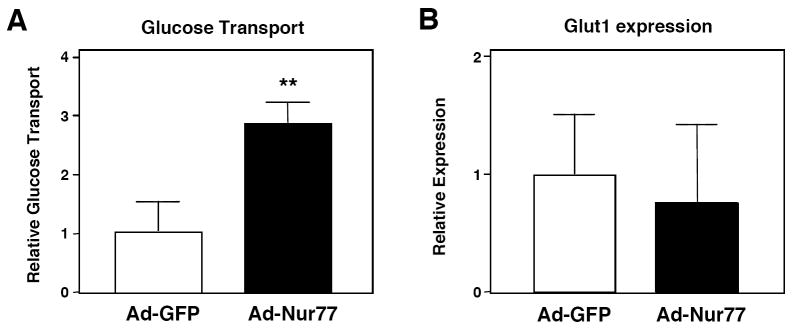
To determine if Nur77-induced upregulation of Glut4 affects cellular glucose uptake, we measured 2-deoxyglucose transport in C2C12 myotubes. Indeed, adenovirus-mediated Nur77 expression increased non-insulin stimulated glucose transport by over 2-fold (Figure 6a). We attribute the increased transport to Glut4 upregulation, as Glut1 mRNA expression was unchanged in response to Nur77 (Figure 6b). Note, C2C12 cells do not undergo significant insulin-stimulated glucose uptake even following ectopic Glut4 expression, and therefore the effect of insulin on Nur77 could not be evaluated using this system (27-29).
Figure 6.
Nur77 enhances glucose uptake in vitro. (a) Differentiated C2C12 cells were infected with Ad-GFP or Ad-Nur77. Glucose transport was determined by 2-[3H]-deoxyglucose uptake at room temperature over 10 minutes. Result is representative of 3 experiments. **p<0.01 (b) Glut1 mRNA expression did not change in response to Nur77 overexpression. Result represents average of 2 experiments.
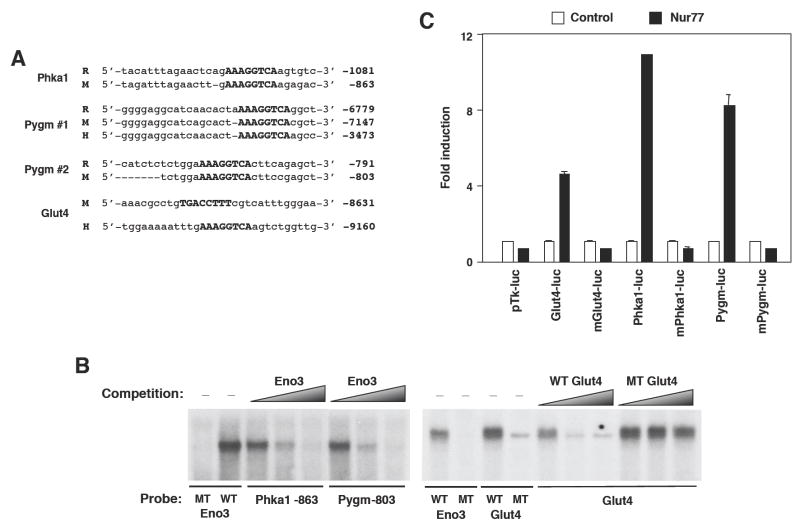
To determine whether muscle metabolic genes are direct targets of Nur77, we examined flanking sequences of these genes for potential Nur77-binding response elements (NBREs). We previously identified enolase 3 as a Nur77 target gene in liver (10). In addition, we now report the identification of consensus Nur77 binding sites (AAAGGTCA), in the promoters of murine Phka1 and Pygm (enzymes involved in glycogenolysis), and Glut4 (Figure 7a). While murine Phka1 and Pygm NBREs are conserved across other species (rat and/or human), the Glut4 NBRE is not conserved. However, the human Glut4 promoter contains a distinct NBRE located 9kb upstream of the transcriptional start site (Figure 7a). 32P-labeled probes spanning the mouse Phka1, Pygm, and Glut4 NBREs bound in vitro-translated Nur77 protein in electromobility shift assays (Figure 7b). The NBRE from the Eno3 gene is shown as a positive control. Increasing concentration of excess unlabeled wildtype, but not mutant probes, effectively competed for probe binding to Nur77, demonstrating sequence-specificity of complex formation.
Figure 7.
Nur77 binds NBREs of several glucose-metabolism genes. (a) Several NBREs (upper case) identified in Phka1 and Pygm are conserved amongst mouse (M), rat (R), and human (H). The murine Glut4 NBRE is not conserved; the NBRE in human Glut4 promoter at position -9160 is shown for comparison. Coordinates represent distance of NBRE from transcriptional start site. (b) EMSA demonstrating Nur77 binding to NBREs of Phka1, Pygm, and Glut4. In vitro-translated Nur77 was used to bind 32P-labeled NBREs. WT = wildtype NBRE, MT = mutant NBRE (AAAGaaTCA). Excess non-labeled Eno3 (left) or Glut4 (right) oligonucleotide was added in increasing concentration (from 5x up to 200x) in competition assay. (c) NBREs of Glut4, Phka1, and Pygm were multimerized and cloned into pTKIII luciferase reporter construct. Co-transfection of MSCV-Nur77 expression plasmid enhanced luciferase activity of 3x-NBRE constructs in HEK293T cells.
Finally, we tested the ability of Nur77 to activate the putative NBREs from glucose metabolic genes in transient transfection assays. NBREs from the murine Phka1, Pygm, and Glut4 promoters were multimerized and cloned upstream of a minimal promoter and luciferase reporter gene. As shown in Figure 7c, cotransfection of a Nur77 expression vector induced expression of wildtype, but not mutant NBREs from these glucose metabolic genes. These results identify the Phka1, Pygm, and Glut4 promoters as direct targets of Nur77.
Discussion
Neuromuscular signaling is important for control of metabolic activity. For example, denervation leads to insulin resistance in skeletal muscle in rats, mainly as a result decreased Glut 4 expression (30-32). Moreover, spinal chord injury in humans leads to insulin resistance due, at least in part, to a loss in muscle mass and Glut4 protein (33). However, the transcription factors that transduce signals from the neuromuscular junction to the nucleus and affect changes in metabolic gene expression are not well defined. We have shown here that the orphan nuclear receptor Nur77 is a potential mediator of innervation-dependent metabolic gene expression. Using two distinct loss-of-function models, shRNA knockdown of Nur77 in rat hindlimb muscle and Nur77-null mice, we demonstrated that loss of Nur77 expression in skeletal muscle inhibits the expression of multiple genes involved in glucose uptake, glycolysis, glycogenolysis, and the glycerophosphate shuttle. Conversely, overexpression of Nur77 in C2C12 muscle cells or denervated rat hindlimb induces the expression of these same genes. These results define a previously unrecognized role for Nur77 in the innervation-dependent expression of metabolic genes in skeletal muscle.
We previously identified NR4A receptors as transcriptional regulators of hepatic gluconeogenesis (10). The present study implicates Nur77 in glucose-utilization in skeletal muscle. One physiologic signal that stimulates both of these pathways is activation of the sympathetic nervous system. Denervation by sciatotomy severs afferent, motor, as well as sympathetic axons. Prior work has shown that the NR4A receptors are involved in the stress response and sympathetic nervous system activation. Nur77 regulates the 21-hydroxylase gene, which encodes an enzyme essential for cortisol biosynthesis (34). Nur77 also negatively regulates glucocorticoid-induced down-regulation of POMC expression, permitting unremitting ACTH synthesis and cortisol production in times of stress (35). We and others have demonstrated that β-adrenergic receptor agonists induce NR4A receptor expression both in vitro and in vivo (18, 24). Thus, the loss of Nur77 expression following denervation of skeletal muscle is consistent with loss of sympathetic tone.
Further evidence supporting a role for Nur77 in sympathetic responses comes from the expression pattern of this transcription factor. Upon activation of the sympathetic nervous system, glucose uptake, glycolysis and glycogenolysis are activated in skeletal muscle to sustain the energy demand. The acute stress response predominantly involves fast-twitch fibers, in which glycogen phosphorylase activity is highest (36). We have shown here that Nur77 is more abundantly expressed in fast-twitch muscles relative to slow-twitch muscles. Depending on intensity, the sympathetic nervous system may also be activated during physical exercise. It is therefore reasonable to suspect that Nur77 may play a role in exercise physiology. In fact, the expression of all three NR4A receptors was reported to be upregulated in vastus lateralis (a fast-twitch muscle) in human subjects three hours after exercise to exhaustion (37). Although the evidence pointing to a link between Nur77 and sympathetic innervation-induced metabolic changes in skeletal muscle is compelling, our data does not preclude the possibility that Nur77 may also regulate contraction (motor-neuron)-induced glucose transport (38) and glycogenolysis (36).
The three NR4A receptors share a high degree of sequence homology (39), and have largely overlapping functions. For instance, all three receptors promote the expression of genes linked to hepatic gluconeogenesis (10). In view of this redundancy, the fact that we observed only modest reductions in the expression of innervation-dependent glucose metabolic genes in Nur77 null mice is not unexpected. The continued expression of Nurr1 and upregulation of NOR1 in Nur77 null mice likely sustains expression of NR4A target genes. Further studies are ongoing to determine whether the two other NR4A receptors regulate expression of these same target genes as Nur77 in muscle. In the future, it will also be of interest to determine the effect of loss of two or even all three NR4A receptors on skeletal muscle function. However, given the early lethality of all NR4A double knockout mice, the use of conditional knockout strategies will likely be required.
In summary, we have identified Nur77 as a novel regulator of glucose metabolism in skeletal muscle whose expression is dependent on innervation. Our findings fit well with prior work showing that NR4A receptors are responsive to sympathetic stimulation and are involved in cortisol production and hepatic gluconeogenesis. Taken together, these observations suggest that Nur77 may coordinate multiple physiologic changes critical to the fight-or-flight response. Further dissection of the role of Nur77 in modulating glucose metabolism in skeletal muscle is expected to broaden our understanding of physiology, and may also offer new pathways for therapeutic intervention in the management of insulin resistance.
Materials and Methods
Cells and Animals
C2C12 cells were maintained in 10% FBS (Omega Scientific) in DMEM at 5% CO2. Cells were differentiated by switching the media to 2% horse serum 1 day after confluence. Male Sprague-Dawley rats (175-200 g) were from Taconic Breeding Laboratory (Germantown, NY). The animals were fed standard chow and kept at constant room temperature (20-22 °C). Rats were anesthetized with pentobarbital sodium (60 mg/kg body wt) by intraperitoneal injection. For denervation, we sectioned the 0.3 cm piece of the sciatic nerve and closed the wound with stitches. For sham operation, we visualized, touched, but did not section the sciatic nerve. We sacrificed the rats 3-6 days after surgery and isolated muscles. Mice were fed a standard chow diet and maintained on a 12-h light-dark cycle and were age- and gender-matched for all experiments. Nur77 null mice were fasted for 12-14 hours prior to sacrifice. Animal studies were performed in accordance with the Boston University and UCLA Animal Research Committees guidelines.
DNA Constructs
Rat primers for PCR were obtained from Invitrogen. We used the following primers for cloning Nur77: Nur77-F: 5’-ccgctcgagcggatggacctggccagcccc-3’, Nur77-R: 5’-cccaagcttgggtcagaaagacaatgtgtc-3’. Nur77 full-length cDNA was cloned by PCR using the primers described above and ligated into pLPCX vector through XhoI and Hind III sites. We designed the shRNA sequence against the DE region of Nur77 as described (18). We used the following oligonucleotides for cloning the Nur77 shRNA targets into pSuper vector: 5’-gatcccgtccctggcttcattgagctttcaagagaagctcaatgaagccagggattttttggaaa-3’, and 5’-agcttttccaaaaaatccctggcttcattgagcttctcttgaaagctcaatgaagccagggacgg-3’. The scrambled shRNA sequence was as described(40) and oligonucleotides used for pSuper vector were: 5’-gatcccggcgcgctttgtaggattcgttcaagagacgaatcctacaaacgcgcgttttttggaaa-3’ and 5’-agcttttccaaaaaacgcgcgtttgtaggattcgtctcttgaacgaatcctacaaagcgcgccgg-3’. For cloning 3x-NBRE constructs, we used the following primers (only sense strand shown): Glut4 WT F – agctttgacctttcgtcatttgggaataaacgcctgtgacctttcgtcatttgggaataaacgcctgtgacctttg; Glut4 MT F – agctttgttctttcgtcatttgggaataaacgcctgtgttctttcgtcatttgggaataaacgcctgtgttctttg; Pygm WT F – agcttaaaggtcacgctaactaatcgcatcagcactaaaggtcacgctaactaatcgcatcagcactaaaggtcag; Pygm MT F – agcttaaagaacacgctaactaatcgcatcagcactaaagaacacgctaactaatcgcatcagcactaaagaacag; Phka1 WT F – agcttaaaggtcaagagacagtctatttagaacttgaaaggtcaagagacagtctatttagaacttgaaaggtcag; Phka1 MT F – agcttaaagaacaagagacagtctatttagaacttgaaagaacaagagacagtctatttagaacttgaaagaacag. The annealed NBRE-oligonucleotides were inserted into pTKIII-Luc vector digested with HindIII and BamHI.
In vivo electroporation
Electroporation into muscle was performed as described (41). We injected pSuper vector (100 μg, 5 μg/μl) encoding the shRNA or Nur77 cDNA (60 μg, 5 μg/μl) in the pLPCX vector into the center of the EDL muscle using a 29G needle. The injection was immediately followed by the application of a pair of needle electrodes attached to the surface of EDL and connected to BTX Electro Square Porator (Model T820). We applied three 50 ms pulses of 200 V/cm at the site of injection, then three more pulses with reverse polarity, and then immediately stitched the incision closed. The other leg of the same animal received appropriate control vectors using the same procedure. We sacrificed the rats 3-6 days after electrotransfer and collected EDL muscles for subsequent analysis.
Membrane preparation and protein analysis
We isolated, minced, and homogenized rat EDL muscle on ice in HES buffer (20 mM HEPES, 250 mM sucrose, 1 mM EDTA, 1 μM aprotinin A, 1 μM pepstatin, 1 μM leupeptin, pH 7.4) using a Polytron homogenizer. After removing debris and non-homogenized tissue by spinning at 2,000 g for 10 minutes, we subjected the supernatant to centrifugation at 9,000 g for 20 minutes. We then spun the supernatant at 180,000 g for 90 minutes, and resuspended the resultant pellet in 50-100 ul PBS with protease inhibitors. For Glut4 blots, we incubated the samples at 37°C for 30 minutes in urea buffer (8 M urea, 5% SDS, 50 mM Tris, 0.1% BPB and 53 mg/ml DTT), and then resolved the proteins by SDS-PAGE. Gels were transferred to polyvinylidene difluoride using a wet transfer protocol. We probed the membranes first with anti-Glut4 monoclonal antibody (1F8) (42) or anti-caveolin 3 monoclonal antibody (BD Transduction labs), then with the appropriate horseradish peroxidase-conjugated secondary antibodies (Sigma). We visualized the signals by enhanced luminal reagents (Perkin Elmer Life Sciences, Boston, MA). Electromobility shift assays were performed as described (7, 17).
Adenoviral infection
Adenoviral vectors expressing GFP and Nur77 were described previously (10). 5 days after initiation of C2C12 cell differentiation, we prepared the adenovirus as follows: we added the adenovirus (at an MOI of 125) to poly-L-lysine hydrobromide (Sigma) 0.5 μg/ml DMEM and incubated the mixture at room temperature for 100 minutes. We then washed the C2C12 cells once carefully with PBS, and then added the adenovirus-poly-L-lysine media (43). Media was refreshed the next day. Cells were harvested with Trizol (Invitrogen) 72 hours after infection.
RT-PCR and Real-Time PCR
RNA was isolated from rat and mouse muscles by homogenizing the tissues in Trizol reagent. cDNA was synthesized as described (7). We analyzed target cDNA levels by PCR in 20 μl reactions containing 10 μl Promega PCR master mix, 1 μM each forward and reverse primers and 1.5 μl of cDNA prepared as described above. The reactions are incubated initially at 94°C for 10 minutes, then cycled the reactions between 25 to 36 cycles with the following condition: 94°C for 30s, 56°C for 1 minute, and 74°C for 1 minute. For real-time PCR, we purchased SybrGreen reagents from Diagenode and followed manufacturer’s instructions. For absolute quantitation of NR4A receptors, we purchased the sense strand of each amplicon as known standards. Results were internally normalized to the signal from 36B4. See Supplemental Tables 1 and 2 for complete primer sequences used in RT-PCR and real-time PCR. Oligonucleotides were purchased from Integrated DNA Technologies.
Microarrays
RNA from differentiated C2C12 cells was prepared three days after adenovirus infection and purified through RNEasy columns (Qiagen). Each condition was done with replicate arrays, each representing 3 pooled wells. cRNA preparation and hybridization to Affymetrix Mouse Genome Arrays 430 v2.0 was performed by the UCLA Microarray Core and data was analyzed using GeneSpring GX7.3.1. We included only genes with raw data signal greater than 500 for at least one condition for analysis. For rat experiments, 10 μg RNA from the control (sham) leg and the denervated leg were prepared for hybridization to Affymetrix GeneChip Rat Genome 230 v2.0. Data analysis was conducted by Boston University Microarray Resource.
Glucose Transport
We performed glucose transport assay as described(44), with the following modifications. We differentiated C2C12 myotubes for 5 days prior to adenovirus infection. Glucose transport assay was done three days later. Cells were incubated with 2-[3H]-deoxyglucose for 10 minutes.
EMSA and Transient Transfection
Electromobility shift assays were performed as described(7, 17). Sequences of the top strand used for EMSA are as follows: Phka1 WT – gatcaacttgaaaggtcaagagac; Phka1 MT – gatcaacttgaaagaacaagagac; Pygm WT – gatctctggaaaaggtcacttccg; Pygm MT – gatctctggaaaagaacacttccg; Glut4 WT – gatccgcctgtgacctttcgtcat; Glut4 MT – gatccgcctgtgttctttcgtcat. We transfected HEK 293T cells in triplicate with or without MSCV-Nur77 expression plasmid, with Lipofectamine (Invitrogen) in accordance to manufacturer’s instructions. Renilla luciferase expression plasmid was co-transfected for normalization of luciferase activity.
Statistics and data analysis
Student t-test was performed to determine statistical significance. All error bars represent standard deviations.
Supplementary Material
Acknowledgments
We thank Dr. P. Cohen and Dr. J. Milbrandt for the Nur77 null mice. We thank Dr. L. Goodyear and members of her lab for advice on in vivo electroporation, and Dr. S. Farmer, for pSuper vector.
Grants: L.C.C. is a fellow of the Pediatric Scientist Development Program (NICHD Grant Award K12-HD00850). P.T. is an Investigator of the Howard Hughes Medical Institute. This work was also supported by NIH grants HL30568 (P.T.) and DK30425 (P.F.P.).
Footnotes
Disclosure statement: The authors have nothing to disclose.
NIH Statement: This is an un-copyedited author manuscript copyrighted by The Endocrine Society. This may not be duplicated or reproduced, other than for personal use or within the rule of “Fair Use of Copyrighted Materials” (section 107, Title 17, U.S. Code) without permission of the copyright owner, The Endocrine Society. From the time of acceptance following peer review, the full text of this manuscript is made freely available by The Endocrine Society at http://www.endojournals.org/. The final copy edited article can be found at http://www.endojournals.org/. The Endocrine Society disclaims any responsibility or liability for errors or omissions in this version of the manuscript or in any version derived from it by the National Institutes of Health or other parties. The citation of this article must include the following information: author(s), article title, journal title, year of publication and DOI.
References
- 1.Zisman A, Peroni OD, Abel ED, Michael MD, Mauvais-Jarvis F, Lowell BB, Wojtaszewski JF, Hirshman MF, Virkamaki A, Goodyear LJ, Kahn CR, Kahn BB. Targeted disruption of the glucose transporter 4 selectively in muscle causes insulin resistance and glucose intolerance. Nat Med. 2000;6:924–8. doi: 10.1038/78693. [DOI] [PubMed] [Google Scholar]
- 2.Hallsten K, Virtanen KA, Lonnqvist F, Sipila H, Oksanen A, Viljanen T, Ronnemaa T, Viikari J, Knuuti J, Nuutila P. Rosiglitazone but not metformin enhances insulin- and exercise-stimulated skeletal muscle glucose uptake in patients with newly diagnosed type 2 diabetes. Diabetes. 2002;51:3479–85. doi: 10.2337/diabetes.51.12.3479. [DOI] [PubMed] [Google Scholar]
- 3.Dela F, Ploug T, Handberg A, Petersen LN, Larsen JJ, Mikines KJ, Galbo H. Physical training increases muscle GLUT4 protein and mRNA in patients with NIDDM. Diabetes. 1994;43:862–5. doi: 10.2337/diab.43.7.862. [DOI] [PubMed] [Google Scholar]
- 4.Kennedy JW, Hirshman MF, Gervino EV, Ocel JV, Forse RA, Hoenig SJ, Aronson D, Goodyear LJ, Horton ES. Acute exercise induces GLUT4 translocation in skeletal muscle of normal human subjects and subjects with type 2 diabetes. Diabetes. 1999;48:1192–7. doi: 10.2337/diabetes.48.5.1192. [DOI] [PubMed] [Google Scholar]
- 5.Gibbs EM, Stock JL, McCoid SC, Stukenbrok HA, Pessin JE, Stevenson RW, Milici AJ, McNeish JD. Glycemic improvement in diabetic db/db mice by overexpression of the human insulin-regulatable glucose transporter (GLUT4) J Clin Invest. 1995;95:1512–8. doi: 10.1172/JCI117823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miles PD, Barak Y, He W, Evans RM, Olefsky JM. Improved insulin-sensitivity in mice heterozygous for PPAR-gamma deficiency. J Clin Invest. 2000;105:287–92. doi: 10.1172/JCI8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laffitte BA, Chao LC, Li J, Walczak R, Hummasti S, Joseph SB, Castrillo A, Wilpitz DC, Mangelsdorf DJ, Collins JL, Saez E, Tontonoz P. Activation of liver X receptor improves glucose tolerance through coordinate regulation of glucose metabolism in liver and adipose tissue. Proc Natl Acad Sci U S A. 2003;100:5419–24. doi: 10.1073/pnas.0830671100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stumvoll M, Haring H. The peroxisome proliferator-activated receptor-gamma2 Pro12Ala polymorphism. Diabetes. 2002;51:2341–7. doi: 10.2337/diabetes.51.8.2341. [DOI] [PubMed] [Google Scholar]
- 9.Lehmann JM, Moore LB, Smith-Oliver TA, Wilkison WO, Willson TM, Kliewer SA. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma) J Biol Chem. 1995;270:12953–6. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 10.Pei L, Waki H, Vaitheesvaran B, Wilpitz DC, Kurland IJ, Tontonoz P. NR4A orphan nuclear receptors are transcriptional regulators of hepatic glucose metabolism. Nat Med. 2006;12:1048–55. doi: 10.1038/nm1471. [DOI] [PubMed] [Google Scholar]
- 11.Knight JB, Eyster CA, Griesel BA, Olson AL. Regulation of the human GLUT4 gene promoter: interaction between a transcriptional activator and myocyte enhancer factor 2A. Proc Natl Acad Sci U S A. 2003;100:14725–30. doi: 10.1073/pnas.2432756100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flaig R, Greschik H, Peluso-Iltis C, Moras D. Structural basis for the cell-specific activities of the NGFI-B and the Nurr1 ligand-binding domain. J Biol Chem. 2005;280:19250–8. doi: 10.1074/jbc.M413175200. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z, Benoit G, Liu J, Prasad S, Aarnisalo P, Liu X, Xu H, Walker NP, Perlmann T. Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature. 2003;423:555–60. doi: 10.1038/nature01645. [DOI] [PubMed] [Google Scholar]
- 14.Tetradis S, Bezouglaia O, Tsingotjidou A. Parathyroid hormone induces expression of the nuclear orphan receptor Nurr1 in bone cells. Endocrinology. 2001;142:663–70. doi: 10.1210/endo.142.2.7926. [DOI] [PubMed] [Google Scholar]
- 15.Arkenbout EK, de Waard V, van Bragt M, van Achterberg TA, Grimbergen JM, Pichon B, Pannekoek H, de Vries CJ. Protective function of transcription factor TR3 orphan receptor in atherogenesis: decreased lesion formation in carotid artery ligation model in TR3 transgenic mice. Circulation. 2002;106:1530–5. doi: 10.1161/01.cir.0000028811.03056.bf. [DOI] [PubMed] [Google Scholar]
- 16.Zetterstrom RH, Solomin L, Jansson L, Hoffer BJ, Olson L, Perlmann T. Dopamine neuron agenesis in Nurr1-deficient mice. Science. 1997;276:248–50. doi: 10.1126/science.276.5310.248. [DOI] [PubMed] [Google Scholar]
- 17.Pei L, Castrillo A, Tontonoz P. Regulation of macrophage inflammatory gene expression by the orphan nuclear receptor Nur77. Mol Endocrinol. 2006;20:786–94. doi: 10.1210/me.2005-0331. [DOI] [PubMed] [Google Scholar]
- 18.Maxwell MA, Cleasby ME, Harding A, Stark A, Cooney GJ, Muscat GE. Nur77 regulates lipolysis in skeletal muscle cells. Evidence for cross-talk between the beta-adrenergic and an orphan nuclear hormone receptor pathway. J Biol Chem. 2005;280:12573–84. doi: 10.1074/jbc.M409580200. [DOI] [PubMed] [Google Scholar]
- 19.Block NE, Menick DR, Robinson KA, Buse MG. Effect of denervation on the expression of two glucose transporter isoforms in rat hindlimb muscle. J Clin Invest. 1991;88:1546–52. doi: 10.1172/JCI115465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duclert A, Piette J, Changeux JP. Influence of innervation of myogenic factors and acetylcholine receptor alpha-subunit mRNAs. Neuroreport. 1991;2:25–8. doi: 10.1097/00001756-199101000-00006. [DOI] [PubMed] [Google Scholar]
- 21.Neville CM, Schmidt M, Schmidt J. Response of myogenic determination factors to cessation and resumption of electrical activity in skeletal muscle: a possible role for myogenin in denervation supersensitivity. Cell Mol Neurobiol. 1992;12:511–27. doi: 10.1007/BF00711232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsay HJ, Schmidt J. Skeletal muscle denervation activates acetylcholine receptor genes. J Cell Biol. 1989;108:1523–6. doi: 10.1083/jcb.108.4.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsujino H, Kondo E, Fukuoka T, Dai Y, Tokunaga A, Miki K, Yonenobu K, Ochi T, Noguchi K. Activating transcription factor 3 (ATF3) induction by axotomy in sensory and motoneurons: A novel neuronal marker of nerve injury. Mol Cell Neurosci. 2000;15:170–82. doi: 10.1006/mcne.1999.0814. [DOI] [PubMed] [Google Scholar]
- 24.Pearen MA, Ryall JG, Maxwell MA, Ohkura N, Lynch GS, Muscat GE. The orphan nuclear receptor, NOR-1, is a target of beta-adrenergic signaling in skeletal muscle. Endocrinology. 2006;147:5217–27. doi: 10.1210/en.2006-0447. [DOI] [PubMed] [Google Scholar]
- 25.Crosson SM, Khan A, Printen J, Pessin JE, Saltiel AR. PTG gene deletion causes impaired glycogen synthesis and developmental insulin resistance. J Clin Invest. 2003;111:1423–32. doi: 10.1172/JCI17975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delibegovic M, Armstrong CG, Dobbie L, Watt PW, Smith AJ, Cohen PT. Disruption of the striated muscle glycogen targeting subunit PPP1R3A of protein phosphatase 1 leads to increased weight gain, fat deposition, and development of insulin resistance. Diabetes. 2003;52:596–604. doi: 10.2337/diabetes.52.3.596. [DOI] [PubMed] [Google Scholar]
- 27.Zhou M, Vallega G, Kandror KV, Pilch PF. Insulin-mediated translocation of GLUT-4-containing vesicles is preserved in denervated muscles. Am J Physiol Endocrinol Metab. 2000;278:E1019–26. doi: 10.1152/ajpendo.2000.278.6.E1019. [DOI] [PubMed] [Google Scholar]
- 28.Kotliar N, Pilch PF. Expression of the glucose transporter isoform GLUT 4 is insufficient to confer insulin-regulatable hexose uptake to cultured muscle cells. Mol Endocrinol. 1992;6:337–45. doi: 10.1210/mend.6.3.1584210. [DOI] [PubMed] [Google Scholar]
- 29.Tortorella LL, Pilch PF. C2C12 myocytes lack an insulin-responsive vesicular compartment despite dexamethasone-induced GLUT4 expression. Am J Physiol Endocrinol Metab. 2002;283:E514–24. doi: 10.1152/ajpendo.00092.2002. [DOI] [PubMed] [Google Scholar]
- 30.Bertelli DF, Ueno M, Amaral ME, Toyama MH, Carneiro EM, Marangoni S, Carvalho CR, Saad MJ, Velloso LA, Boschero AC. Reversal of denervation-induced insulin resistance by SHIP2 protein synthesis blockade. Am J Physiol Endocrinol Metab. 2003;284:E679–87. doi: 10.1152/ajpendo.00345.2002. [DOI] [PubMed] [Google Scholar]
- 31.Henriksen EJ, Rodnick KJ, Mondon CE, James DE, Holloszy JO. Effect of denervation or unweighting on GLUT-4 protein in rat soleus muscle. J Appl Physiol. 1991;70:2322–7. doi: 10.1152/jappl.1991.70.5.2322. [DOI] [PubMed] [Google Scholar]
- 32.Coderre L, Monfar MM, Chen KS, Heydrick SJ, Kurowski TG, Ruderman NB, Pilch PF. Alteration in the expression of GLUT-1 and GLUT-4 protein and messenger RNA levels in denervated rat muscles. Endocrinology. 1992;131:1821–5. doi: 10.1210/endo.131.4.1396328. [DOI] [PubMed] [Google Scholar]
- 33.Hjeltnes N, Galuska D, Bjornholm M, Aksnes AK, Lannem A, Zierath JR, Wallberg-Henriksson H. Exercise-induced overexpression of key regulatory proteins involved in glucose uptake and metabolism in tetraplegic persons: molecular mechanism for improved glucose homeostasis. Faseb J. 1998;12:1701–12. doi: 10.1096/fasebj.12.15.1701. [DOI] [PubMed] [Google Scholar]
- 34.Wilson TE, Mouw AR, Weaver CA, Milbrandt J, Parker KL. The orphan nuclear receptor NGFI-B regulates expression of the gene encoding steroid 21-hydroxylase. Mol Cell Biol. 1993;13:861–8. doi: 10.1128/mcb.13.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okabe T, Takayanagi R, Adachi M, Imasaki K, Nawata HMS. Nur77, a member of the steroid receptor superfamily, antagonizes negative feedback of ACTH synthesis and secretion by glucocorticoid in pituitary corticotrope cells. J Endocrinol. 1998;156:169–75. doi: 10.1677/joe.0.1560169. [DOI] [PubMed] [Google Scholar]
- 36.Richter EA, Ruderman NB, Gavras H, Belur ER, Galbo H. Muscle glycogenolysis during exercise: dual control by epinephrine and contractions. Am J Physiol. 1982;242:E25–32. doi: 10.1152/ajpendo.1982.242.1.E25. [DOI] [PubMed] [Google Scholar]
- 37.Mahoney DJ, Parise G, Melov S, Safdar A, Tarnopolsky MA. Analysis of global mRNA expression in human skeletal muscle during recovery from endurance exercise. Faseb J. 2005;19:1498–500. doi: 10.1096/fj.04-3149fje. [DOI] [PubMed] [Google Scholar]
- 38.Jessen N, Goodyear LJ. Contraction signaling to glucose transport in skeletal muscle. J Appl Physiol. 2005;99:330–7. doi: 10.1152/japplphysiol.00175.2005. [DOI] [PubMed] [Google Scholar]
- 39.Martinez-Gonzalez J, Badimon L. The NR4A subfamily of nuclear receptors: new early genes regulated by growth factors in vascular cells. Cardiovasc Res. 2005;65:609–18. doi: 10.1016/j.cardiores.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 40.Zhang L, Fogg DK, Waisman DM. RNA interference-mediated silencing of the S100A10 gene attenuates plasmin generation and invasiveness of Colo 222 colorectal cancer cells. J Biol Chem. 2004;279:2053–62. doi: 10.1074/jbc.M310357200. [DOI] [PubMed] [Google Scholar]
- 41.Cleasby ME, Davey JR, Reinten TA, Graham MW, James DE, Kraegen EW, Cooney GJ. Acute bidirectional manipulation of muscle glucose uptake by in vivo electrotransfer of constructs targeting glucose transporter genes. Diabetes. 2005;54:2702–11. doi: 10.2337/diabetes.54.9.2702. [DOI] [PubMed] [Google Scholar]
- 42.James DE, Brown R, Navarro J, Pilch PF. Insulin-regulatable tissues express a unique insulin-sensitive glucose transport protein. Nature. 1988;333:183–5. doi: 10.1038/333183a0. [DOI] [PubMed] [Google Scholar]
- 43.Orlicky DJ, Schaack J. Adenovirus transduction of 3T3-L1 cells. J Lipid Res. 2001;42:460–6. [PubMed] [Google Scholar]
- 44.Michael LF, Wu Z, Cheatham RB, Puigserver P, Adelmant G, Lehman JJ, Kelly DP, Spiegelman BM. Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1. Proc Natl Acad Sci U S A. 2001;98:3820–5. doi: 10.1073/pnas.061035098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.