Abstract
The oxidative folding of frog onconase (ONC), a member of the ribonuclease A family, was examined and shows markedly different behavior compared to its structural homologue bovine pancreatic ribonuclease A (RNase A) under similar conditions. Application of a reduction pulse (using a small amount of reduced dithiothreitol) during the oxidative regeneration of ONC indicated the survival of the native protein along with three other (structured) species, I1, I2 and I3, with the rest of the unstructured species being converted to fully reduced protein. Mass spectrometry indicates that I1 has two disulfide bonds, whereas I2 and I3 have three disulfide bonds each. A disulfide mapping method, based on cyanylation, was used to identify I2 and I3 as des-[30–75] and des-[19–68], respectively. On enzymatic digestion using trypsin, I1 was identified as des-[19–68, 30–75]. Differences in the intermediates that are generated during the oxidative folding of the two structural homologues, RNase A and ONC, demonstrate that regenerative pathways are not necessarily influenced by tertiary structure. This indicates that the lack of a disulfide bond in ONC, analogous to the (65–72) disulfide bond in RNase A which plays an important role in its oxidative regeneration, does not adversely affect the oxidative folding of ONC.
Keywords: onconase, oxidative folding, RNase A, structured intermediate
Introduction
Onconase (ONC; registered trademark of Alfacell Corp., Bloomfield, NJ, USA), a frog ribonuclease found in the oocytes and early embryos of the northern leopard frog (Rana pipiens) (Ardelt et al. 1991), has four disulfide bonds and is structurally homologous to bovine pancreatic ribonuclease A (RNase A) (Wlodawer et al. 1988; Mosimann et al. 1994; Leland and Raines 2001). The frog variant demonstrates cytostatic and cytotoxic characteristics toward certain types of tumor cells (Boix et al. 1996; Leland et al. 1998; Leland et al. 2000), can degrade tRNAs selectively (Saxena et al. 2002) and is currently in Phase IIIb clinical trials for the treatment of asbestos-related lung cancer (http://www.alfacell.com/clinical_trials/mm_trials.htm). Ongoing effort in various laboratories is devoted to understand and improve the biomedical potential of ONC, and has resulted in the discovery of several new and interesting properties of this macromolecule (Newton et al. 1998, 2001; Welker et al. 2007).
Research in our laboratory is devoted to determining the intramolecular interactions that govern the oxidative folding of multi-disulfide-containing proteins (Rothwarf and Scheraga 1993; Rothwarf et al. 1998a; Narayan et al. 2000; Wedemeyer et al. 2000;Welker et al. 2001a, 2001b,2004). The ultimate aim was to be able to predict the structures of proteins from their amino acid sequences, which will aid in their applications in medicine and biotechnology. In this context, studies of the intermediates that accumulate during the folding pathway are useful in determining key interactions that allow for the formation and stabilization of those intermediates, and of the folded protein. Knowledge of such interactions is essential for the design of more potent mimics of the naturally occurring biomolecule, and is also required to improve the yields during the oxidative folding of recombinant native and mutant variants that are created for biotechnological and therapeutic purposes.
To understand the factors influencing the oxidative folding pathways of multi-disulfide-containing proteins, we have carried out in-depth studies of the regeneration of RNase A and its three-disulfide-containing mutants (Rothwarf and Scheraga 1993; Iwaoka et al. 1998; Rothwarf et al. 1998a; Xu and Scheraga 1998). Our work, together with research in other laboratories, has demonstrated that multi-disulfide-containing proteins regenerate by first forming unstructured disulfide-containing intermediates (Weissman and Kim 1991; van den Berg et al. 1999; Narayan et al. 2000; Chang and Li 2002). The rate determining steps in most of these cases involve the formation of a structured intermediate, or the native protein, either through reshuffling-coupled conformational folding reactions (such as the 3S→3S* step in RNase A (Rothwarf et al. 1998b) or through oxidation-coupled conformational folding reactions (such as the 2S→N [or 3S*] step in hirudin (Thannhauser et al. 1997) and in the three-disulfide-containing mutants of RNase A (Iwaoka et al. 1998; Xu and Scheraga 1998). The formation of the remainder of the disulfide bonds in a structured intermediate is more rapid in this kinetic mechanism and, thus, the formation of such a structured intermediate plays an important role in the overall oxidative folding pathway.
In this study, we have examined the oxidative folding of ONC and the effects of any structured intermediates in the early stages of regeneration on the oxidative folding. We have also identified the disulfide-bonded residues in the oxidative folding intermediates. The results obtained are compared with the oxidative folding pathways in the regeneration of RNase A, for which a large amount of data already exists (Rothwarf and Scheraga 1993; Iwaoka et al. 1998; Rothwarf et al. 1998a, 1998b; Xu and Scheraga 1998; Narayan et al. 2000; Wedemeyer et al. 2000;Welker et al. 2001a, 2001b,2004), and with the reductive unfolding pathways of RNase A and ONC (Narayan et al. 2004). Furthermore, we use these structural homologues to determine the influence of three-dimensional structure on the oxidative folding pathways of multi-disulfide-containing proteins. Finally, the impact of the absence of the (65–72) disulfide bond in the oxidative folding of ONC is discussed, given that this disulfide bond plays a major role in the regeneration of RNase A (Xu et al. 1996, 2004; Iwaoka and Juminaga et al. 1998; Rothwarf et al. 1998a, 1998b).
Materials and methods
Materials
WT-ONC cDNA in a pET11 expression vector was kindly provided by R.J. Youle (Boix et al. 1996). This cDNA was amplified by PCR and cloned to a pET22b(+) vector in-frame with the pelB signal sequence without the starting methionine residue. WT-ONC (Boix et al. 1996) was expressed in BL21 (DE3) cells and purified as described earlier for RNase A (Laity et al. 1993, 1997; Narayan et al. 2004), and the level of expression was comparable with that of an independent expression of WT-ONC from another wild-type plasmid (pONC) which was kindly provided by R.T. Raines (Leland et al. 1998). The conversion of the leading glutamine to pyroglutamic acid was made from the uncyclized protein by dialyzing the folded protein against 100 mM phosphate buffer, pH 7.0, at room temperature for 2 days. The protein was purified by cation-exchange HPLC as described earlier (Laity et al. 1993, 1997), and lyophilized. The conversion of the leading glutamine to pyroglutamic acid was verified by MALDI-TOF mass spectrometry (Cornell University Life Sciences Core Laboratories Center Proteomics and Mass Spectrometry Facility).
AEMTS (>99% pure) was purchased from Anatrace and used without further purification. DTTox, DTTred, CDAP, methylamine HCl and TCEP were obtained from Sigma and used without further purification. Trypsin Gold, mass spectroscopy grade, was purchased from Promega in vials of 100 μg and was dissolved in 50 mM acetic acid, pH 3, to a concentration of 1 mg/ml. Aliquots (10 μl) of this stock solution were stored frozen at −70°C. Such aliquots were not used after three freeze/thaw cycles. All other chemicals were of the highest grade commercially available.
Preparation of reduced ONC
Lyophilized ONC was dissolved in buffer (pH 8, 25 mM Tris–HCl, 3 mM EDTA) containing 6 M GdnHCl and 100 mM DTTred at a concentration of 2 mg/ml and allowed to incubate at room temperature for a period of 2 h under humidified argon. The reducing agent and salts were removed from the reduced protein by using a G25 size-exclusion column (50 mM acetic acid as the running buffer). Water and acetic acid were removed by lyophilization. Lyophilized, reduced-ONC (R-ONC) was stored in 2 mg/ml aliquots in a 3 mM acetic acid buffer (pH 3), frozen at −70°C, until further use.
Oxidative folding of R-ONC
Oxidative folding of ONC was initiated by introducing an aliquot of R-ONC into a solution containing 25 mM DTTox (final pH 8.0, 100 mM Tris, 3 mM EDTA, 25°C), which was previously purged of oxygen by humidified argon, to a final concentration of 0.2 mg/ml of protein. The reaction mixture was kept under humidified argon for the entire time required to populate the oxidative folding intermediates.
Aliquots of the reaction mixture were withdrawn periodically, and free thiols in each aliquot were blocked immediately with AEMTS. The blocking was performed by adding 100 μl of a blocking buffer (pH 8.7, 2M Tris, 20 mM EDTA) with a sufficient amount of AEMTS to achieve a 100-fold molar excess over the thiol content, and was allowed to proceed for 2 min, then stopped by adding glacial acetic acid; every sample was immediately desalted on a G25 column, with 50 mM acetic acid as a running buffer. The intermediate species populated during oxidative folding were separated from the AEMTS blocked mixture by HPLC using a TOSOH Biosciences TSKgel™ column SP-5PW 7.5 cm × 7.5 mm, analytical strong cation-exchange (SCX) column with a salt gradient (0–480 mM NaCl in 120 min). The mobile phase was 25 mM HEPES with 2 mM EDTA buffer, pH 8.0.
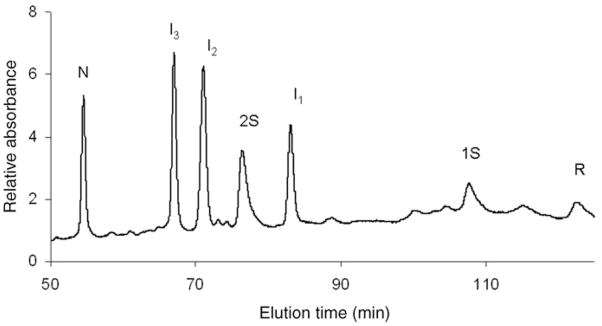
In a previous study, some aliquots were subjected to a reduction pulse (Rothwarf et al. 1998a) by the addition of 5 mM DTTred for a period of 2 min at 25°C before being blocked with AEMTS. Three chromatographic intermediates, namely as I1, I2 and I3, were observed (Welker et al. 2004); their survival during a reduction pulse implies that they are structured. These chromatographic intermediates are reproduced in Fig. 1. According to the mass spectrometric results in Table I, I2 and I3 each has three disulfide bonds, and I1 has two disulfide bonds. Favorable conditions were established to populate each of these intermediates in large enough quantities to determine their disulfide connectivities.
Fig. 1.
SCX-HPLC separation of the species involved in the oxidative folding of ONC at 25 mM DTTox, pH 8.0, 25°C, 0.35 mg/ml ONC, 50 min after the start of regeneration. The absorbance at 280 nm was monitored.
Table I.
Molecular weights of native ONC and its AEMTS-blocked intermediates, I1, I2 and I3
| Observed chromatographic peaks of Fig. 1 | Experimental MW of ONC species in Fig. 1, blocked with AEMTSa | Theoretical MW of ONC species blocked with AEMTS | Number of disulfide bonds in each species in Fig. 1 |
|---|---|---|---|
| N | 11 820b | 11 819b,c | 4 |
| I3 | 11 970 | 11 971 | 3 |
| I2 | 11 971 | 11 971 | 3 |
| I1 | 12 121 | 12 123 | 2 |
| R | 12 427 | 12 427 | 0 |
Each AEMTS adduct adds 76 Da to the molecular weight of the native protein.
Mass measurements were made on a MALDI-TOF MS instrument.
N cannot be blocked with AEMTS.
Populating the oxidative folding intermediates for disulfide mapping
To optimally populate I1, oxidative folding was initiated and allowed to proceed for 15 min, before being blocked with AEMTS, and isolated from an SCX-HPLC separation, as described in ‘Oxidative folding of R-ONC’ section. After a fraction from the HPLC column containing I1 was collected, the pH of the solution was reduced to 3, and the sample was frozen at −70°C until further use. NaCl and buffer salts were removed by RP-HPLC using a 25 cm × 4.6 mm SULPELCO Discovery® BIO Wide Pore C18, 5 μm particle size column with a 0.1% TFA–water/0.1% TFA–acetonitrile mobile phase with a constant gradient of 0% acetonitrile to elute salts, followed by a constant gradient of 80% acetonitrile to elute I1. After collecting AEMTS-blocked I1 from RP-HPLC, the collected fraction was frozen by allowing the vessel to contact dry ice, prior to lyophilization (for removal of water, acetonitrile and TFA). The lyophilized powder was re-dissolved at pH 3 in 3 mM acetic acid buffer and frozen at −70°C until subjected to enzymatic digestion.
I2 and I3 were optimally populated by allowing the oxidative folding reaction to proceed for 60 min. Then, the reaction was stopped by reducing the pH to 3 by adding 100 μl of glacial acetic acid. They were then separated by RP-HPLC using a 25 cm × 4.6 mm SULPELCO Discovery® BIO Wide Pore C18, 5 μm particle size column with 0.1% TFA–water/0.1% TFA–acetonitrile mobile phase and a gradient of 30–45% acetonitrile over 60 min. After I2 and I3 were collected in separate fractions, each fraction was frozen by allowing the vessel to contact dry ice and lyophilized to remove the TFA, water and acetonitrile. Unblocked I2 and I3 were stored frozen in solution either as a collected fraction from RP-HPLC or reconstituted from the lyophilized form in 3 mM acetic acid at −70°C.
Free cysteines in I2 and I3 are unblocked during their separation by RP-HPLC because the mapping method used (Wu and Watson 1997) requires that the cysteines, which eventually must be cyanylated, are free, i.e. not blocked with AEMTS. To verify that the peaks separated by RP-HPLC were I2 and I3, each separate fraction containing a single moiety, corresponding to I2 or I3, was lyophilized, reconstituted in 3 mM acetic acid over ice for 15 min and blocked with AEMTS, as described in ‘Oxidative folding of R-ONC’ section. The separate fractions were injected individually into the SCX-HPLC column and analyzed by the same method described in ‘Oxidative folding of R-ONC’ section. Each of the fractions analyzed separately corresponded to the intermediates I2 and I3 according to the SCX-HPLC chromatogram shown in Fig. 1.
Populating the oxidative folding intermediates by reductive unfolding for disulfide mapping
In order to map each oxidative folding intermediate unambiguously, the connectivity of the disulfide bonds had to be verified. Since these intermediates show evidence of native-like structure, it seems that the intermediates have native disulfide bonds. In order to verify this, each intermediate was populated both by reductive unfolding of native ONC (N-ONC) and by oxidative folding of R-ONC, and the cysteines not involved in disulfide bonds were identified in both cases. The disulfide bond connectivity could be verified as native only when the two mass spectra of each intermediate are the same, whether it was populated from N-ONC or R-ONC. Co-elution tests were carried out to verify that an oxidative folding intermediate was also populated by reductive unfolding.
I2 and I3 were populated by reductive unfolding by introducing an aliquot of N-ONC into to a solution containing 6.6 mM TCEP (pH 3.1, 0.1 M citric acid, 6 M GdnHCl, 2 mM EDTA, 25°C) to a final concentration of 0.3 mg/ml for 1 h. In previous work (Narayan et al. 2004; Xu et al. 2004), only I2 was observed upon reduction of native protein (25°C, pH 8), in the absence of denaturant. Therefore, to access the disulfide bond to be reduced to populate I3, 6 M GdnHCl had to be added to expose all of the disulfide bonds in N-ONC. After 1 h, in the presence of GdnHCl, unblocked I2 and I3 could be separated by the same method and conditions used to separate the intermediates described in ‘Populating the oxidative folding intermediates for disulfide mapping’ section, i.e. a brief reduction time at a low concentration of TCEP. I2 and I3, populated by reductive unfolding and blocked with AEMTS, could thus be separated. After N-ONC was permitted to equilibrate in the reaction mixture for 1 h, the mixture was diluted by one-half with 100 mM Tris–HCl, 2 mM EDTA buffer, pH 8.3, and immediately added to a solution containing AEMTS, as described in ‘Oxidative folding of R-ONC’ section. After 2 min, the resulting mixture was desalted on a G25 column with 50 mM acetic acid as a running buffer. AEMTS-blocked I2 and I3 could be separated by SCX-HPLC, using the method described in ‘Oxidative folding of R-ONC’ section.
I1 was also populated by first reducing N-ONC to I2 and isolating unblocked I2, then populating I1 by reducing unblocked I2. Unblocked I2 was most efficiently populated from reductive unfolding by introducing an aliquot of N-ONC to a solution containing 10 mM DTTred (pH 8.0, 100 mM Tris, 2 mM EDTA) to a final concentration of 1 mg/ml for 1 h. Glacial acetic acid was added to stop the reduction reaction. The solution was injected into an RP-HPLC column, and unblocked I2 was separated and collected by the method described in ‘Populating the oxidative folding intermediates for disulfide mapping’ section. The fraction containing unblocked I2 was frozen by allowing the vessel to contact dry ice and lyophilized to remove water, acetonitrile and TFA. The lyophilized powder was re-dissolved in 3 mM acetic acid buffer, pH 3, at a concentration of 7.5 mg/ml of unblocked I2 stock solution. A 20 μl aliquot was removed from this stock solution and added to a reducing buffer containing 6.6 mM TCEP (pH=3.1, 6 M GdnHCl, 0.1 M Tris, 2 mM EDTA) to obtain a final protein concentration of 0.3 mg/ml, and sufficient amount of I1 was produced after 45 min. In order to block I1 with AEMTS, the mixture was treated in a similar manner as for the AEMTS-blocking of I2 and I3 and separated on an SCX-HPLC column, as described previously.
Wu–Watson cyanylation and base cleavage method employed for disulfide bond connectivity identification in I2 and I3
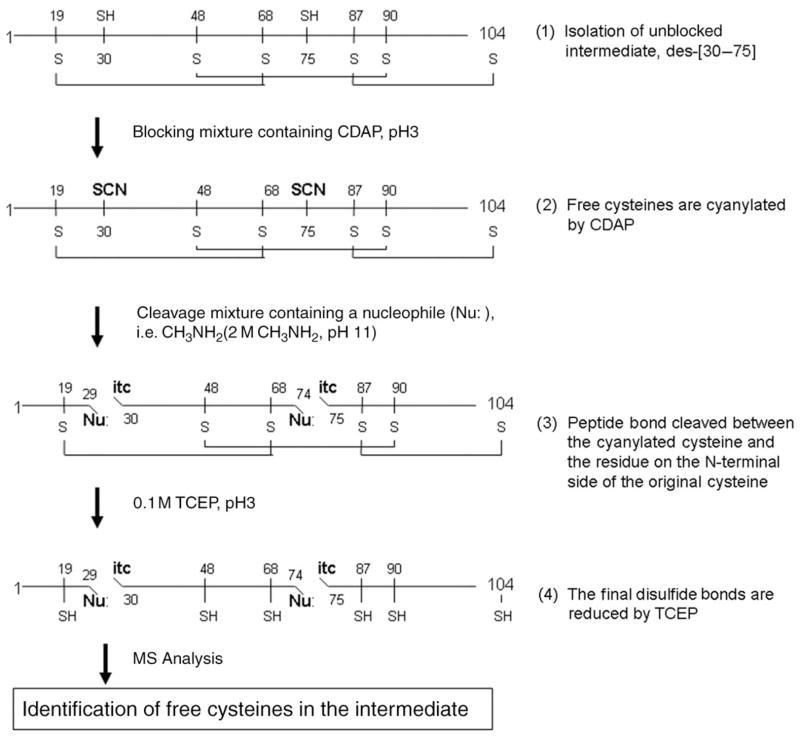
Location of free cysteines in I2 and I3 was achieved with a method developed by Wu and Watson (Wu and Watson 1997). This method makes use of a nucleophilic attack by a base on the carbonyl carbon of the residue located on the N-terminal side of a cyanylated cysteine, resulting in cleavage of the peptide bond (Fig. 2). Following the base-induced cleavage, the free amino group of the cyanylated cysteine residue performs a nucleophilic attack on the cyanyl group and forms a five-member ring, abbreviated as –itc (Wu and Watson 1998). The cysteines not involved in a disulfide-bond in an oxidative folding intermediate are cyanylated by CDAP, and the free cyteines can be identified by the size of the fragments produced by the cleavage.
Fig. 2.
The mapping method developed by Wu and Watson (1997) applied to a three-disulfide-bond-containing intermediate of ONC; this scheme is illustrated with I2 as an example. The cysteines in ONC are numbered, and the native disulfide bonds are connected. The locations of the free cysteines are identified by the sizes of the fragments produced by the base cleavage.
After fractions of unblocked I2 and I3 were separated and collected individually from RP-HPLC and lyophilized, the dried fractions were treated with a cyanylation buffer. For the cyanylation of I2, a 50 μl mixture containing 0.1 M CDAP (pH 3, 0.1 M citric acid) was added to the lyophilized powder for 15 min at 25°C. For the cyanylation of I3, a 25 μl mixture containing 0.2 M CDAP (pH 3, 0.1 M citric acid, 6 M GdnHCl) was added to the lyophilized powder for 15 min at 25°C. Cyanylation of both samples was quenched by freezing them. CDAP, GdnHCl and buffer salts were removed from each fraction by RP-HPLC, using the same method to remove salts described in ‘Populating the oxidative folding intermediates for disulfide mapping’ section. The collected intermediates were frozen by allowing the vessel to contact dry ice and lyophilized. The conditions for cyanylating I2 and I3 are different because of the different reactivity of the free cysteines in both intermediates; i.e. the free cysteines in I3 are less reactive than those in I2 and, as a result, a higher concentration of CDAP and the presence of 6 M GdnHCl are required for complete blocking. Reshuffling and incomplete cyanylation was observed if these two procedures were not adopted in cyanylating I3.
Fragmentation was carried out with a 50 μl mixture containing 2 M methylamine (pH 11, 6 M GdnHCl, 0.1 M phosphate) at 2°C for 10 min. Before the fragmentation mixture was added to the dried cyanylated intermediates, the fragmentation mixture was equilibrated for 5 min at 2°C. After 10 min, the fragmentation was stopped by adding 40 μl of formic acid, and the disulfide bonds in each intermediate were reduced by adding 800 μl of 0.1 M TCEP (pH 3.1, 0.1 M citric acid) for 40 min at 37°C.
Trypsin digestion of AEMTS-blocked I1
Tryptic digestion, without the addition of GdnHCl, was used to produce peptides, identifiable by mass spectroscopy, for the identification of I1. The peptides were compared to those produced from tryptic digestion of a completely reduced and AEMTS-blocked species of ONC, RB-ONC.
The conditions for enzymatic digestion were 37°C and pH 8.0 (0.1 M Tris–HCl, 2 mM EDTA). The concentration of the protein was 0.5 mg/ml, and a sufficient amount of trypsin was added to form an enzyme:I1 mass ratio of 1:25. The digestion was stopped by adding formic acid after 1 h.
Obtaining mass spectra for each intermediate
Each sample was desalted using a Sep-Pak reversed-phase Solid-Phase Extraction procedure before being loaded onto a 4700 Proteomics Analyzer (Applied Biosystems) MALDI-TOF mass spectrometer. Peptides were analyzed in both linear and reflector modes.
Population of I1 from I3
I1 was populated from I3 first by reducing N-ONC to I3, isolating unblocked I3 from RP-HPLC, and lyophilizing and storing the re-dissolved protein in 3 mM acetic acid, as discussed in ‘Populating the oxidative folding intermediates by reductive unfolding for disulfide mapping’ section. For the reduction of I3, this unblocked intermediate was added to a reducing buffer containing 0.1 M TCEP (pH 3.1, 0.1 M citric acid) to a final protein concentration of 0.1 mg/ml, and I1 was produced after 30 min. I3 and I1 were separated by RP-HPLC according to ‘Populating the oxidative folding intermediates for disulfide mapping’ section.
Results
Oxidative folding of ONC
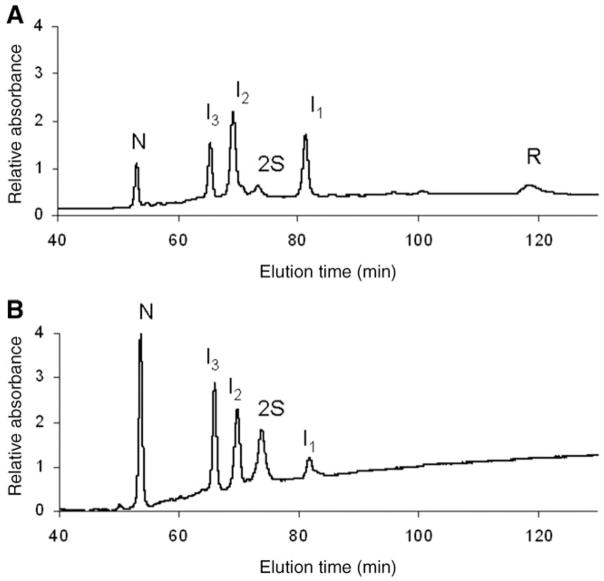
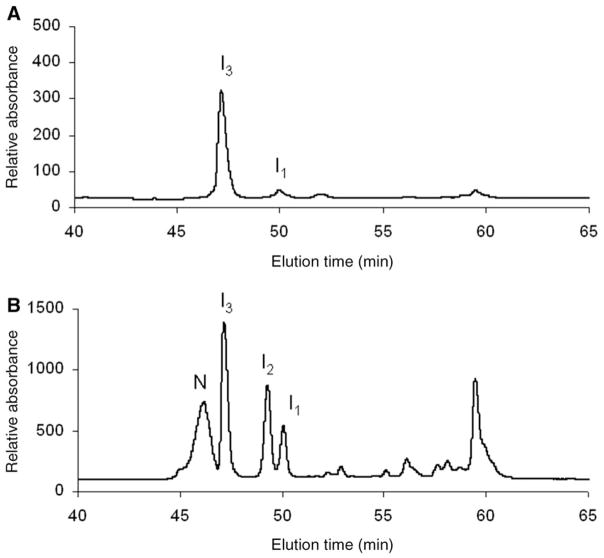
Figure 3A and B are typical HPLC chromatograms showing the oxidative regeneration of N-ONC from its reduced form as a function of time, 15 and 115 min, respectively (using 25 mM DTTox). During the course of oxidative folding, several intermediates are visible, besides the native (N) in Fig. 3A and B and fully-reduced (R) protein in Fig. 3A. After only 15 min, almost all of R and 1S are consumed. This is much different from the oxidative folding behavior of RNase A, in which R and 1S are in an identifiable pre-equilibrium in which they each constitute 10 and 20%, respectively, of the intermediate species over a number of hours under similar redox conditions (Rothwarf and Scheraga 1993). This shows that strong interactions play a role early in the oxidative folding pathway of ONC, and drive formation of the early forming intermediate, I1. According to Fig. 3B, 40% of N is regenerated after 115 min. Compared with RNase A, this degree of folding in such a short amount of time is observed only with 200 mM DTTox, eight times the amount used for ONC. Another key difference is that the intermediates, I1, I2 and I3, are present to a large extent, 10–30% over the course of the oxidative folding process, compared with RNase A. The stable intermediates in RNase A, des-[65–72] and des-[40–95] were populated to no more than 10% (Rothwarf et al. 1998b) under conditions much more oxidizing than the ones used for ONC in Fig. 3. Also, the reactivities of I2 and I3 along the oxidative folding pathway of ONC appear to be different from each other. In Fig. 3A, I2 is produced to a larger extent than I3, whereas in Fig. 3B more I2 is being turned over than I3. Further work (in progress) would determine how their different reactivities play a role in the oxidative folding mechanism.
Fig. 3.
Typical HPLC chromatograms showing the oxidative regeneration of ONC using DTTox (25 mM) at pH 8.0 and 25°C (100 mM Tris–HCl, 3 mM EDTA) at an R-ONC concentration of 0.2 mg/ml. (A) 15 min and (B) 115 min after the start of the regeneration. The samples were blocked with AEMTS before injection onto the HPLC, as described in ‘Oxidative folding of R-ONC’ section. The absorbance at 280 nm was monitored.
Production of oxidative folding intermediates by reductive unfolding
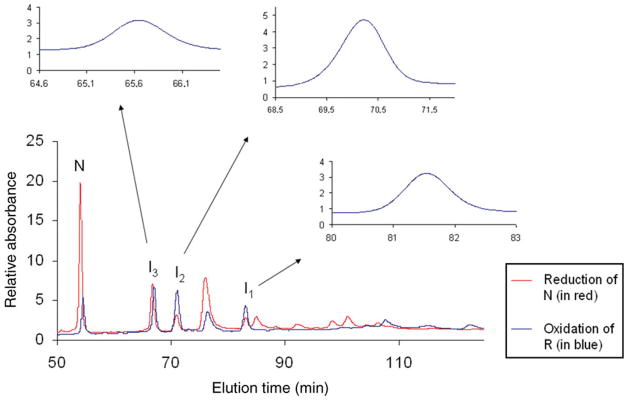
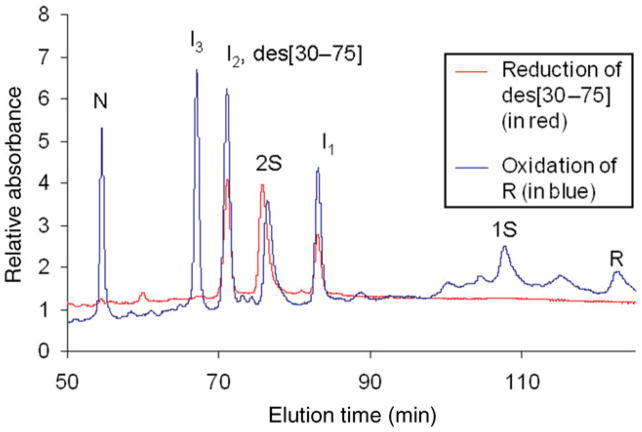
The oxidative folding intermediates produced by the reductive unfolding of N and subsequently blocked with AEMTS are shown in Fig. 4. The chromatogram for this separation is superimposed on the chromatogram for separation of the AEMTS-blocked intermediates produced by oxidative folding. To show that the intermediates from reduction and oxidation are the same species, a co-elution test was carried out and the results (in the inset of Fig. 4) indicated that they are the same species. As pointed out in ‘Populating the oxidative folding intermediates by reductive unfolding for disulfide mapping’ section, only I2 is observed in the reductive unfolding pathway in ONC; this is attributed to the highly exposed (30–75) disulfide bond in N-ONC which is preferentially reduced to form I2 (Narayan et al. 2004; Xu et al. 2004). Also, 6 M GdnHCl was required to expose the other disulfide bonds to produce I3 in reductive unfolding.
Fig. 4.
Superposition of the chromatograms of the oxidative folding intermediates produced by reduction of N-ONC and by oxidation of R-ONC. The conditions for reduction were: 25°C, 6.6 mM TCEP, pH 3, 6 M GdnHCl, 0.3 mg/ml N-ONC. The conditions for oxidation were: 25°C, 25 mM DTTox, pH 8.0, 0.35 mg/ml R-ONC. To confirm that I1, I2 and I3 were produced from both N-ONC and R-ONC, co-elution tests were carried out, and the results are shown as insets in these chromatograms. Each intermediate was collected from either oxidative folding or reductive unfolding experiments, mixed together and, when re-injected into the HPLC, eluted as one peak. The absorbance at 280 nm was monitored.
In order to produce I1 by reductive unfolding for trypsin digestion, the production of I1 from I2 is shown in Fig. 5. I2 was used because it was shown not to reshuffle under the conditions for reduction, and the yield is much higher for I1 than a reduction from N when compared with Fig. 4 (R.F. Gahl and H.A. Scheraga, in preparation). In addition, I1 was produced by reducing unblocked I3, as shown in Fig. 6. I2 and I3 have three disulfides each, and the reduction of these two intermediates can possibly generate three two-disulfide-bond-containing species. In Figs 5 and 6, only two peaks are visible between I2 and I3 and the reduced ONC. It is possible that two unstructured 2S species are eluted at the same time or one 2S species is not produced abundantly and thus only two peaks are detected in the chromatogram besides the original intermediate and the reduced form. The conditions to produce I1 from I3 (0.1 M TCEP) are different than those used to produce I1 from I2 (6.6 mM TCEP, 6 M GdmCl). This is a reflection of the relative reactivities around the (30–75) disulfide bond in I3 and the (19–68) disulfide bond in I2. The (19–68) disulfide must be buried in des-[30–75] because GdnHCl was required to expose it for reduction. Also, the (30–75) disulfide bond was exposed in des-[19–68] but is not as reactive as when it is reduced in N-ONC (Narayan et al. 2004; Xu et al. 2004). These interactions will have implications for determining the oxidative folding pathway of ONC (R.F. Gahl and H.A. Scheraga, in preparation).
Fig. 5.
Chromatogram for the production of I1 from isolated and unblocked I2 (des-[30–75]) after 1 h. The conditions were: 25°C, 6.6 mM TCEP, pH 3, 6 M GdnHCl, 0.3 mg/ml unblocked I2. The absorbance at 280 nm was monitored. The chromatogram shown in Fig. 1 is superimposed for comparison.
Fig. 6.
(A) I3 can be reduced with 0.1 M TCEP (pH 3.1, 0.1 M citric acid, 0.1 mg/ml protein, 25°C) to form I1 after 40 min, and separated by RP-HPLC as described in ‘Populating the oxidative folding intermediates for disulfide mapping’ section. (B) Separation of all the oxidative folding intermediates produced with 25 mM DTTox after 30 min (0.35 mg/ml protein, 25°C, pH 8.0, 0.1 M Tris) using the same RP-HPLC method. The absorbance at 210 nm was monitored.
I2 and I3 are intermediates with three disulfide bonds, whereas I1 has two. In ‘Populating the oxidative folding intermediates by reductive unfolding for disulfide mapping’ section, it was demonstrated that one disulfide bond was reduced in I2 to produce I1. Since ONC is a four disulfide-bond-containing protein, and if I1 could also be populated by another intermediate with three disulfide bonds, namely I3, then I1 must lack the disulfide bonds missing in both I2 and I3. This is indeed the case, and the results are discussed in ‘Identity of I1, I2 and I3’ section.
Methodology to identify the oxidative folding intermediates, I1, I2 I3
In order to identify the oxidative folding intermediates, i.e. map the disulfide linkages, two questions must be answered: (i) which particular cysteine is involved in a disulfide bond? (ii) If it is, what is its connectivity? A method developed by Watson and co-workers (Wu and Watson 1997) was adopted to identify the free cysteines in I2 and I3. Tryptic digestion was used to identify the free cysteines in I1. While each of these methods for mapping the intermediates determines whether a cysteine is involved in a disulfide bond or not, the disulfide-bond connectivity still had to be determined. To determine whether the connectivity of the disulfide bonds in the oxidative folding intermediates is native, the intermediates are also populated by reductive unfolding of N-ONC whose disulfide bonds are known from NMR (Gorbatyuk et al. 2004) and crystallography (Mosimann et al. 1994). The oxidative folding intermediates produced by reductive unfolding should have native disulfide bonds if they are not preceded by a species that reshuffles during the reduction. To eliminate reshuffling, which could produce non-native disulfide bonds, the reductive unfolding was performed at pH 3 with TCEP. To ensure access to all of the disulfide bonds in ONC, the reduction was performed in the presence of 6 M GdnHCl.
Identity of I1, I2 and I3
Table II shows the possible fragments that are produced using the method developed by Wu and Watson (1997). The assignment of the disulfides to I3 is unambiguous. All three peaks in the mass spectrum corresponding to des-[19–68] are present, and the peaks corresponding to other ‘des’ species are absent. The same is true for the assignment to I2 as des-[30–75].
Table II.
Results of fragmentation of I2 and I3 using the method of Wu and Watson (1997)
|
|
|
||
| 3516.6b (1–29-Nu:, 3516.6c) | 3516.6 (1–29-Nu:, 3516.6) | ||
| 5079.8 (itc-30–74-Nu:, 5078.8) | 5079.2 (itc-30–74-Nu:, 5078.8) | ||
| 3343.6 (itc-75–104, 3343.6) | 3343.5 (itc-75–104, 3343.6) | ||
|
| |||
|
|
|
||
|
| |||
| 2241.1 (1–18-Nu:, −2241.5) | 2241.1 (1–18-Nu:, 2241.5) | ||
| 5600.0 (itc-19–67-Nu:, 5596.5) | 5590.8 (itc-19–67-Nu:, 5596.5) | ||
| 4107.4 (itc-68–104, 4103.8) | 4103.2 (itc-68–104, 4103.8) | ||
The superscripts ‘red’ and ‘ox’ refer to an intermediate produced by reductive unfolding and oxidative folding, respectively. The abbreviations –itc and –Nu: refer to the same modifications of the intermediates described in Fig. 2; these modifications are taken into account in determining the masses of the fragments. The fragments in each of the three rows identify the positions of the free cysteines. Since the same fragments are observed in fragmenting an intermediate produced by reductive unfolding or by oxidative folding and the intermediates from oxidative folding and reductive unfolding co-elute, the disulfide bonds in the intermediates are native.
I2 is des-[30–75].
The numbers in these columns are the masses obtained by Mass Spectroscopy.
The theoretical masses for each of the fragments.
I3 is des-[19–68].
The peptides of I1 generated by trypsin that identify it as des-[19–68, 30–75] are listed in Table III. The peptides from I1 populated by oxidative folding ( ) and reductive unfolding ( ) are compared to the peptides generated by RB-ONC under the same conditions, and the two disulfide bonds were identified as Cys87–Cys104 and Cys48–Cys90.
Table III.
Results from tryptic digestion of RB-ONC, , and
| Mass of peptides | Location of peptides in ONC | Cys state | RB-ONC |
|
|
||
|---|---|---|---|---|---|---|---|
| 1994.9 (1994.9) | 86–104 | C87-C104, C90-SH | No | Yes | Yes | ||
| 2004.8 (2004.8) | 16–31 | C19blo, C30blo | Yes | No | No | ||
| 2221.9 (2221.9) | 86–104 | C87blo, C90blo, C104blo | Yes | No | No | ||
| 2426.1 (2426.1) | 46–49…86–104 | C48-C90, C87-C104 | No | Yes | Yes | ||
| 2527.1 (2527.1) | 56–76 | C68blo, C75blo | Yes | No | No | ||
| 5561.8 (5562.1) | 1–45 | C19blo, C30blo | Yes | Yes | Yes |
The superscript ‘red’ refers to the population produced by reductive unfolding, and the superscript ‘ox’ refers to the population produced by oxidative folding. Cysteines in a fragment blocked with an group from AEMTS are indicated as CXXblo, or in a disulfide bond as CXX-CXX, and unblocked as CXX-SH. The numbers in the first part of column 1 are the experimental masses, and those in parentheses are the theoretical masses of the fragments. Since the same fragments are produced from AEMTS-blocked I1 populated by either reductive unfolding or oxidative folding, I1 must have native disulfide bonds.
‘Yes’ and ‘No’ indicate that the particular peptide is, or is not, present in the mass spectrum.
I1 is des-[19–68, 30–75].
The corresponding mass spectra to produce Tables II and III are provided in the Supplementary Material (Supplementary data are available at PEDS online).
Discussion
The oxidative folding of ONC with DTTox has been examined at 25°C (pH 8). Three peaks that contain intermediates with disulfide bonds (I1, I2 and I3) are found to survive a reduction pulse, indicating that the intermediates constituting these peaks have some degree of structure that protects their disulfides from a reducing agent. Further studies (in preparation), including how they are involved in the oxidative folding pathway, will provide insight into the process by which ONC obtains its native structure from its completely reduced form. Preliminary oxidative folding data indicate that I1 is formed and consumed early in the regeneration process. While I2 and I3 are produced after I1 first appears, I2 is consumed more quickly before the end of the regeneration; I3 reacts more slowly relative to I2 and, therefore, is not as reactive and not readily oxidized to N.
The formation of such a structured intermediate (I1) in the early stages of ONC folding is compatible with the enhanced rate of regeneration. Once a structured intermediate is formed, its disulfides are sequestered from reduction and reshuffling reactions by its remaining cysteines or by an internal redox agent. Additionally, the remaining cysteines are aligned in native fashion and facilitate formation of the native disulfide bonds (Laity et al. 1997). Thus, I1 would be ideally poised to be oxidized to a structured three-disulfide-containing intermediate that can then be oxidized easily to N (2S*→3S*→N). The current work is being devoted to determine how these reactions fit into an overall oxidative folding mechanism.
Although RNase A and ONC are structurally very similar, our results indicate that there are significant differences in their oxidative folding pathways. The oxidative folding of RNase A at 25°C is characterized by the presence of two stable, three-disulfide-containing on-pathway intermediates, namely des-[40–95] and des-[65–72] (Rothwarf et al. 1998a). Furthermore, no stable two-disulfide intermediate(s) have been found to exist in the RNase A folding pathway whereas, in ONC, the presence of a stable, two-disulfide-containing intermediate (I1) is clearly evident. It is clear that the nature of the intermediates produced in these regenerative oxidative pathways is very different. This indicates that the intramolecular interactions that stabilize their respective intermediates during the oxidative folding process differ, and yet are capable of giving rise to a very similar three-dimensional fold in the final stage of the folding process. By identifying the interactions that give rise to the different folding pathways in future studies, more information can be gained about the interactions that drive the folding process.
Finally, the oxidative folding of ONC is of special interest because it lacks the (65–72) disulfide bond that plays a major role in the oxidative folding of RNase A. In the unstructured intermediates of RNase A, this bond is present to an extent of ~40% of all the disulfide bonds in the 1S and 2S ensembles (Xu et al. 1996; Volles et al. 1999), and the structured intermediate lacking this disulfide bond (des-[65–72]) is formed to a minor extent (~20%) in the rate-determining step of its regeneration (Rothwarf et al. 1998b). It is, therefore, of interest that the oxidative folding of ONC is faster than that of RNase A, despite the absence of the analog of the (65–72) disulfide bond in ONC. Such a disulfide bond analog does not appear in the native structure of ONC. ONC has two cysteines, Cys68 and Cys75, the same distance apart along the peptide backbone as the (65–72) disulfide bond in RNase A. However, in N-ONC, Cys68 is involved in a disulfide bond with Cys19, and Cys75 is involved in a disulfide bond with Cys30. Each of these disulfide bonds are 39 and 45 residues apart, respectively, and their formation incurs a greater loss in entropy for the polypeptide than forming a disulfide bond seven residues apart. However, the formation of the 19–68 and 30–75 disulfide bonds may be driven by enthalpically favorable interactions of the side chains. A similar preference to form a disulfide by enthalpic interactions is seen in the case of RNase A. Cys58 and Cys65 are seven residues apart, but Cys65 forms a disulfide bond with Cys72 because the (65–72) bond is a native disulfide bond, and more favorable enthalpic interactions promote this disulfide preference. Cys58 forms a native disulfide bond with Cys110.
In conclusion, we have demonstrated that the oxidative folding pathway of ONC is significantly different from that of its structural homologue, RNase A. Early-forming, structured intermediates can significantly accelerate the oxidative folding rate. Further experiments are in progress to identify the intramolecular interactions that account for the increased rate and formation of the structured intermediates.
Acknowledgments
We thank Dr. Ervin Welker for assistance in the expression of ONC and for helpful comments during the course of this work and also thank Robert Sherwood at the Cornell University Life Sciences Core Laboratories Center Proteomics and Mass Spectrometry Facility for obtaining the mass spectra.
Funding
National Institute of General Medical Sciences of the National Institutes of Health (Grant no. GM-24893).
Abbreviations
- ONC
frog onconase (Rana pipiens)
- R-ONC
onconase with all its disulfide bonds reduced
- RB-ONC
onconase with all its cysteines blocked with AEMTS and tagged with an group
- RNase A
bovine pancreatic ribonuclease A
- AEMTS
2-aminoethylmethylthiosulfonate
- CDAP
1-cyano-4-dimethylamino-pyridinium tetrafluoroborate
- DTTox and DTTred
oxidized and reduced dithiothreitol, respectively
- TCEP
tris(2-carboxyethyl)phosphine hydrochloride
- GdnHCl
guanidine hydrochloride
- Tris
tris(hydroxymethyl)aminomethane
- HEPES
N-(2-hydroxyethyl)piperazine-N′-(2-ethanesulfonic acid)
- TFA
trifluoroacetic acid
- SCX
strong cation exchange
- RP
reversed phase
- HPLC
high-performance liquid chromatography
- des-[a–b, c–d]
a species that contains all native disulfide bonds except for the a–b and c–d disulfide bonds
- nS
an ensemble of disulfide-containing intermediates each having n disulfide bonds
- nS*
a single species containing n disulfide bonds
- N
native onconase
- –itc
iminothiazolidinyl carboxyl residue
- MALDI-TOF
matrix-assisted laser desorption/ionization time-of-flight
References
- Ardelt W, Mikulski SM, Shogen K. J Biol Chem. 1991;266:245–251. [PubMed] [Google Scholar]
- Boix E, Wu Y, Vasandani VM, Saxena SK, Ardelt W, Ladner J, Youle RJ. J Mol Biol. 1996;257:992–1007. doi: 10.1006/jmbi.1996.0218. [DOI] [PubMed] [Google Scholar]
- Chang JY, Li L. Biochemistry. 2002;41:8405–8413. doi: 10.1021/bi020169k. [DOI] [PubMed] [Google Scholar]
- Gorbatyuk VY, Tsai CK, Chang CF, Huang T. J Biol Chem. 2004;279:5772–5780. doi: 10.1074/jbc.M311233200. [DOI] [PubMed] [Google Scholar]
- Iwaoka M, Juminaga D, Scheraga HA. Biochemistry. 1998;37:4490–4501. doi: 10.1021/bi9725327. [DOI] [PubMed] [Google Scholar]
- Laity JH, Shimotakahara S, Scheraga HA. Proc Natl Acad Sci USA. 1993;90:615–619. doi: 10.1073/pnas.90.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laity JH, Lester CC, Shimotakahara S, Zimmerman DE, Montelione GT, Scheraga HA. Biochemistry. 1997;36:12683–12699. doi: 10.1021/bi970878b. [DOI] [PubMed] [Google Scholar]
- Leland PA, Raines RT. Chem Biol. 2001;8:405–413. doi: 10.1016/s1074-5521(01)00030-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leland PA, Schultz LW, Kim BM, Raines RT. Proc Natl Acad Sci USA. 1998;95:10407–10412. doi: 10.1073/pnas.95.18.10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leland PA, Staniszewski KE, Kim BM, Raines RT. FEBS Lett. 2000;477:203–207. doi: 10.1016/s0014-5793(00)01804-4. [DOI] [PubMed] [Google Scholar]
- Mosimann SC, Ardelt W, James MNG. J Mol Biol. 1994;236:1141–1153. doi: 10.1016/0022-2836(94)90017-5. [DOI] [PubMed] [Google Scholar]
- Narayan M, Welker E, Wedemeyer WJ, Scheraga HA. Acc Chem Res. 2000;33:805–812. doi: 10.1021/ar000063m. [DOI] [PubMed] [Google Scholar]
- Narayan M, et al. J Mol Biol. 2004;338:795–809. doi: 10.1016/j.jmb.2004.03.014. [DOI] [PubMed] [Google Scholar]
- Newton DL, Boque L, Wlodawer A, Huang CY, Rybak SM. Biochemistry. 1998;37:5173–5183. doi: 10.1021/bi972147h. [DOI] [PubMed] [Google Scholar]
- Newton DL, Hansen HJ, Mikulski SM, Goldenberg DM, Rybak SM. Blood. 2001;97:528–535. doi: 10.1182/blood.v97.2.528. [DOI] [PubMed] [Google Scholar]
- Rothwarf DM, Li YJ, Scheraga HA. Biochemistry. 1998a;37:3760–3766. doi: 10.1021/bi972822n. [DOI] [PubMed] [Google Scholar]
- Rothwarf DM, Li YJ, Scheraga HA. Biochemistry. 1998b;37:3767–3776. doi: 10.1021/bi972823f. [DOI] [PubMed] [Google Scholar]
- Rothwarf DM, Scheraga HA. Biochemistry. 1993;32:2671–2679. doi: 10.1021/bi00061a027. [DOI] [PubMed] [Google Scholar]
- Saxena SK, Sirdeshmukh R, Ardelt W, Mikulski SM, Shogen K, Youle RJ. J Biol Chem. 2002;277:15142–15146. doi: 10.1074/jbc.M108115200. [DOI] [PubMed] [Google Scholar]
- Thannhauser TW, Rothwarf DM, Scheraga HA. Biochemistry. 1997;36:2154–2165. doi: 10.1021/bi962340w. [DOI] [PubMed] [Google Scholar]
- Volles MJ, Xu X, Scheraga HA. Biochemistry. 1999;38:7284–7293. doi: 10.1021/bi990570f. [DOI] [PubMed] [Google Scholar]
- van den Berg B, Chung EW, Robinson CV, Mateo PL, Dobson CM. EMBO J. 1999;18:4794–4803. doi: 10.1093/emboj/18.17.4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedemeyer WJ, Welker E, Narayan M, Scheraga HA. Biochemistry. 2000;39:4207–4216. doi: 10.1021/bi992922o. [DOI] [PubMed] [Google Scholar]
- Weissman JS, Kim PS. Science. 1991;253:1386–1393. doi: 10.1126/science.1716783. [DOI] [PubMed] [Google Scholar]
- Welker E, Narayan M, Wedemeyer WJ, Scheraga HA. Proc Natl Acad Sci USA. 2001a;98:2312–2316. doi: 10.1073/pnas.041615798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker E, Wedemeyer WJ, Narayan M, Scheraga HA. Biochemistry. 2001b;40:9059–9064. doi: 10.1021/bi010409g. [DOI] [PubMed] [Google Scholar]
- Welker E, Hathaway L, Scheraga HA. J Am Chem Soc. 2004;126:3270–3721. doi: 10.1021/ja031658q. [DOI] [PubMed] [Google Scholar]
- Welker E, Hathaway L, Xu G, Narayan M, Pradeep L, Shin HC, Scheraga HA. Biochemistry. 2007;46:5485–5493. doi: 10.1021/bi602495a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodawer A, Svensson LA, Sjölin L, Gilliland GL. Biochemistry. 1988;27:2705–2717. doi: 10.1021/bi00408a010. [DOI] [PubMed] [Google Scholar]
- Wu J, Watson JT. Prot Sci. 1997;6:391–398. doi: 10.1002/pro.5560060215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Watson JT. Anal Biochem. 1998;258:268–276. doi: 10.1006/abio.1998.2596. [DOI] [PubMed] [Google Scholar]
- Xu X, Scheraga HA. Biochemistry. 1998;37:7561–7571. doi: 10.1021/bi980086x. [DOI] [PubMed] [Google Scholar]
- Xu X, Rothwarf DM, Scheraga HA. Biochemistry. 1996;35:6406–6417. doi: 10.1021/bi960090d. [DOI] [PubMed] [Google Scholar]
- Xu G, Narayan M, Welker E, Scheraga HA. Biochemistry. 2004;43:3246–3254. doi: 10.1021/bi036215d. [DOI] [PubMed] [Google Scholar]