Abstract
Hereditary haemorrhagic telangiectasia (HHT) is an autosomal dominant disorder characterised by epistaxis, telangiectases, and multiorgan vascular dysplasia. The two major types of disease, HHT1 and HHT2, are caused by mutations in the ENG (endoglin) and ACVRL1 genes, respectively. The corresponding endoglin and ALK‐1 proteins are specific endothelial receptors of the transforming growth factor β superfamily essential for maintaining vascular integrity. Many mutations have been identified in ENG and ACVRL1 genes and support the haploinsufficiency model for HHT. Two more genes have recently been implicated in HHT: MADH4 mutated in a combined syndrome of juvenile polyposis and HHT (JPHT), and an unidentified HHT3 gene linked to chromosome 5. Current knowledge on the genetics of HHT is summarised, including the pathways that link the genes responsible for HHT and the potential mechanisms underlying the pathogenesis of the disease.
Keywords: ACVRL1, ALK‐1, ENG, hereditary haemorrhagic telangiectasia, vascular disorders
Hereditary haemorrhagic telangiectasia (HHT) or Rendu–Osler–Weber syndrome is an autosomal dominant disorder characterised primarily by epistaxis, telangiectases, and multiorgan vascular dysplasia. A minimum prevalence rate of HHT is estimated to be 1 in 10 0001 and higher in some geographically isolated regions. Thus, for example, the incidence in the Akita prefecture of northern Japan is estimated to be 1:5000 to 1:8000,2 roughly comparable with those reported in the Fyn County of Denmark (1:1641 to 1:7246)3 and other European and US populations. A higher prevalence (1:2351) is reported in the Haut Jura region of France.4,5,6 The Afro‐Caribbean population of the Netherlands Antilles has a prevalence of 1 in 1331 inhabitants, the highest known in the world.7
Individuals with HHT present with a wide range of symptoms and there is great variability in the clinical manifestations between families and among members of the same family. Spontaneous recurrent nosebleeds from telangiectasia of the nasal mucosa is the presenting sign in more than 90% of HHT patients.8 The severity and frequency of nosebleeds generally increases with age and can lead to chronic anaemia and blood transfusion requirement. Multiple telangiectases on the face, lips, oral cavity, nose, and fingers are common.6 Telangiectases can also develop in the gastrointestinal tract, particularly in the stomach and small bowel of older patients, who present with gastrointestinal haemorrhage and iron deficiency anaemia, usually in their fifth or sixth decades of life.6 Liver involvement is now more widely recognised and reported in up to 40% of HHT patients.9 It is usually asymptomatic in up to 50% of the affected individuals and reflects the presence of multiple intrahepatic telangiectases leading to the formation of shunts between the major vessels of the liver (from the hepatic artery to either portal or hepatic veins and from the portal vein to hepatic vein or vena cava).9,10
HHT patients may have arteriovenous malformations (AVM) in the pulmonary and cerebral/spinal circulation. Pulmonary arteriovenous malformations (PAVM) have been reported in up to 50% of patients and are caused by a direct connection between the pulmonary artery and the pulmonary vein, bypassing the capillary bed.6,11 This left to right shunting of blood by PAVM can lead to hypoxaemia, stroke, and brain abscess.12,13,14 Cerebral involvement may be associated with telangiectases, cerebral arteriovenous malformations (CAVM), aneurysms, or cavernous angiomas and can lead to seizures and life threatening or disabling haemorrhagic stroke.15,16
The clinical diagnosis of HHT is generally made according to the established Curaçao criteria.17 An individual is considered to have HHT if three of the following four diagnostic criteria are met: recurrent spontaneous epistaxis; mucocutaneous telangiectasia; visceral involvement such as pulmonary and cerebral/spinal AVMs, gastrointestinal bleeding or intrahepatic shunting; and a family history of HHT. The presence of two criteria warrants a possible or suspected diagnosis, while a single criterion renders the diagnosis unlikely. It should be noted that some features of HHT, such as epistaxis or gastrointestinal bleeding, are common in the general population and may occur in other conditions. Many signs of disease are also age dependent and do not manifest until later in life; therefore clinical criteria should be considered carefully, particularly in children with sporadically occurring disease.
More than 100 years after the initial recognition of the clinical entity, HHT is now associated with mutations in genes implicated in the mediation of transforming growth factor β (TGFβ) effects in endothelial cells. The targeted genes are predominantly expressed on vascular endothelium and define an endothelium specific pathway. Endoglin, coded for by the HHT1 gene, is a co‐receptor for TGFβ1 and TGFβ3 isoforms, while ALK‐1, encoded by the HHT2 gene, is an alternate type I serine–threonine kinase receptor which signals through Smad1/5. MADH4 gene, which is mutated in the combined syndrome of juvenile polyposis and HHT,18 codes for Smad4, the common Smad implicated in TGFβ signalling and present in all cell types.19 The nature of the putative HHT3 gene, recently linked to chromosome 5,20 remains to be determined, as well as its relation, if any, to members to the TGFβ superfamily.
Our review will summarise the current knowledge on the genetics of HHT, focusing on HHT1 and HHT2 and the potential mechanisms underlying disease pathogenesis.
Genetics of HHT and related disorders
HHT is a genetically heterogeneous disorder, and linkage studies have mapped it to regions 9q33‐q34.1 on chromosome 9 (HHT type 1) and 12q11‐q14 on chromosome 12 (HHT type 2). HHT type 1 (HHT1; OMIM 187300) is caused by mutations in the ENG (endoglin) gene, whereas HHT type 2 (HHT2; OMIM 600376) is caused by mutations in the ACVRL1 (activin receptor‐like kinase 1 or ALK‐1) gene. About 20% of HHT families remain unresolved after mutation analysis of these two genes, suggesting that other genes may be implicated. Indeed, a new locus for HHT (HHT3) was recently mapped to chromosome 5, though the causative gene remains unidentified.20 A subset of patients with a combined syndrome of juvenile polyposis and HHT (JPHT; OMIM 175050) harbour mutations in the MADH4 gene.18
ENG gene: structure, mutations, and polymorphisms
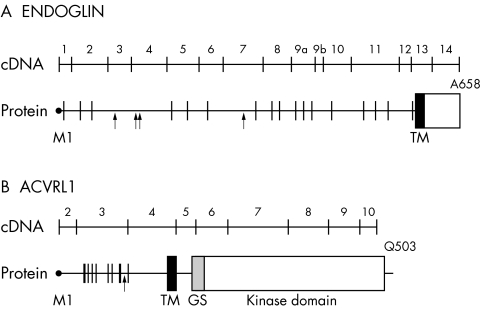
Linkage analysis first mapped HHT to chromosome 9q33‐q34.1,21,22 where endoglin was previously mapped.23 The chromosomal location of endoglin and its expression pattern and function led to its testing and confirmation as the disease associated gene (HHT1).24 A cDNA encoding endoglin, a type I integral membrane glycoprotein, was isolated in 1990.25 The protein exists as a covalently linked homodimer of Mr = 180 000, comprising polypeptide chains of Mr = 68 051 and N‐linked and O‐linked glycans. The 17 cysteine residues and the generated intra‐ and interchain bonds suggest that folding is tightly regulated; this is supported by the findings that most endoglin mutations lead to structural instability and loss of protein function. The extracellular region of endoglin, where all mutations have been found to date, consists of 561 amino acids, with a short hydrophobic stretch of 17 amino acids separating the regions rich in N‐linked (residues 63, 96, 109, and 282 from the N‐terminal) and O‐linked (residues 311 to 551) glycosylation sites (fig 1A). A hydrophobic region of 25 amino acids spans the plasma membrane, and the cytoplasmic tail is 47 residues long, rich in serine and threonine, and heavily phosphorylated, predominantly on serine residues.26
Figure 1 Schematic diagram of the cDNA and protein of endoglin (A) and ALK‐1 (B). The exon–intron boundaries are indicated on the protein; ATG initiation codon corresponds to base pair (bp) 1 and M1 of the leader peptide; the last codon is also indicated. In the polypeptide structure, vertical lines illustrate the position of the cysteine residues, while arrows indicate the potential N‐linked glycosylation sites. GS, glycine/serine‐rich domain; TM, transmembrane domain.
A comprehensive review of all published reports reveals 155 different ENG mutations (table 1 and the references cited). Their overall distribution and frequency is illustrated in fig 2A. Mutations to date were found in exons 1 to 12 (coding for the extracellular domain) and are of all types: deletions (n = 51), missense (n = 31), splice sites (n = 21), insertions (n = 25), nonsense (n = 23), and indels (n = 4). The total number of mutations per exon is similar except for smaller numbers in exons 1, 9b, and 12, and none in exons 13 and 14, coding for the transmembrane and cytoplasmic domains, respectively (fig 2A).
Table 1 Summary of known ENG mutations.
| No | Location | Type | Nucleotide change | Consequence | Refs |
|---|---|---|---|---|---|
| 1 | Ex 1 | M | c.1A→G | p.M1V | 27 |
| 2 | Ex 1 | M | c.2T→C | p.M1T | 28, 29 |
| 3 | Ex 1 | M | c.2T→G | p.M1R | 30 |
| 4 | Ex 1 | M | c.23T→C | p.L8P | 30 |
| 5 | Ex1 | D | c.63delC | p.T22fs | 31, 32 |
| 6 | Int 1 | Sp | c.67+1G→A | Ex 1 skip, p.M1fs | 27, 33 |
| 7 | Int 1 | Sp | c.68G→A (c.68–1G→A)* | Ex 2 del, p.S23fs | 34* |
| 8 | Int 1 | D | c.68_219del | Ex 2 skip, p.S23fs | 34, 35 |
| 9 | Ex 2 | N | c.90T→A | p.C30X | 36 |
| 10 | Ex 2 | M | c.95T→G | p.L32R | 37 |
| 11 | Ex 2 | D | c.115delA | p.R39fs | 35 |
| 12 | Ex 2 | N | c.121G→T | p.E41X | 35 |
| 13 | Ex 2 | N | c.142C→T | p.Q48X | 27 |
| 14 | Ex 2 | M | c.145G→T | p.V49F | 30 |
| 15 | Ex 2 | M | c.155G→T | p.G52V | 29, 38 |
| 16 | Ex 2 | M | c.155G→A | p.G52D | 37 |
| 17 | Ex 2 | M | c.157T→C | p.C53R | 16, 28, 29, 38 |
| 18 | Ex 2 | N | c.159C→A (c.157C→A)* | p.C53X | 27* |
| 19 | Ex 2 | I | c.195_196insT | p.H65fs | 30 |
| 20 | Int 2 | Sp | c.219+5G→A | Ex 2 skip, p.S23fs | 35 |
| 21 | Ex 3 | N | c.229C→T | p.Q77X | 32 |
| 22 | Ex 3 | N | c.247C→T | p.Q83X | 27 |
| 23 | Ex 3 | I | c.258–276insCAAAATGGCACCTGGCCC | p.K85_Q86insQNGTWP | 39 |
| 24 | Ex 3 | N | c.277C→T | p.R93X (p.R83X)* | 27, 28, 39* |
| 25 | Ex 3 | M | c.320T→G | p.L107R | 30 |
| 26 | Ex 3 | D | c.332_338delCCCTGGG | p.A111fs | 27 |
| 27 | Ex 3 | N | c.360C→A | p.Y120X | 39 |
| 28 | Int 3 | Sp | c.360+1G→A | Ex 3 skip, p.G74_Y120del | 16, 27, 28, 35, 40 |
| 29 | Int 3 | Sp | c.360+1G→C | Ex 3 skip, p.G74_Y120del | 2 |
| 30 | Int 3 | Sp | c.360+4A→G | Ex 3 skip, p.G74_Y120del | 34 |
| 31 | Int 3 | Sp | c.360+5G→A | splice defect | 27 |
| 32 | Int 3 | Sp | c.361–2A→G | splice defect | 34, 39 |
| 33 | Ex 4 | M | c.374T→A | p.V125D | 37 |
| 34 | Ex 4 | M | c.447G→C | p.W149C | 29, 41 |
| 35 | Ex 4 | D | c.461delG | p.G154fs | 30 |
| 36 | Ex 4 | M | c.479C→A | p.A160D | 37, 42 |
| 37 | Ex 4 | M | c.494C→T | p.P165L | 35 |
| 38 | Ex 4 | D | c.496delC | p.Q166fs | 30 |
| 39 | Ex 4 | I | c.497_498insC | p.Q166fs | 27 |
| 40 | Ex 4 | N | c.511C→T | p.R171X | 30, 32, 34, 36, 43 |
| 41 | Ex 4 | Sp | c.523G→C, c.361_523del | Ex 4 skip, p.N121fs | 35 |
| 42 | Int 4 | Sp | c.524–2A→G | Ex 4 skip, pN121fs | 29 |
| 43 | Int 4 | D | c.524_689del | Ex 5 del, p.A175fs | 35 |
| 44 | Ex 5 | I | c.562_563insC | p.Q188fs | 35 |
| 45 | Ex 5 | D | c.576_596del21bp | p.R192_P198del | 34 |
| 46 | Ex 5 | I | c.577_578insGC | p.T193fs | 27 |
| 47 | Ex 5 | M | c.581T→C | p.L194P | 35 |
| 48 | Ex 5 | I | c.580_581insACGC | p.L194fs | 28 |
| 49 | Ex 5 | D | c.586_604delTGGCGGCCGCGTACTCCAG | p.W196fs | 31 |
| 50 | Ex 5 | M | c.586T→C | p.W196R | 16 |
| 51 | Ex 5 | N | c.587G→A | p.W196X | 28, 44 |
| 52 | Ex 5 | N | c.588G→A | p.W196X | 35 |
| 53 | Ex 5 | D | c.591_619del29bp | p.P198fs | 32 |
| 54 | Ex 5 | I | c.596_597insCG | p.R199fs | 16 |
| 55 | Ex 5 | D | c.619_621delTGC | p.C207del | 30 |
| 56 | Ex 5 | D | c.640_643delGGCC | p.G214fs | 16 |
| 57 | Ex 5 | D | c.657_658delCA | p.I220fs | 45 |
| 58 | Ex 5 | M | c.662T→C | p.L221P | 38 |
| 59 | Ex 5 | D | c.682_686delTCGGC | p.S228fs | 46 |
| 60 | Int 5 | Sp | c.689+2T→C | Ex 6 skip, p.G230fs | 27, 30 |
| 61 | Int 5 | Sp | c.689–2A→T | Ex 6 skip, p.G230fs | 28 |
| 62 | Ex 6 | D | c.694_699delCGGACG | p.R232_T233del | 30 |
| 63 | Ex 6 | I | c.701_702insACGG | p.V234fs | 27 |
| 64 | Ex 6 | N | c.715G→T | p.E239X | 31 |
| 65 | Ex 6 | D | c.733delG | p.G245fs | 27 |
| 66 | Ex 6 | D | c.736delG | p.D246fs | 35 |
| 67 | Ex 6 | I | c.766_767insC | p.P256fs | 27 |
| 68 | Ex 6 | I | c.772_773insC | p.Y258fs | 30 |
| 69 | Ex 6 | M | c.781T→C | p.W261R | 27 |
| 70 | Ex 6 | N | c.782G→A | p.W261X | 31, 37 |
| 71 | Ex 6 | D | c.785_789delTCATC | p.L262fs | 30 |
| 72 | Ex 6 | D | c.787_789delATC | p.I263del | 27, 30 |
| 73 | Ex 6 | M | c.788T→C | p.I263T | 30 |
| 74 | Ex 6 | M | c.790G→A | p.D264N | 27 |
| 75 | Int 6 | Indel | c.817_1134del;insTAAAC (g.IVS6+140_IVS8+20delTAACCins)* | p.T273_A378del | 35* |
| 76 | Ex 7 | InDel | c.820_826delACTGGAGinsTTGAAGGTCTTTCCAGAGAAAAAC | p.T274fs | 28 |
| 77 | Ex 7 | I | c.828_829insA | p.Y277fs | 2 |
| 78 | Ex 7 | N | c.831C→G | p.Y277X | 24 |
| 79 | Ex 7 | D | c.882_920del39bp | p.T295_N307del | 24 |
| 80 | Ex 7 | InDel | c.887_918del32bp;c.919_920 insCAAGCTCCCAG | p.Q297fs | 27 |
| 81 | Ex 7 | D | c.893delG | p.G298fs | 30 |
| 82 | Ex 7 | I | c.904_905insGG | p.E302fs | 35 |
| 83 | Ex 7 | D | c.909_929del21bp | p.R304_I310del | 32 |
| 84 | Ex 7 | M | c.917T→C | p.L306P | 27, 29 |
| 85 | Ex 7 | M | c.932T→G | p.V311G | 47 |
| 86 | Ex 7 | I | c.967_968insT | p.V323fs | 34, 46 |
| 87 | Ex 7 | M | c.991G→A | p.G331S | 27 |
| 88 | Int 7 | D | c.993–25_1120del152 | p.G332fs | 30 |
| 89 | Ex 8 | D | c.993_1134del | Ex 8del, p.G332fs | 34 |
| 90 | Ex 8 | D | c.995delG | p.G332fs | 27 |
| 91 | Ex 8 | I | c.1048_1049dupTT | p.T349fs | 30 |
| 92 | Ex 8 | N | c.1050T→A | p.C350X | 24, 44 |
| 93 | Ex 8 | N | c.1078C→T | p.Q360X | 27 |
| 94 | Ex 8 | D | c.1078_1081delCAGA | p.G360fs | 29 |
| 95 | Ex 8 | M | c.1088G→A | p.C363Y | 37 |
| 96 | Ex 8 | D | c.1089_1090delTG | p.C363fs | 28 |
| 97 | Ex 8 | I | c.1111_1112insG | p.V371fs | 28, 40, 44 |
| 98 | Ex 8 | D | c.1111_1133del | p.V371fs | 16, 30 |
| 99 | Ex 8 | I | c.1117_1118insT | p.K373fs | 27 |
| 100 | Ex 8 | D | c.1120_1123delAAAG | p.K374fs | 2 |
| 101 | Ex 8 | M | c.1121_1122AA→GC | p.K374S | 27 |
| 102 | Ex 8 | I | c.1122_1123insA | p.E375fs | 35 |
| 103 | Ex 8 | N | c.1123G→T | p.E375X | 31 |
| 104 | Int 8 | Sp | c.1134G→A | splice defect, p.A378A | 27 |
| 105 | Int 8 | Sp | c.1134+1G→A | Ex 8 skip, p.G332fs | 34, 39 |
| 106 | Int 8 | Sp | c.1134+1G→C | Ex 8 skip, p.G332fs | 31 |
| 107 | Int 8 | I | c.1134_1135ins220_1134 | p.Ala378_His379insGly74_Ala378 | 34, 41 |
| 108 | Int 8 | D | c.1135_1977del | Ex 8–14del p.H379_A658del | 34, 35 |
| 109 | Ex 9a | I | c.1142_1143insT | p.K381fs | 27 |
| 110 | Ex 9a | M | c.1146C→G | p.C382W | 16 |
| 111 | Ex 9a | D | c.1165–1167delTTC | p.F389del | 39 |
| 112 | Ex 9a | D | c.1186delG | p.A396fs | 28 |
| 113 | Ex 9a | D | c.1195delA | p.R399fs | 16 |
| 114 | Ex 9a | D | c.1199delG | p.G400fs | 30, 35 |
| 115 | Ex 9a | D | c.1206delG | p.K402fs | 29 |
| 116 | Ex 9a | I | c.1213_1214ins11bp | p.L405fs | 30 |
| 117 | Ex 9a | M | c.1220G→A | p.S407N | 37 |
| 118 | Ex 9a | D | c.1231_1233delAGC | p.S411del | 37 |
| 119 | Ex 9a | M | c.1234T→A | p.C412S | 30 |
| 120 | Ex 9a | M | c.1238G→T | p.G413V | 27, 33 |
| 121 | Ex 9a | D | c.1255delA | p.S419fs | 27 |
| 122 | Ex 9a | D | c.1267delA | p.N423fs | 29, 31 |
| 123 | Ex 9b | Sp | c.1310delG | splice defect | 27 |
| 124 | Ex 9b | Sp | c.1311G→C | splice defect, p.R437R | 29 |
| 125 | Ex 9b | Sp | c.1311G→A | splice defect, p.R437R | 27 |
| 126 | Int 9b | Sp | c.1311+2T→A | Ex 9b skip, p.A425fs | 28 |
| 127 | Ex 10 | I | c.1317_1318insA | p.V440fs | 27 |
| 128 | Ex 10 | D | c.1334delT | p.M445fs | 46 |
| 129 | Ex 10 | D | c.1346_1347delCT | p.S449fs | 27, 30, 35 |
| 130 | Ex 10 | D | c.1347_1350delTTTC | p.F450fs | 28 |
| 131 | Ex 10 | I | c.1361_1362insT | p.Y455fs | 30 |
| 132 | Ex 10 | I | c.1392_1393insC | p.N465fs | 30 |
| 133 | Ex 10 | D | c.1410delG | p.G470fs | 30 |
| 134 | Ex 10 | N | c.1414C→T | p.Q472X | 40, 44 |
| 135 | Ex 10 | InDel | c.1415_1417delAGAinsGT | p.Q472fs | 28, 37 |
| 136 | Ex 10 | Sp | c.1428G→A | splice defect | 30 |
| 137 | Ex 11 | D | c.1432_1433delAG | p.R478fs | 29 |
| 138 | Ex 11 | D | c.1437_1438delGT | p.V479fs | 27 |
| 139 | Ex 11 | N | c.1469T→G | p.L490X | 30 |
| 140 | Ex 11 | I | c.1470_1471insA | p.D491fs | 30, 31, 37 |
| 141 | Ex 11 | M | c.1510G→A | p.V504M | 31, 39 |
| 142 | Ex 11 | M | c.1522C→T | p.Q508X | 30 |
| 143 | Ex 11 | D | c.1550_1551delTG | p.V517fs | 44 |
| 144 | Ex 11 | D | c.1553_1554delGC | p.S518fs | 24 |
| 145 | Ex 11 | D | c.1609delT | p.Y537fs | 30 |
| 146 | Ex 11 | N | c.1611C→A | p.Y537X | 27 |
| 147 | Ex 11 | I | c.1623_1624insA | p.P542fs | 30 |
| 148 | Ex 11 | D | c.1630delA | p.T544fs | 31 |
| 149 | Ex 11 | D | c.1655delC | p.A552fs | 29, 44 |
| 150 | Ex 11 | D | c.1672_1684delGGGTCTCAAGACC | p.G558fs | 37 |
| 151 | Ex 11 | N | c.1678C→T(c.1778C→T)* | p.Q560X | 27* |
| 152 | Ex 11 | N | c.1684C→T | p.Q562X | 27 |
| 153 | Ex 12 | D | c.1689_1699delAGTCCATAGGA | p.E563fs | 37 |
| 154 | Ex 12 | N | c.1715T→A | p.L572X | 37 |
| 155 | Ex 12 | Sp | c.1741_1742insTCTG | p.G581fs | 31 |
| Non‐disease associated variants | |||||
| Ex 1 | M | c.14C→T | p.T5M | 31, 32, 34 | |
| Ex 4 | M | c.392C→T | p.P131L | 27, 31, 35, 39 | |
| Ex 5 | M | c.572G→A | p.G191D | 30, 31 | |
| Ex 5 | M | c.590G→A | p.R197Q | 31 | |
| Ex 8 | M | c.1055C→T | p.P352L | 32 | |
| Ex 8 | M | c.1096G→C | p.D366H | 31, 32, 48 | |
| Ex 13 | M | c.1794T→G | p.I575T | 32 | |
Mutations as described in original* article are in parentheses.
D, deletion; Ex, exon; I, insertion; InDel, insertion+deletion; Int, intron; M, missense; N, nonsense; Sp, splice site.
Figure 2 Distribution and frequency of mutations in the (A) ENG and (B) ALK‐1 genes.
The vast majority (80%) of mutations of the ENG gene identified in HHT1 patients lead to premature stop codons and truncated polypeptides (table 1). It was initially proposed that these mutant proteins could be secreted locally and exert a dominant negative effect by disrupting normal endoglin function.24,44 However, expression analysis of mutant endoglin proteins showed that they are rarely detectable and if expressed, only as transient species that do not reach the cell surface.34,37,40 It has been demonstrated that most mutations leading to frame shift and truncation probably result in nonsense mediated decay and therefore reduced mRNA levels and very unstable mutant proteins.49 Twenty one mutations (14%) lead to splice site defects including six in exonic sequences. Three of the 31 missense mutations affect the ATG start codon and are predicted to lead to null alleles (table 1). The remaining 28 missense mutations are distributed as shown in fig 2A, none being present in exons 9b, 10, and 12.
Several single base pair ENG substitutions altering an amino acid, and previously described as disease causing mutations, are now recognised as polymorphic variants. These include p.T5M in the leader peptide, p.G191D, p.R197Q, p.P352L, p.D366H and p.I575T (table 1). More recently, p.P131L was reported by several investigators as a disease causing mutation.27,31,35,39 However, this variant has been observed in non‐affected individuals and an additional disease causing mutation was found in some of the affected patients with this polymorphism.
Some families suspected of HHT1 remain unresolved after analysis, possibly because of insufficient sensitivity of the methods applied, the complexity of mutations, or their location in ENG regulatory regions.34,35 A human genomic clone containing the 5′‐flanking region of the ENG gene was used to identify the promoter region with two GC‐rich boxes (−5 to +16 and −47 to −29) and an Sp1 site (−37) near the initiation site, but no consensus TATA and CAAT boxes,50 a feature common to members of the TGFβ superfamily. Various consensus sites for GATA—ets, AP‐2, NFκB, Mad, as well as TGFβ, glucocorticoid, vitamin D, and oestrogen responsive elements—were found to be located upstream of the translation initiation codon. The report confirmed that the ENG promoter activity was stimulated by TGFβ1.50 A subsequent study showed that mutation of the Sp1 binding sequence abolished the basal activity of the promoter and speculated that such mutations would lead to HHT1.51 Further studies are necessary to determine if such promoter mutations occur in HHT and if so, what are their effects on the protein function.
Haploinsufficiency as a mechanism for HHT1
Analysis of endoglin protein levels in affected patients strongly supports haploinsufficiency and the associated reduced levels of functional protein as the underlying cause of HHT1. Our results are based on the estimation (by metabolic labelling and immunoprecipitation) of the amount of newly synthesised protein in patient samples expressed relative to those of the control samples included in every experiment.28,35,38,40 Table 2 shows that the age distribution did not differ significantly in the patient groups analysed. However, the distribution of endoglin level in peripheral blood monocytes of patients with characterised ENG mutations (n = 109) was significantly lower (48% v 88.5%, respectively) than in the control group (n = 84), which was also analysed relative to an internal experimental control sample.
Table 2 Summary of endoglin analysis in peripheral blood activated monocytes of patients with hereditary haemorrhagic telangiectasia (HHT).
| Group | Mutation confirmed | n | Age (years) | Endoglin levels (%) | ||
|---|---|---|---|---|---|---|
| Median | Range (25–75%) | Median | Range (25–75%) | |||
| HHT1 | ENG | 109 | 42.4 | 21.3 | 48.0* | 19.0 |
| HHT2 | ALK‐1 | 61 | 42.2 | 22.3 | 91.0 | 37.0 |
| Control | None | 84 | 37.9 | 38.9 | 88.5 | 29.5 |
HHT1 and HHT2 refer to individuals with a confirmed mutation. The control group is composed of spouses of affected members, unrelated individuals, and non‐affected family members as confirmed by the absence of mutation. The endoglin level present on activated monocytes is expressed relative to a normal control ran in each experiment.
The two sided pair normal approximation obtained from the two sample Wilcoxon test is reported: *p<0.001 relative to HHT2 and control groups (level of significance α = 0.008).
Human umbilical vein endothelial cells (HUVEC) from newborn infants of HHT families were also studied for endoglin expression.28,35,37,38,40 Table 3 shows that the distribution of endoglin levels on HUVEC from newborn infants with an ENG mutation was significantly different from control, with a median value of 45%. The group of newborns found not to carry the familial ENG mutation had endoglin levels similar to those of controls (median 98%). Thus endoglin functional levels on peripheral blood monocytes and endothelial cells of individuals with ENG mutations are significantly reduced and must predispose to the clinical manifestations of HHT1.
Table 3 Levels of endoglin and ALK‐1 in human umbilical vein endothelial cells of newborns from families with hereditary haemorrhagic telangiectasia (HHT).
| HHT type | Mutation status of neonate | n | Endoglin level (%) | n | ALK‐1 level (%) | ||
|---|---|---|---|---|---|---|---|
| Median | Range (25–75%) | Median | Range (25–75%) | ||||
| HHT1 | Yes (ENG) | 30 | 45.0* | 13.0 | 7 | 94.0 | 18.0 |
| HHT1 | None | 18 | 98.0 | 17.0 | 3 | 92.0 | 23.0 |
| HHT2 | Yes (ALK‐1) | 8 | 98.5 | 25.5 | 4 | 60.5† | 24.5 |
| HHT2 | None | 6 | 106.5 | 31.0 | 3 | 96.0 | 3.0 |
*The distribution of endoglin levels in the group with ENG mutations was significantly different from all other groups. The two sided pair normal approximation was obtained from the two sample Wilcoxon test for comparison to the non‐affected HHT1 group (p<0.001), while the t test was used for comparison to the groups with and without an ALK‐1 mutation (p = 0.0002 in both cases).
†The distribution of ALK‐1 levels for the group with an ALK‐1 mutation was p = 0.03 (by t test) when compared with the combined HHT1 groups or with the last group (level of significance α = 0.01)
Some large deletions and insertions as well as some splice site mutations causing in‐frame deletions or insertions of several exons were shown to produce detectable mutants.40,41 However, such mutant proteins were only observed by metabolic labelling and represent intra‐cytoplasmic, transient, and unstable proteins that would not be functional. The missense mutants were shown to be expressed as partially glycosylated precursor proteins with a lower molecular weight (80 000 kDa v 90 000 kDa for the fully processed monomer).37 These mutants are probably misfolded and consequently unable to form heterodimers with normal endoglin and reach the cell surface. Thus the missense mutants studied to date are expressed intracellularly and cannot interfere with the normal function of the cell surface endoglin protein. These data overwhelmingly support the view that ENG mutations act as null alleles, resulting in haploinsufficiency as the underlying mechanism of HHT1.
ACVRL1 gene: structure, mutations, and polymorphisms
The second HHT locus, HHT2, was mapped to chromosome 12q1352,53 and the candidate gene identified as ACVRL1.54 The ACVRL1 gene spans more than 15 kb of genomic DNA and the cDNA encodes a protein of 503 amino acids.55,56 The coding region is contained within nine exons, the start codon being in exon 2 and the termination codon in exon 10 (fig 1B). All introns follow the GT–AG rule except for intron 6, which has a TAGgcaag 5′ splice junction. Two descriptions of the 5′ untranslated sequence of ACVRL1 have been published.56,57 In the first variant, the 5′UTR sequence is part of exon 2, while the second variant arises from the splicing of exon 1 and joining to a consensus junction 7 bp upstream of the start codon in exon 2; the remainder of the sequences is identical.
ALK‐1 is a type I cell surface receptor of the TGFβ superfamily of ligands, which was shown to bind TGFβ and to mediate is effects through Smad 1,5, and 8. It shares with other type I receptors a relatively high degree of similarity in serine/threonine kinase subdomains, a glycine/serine‐rich (GS) region preceding the intracellular kinase domain and a short C‐terminal tail (fig 1B).57,58 ALK‐1 contains 10 conserved cysteine residues and a potential N‐linked glycosylation site in the extracellular domain. The intracellular part of ALK‐1 consists almost entirely of a kinase domain containing 12 subdomains with highly conserved residues.59,60
To date, 123 mutations in the ACVRL1 gene have been reported. Unlike ENG, more than half (53%) of the mutations identified in ACVRL1 are missense substitutions (n = 65) (table 4). The remaining mutations include 24 deletions, 13 insertions, 16 nonsense, one indel, and four splice site mutations (table 4). Twenty five mutations (20%) were identified in the extracellular domain, six (5%) in the transmembrane domain, and 92 (75%) in the intracellular domain. The frequency of mutations is highest in exons 8, 7, and 3, accounting for 65% of all reported mutations (fig 2B).
Table 4 Summary of known ALK‐1 mutations.
| Location | Type | Nucleotide change | Consequence | References | |
|---|---|---|---|---|---|
| 1 | Ex 2 | D | c.37delC | p.L13fs | 46, 61 |
| 2 | Int 2 | Sp | c.61+10G→A | unknown | 30 |
| 3 | Ex 3 | D | c.83delG | p.R28fs | 27 |
| 4 | Ex 3 | D | c.86delG | p.G29fs | 60, 62 |
| 5 | Ex 3 | I | c.139_140insG | p.G48fs | 63 |
| 6 | Ex 3 | M | c.142G→A | p.G48R | 30 |
| 7 | Ex 3 | M | c.143G→A | p.G48E | 39 |
| 8 | Ex 3 | M | c.143G→A, 145delG, 146_147insT | p.G48E,A49P | 64 |
| 9 | Ex 3 | I | c.144_145insG | p.A49fs | 63, 65 |
| 10 | Ex 3 | D | c.145delG | p.A49fs | 65 |
| 11 | Ex 3 | M | c.150G→T | p.W50C | 55, 62–64 |
| 12 | Ex 3 | N | c.150G→A | p.W50X | 27 |
| 13 | Ex 3 | M | c.152G→A | p.C51Y | 63, 65 |
| 14 | Ex 3 | N | c.172G→T | p.E58X | 65 |
| 15 | Ex 3 | D | c.190delC | p.Q64fs | 27 |
| 16 | Ex 3 | M | c.199C→T | p.R67W | 31, 65 |
| 17 | Ex 3 | M | c.200G→A | p.R67Q | 55 |
| 18 | Ex 3 | D | c.203delG | p.G68fs | 27 |
| 19 | Ex 3 | M | c.231C→G | p.C77W | 63 |
| 20 | Ex 3 | I | c.237_238insG | p.R80fs | 60 |
| 21 | Ex 3 | I | c.243_244dupC | p.T82fs | 30 |
| 22 | Ex 3 | M | c.286A→G | p.N96D | 63 |
| 23 | Ex 3 | D | c.289_294delCACAAC | p.H97_N98del | 65 |
| 24 | Ex 3 | D | c.301_307delCTGGTGC | p.L101fs | 30 |
| 25 | Ex 4 | N | c.352C→T | p.Q118X | 27 |
| 26 | Ex 4 | I | c.372_373insCC | p.P125fs | 27 |
| 27 | Ex 4 | M | c.383C→A | p.A128D | 31 |
| 28 | Ex 4 | D | c.400delG | p.A134fs | 63 |
| 29 | Ex 4 | D | c.406_409delGGTG | p.G136fs | 63 |
| 30 | Ex 4 | N | c.423G→A | p.W141X | 55, 60 |
| 31 | Ex 4 | N | c.430C→T | p.R144X | 30, 37, 60 |
| 32 | Ex 4 | N | c.439C→T | p.Q147X | 27 |
| 33 | Ex 4 | N | c.475G→T | p.E159X | 55 |
| 34 | Ex 4 | D | c.510delC | p.G170fs | 30 |
| 35 | Ex 5 | M | c.536A→C | p.D179A | 46 |
| 36 | Ex 5 | I | c.625_626insTG | Splice defect, p.G209fs | 27 |
| 37 | Int5 | Sp | c.626–6del14bp | Unknown | 27 |
| 38 | Ex 6 | M | c.632G→A | p.G211D | 16, 46 |
| 39 | Ex 6 | M | c.643G→A | p.E215K | 30 |
| 40 | Ex 6 | D | c.664_668delCACGG | p.H222X | 30 |
| 41 | Ex 6 | M | c.667G→C | p.G223R | 30 |
| 42 | Ex 6 | D | c.682delG | p.V228fs | 30 |
| 43 | Ex 6 | M | c.686A→G | p.K229R | 30 |
| 44 | Ex 6 | D | c.694_696delCTC | p.S232del | 30, 54, 55, 64 |
| 45 | Ex 6 | D | c.704delA | p.D235fs | 30 |
| 46 | Ex 6 | D | c.759_761delCGA | p.D254del | 61 |
| 47 | Int 6 | Sp | c.773–2A→G | splice defect | 30 |
| 48 | Ex 7 | D | c.810_822delCACGCAGCTGTGG | p.S271fs | 65 |
| 49 | Ex 7 | M | c.818T→C | p.L273P | 31 |
| 50 | Ex 7 | M | c.827T→C | p.I276T | 31 |
| 51 | Ex 7 | I | c.838_846dupCACGAGCAC (c.1120_1129dupCACGAGCAC)* | p.H280_H282dupHEH (Y379_380dupHEH)* | 39* |
| 52 | Ex 7 | M | c.851C→T | p.S284F | 31 |
| 53 | Ex 7 | M | c.853C→T | p.L285F | 30 |
| 54 | Ex 7 | N | c.858C→A | p.Y286X | 27, 65 |
| 55 | Ex 7 | I | c.864_865insT | p.L289fs | 55 |
| 56 | Ex 7 | M | c.913T→C | p.S305P | 31 |
| 57 | Ex 7 | M | c.916G→C | p.A306P | 30 |
| 58 | Ex 7 | N | c.924C→A | p.C308X | 55 |
| 59 | Ex 7 | M | c.925G→A | p.G309S | 27 |
| 60 | Ex 7 | M | c.940C→T | p.H314Y | 30 |
| 61 | Ex 7 | N | c.961C→T | p.Q321X | 27 |
| 62 | Ex 7 | D | c.972delA | p.P324fs | 30 |
| 63 | Ex 7 | M | c.986G→A | p.R329H | 60 |
| 64 | Ex 7 | M | c.988G→T | p.D330Y | 27, 65 |
| 65 | Ex 7 | M | c.988G→A | p.D330N | 27 |
| 66 | Ex 7 | M | c.998G→T | p.S333I | 55, 62, 64 |
| 67 | Ex 7 | InDel | c.1000_1005delCGCAATinsG | p.R334fs | 16 |
| 68 | Ex 7 | M | c.1010T→C | p.L337P | 30 |
| 69 | Ex 7 | M | c.1023C→G | p.N341K | 16 |
| 70 | Ex 7 | M | c.1031G→A | p.C344Y | 46, 64 |
| 71 | Ex 7 | M | c.1031G→T | p.C344F | 64, 65 |
| 72 | Ex 7 | M | c.1039G→C | p.A347P | 30 |
| 73 | Ex 7 | D | c.1042delG | p.D348fs | 27 |
| 74 | Ex 7 | Sp (M) | c.1048G→C | Splice defect, p.G350fs (p.G350R) | 27 |
| 75 | Ex 8 | M | c.1054G→C | p.A352P | 65 |
| 76 | Ex 8 | D | c.1061_1068del8bp | p.M354fs | 27 |
| 77 | Ex 8 | I | c.1062_1080dupGCACTCACAGGGCAGCGAT | p.Y361fs | 65 |
| 78 | Ex 8 | N | c.1069C→T | p.Q357X | 31 |
| 79 | Ex 8 | D | c.1071delG | p.Q357fs | 27 |
| 80 | Ex 8 | D | c.1107_1108delAG | p.R369fs | 27 |
| 81 | Ex 8 | I | c.1112_1113insG | p.G371fs | 30, 62, 64 |
| 82 | Ex 8 | M | c.1120C→T (c.1402C→T)* | p.R374W | 27, 31, 39*, 43 |
| (p.R354W)* | 46, 55, 60, 66, 67 | ||||
| 83 | Ex 8 | D | c.1120_1137del18bp | p.R374_E379del | 27 |
| 84 | Ex 8 | M | c.1121G→A | p.R374Q | 27, 30, 56, 62 |
| 85 | Ex 8 | M | c.1123T→C | p.Y375H | 60, 62 |
| 86 | Ex 8 | I | c.1125_1126insGTAC | p.M376fs | 30 |
| 87 | Ex 8 | M | c.1126A→G | p.M376V | 30 |
| 88 | Ex 8 | M | c.1127T→G | p.M376R | 54, 55 |
| 89 | Ex 8 | M | c.1133C→T | p.P378L | 65 |
| 90 | Ex 8 | M | c.1133C→A | p.P378H | 31 |
| 91 | Ex 8 | M | c.1135G→A | p.E379K (p.N379L)* | 30, 39* |
| 92 | Ex 8 | M | c.1139T→G | p.V380G | 27 |
| 93 | Ex 8 | M | c.1157G→A | p.R386H | 31 |
| 94 | Ex 8 | N | c.1171G→T | p.E391X | 60 |
| 95 | Ex 8 | M | c.1189G→A | p.D397N | 27 |
| 96 | Ex 8 | M | c.1190A→G | p.D397G | 30 |
| 97 | Ex 8 | M | c.1193T→A (c.1475T→A)* | p.I398N | 39*, 67 |
| 98 | Ex 8 | M | c.1196G→C | p.W399S | 46 |
| 99 | Ex 8 | M | c.1199C→A | p.A400D | 65 |
| 100 | Ex 8 | M | c.1204G→A | p.G402S | 16 |
| 101 | Ex 8 | M | c.1218G→C | p.W406C | 27 |
| 102 | Ex 8 | M | c.1221G→T | p.E407D | 62, 64 |
| 103 | Ex 8 | M | c.1231C→T | p.R411W | 27, 30, 31, 60–62, 68 |
| 104 | Ex 8 | M | c.1232G→A | p.R411Q (p.R411N)* | 30, 39*, 46, 54, 55, 60, 62, 69 |
| 105 | Ex 8 | M | c.1232G→C | p.R411P | 30 |
| 106 | Ex 9 | M | c.1270C→A | p.P424T | 55 |
| 107 | Ex 9 | M | c.1270C→T | p.P424S | 31 |
| 108 | Ex 9 | M | c.1271C→T | p.P424L | 27 |
| 109 | Ex 9 | M | c.1275C→G | p.F425L | 30 |
| 110 | Ex 9 | M | c.1297C→T | p.P433S | 27 |
| 111 | Ex 9 | D | c.1299delC | p.P433fs | 55, 62 |
| 112 | Ex 9 | M | c.1313T→C | p.M438T | 27 |
| 113 | Ex 9 | M | c.1321G→A | p.V441M | 31 |
| 114 | Ex 9 | M | c.1355C→T | p.P452L | 31 |
| 115 | Ex 10 | N | c.1385C→G | p.S462X | 66 |
| 116 | Ex 10 | N | c.1408G→T | p.E470X | 27 |
| 117 | Ex 10 | I | c.1428_1429dupC | p.S477fs | 30 |
| 118 | Ex 10 | N | c.1435C→T | p.R479X | 27, 30, 31, 66 |
| 119 | Ex 10 | M | c.1436G→T | p.R479L | 30 |
| 120 | Ex 10 | M | c.1450C→T | p.R484W | 27, 30, 61 |
| 121 | Ex 10 | I | c.1450C→T, 1450_1451insG | p.R484Wfs | 66 |
| 122 | Ex 10 | M | c.1460A→C | p.K487T | 46 |
| 123 | Ex 10 | N | c.1468C→T | p.Q490X | 61 |
| Non‐disease associated variants | |||||
| Ex 10 | M | c.1445C→T | p.A482V | 27, 30, 70 | |
Mutations as described in original article* are in parentheses.
D, deletion; Ex, exon; I, insertion; InDel, insertion/deletion; Int, intron; M, missense; N, nonsense; Sp, Splice site.
Twenty two mutations (18%) were identified in exon 3 which codes for the extracellular domain; 13 of these changes lead to formation of premature stop codons. Three missense mutations (p.G48R; p.G48E; p.G48E, A49P) alter G48, which is conserved in man, mouse, rat, chicken, and cow sequences. This last missense mutation is a result of a complex rearrangement (G143A substitution, deletion of G145, and insertion of T147) in the coding region of ACVRL1. HUVEC from a newborn infant with such a mutation had reduced levels of ALK‐1 protein as measured by metabolic labelling and flow cytometry.64 Another missense mutation, p.W50C, was shown to abrogate the signalling activity when introduced into the extracellular domain of the ALK‐1/TβRI chimera.71 This loss of signalling activity in COS cells and the reduction of ALK‐1 expression in HUVEC probably reflects low levels of expression of the unstable mutant protein.64 A p.C51Y missense substitution affects another highly conserved cysteine at position 51. There is also a report of a conserved cysteine, C77, mutated to tryptophan.63 Arginine at position 67 is a source of two missense substitutions, p.R67W and p.R67Q,31,55,65 and is conserved in man, mouse, rat, and zebra fish. Another substitution, p.N96D, affects an asparagine residue that is conserved in man, mouse, rat, chicken, cow, and zebra fish. Ten of the mutations affect codons G48‐A49‐W50‐C51 and this may be considered a “hot spot”. Various other residues in the kinase domain—for example, R374‐V380, R411, P424, and so on—are also hot spots.
Five of the six mutations reported in the short transmembrane region lead to formation of premature stop codons owing to nonsense and frame shift mutations (table 4). The resulting proteins would be severely truncated and lacking an intracellular signalling kinase domain. A single missense mutation (p.A128D) changes a poorly conserved non‐polar, weakly hydrophobic residue into a polar residue.
Type I receptors have a highly conserved GS motif (SGSGSGLP) in the cytoplasmic juxtamembrane region immediately preceding the kinase domain. This domain plays an important role in intracellular signalling, as its deletion abolishes the ability of TβRI to undergo phosphorylation and to mediate TGFβ dependent responses.72 Single substitutions in the GS domain have also been shown to cause loss of function or gain of function mutations in TβRI.73,74 Only two mutations are reported in exon 5. The first, an A to C substitution at position 536 (p.D179A), was detected in a patient with primary pulmonary hypertension and no clinical or family history of HHT.46 Functional studies with a GFP tagged mutant construct showed localisation of this protein at the cell surface, but structural modelling revealed a loss of a hydrogen bond with arginine 252, a residue critical in GS–kinase interaction. The second mutation is an insertion, c.625_626insTG, which leads to a splice defect.
Most ACVRL1 mutations (75%) are found in the intracellular kinase domain (table 4). Of these 92 mutations, premature stop codons occur as a result of insertions/deletions (n = 23), nonsense mutations (n = 11), and splice site mutations (n = 3). Another 55 mutations were caused by missense substitutions. Homology modelling of ALK‐1 kinase domain was previously used to determine any possible structural or functional consequences resulting from missense substitutions.60 The vast majority of missense mutations in the kinase domain occur at residues that are conserved not only among ALK‐1 in different species but also among the different type I receptors. Modelling suggested that they cause alterations in the polarity, charge, hydrophobicity, or size of the substituted amino acid and probably have structural effects creating misfolded unstable proteins.
ACVRL1 mutations were also identified in a rare group of patients with HHT who developed pulmonary hypertension.61 These patients had vascular dilatations and AVMs characteristic of HHT, as well as occlusive arteriopathy typical of primary pulmonary hypertension. To date, at least 15 ACVRL1 mutations have been identified in patients with HHT related pulmonary hypertension.46,61,66 Of these, six lead to formation of premature stop codons and eight alter highly conserved amino acids within the functional kinase domain of ALK‐1.
Some polymorphisms in the ACVRL1 gene have been published.31 Of interest is the p.A482V (c.1445C→T) variant first reported in a patient's pituitary adenoma and control leucocyte DNA.70 Both samples were heterozygous for the observed substitution; however, the patient did not have HHT, as evidenced from clinical data and family history. The presence of the same variant in another two unrelated patients—one with an ENG and another with an ACVRL1 mutation—suggested that it might be a polymorphic variant27 rather than a deleterious substitution, and this was supported by the structural analysis of the ALK‐1 protein (data not shown). Another patient with a confirmed diagnosis of HHT carried this variant but further studies on this family are needed to ascertain the relevance to disease.30
Analysis of ALK‐1 and endoglin protein levels in HHT2
It therefore appears that most ACVRL1 mutations lead to unstable and non‐functional mutant proteins, supporting haploinsufficiency as a predominant model of HHT2. Table 3 reports several HUVEC samples where ALK‐1 protein levels were measured. Three of these mutants (G48E, A49P; W50C; S333I) had reduced levels of ALK‐1 relative to control.64 The fourth mutant, with deletion of S232, had an ALK‐1 level of >85%; this residue is in the ATP binding site of the kinase domain so its loss would lead to a non‐functional protein.64
The lack of monoclonal antibodies to ALK‐1 and the very low levels of protein expressed on activated monocytes present a difficulty in the routine assessment of the ALK‐1 levels in HHT patients. As endoglin and ALK‐1 are both specialised endothelial TGFβ receptors associated with HHT, we determined whether endoglin levels might be reduced in HHT2 patients. Table 2 shows that the level of newly synthesised endoglin protein as measured by metabolic labelling, is not altered in HHT2 patients (n = 61) with a confirmed molecular diagnosis (91%, v 88.5% in controls). These results differ from a recent study of a limited number of patients showing that endoglin steady state surface levels, assessed by flow cytometry in peripheral blood activated monocytes, were reduced in patients with either HHT1 (one of two affected family members) or HHT2 (three of six affected members).43 Furthermore, it was suggested that endoglin levels were lower in more severely affected HHT1 and HHT2 patients, and also decreased with age. The analysis of our data with respect to age and disease severity does not support these findings.
Table 3 shows that the eight HUVEC samples from newborn infants with ACVRL1 mutations have an endoglin level distribution similar to that of the control group (98.5% v 106.5%) and clearly distinct from that of the neonates with an ENG mutation (98.5% v 45%). Thus our studies detect reduced levels of ALK‐1 in HUVEC of infants with mutations in the ACVRL1 gene but normal levels of endoglin in the same HUVEC samples and in peripheral blood monocytes of HHT2 patients.
MADH4 mutations in combined syndrome of juvenile polyposis and HHT (JPHT)
The presence of both juvenile polyposis and HHT in an affected individual defines the syndrome of juvenile polyposis and HHT (JPHT).18 First reports of the coexistence of an autosomal dominant juvenile gastrointestinal polyposis and PAVMs with digital clubbing date back to the early 1980s.75,76,77 Juvenile polyposis, a predisposing factor in gastrointestinal malignancy, was subsequently shown to be associated with mutations in either MADH4 or BMPRIA.78,79 In 2004, Gallione et al described patients from seven unrelated families meeting the diagnostic criteria of both juvenile polyposis and HHT and carrying mutations in the MADH4 gene, but none in ENG or ACVRL1 genes.18 These mutations were detected in the region of the MADH4 gene coding for the highly conserved carboxyl terminus and comprise four missense mutations (c.1054G→A, c.1081C→G, c.1157G→A, c.1598T→G), one nonsense mutation (c.1600C→T), and two frameshift mutations (c.1594delG, c.1612del14) in exons 8, 9, and 11.18 Three of these occurred de novo: c.1081C→G and c.1594delG, identified each in a single affected individual, and the third mutation, c.1157G→A, detected in a proband and his similarly affected offspring but not in the parents, who had no signs or symptoms of either disorder. The presence of the de novo mutations confirmed that mutations in MADH4 are the likely cause of JPHT. The severity (pulmonary and hepatic AVMs, cerebral involvement) and often early onset of HHT symptoms in these patients argue in favour of systemic screening for visceral manifestations in juvenile polyposis patients with MADH4 mutations.18
Role of HHT gene products in TGFβ signalling
TGFβ is a member of a large family of proteins that exhibit many biological effects including regulation of cellular proliferation, differentiation, migration, and extracellular matrix formation. Members of the TGFβ superfamily include the structurally related cytokines, TGFβs, activins and bone morphogenetic proteins (BMP), which exert their biological effects through binding to heteromeric complexes containing two different transmembrane serine/threonine kinases, known as type I (RI) and type II (RII) receptors. Five type II and seven type I receptors are known in vertebrates. Upon ligand binding to RII, RI is recruited, phosphorylated in the GS domain, and activated by the constitutively active RII. Each member of the TGFβ superfamily binds characteristic type I and type II receptors. The activated RI then transmits intracellular signal to the nucleus by phosphorylating members of the receptor regulated R‐Smad proteins (Smad1, Smad2, Smad3, Smad 5, Smad8). ALK‐1, and the BMP type I receptors ALK‐2 (ActRI), ALK‐3 (BMPRIA) and ALK‐6 (BMPRIB) phosphorylate Smad1, Smad5, and Smad8, while TGFβ and activin type I receptors ALK‐5 (TβRI) and ALK‐4 (ActRIB) phosphorylate Smad2 and Smad3. The specificity of the interaction between the RI receptors and the R‐Smads is determined by a few conserved amino acid residues in the L45 loop of the small lobe of the RI kinase and the L3 loop of the MH2 domain of R‐Smads.72,80,81,82 Activated R‐Smads then bind Smad4 and translocate to the nucleus where they exert their effects by controlling gene expression through interactions with transcription factors, co‐activators, and co‐repressors. Two more Smads, Smad6 and Smad7, known as inhibitory Smads (I‐Smads), control TGFβ superfamily signalling. Specifically, activin/TGFβ signalling induces Smad7 expression, while BMPs induce Smad6 activation. A more detailed description of the TGFβ/Smad signalling pathways can be found elsewhere.83,84,85
In most cell types TGFβ signals through ALK‐5, while in endothelial cells it can also signal through ALK‐1.86,87 The ALK‐5 pathway is mediated by Smads 2/3, while the ALK‐1 pathway is mediated through Smads1/5/8.87,88,89 This dual route increases the complexity of TGFβ signalling in the endothelium and suggests that endoglin and ALK‐1 are endothelial specific receptors essential for vascular functions. ALK‐1 is indeed expressed predominantly in endothelial cells but is also found at epithelial–mesenchymal cell interaction sites.90 Endoglin is expressed at high levels on vascular endothelial cells and syncytiotrophoblast of full term human placenta, as well as transiently on extravillous cytotrophoblasts and on the cardiac endocardium during development.25,91,92 It is an accessory protein that interacts with multiple heteromeric receptor complexes containing TGFβs, activins, and BMPs.93 In endothelial cells, endoglin can also interact with ALK‐1 and ALK‐5 in the absence of TGFβ.64,94 In addition, endoglin has been shown to physically associate with ALK‐1, potentiate ALK‐1/Smad1 dependent signalling, and inhibit ALK‐5/Smad3 pathway in COS transfected cells.95 Though most studies of endoglin have focused on its binding to TGFβ1 and TGFβ3 and its ability to regulate responses to these ligands and form heteromeric complexes with TβRI and TβRII, its role in the TGFβ receptor complex of endothelial cells has not been fully elucidated.96,97 In murine embryonic endothelial cells (MEEC) devoid of endoglin through siRNA knockdown, Lebrin et al reported the requirement of endoglin in the TGFβ dependent activation of ALK‐1 and subsequent Smad1/5/8 signalling but decreased ALK‐5 levels.98 In contrast, more recent data using MEEC derived from Eng null embryos show that endoglin is not required for the TGFβ dependent activation of Smad1/5/8 pathway and probably controls the levels of cell surface receptors and their binding characteristics.99 Both reports indicate that endoglin is not required for TGFβ dependent activation of Smad2/3; however, future studies are needed to elucidate specific molecular mechanisms by which endoglin might regulate TGFβ receptor expression and contribute to endothelial cell function.
TGFβ is a multifunctional protein that plays an important role in angiogenesis and vascular remodelling. It has been shown to modify cell function depending on in vivo and in vitro conditions and the cell type.86,100,101 TGFβ regulates endothelial cell function by either stimulating or inhibiting their proliferation through a fine balance between ALK‐1 and ALK‐5 signalling.89 However, ALK‐5 kinase activity and the TβRII receptor are also required for optimal ALK‐1 activation,86 suggesting that ALK‐1 and ALK‐5 form a heteromeric receptor complex with TβRII and signal through the Smad1/5 pathway. The TGFβ/ALK‐5 pathway leads to inhibition of cell migration and proliferation, whereas the TGFβ/ALK‐1 pathway induces endothelial cell migration and proliferation.89 A specific gene, Id1, was found to mediate the TGFβ/ALK‐1 induced (and Smad dependent) migration, while induction of plasminogen activator inhibitor‐1 (PAI‐1) by activated ALK‐5 may contribute to the TGFβ induced maturation of blood vessels.101
Smad4 forms heteromeric complexes with TGFβ/activin activated Smad2 and Smad3 and with BMP activated Smad1, Smad5, and Smad8. It binds DNA in response to a TGFβ ligand induced intracellular signalling cascade. SMAD4 is deleted or mutated during tumorigenesis in many human tumours. Some of these mutations occur in the N‐terminal part of the protein, the Mad homology 1 (MH1) region, which shows sequence specific DNA binding. The MH2 domain of Smad4 appears to be responsible for homo‐oligomerisation of Smad4 trimers and hetero‐oligomerisation between Smad4 and R‐Smad trimers.102 Smad4 is not phosphorylated by R‐Smads and is required for the formation of functional transcriptional complexes.103 Regulation of TGFβ family signalling and transcription thus occurs through the recruitment of the Smad complex, while the Smad interaction with various co‐activators and co‐repressors may determine the outcome of signalling events, also dependent on the relative levels and activities of these proteins.104 As many of the Smad co‐factors, co‐activators, and co‐repressors are known to be involved in other signalling pathways, it would be interesting to determine the relations and possible influence of these other pathways on the outcome of the TGFβ responses.
Eng and ACVRl1 in vascular pathology
Various human syndromes and diseases, both hereditary and spontaneous, have been attributed to mutations in members of the TGFβ superfamily. For the purpose of this review we will focus on the genetic disorders associated with ENG and ACVRL1 genes (fig 3) as other disorders have been extensively described elsewhere.
Figure 3 Model of diseases associated with ENG, ACVRL1, and MADH4 mutations. The type II (RII) and type I (RI) receptors are related serine/threonine kinases which form high affinity complexes upon ligand binding. In endothelial cells, TGFβ1 and TGFβ3 isoforms bind the RII receptor (TβRII), and an RI receptor is then recruited (ALK‐5 or ALK‐1, which phosphorylate Smad2/3 or Smad1/5, respectively). Endoglin interacts with either of these receptor complexes. In the case of BMP‐2, BMP‐4, or BMP‐6, the RI receptors (ALK‐3 and ALK‐6) first bind the ligand, engage BMPRII receptor and transit the signal through Smad1/5. Smad4 represents a common Smad through which signals from different receptors converge and translocate to the nucleus to regulate transcriptional responses. Mutations targeting different components (red) of the TGFβ signalling pathway contribute to several human disorders (in parentheses). Mutations in the endoglin gene are known to cause HHT1 and probably pulmonary arterial hypertension. ALK‐1 mutations are associated with HHT2 and PPH, the later being also caused by BMPRII mutations. Mutations in ALK‐3 also cause juvenile polyposis, while mutations in MADH4 coding for Smad4 lead to a combined syndrome of juvenile polyposis and HHT. BMP, bone morphogenetic protein; HHT, hereditary haemorrhagic telangiectasia; JPHT, syndrome of juvenile polyposis and hereditary haemorrhagic telangiectasia; PAH, pulmonary arterial hypertension; PPH, primary pulmonary hypertension; TGF, transforming growth factor
Mutations in ENG and ACVRL1 genes of the TGFβ superfamily cause HHT1 and HHT2, respectively. The two types of HHT are difficult to distinguish clinically, as all reported manifestations are known to occur in both disease types and show significant inter‐ and intrafamilial variations. HHT2 has a later onset and lower penetrance, while PAVMs are reported to be much more common in HHT1.24,36 Several studies reported that patients with HHT1 are at a higher risk of developing PAVMs and CAVMs than those with HHT2.28,36,62,105 gastrointestinal bleeding and liver involvement are also present in both groups but might be more common in HHT2 families.62,65
As vascular lesions are associated with both HHT1 and HHT2, it remains to be determined how a reduction in endoglin or ALK‐1 predisposes to HHT, and what causes vascular lesions to develop selectively in limited vascular beds. Endoglin null mice die at mid‐gestation (days E10.5–11.5) from angiogenic and cardiovascular defects.106,107,108 The failure in endothelial remodelling was also noted in yolk sac, indicating that endoglin is required for TGFβ1 signalling during both extraembryonic and embryonic vascular development and is critical for both angiogenesis and heart valve formation.109 Angiogenesis, the process of new vessel formation from pre‐existing blood vessels, consists of activation and resolution phases. During the activation phase, endothelial cells degrade the perivascular membrane and invade and migrate into the extracellular space, where they proliferate and form a lumen. In the resolution or maturation phase, cells stop migrating and proliferating and reconstitute the basement membrane, generating a new vessel. The role of ALK‐1 in angiogenesis remains controversial and not clearly defined. In one study, it was shown that transfection of a constitutively active form of ACVRL1 inhibits the proliferation and migration of endothelial cells by upregulating p21 and downregulating c‐myc protooncogene.110 Similarly, an increased number of endothelial cells within the dilated vessels of a zebra fish Acvrl1 mutant, vbg, supports the role of Acvrl1 in the inhibition of endothelial cell proliferation and in maintaining vascular integrity.111 Contrary to these reports, Goumans et al89 showed that ACVRL1 signalling promotes endothelial cell migration and proliferation by upregulating Id1 through transcriptional repression of thrombospondin‐1, an inhibitor of angiogenesis.89,101 The predominant expression of Acvrl1 was also noted in the developing arterial endothelium of Acvrl1 null mouse line and in newly forming arterial vessels during wound healing and tumour angiogenesis, pointing to the role of Acvrl1 in the resolution phase rather than the activation phase of angiogenesis.112 It is difficult to account for these discrepancies in the angiogenic role of ALK‐1 reported by different research groups; however, they could partly reflect the intrinsic differences between the cell lines used and the experimental or culture conditions.
Endoglin heterozygous mice can develop signs of HHT such as nosebleeds, telangiectases, dilated thin walled vessels and even cerebral AVMs and other complications associated with HHT,107,113,114 thus serving as an animal model of HHT1. However, our results show that some strains appear to be more affected than others, suggesting that modifier genes and even epigenetic factors contribute to the disease heterogeneity.114 Similarly, Acvrl1 null mice are embryonic lethal and exhibit defective vascular remodelling. The Acvrl1 null embryos showed few well defined capillary vessels, severely dilated major blood vessels, and suspected AVMs.87,115 Reminiscent of patients with HHT, Acvrl1 heterozygous mice, with age, developed vascular lesions in the skin, oral cavity, lung, brain, liver, spleen, and intestine.116 Some of these mice had grossly enlarged liver, leading to high output cardiac failure and pulmonary hypertension secondary to presumed hepatic AVMs, reminiscent of the cardiac pathology reported in HHT, particularly HHT2.116 These findings suggest that ACVRL1 is required for developing distinct arterial and venous vascular beds, as its lack might result in loss of anatomical, molecular, and functional distinctions between arteries and veins. It is also becoming more evident that ENG and ACVRL1 defects have a drastic affect on blood vessel development and angiogenesis, but that environmental factors such as vascular stress must trigger a vascular endothelium weakened by reduced expression of these essential endothelial specific TGFβ receptors. Other factors such as inflammation or the products of modifier genes are likely to contribute to disease progression.114,117
Pulmonary hypertension has emerged as a rare but important complication of HHT and there is probably some molecular and mechanistic overlap between these conditions. Mutations in either ACVRL1 or BMPRII genes predispose to a pulmonary hypertension syndrome characterised by obstruction of precapillary pulmonary arteries and leading to sustained elevation of pulmonary artery pressure, right ventricular failure, and death.118,119 Both BMPRII and ACVRL1 mediate BMP and TGFβ effects, respectively; signalling acts through Smads1/5/8, suggesting that related downstream genes might be affected in both disorders. For instance, mice with homozygous deletion of Smad6, an inhibitor of the BMP pathway, have multiple cardiovascular abnormalities and raised blood pressure.120 Smad6 was predominantly expressed in the heart and blood vessels, suggesting that it is important in the homeostasis of the cardiovascular system and tissue specific modulation of TGFβ superfamily signalling in vivo. However, further studies are required to examine the relevance of these findings and the role of I‐Smads and downstream transcriptional factors in the pathogenesis of primary pulmonary hypertension or HHT.
In HHT, pulmonary hypertension has typically been described as a consequence of a high blood flow through large AVMs. For instance, Harrison et al described two such patients harbouring ENG mutations: one had PAVM and developed pulmonary hypertension secondary to thromboembolic disease; the other had both lung and liver AVMs, and developed pulmonary hypertension caused by high blood flow through the extensive liver AVMs.46 In the same study, an individual with HHT, pulmonary hypertension, and a history of exposure to appetite suppressants was reported to carry a mutation in the ACVRL1 gene.46 Another case of appetite suppressant (dexfenfluramine) associated pulmonary arterial hypertension has been described in a patient with HHT1.120 Appetite suppressants are known risk factors for the development of pulmonary hypertension; however, their mechanism of action in individuals with genetic defects in BMPRII, ALK‐1 and ENG genes remains to be determined.
An imbalance in the regulation of TGFβ/BMP mediated endothelial pathways, caused by mutations in ALK‐3 (BMPR1A) or Smad4, can also lead to juvenile polyposis, while mutations in Smad4 can be associated with HHT in patients with juvenile polyposis (fig 3).78,79 Mutations in Smads are usually associated with cancers, particularly those of the colon and gastrointestinal tract. Three of the Smad genes—Smad2, Smad4, and Smad7—are closely clustered at 18q21.1, a region that is often deleted in human cancers. For instance Smad4, initially identified as DPC (deleted in pancreatic cancer), is mutated in up to 50% of pancreatic carcinomas and a third of colorectal cancers.122 This testifies to the crucial role of these pathways in the maintenance of integrity in endothelium and epithelium. It also implies crosstalk between TGFβ/BMP pathways and the tight regulation needed to avoid the many pathologies arising from imbalance in receptor or Smad levels.
Conclusions and future prospects
Numerous data strongly support haploinsufficiency as the mechanism responsible for HHT and indicate that disease heterogeneity cannot be explained by the position and type of mutations. Mutations in ENG or ACVRL1 genes thus result in a significant reduction in the level of functional endoglin and ALK‐1 proteins and to dysregulation of the TGFβ signalling pathways. Such alterations in these endothelial receptors appear to weaken the vascular endothelium and predispose to the formation of focal vascular lesions that might be more frequent in patients with modifier genes. However, it is likely that vascular stress triggers the formation of focal lesions. It has been suggested that local tissue inflammation or endothelial cell injury, perhaps caused by hypoxia or haemodynamic changes, could act as triggers.87,123 Further investigations in the animal models are required to establish the potential mechanisms of induction of HHT vascular lesions by haemodynamic changes.
The identification of Smad4 as a potential target gene for HHT in a selected group of patients with familial polyposis raises the possibility that other genes can lead to HHT associated with specific clinical manifestations. It also suggests that HHT patients with no detected ACVRL1 or ENG mutations should be screened for potential gastrointestinal complications and analysed for MADH4 mutations. Mutations in patients with both HHT and other disorders (such as primary pulmonary hypertension and juvenile polyposis) point to the complexity and intricate interaction between members of the TGFβ superfamily and their importance in maintaining homeostasis. The report of a locus for HHT3 in a family with PAVMs20 should ultimately lead to the identification of the third HHT gene and contribute to our understanding of HHT and its underlying mechanisms. Extensive genotype/phenotype studies might yield a better characterisation of the types of HHT and their specific clinical manifestations, such as higher prevalence of PAVMs in HHT1, primary pulmonary hypertension in association with HHT2, and polyposis in HHT patients with MADH4 mutations.
The biological effects of TGFβ are extremely varied, dose dependent, and differ according to the type and environment of the target cell.124 Identifying novel Smad partners and regulators is crucial for understanding TGFβ function. Further studies should determine if selective activation of Smads, including the I‐Smads, might account for the different angiogenic responses observed in conditions such as HHT with a dysregulation of TGFβ responses.
Abbreviations
AVM - arteriovenous malformation
BMP - bone morphogenetic protein
CAVM - cerebral arteriovenous malformations
HHT - hereditary haemorrhagic telangiectasia
HUVEC - human umbilical vein endothelial cell
JPHT - syndrome of juvenile polyposis and hereditary haemorrhagic telangiectasia
MEEC - murine embryonic endothelial cells
PAVM - pulmonary arteriovenous malformation
TGF - transforming growth factor
Footnotes
Conflicts of interest: none declared
References
- 1.Guttmacher A E, Marchuk D A, White R I. Hereditary hemorrhagic telangiectasia. N Engl J Med 1995333918–924. [DOI] [PubMed] [Google Scholar]
- 2.Dakeishi M, Shioya T, Wada Y, Shindo T, Otaka K, Manabe M, Nozaki J, Inoue S, Koizumi A. Genetic epidemiology of hereditary haemorrhagic telangiectasia in a local community in the northern part of Japan. Hum Mutat 200219140–148. [DOI] [PubMed] [Google Scholar]
- 3.Kjeldsen A D, Vase P, Green A. Hereditary haemorrhagic telangiectasia: a population‐based study of prevalence and mortality in Danish patients. J Intern Med 199924531–39. [DOI] [PubMed] [Google Scholar]
- 4.Plauchu H, Bideau A. Epidemiologie et constitution d'un registre de population a propos d'une concentration geographique d'une maladie hereditaire rare. Population 19844–5765–786. [Google Scholar]
- 5.Bideau A, Plauchu H, Jacquard A, Robert J M, Desjardins B. Genetic aspects of Rendu‐Osler disease in Haut‐Jura: convergence of methodological approaches of historic demography and medical genetics. J Genet Hum 198028127–147. [PubMed] [Google Scholar]
- 6.Plauchu H, de Chadarevian J P, Bideau A, Robert J M. Age‐related clinical profile of hereditary haemorrhagic telangiectasia in an epidemiologically recruited population. Am J Med Genet 198932291–297. [DOI] [PubMed] [Google Scholar]
- 7.Westermann C J, Rosina A F, De Vries V, de Coteau P A. The prevalence and manifestations of hereditary haemorrhagic telangiectasia in the Afro‐Caribbean population of the Netherlands Antilles: a family screening. Am J Med Genet 2003116324–328. [DOI] [PubMed] [Google Scholar]
- 8.Assar O S, Friedman C M, White R I. The natural history of epistaxis in hereditary haemorrhagic telangiectasia. Laryngoscope 1991101977–980. [DOI] [PubMed] [Google Scholar]
- 9.Buscarini E, Danesino C, Plauchu H, de Fazio C, Olivieri C, Brambilla G, Menozzi F, Reduzzi L, Blotta P, Gazzaniga P, Pagella F, Grosso M, Pongiglione G, Cappiello J, Zambelli A. High prevalence of hepatic focal nodular hyperplasia in subjects with hereditary haemorrhagic telangiectasia. Ultrasound Med Biol 2004301089–1097. [DOI] [PubMed] [Google Scholar]
- 10.Garcia‐Tsao G, Korzenik J R, Young L, Henderson K J, Jain D, Byrd B, Pollak J S, White R I. Liver disease in patients with hereditary haemorrhagic telangiectasia. N Engl J Med 2000343931–936. [DOI] [PubMed] [Google Scholar]
- 11.Nanthakumar K, Graham A T, Robinson T I, Grande P, Pugash R A, Clarke J A, Hutchison S J, Mandzia J L, Hyland R H, Faughnan M E. Contrast echocardiography for detection of pulmonary arteriovenous malformations. Am Heart J 2001141243–246. [DOI] [PubMed] [Google Scholar]
- 12.Ference B A, Shannon T M, White R I, Zawin M, Burdge C M. Life‐threatening pulmonary hemorrhage with pulmonary arteriovenous malformations and hereditary haemorrhagic telangiectasia. Chest 19941061387–1390. [DOI] [PubMed] [Google Scholar]
- 13.Shovlin C L, Winstock A R, Peters A M, Jackson J E, Hughes J M. Medical complications of pregnancy in hereditary haemorrhagic telangiectasia. Q J Med 199588879–887. [PubMed] [Google Scholar]
- 14.Kjeldsen A D, Oxhoj H, Andersen P E, Green A, Vase P. Prevalence of pulmonary arteriovenous malformations (PAVMs) and occurrence of neurological symptoms in patients with hereditary haemorrhagic telangiectasia (HHT). J Intern Med 2000248255–262. [DOI] [PubMed] [Google Scholar]
- 15.Fulbright R K, Skudlarski P, Lacadie C M, Warrenburg S, Bowers A A, Gore J C, Wexler B E. Functional MR imaging of regional brain responses to pleasant and unpleasant odors. Am J Neuroradiol 1998191721–1726. [PMC free article] [PubMed] [Google Scholar]
- 16.Bayrak‐Toydemir P, Mao R, Lewin S, McDonald J. Hereditary haemorrhagic telangiectasia: an overview of diagnosis and management in the molecular era for clinicians. Genet Med 20046175–191. [DOI] [PubMed] [Google Scholar]
- 17.Shovlin C L, Guttmacher A E, Buscarini E, Faughnan M E, Hyland R H, Westermann C J, Kjeldsen A D, Plauchu H. Diagnostic criteria for hereditary haemorrhagic telangiectasia (Rendu‐Osler‐Weber syndrome). Am J Med Genet 20009166–67. [DOI] [PubMed] [Google Scholar]
- 18.Gallione C J, Repetto G M, Legius E, Rustgi A K, Schelley S L, Tejpar S, Mitchell G, Drouin E, Westermann C J, Marchuk D A. A combined syndrome of juvenile polyposis and hereditary haemorrhagic telangiectasia associated with mutations in MADH4 (SMAD4). Lancet 2004363852–859. [DOI] [PubMed] [Google Scholar]
- 19.Sirard C, de la Pompa J L, Elia A, Itie A, Mirtsos C, Cheung A, Hahn S, Wakeham A, Schwartz L, Kern S E, Rossant J, Mak T W. The tumor suppressor gene Smad4/Dpc4 is required for gastrulation and later for anterior development of the mouse embryo. Genes Dev 199812107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cole S, Begbie M, Wallace G, Shovlin C. A new locus for hereditary haemorrhagic telangiectasia (HHT3) maps to chromosome 5. J Med Genet 200542577–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDonald M T, Papenberg K A, Ghosh S, Glatfelter A A, Biesecker B B, Helmbold E A, Markel D S, Zolotor A, McKinnon W C, Vanderstoep J L.et al A disease locus for hereditary haemorrhagic telangiectasia maps to chromosome 9q33–34. Nat Genet 19946197–204. [DOI] [PubMed] [Google Scholar]
- 22.Shovlin C L, Hughes J M, Tuddenham E G, Temperley I, Perembelon Y F, Scott J, Seidman C E, Seidman J G. A gene for hereditary haemorrhagic telangiectasia maps to chromosome 9q3. Nat Genet 19946205–209. [DOI] [PubMed] [Google Scholar]
- 23.Fernandez‐Riuz E, St‐Jacques S, Bellon T, Letarte M, Bernabeu C. Assignment of the human endoglin gene (END) to 9q34→qter. Cytogenet Cell Genet 199364204–207. [DOI] [PubMed] [Google Scholar]
- 24.McAllister K A, Grogg K M, Johnson D W, Gallione C J, Baldwin M A, Jackson C E, Helmbold E A, Markel D S, McKinnon W C, Murrell J.et al Endoglin, a TGF‐β binding protein of endothelial cells, is the gene for hereditary haemorrhagic telangiectasia type 1. Nat Genet 19948345–351. [DOI] [PubMed] [Google Scholar]
- 25.Gougos A, Letarte M. Primary structure of endoglin, an RGD‐containing glycoprotein of human endothelial cells. J Biol Chem 19902658361–8364. [PubMed] [Google Scholar]
- 26.Lastres P, Martin‐Perez J, Langa C, Bernabeu C. Phosphorylation of the human transforming‐growth‐factor‐beta‐binding protein endoglin. Biochem J 1994301765–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Letteboer T G, Zewald R A, Kamping E J, de Haas G, Mager J J, Snijder R J, Lindhout D, Hennekam F A, Westermann C J, Ploos van Amstel J K. Hereditary haemorrhagic telangiectasia: ENG and ALK‐1 mutations in Dutch patients. Hum Genet 20051168–16. [DOI] [PubMed] [Google Scholar]
- 28.Cymerman U, Vera S, Pece‐Barbara N, Bourdeau A, White R I, Dunn J, Letarte M. Identification of hereditary haemorrhagic telangiectasia type 1 in newborns by protein expression and mutation analysis of endoglin. Pediatr Res 20004724–35. [DOI] [PubMed] [Google Scholar]
- 29.Gallione C J, Klaus D J, Yeh E Y, Stenzel T T, Xue Y, Anthony K B, McAllister K A, Baldwin M A, Berg J N, Lux A, Smith J D, Vary C P, Craigen W J, Westermann C J, Warner M L, Miller Y E, Jackson C E, Guttmacher A E, Marchuk D A. Mutation and expression analysis of the endoglin gene in hereditary haemorrhagic telangiectasia reveals null alleles. Hum Mutat 199811286–294. [DOI] [PubMed] [Google Scholar]
- 30.Lesca G, Plauchu H, Coulet F, Lefebvre S, Plessis G, Odent S, Riviere S, Leheup B, Goizet C, Carette M F, Cordier J F, Pinson S, Soubrier F, Calender A, Giraud S. French Rendu‐Osler Network. Molecular screening of ALK1/ACVRL1 and ENG genes in hereditary haemorrhagic telangiectasia in France. Hum Mutat 200423289–299. [DOI] [PubMed] [Google Scholar]
- 31.Abdalla S A, Cymerman U, Rushlow D, Chen N, Stoeber G, Lemire E, Letarte M. Novel mutations and polymorphisms in genes causing hereditary haemorrhagic telangiectasia. Hum Mutat 200525320–321. [DOI] [PubMed] [Google Scholar]
- 32.Lastella P, Sabba C, Lenato G M, Resta N, Lattanzi W, Gallitelli M, Cirulli A, Guanti G. Endoglin gene mutations and polymorphisms in Italian patients with hereditary haemorrhagic telangiectasia. Clin Genet 200363536–540. [DOI] [PubMed] [Google Scholar]
- 33.Gallione C J, Scheessele E A, Reinhardt D, Duits A J, Berg J N, Westermann C J, Marchuk D A. Two common endoglin mutations in families with hereditary haemorrhagic telangiectasia in the Netherlands Antilles: evidence for a founder effect. Hum Genet 200010740–44. [DOI] [PubMed] [Google Scholar]
- 34.Shovlin C, Hughes J, Scott J, Seidman C, Seidman J. Characterization of endoglin and identification of novel mutations in hereditary haemorrhagic telangiectasia. Am J Hum Genet 19976168–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cymerman U, Vera S, Karabegovic A, Abdalla S, Letarte M. Characterization of 17 novel endoglin mutations associated with hereditary haemorrhagic telangiectasia. Hum Mutat 200321482–492. [DOI] [PubMed] [Google Scholar]
- 36.Berg J, Porteous M, Reinhardt D, Gallione C, Holloway S, Umasunthar T, Lux A, McKinnon W, Marchuk D, Guttmacher A. Hereditary haemorrhagic telangiectasia: a questionnaire based study to delineate the different phenotypes caused by endoglin and ALK‐1 mutations. J Med Genet 200340585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paquet M E, Pece‐Barbara N, Vera S, Cymerman U, Karabegovic A, Shovlin C, Letarte M. Analysis of several endoglin mutants reveals no endogenous mature or secreted protein capable of interfering with normal endoglin function. Hum Mol Genet 2001101347–1357. [DOI] [PubMed] [Google Scholar]
- 38.Pece‐Barbara N, Cymerman U, Vera S, Marchuk D A, Letarte M. Expression analysis of four endoglin missense mutations suggests that haploinsufficiency is the predominant mechanism for hereditary haemorrhagic telangiectasia type I. Hum Mol Genet 199982171–2181. [DOI] [PubMed] [Google Scholar]
- 39.Brusgaard K, Kjeldsen A D, Poulsen L, Moss H, Vase P, Rasmussen K, Kruse T A, Horder M. Mutations in endoglin and in activin receptor‐like kinase 1 among Danish patients with hereditary haemorrhagic telangiectasia. Clin Genet 200466556–561. [DOI] [PubMed] [Google Scholar]
- 40.Pece N, Vera S, Cymerman U, White R I J, Wrana J L, Letarte M. Mutant endoglin in hereditary haemorrhagic telangiectasia type I is transiently expressed intracellularly and is not a dominant negative. J Clin Invest 19971002568–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bourdeau A, Cymerman U, Paquet M E, Meschino W, McKinnon W C, Guttmacher A E, Becker L, Letarte M. Endoglin expression is reduced in normal vessels but still detectable in arteriovenous malformations of patients with hereditary haemorrhagic telangiectasia type 1. Am J Pathol 2000156911–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamaguchi H, Azuma H, Shigekiyo T, Inoue H, Saito S. A novel missense mutation in the endoglin gene in hereditary haemorrhagic telangiectasia. Thromb Haemost 199777243–247. [PubMed] [Google Scholar]
- 43.Sanz‐Rodriguez F, Fernandez‐L A, Zarrabeitia R, Perez‐Molino A, Ramirez J R, Coto E, Bernabeu C, Botella L. Mutation analysis in Spanish patients with hereditary haemorrhagic telangiectasia: deficient endoglin up‐regulation in activated monocytes. Clin Chem 2004502003–2011. [DOI] [PubMed] [Google Scholar]
- 44.McAllister K A, Baldwin M A, Thukkani A K, Gallione C J, Berg J N, Porteus M E, Guttmacher A E, Marchuk D A. Six novel mutations in the endoglin gene in hereditary haemorrhagic telangiectasia type 1 suggest a dominant‐negative effect of receptor function. Hum Mol Genet 199541983–1985. [DOI] [PubMed] [Google Scholar]
- 45.Bourdeau A, Faughnan M E, Letarte M. Endoglin‐deficient mice, a unique model to study hereditary haemorrhagic telangiectasia. Trends Cardiovasc Med 200010279–285. [DOI] [PubMed] [Google Scholar]
- 46.Harrison R E, Flanagan J A, Sankelo M, Abdalla S A, Rowell J, Machado R D, Elliott C G, Robbins I M, Olschewski H, McLaughlin V, Gruenig E, Kermeen F, Halme M, Raisanen‐Sokolowski A, Laitinen T, Morrell N W, Trembath R C. Molecular and functional analysis identifies ALK‐1 as the predominant cause of pulmonary hypertension related to hereditary haemorrhagic telangiectasia. J Med Genet 200340865–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karabegovic A, Shinawi M, Cymerman U, Letarte M. No live individual homozygous for a novel endoglin mutation was found in a consanguineous Arab family with hereditary haemorrhagic telangiectasia. J Med Genet 200441e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin W D, Tsai F J, Lee C C, Wu J Y. Identification of a polymorphism (D366H) in the endoglin gene in Chinese. Hum Mutat 200015583. [DOI] [PubMed] [Google Scholar]
- 49.Frischmeyer P A, Dietz H C. Nonsense‐mediated mRNA decay in health and disease. Hum Mol Genet 199981893–1900. [DOI] [PubMed] [Google Scholar]
- 50.Ríus C, Smith J D, Almendro N, Langa C, Botella L M, Marchuk D A, Vary C P H, Bernabeu C. Cloning of the promoter region of human endoglin, the target gene for hereditary haemorrhagic telangiectasia type 1. Blood 1998924677–4690. [PubMed] [Google Scholar]
- 51.Botella L M, Sanchez‐Elsner T, Rius C, Corbi A, Bernabeu C. Identification of a critical Sp1 site within the endoglin promoter and its involvement in the transforming growth factor‐beta stimulation. J Biol Chem 200127634486–34494. [DOI] [PubMed] [Google Scholar]
- 52.Vincent P, Plauchu H, Hazan J, Faure S, Weissenbach J, Godet J. A third locus for hereditary haemorrhagic telangiectasia maps to chromosome 12q. Hum Mol Genet 19954945–949. [DOI] [PubMed] [Google Scholar]
- 53.Johnson D W, Berg J N, Gallione C J, McAllister K A, Warner J P, Helmbold E A, Markel D S, Jackson C E, Porteus M E, Marchuk D A. A second locus for hereditary hemorrhagic telangiectasia maps to chromosome 12. Genome Res 1995521–28. [DOI] [PubMed] [Google Scholar]
- 54.Johnson D W, Berg J N, Baldwin M A, Gallione C J, Marondel I, Yoon S J, Stenzel T T, Speer M, Pericak‐Vance M A, Diamond A, Guttmacher A E, Jackson C E, Attisano L, Kucherlapati R, Porteous M E, Marchuk D A. Mutations in the activin receptor‐like kinase 1 gene in hereditary haemorrhagic telangiectasia type 2. Nat Genet 199613189–195. [DOI] [PubMed] [Google Scholar]
- 55.Berg J N, Gallione C J, Stenzel T T, Johnson D W, Allen W P, Schwartz C E, Jackson C E, Porteous M E, Marchuk D A. The activin receptor‐like kinase 1 gene: genomic structure and mutations in hereditary haemorrhagic telangiectasia type 2. Am J Hum Genet 19976160–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.ten Dijke P, Ichijo H, Franzen P, Schulz P, Saras J, Toyoshima H, Heldin C H, Miyazono K. Activin receptor‐like kinases: a novel subclass of cell‐surface receptors with predicted serine/threonine kinase activity. Oncogene 199382879–2887. [PubMed] [Google Scholar]
- 57.Attisano L, Carcamo J, Ventura F, Weis F M, Massague J, Wrana J L. Identification of human activin and TGF beta type I receptors that form heteromeric kinase complexes with type II receptors. Cell 199375671–680. [DOI] [PubMed] [Google Scholar]
- 58.ten Dijke P, Yamashita H, Ichijo H, Franzen P, Laiho M, Miyazono K, Heldin C H. Characterization of type I receptors for transforming growth factor‐beta and activin. Science 1994264101–104. [DOI] [PubMed] [Google Scholar]
- 59.Hanks S K, Quinn A M, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 198824142–52. [DOI] [PubMed] [Google Scholar]
- 60.Abdalla S A, Cymerman U, Johnson R M, Deber C M, Letarte M. Disease‐associated mutations in conserved residues of ALK‐1 kinase domain. Eur J Hum Genet 200311279–287. [DOI] [PubMed] [Google Scholar]
- 61.Trembath R C, Thomson J R, Machado R D, Morgan N V, Atkinson C, Winship I, Simonneau G, Galie N, Loyd J E, Humbert M, Nichols W C, Morrell N W, Berg J, Manes A, McGaughran J, Pauciulo M, Wheeler L. Clinical and molecular genetic features of pulmonary hypertension in patients with hereditary haemorrhagic telangiectasia. N Engl J Med 2001345325–334. [DOI] [PubMed] [Google Scholar]
- 62.Abdalla S A, Geisthoff U W, Bonneau D, Plauchu H, McDonald J, Kennedy S, Faughnan M E, Letarte M. Visceral manifestations in hereditary haemorrhagic telangiectasia type 2. J Med Genet 200340494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klaus D J, Gallione C J, Anthony K, Yeh E Y, Yu J, Lux A, Johnson D W, Marchuk D A. Novel missense and frameshift mutations in the activin receptor‐like kinase‐1 gene in hereditary haemorrhagic telangiectasia. Mutations in brief no. 164. Hum Mutat 199812137. [DOI] [PubMed] [Google Scholar]
- 64.Abdalla S A, Pece‐Barbara N, Vera S, Tapia E, Paez E, Bernabeu C, Letarte M. Analysis of ALK‐1 and endoglin in newborns from families with hereditary haemorrhagic telangiectasia type 2. Hum Mol Genet 200091227–1237. [DOI] [PubMed] [Google Scholar]
- 65.Olivieri C, Mira E, Delù G, Pagella F, Zambelli A, Malvezzi L, Buscarini L, Danesino C. Identification of 13 new mutations in the ACVRL1 gene in a group of 52 unselected Italian patients affected by hereditary haemorrhagic telangiectasia. J Med Genet 200239e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abdalla S A, Gallione C J, Barst R J, Horn E M, Knowles J A, Marchuk D A, Letarte M, Morse J H. Primary pulmonary hypertension in families with hereditary haemorrhagic telangiectasia. Eur Respir J 200423373–377. [DOI] [PubMed] [Google Scholar]
- 67.Kjeldsen A D, Brusgaard K, Poulsen L, Kruse T, Rasmussen K, Green A, Vase P. Mutations in the ALK‐1 gene and the phenotype of hereditary haemorrhagic telangiectasia in two large Danish families. Am J Med Genet 200198298–302. [DOI] [PubMed] [Google Scholar]
- 68.Zhang G, Yi Y, Peng H, Shen J, Xie D, He X. Clinical phenotypes, ALK1 gene mutation and level of related plasma proteins in Chinese hereditary haemorrhagic telangiectasia. Chin Med J 2004117808–812. [PubMed] [Google Scholar]
- 69.Lin W, Wu J, Hsu H, Tsai F, Lee C, Tsai C. Mutation analysis of a family with hereditary haemorrhagic telangiectasia associated with hepatic arteriovenous malformation. J Formos Med Assoc 2001100817–819. [PubMed] [Google Scholar]
- 70.D'Abronzo F H, Swearingen B, Klibanski A, Alexander J M. Mutational analysis of activin/transforming growth factor‐beta type I and type II receptor kinases in human pituitary tumors. J Clin Endocrinol Metab 1999841716–1721. [DOI] [PubMed] [Google Scholar]
- 71.Lux A, Attisano L, Marchuk D A. Assignment of transforming growth factor beta1 and beta3 and a third new ligand to the type I receptor ALK‐1. J Biol Chem 19992749984–9992. [DOI] [PubMed] [Google Scholar]
- 72.Feng X H, Derynck R. A kinase subdomain of transforming growth factor‐beta (TGF‐beta) type I receptor determines the TGF‐beta intracellular signaling specificity. EMBO J 1997163912–3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wieser R, Wrana J L, Massague J. GS domain mutations that constitutively activate T beta R‐I, the downstream signaling component in the TGF‐beta receptor complex. EMBO J 1995142199–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Saitoh M, Nishitoh H, Amagasa T, Miyazono K, Takagi M, Ichijo H. Identification of important regions in the cytoplasmic juxtamembrane domain of type I receptor that separate signaling pathways of transforming growth factor‐beta. J Biol Chem 19962712769–2775. [DOI] [PubMed] [Google Scholar]
- 75.Cox K L, Frates R C, Wong A, Gandhi G. Hereditary generalized juvenile polyposis associated with pulmonary arteriovenous malformation. Gastroenterology 1980781566–1570. [PubMed] [Google Scholar]
- 76.Conte W J, Rotter J I, Schwartz A G, Congleton J E. Hereditary generalized juvenile polyposis, areteriovenous malformations and colonic carcinoma [abstract]. Clin Res 19823093A [Google Scholar]
- 77.Baert A L, Casteels‐Van Daele M, Broeckx J, Wijndaele L, Wilms G, Eggermont E. Generalized juvenile polyposis with pulmonary arteriovenous malformations and hypertrophic osteoarthropathy. Am J Roentgenol 198314661–662. [DOI] [PubMed] [Google Scholar]
- 78.Howe J R, Roth S, Ringold J C, Summers R W, Jarvinen H J, Sistonen P, Tomlinson I P, Houlston R S, Bevan S, Mitros F A, Stone E M, Aaltonen L A. Mutations in the SMAD4/DPC4 gene in juvenile polyposis. Science 19982801086–1088. [DOI] [PubMed] [Google Scholar]
- 79.Howe J R, Bair J L, Sayed M G, Anderson M E, Mitros F A, Petersen G M, Velculescu V E, Traverso G, Vogelstein B. Germline mutations of the gene encoding bone morphogenetic protein receptor 1A in juvenile polyposis. Nat Genet 200128184–187. [DOI] [PubMed] [Google Scholar]
- 80.Chen Y G, Hata A, Lo R S, Wotton D, Shi Y, Pavletich N, Massague J. Determinants of specificity in TGF‐beta signal transduction. Genes Dev 1998122144–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huse M, Chen Y G, Massague J, Kuriyan J. Crystal structure of the cytoplasmic domain of the type I TGF beta receptor in complex with FKBP12. Cell 199996425–436. [DOI] [PubMed] [Google Scholar]
- 82.Lo R S, Chen Y G, Shi Y, Pavletich N P, Masague J. The L3 loop: a structural motif determining specific interactions between SMAD proteins and TGF‐beta receptors. EMBO J 199817996–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.ten Dijke P, Hill C S. New insights into TGF‐beta‐Smad signalling. Trends Biochem Sci 200429265–273. [DOI] [PubMed] [Google Scholar]
- 84.Mehra A, Wrana J L. TGF‐beta and the Smad signal transduction pathway. Biochem Cell Biol 200280605–622. [DOI] [PubMed] [Google Scholar]
- 85.Chang H, Brown C W, Matzuk M M. Genetic analysis of the mammalian transforming growth factor‐beta superfamily. Endocr Rev 200223787–823. [DOI] [PubMed] [Google Scholar]
- 86.J M, Lebrin F, Valdimarsdottir G. Controlling the angiogenic switch: a balance between two distinct TGF‐beta receptor signaling pathways. Trends Cardiovasc Med 200313301–307. [DOI] [PubMed] [Google Scholar]
- 87.Oh S, Seki T, Goss K, Imamura T, Yi Y, Donahoe P, Li L, Miyazono K, ten Dijke P, Kim S, Li E. Activin receptor‐like kinase 1 modulates transforming growth factor‐beta 1 signaling in the regulation of angiogenesis. Proc Natl Acad Sci USA 2000972626–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen Y, Massague J. Smad1 recognition and activation by the ALK1 group of transforming growth factor‐beta family receptors. J Biol Chem 19992743672–3677. [DOI] [PubMed] [Google Scholar]
- 89.Goumans M J, Valdimarsdottir G, Itoh S, Rosendahl A, Sideras P, ten Dijke P. Balancing the activation state of the endothelium via two distinct TGF‐β type I receptors. EMBO J 2002211743–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Roelen B A, van Rooijen M A, Mummery C L. Expression of ALK‐1, a type 1 serine/threonine kinase receptor, coincides with sites of vasculogenesis and angiogenesis in early mouse development. Dev Dyn 1997209418–430. [DOI] [PubMed] [Google Scholar]
- 91.St‐Jacques S, Cymerman U, Pece N, Letarte M. Molecular characterization and in situ localization of murine endoglin reveal that it is a transforming growth factor‐β binding protein of endothelial and stromal cells. Endocrinology 19941342645–2657. [DOI] [PubMed] [Google Scholar]
- 92.Qu R, Silver M M, Letarte M. Distribution of endoglin in early human development reveals high levels on endocardial cushion tissue mesenchyme during valve formation. Cell Tissue Res 1998292333–343. [DOI] [PubMed] [Google Scholar]
- 93.Barbara N P, Wrana J L, Letarte M. Endoglin is an accessory protein that interacts with the signaling receptor complex of multiple members of the transforming growth factor‐beta superfamily. J Biol Chem 1999274584–594. [DOI] [PubMed] [Google Scholar]
- 94.Guerrero‐Esteo M, Sanchez‐Elsner T, Letamendia A, Bernabeu C. Extracellular and cytoplasmic domains of endoglin interact with the transforming growth factor‐beta receptors I and II. J Biol Chem 200227729197–29209. [DOI] [PubMed] [Google Scholar]
- 95.Blanco F, Santibanez J, Guerrero‐Esteo M, Langa C, Vary C, Bernabeu C. Interaction and functional interplay between endoglin and ALK‐1, two components of the endothelial transforming growth factor‐beta receptor complex. J Cell Physiol. Published Online First: 8 February 2005. doi:10.1002/jcp.20311 [DOI] [PubMed]
- 96.Cheifetz S, Bellon T, Cales C, Vera S, Bernabeu C, Massague J, Letarte M. Endoglin is a component of the transforming growth factor‐beta receptor system in human endothelial cells. J Biol Chem 199226719027–19030. [PubMed] [Google Scholar]
- 97.Letamendia A, Lastres P, Botella L M, Raab U, Langa C, Velasco B, Attisano L, Bernabeu C. Role of endoglin in cellular responses to transforming growth factor‐beta. A comparative study with betaglycan. J Biol Chem 199827333011–33019. [DOI] [PubMed] [Google Scholar]
- 98.Lebrin F, Goumans M J, Jonker L, Carvalho R L, Valdimarsdottir G, Thorikay M, Mummery C, Arthur H M, ten Dijke P. Endoglin promotes endothelial cell proliferation and TGF‐β/ALK‐1 signal transduction. EMBO J 2004234018–4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pece‐Barbara N, Vera S, Kathirkamathamby K, Liebner S, Di Guglielmo G, Dejana E, Wrana J, Letarte M. Endoglin null endothelial cells proliferate faster, and are more responsive to TGF‐β1 with higher affinity receptors and an activated ALK‐1 pathway. J Biol Chem. (in press) [DOI] [PubMed]
- 100.Massague J, Blain S W, Lo R S. TGF‐β signaling in growth control, cancer and heritable disorders. Cell 2000103295–309. [DOI] [PubMed] [Google Scholar]
- 101.Ota T, Fujii M, Sugizaki T, Ishii M, Miyazawa K, Aburatani H, Miyazono K. Targets of transcriptional regulation by two distinct type I receptors for TGF‐β in human umbilical vein endothelial cells. J Cell Physiol 2002193299–318. [DOI] [PubMed] [Google Scholar]
- 102.Wu G, Chen Y G, Ozdamar B, Gyuricza C A, Chong P A, Wrana J L, Massague J, Shi Y. Structural basis of Smad2 recognition by the Smad anchor for receptor activation. Science 200028792–97. [DOI] [PubMed] [Google Scholar]
- 103.Liu F, Pouponnot C, Massague J. Dual role of the Smad4/DPC tumor suppressor in TGF‐β‐inducible transcriptional responses. Genes Dev 1997113157–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Massague J, Wotton D. Transcriptional control by the TGF‐β/Smad signaling system. EMBO J 2000191745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Morgan T, McDonald J, Anderson C, Ismail M, Miller F, Mao R, Madan A, Barnes P, Hudgins L, Manning M. Intracranial hemorrhage in infants and children with hereditary haemorrhagic telangiectasia (Osler‐Weber‐Rendu syndrome). Pediatrics 2002109E12. [DOI] [PubMed] [Google Scholar]
- 106.Li D Y, Sorensen L K, Brooke B S, Urness L D, Davis E C, Taylor D G, Boak B B, Wendel D P. Defective angiogenesis in mice lacking endoglin. Science 19992841534–1537. [DOI] [PubMed] [Google Scholar]
- 107.Bourdeau A, Dumont D J, Letarte M. A murine model of hereditary haemorrhagic telangiectasia. J Clin Invest 19991041343–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Arthur H M, Ure J, Smith A J, Renforth G, Wilson D I, Torsney E, Charlton R, Parums D V, Jowett T, Marchuk D A, Burn J, Diamond A G. Endoglin, an ancillary TGF‐β receptor, is required for extraembryonic angiogenesis and plays a key role in heart development. Dev Biol 200021742–53. [DOI] [PubMed] [Google Scholar]
- 109.Carvalho R L, Jonker L, Goumans M J, Larsson J, Bouwman P, Karlsson S, ten Dijke P, Arthur H, Mummery C. Defective paracrine signalling by TGF‐β in yolk sac vasculature of endoglin mutant mice: a paradigm for hereditary haemorrhagic telangiectasia. Development 20041316237–6247. [DOI] [PubMed] [Google Scholar]
- 110.Lamouille S, Mallet C, Feige J J, Baily S. Activin receptor‐like kinase 1 is implicated in the maturation phase of angiogenesis. Blood 20021004495–4501. [DOI] [PubMed] [Google Scholar]
- 111.Roman B L, Pham V N, Lawson N D, Kulik M, Childs S, Lekven A C, Garrity D M, Moon R T, Fishman M C, Lechleider R J, Weinstein B M. Disruption of acvrl1 increases endothelial cell number in zebrafish cranial vessels. Development 20021293009–3019. [DOI] [PubMed] [Google Scholar]
- 112.Seki T, Yun J, Oh S. Arterial endothelium‐specific activin receptor‐like kinase 1 expression suggests its role in arterialization and vascular remodeling. Circulation Res 200393682–689. [DOI] [PubMed] [Google Scholar]
- 113.Satomi J, Mount R J, Toporsian M, Paterson A D, Wallace M C, Harrison R V, Letarte M. Cerebral vascular abnormalities in a murine model of hereditary haemorrhagic telangiectasia. Stroke 200334783–789. [DOI] [PubMed] [Google Scholar]
- 114.Bourdeau A, Faughan M, McDonald M L, Paterson A D, Wanless I R, Letarte M. Potential role of modifier genes influencing TGF‐β1 levels in the development of vascular defects in endoglin heterozygous mice with hereditary hemorrhagic telangiectasia. Am J Pathol 20011802011–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Urness L D, Sorensen L K, Li D Y. Arteriovenous malformations in mice lacking activin receptor‐like kinase‐1. Nat Genet 200026328–331. [DOI] [PubMed] [Google Scholar]
- 116.Srinivasan S, Hanes M, Dickens T, Porteous M, Oh S, Hale L, Marchuk D. A mouse model for hereditary haemorrhagic telangiectasia (HHT) type 2. Hum Mol Genet 200312473–482. [DOI] [PubMed] [Google Scholar]
- 117.Torsney E, Charlton R, Parums D, Collis M, Arthur H M. Inducible expression of human endoglin during inflammation and wound healing in vivo. Inflamm Res 200251464–470. [DOI] [PubMed] [Google Scholar]
- 118.Rubin L J. Primary pulmonary hypertension. N Engl J Med 1997336111–117. [DOI] [PubMed] [Google Scholar]
- 119.Machado R D, Pauciulo M W, Thomson J R, Lane K B, Morgan N V, Wheeler L, Phillips J A, 3rd, Newman J, Williams D, Galie N, Manes A, McNeil K, Yacoub M, Mikhail G, Rogers P, Corris P, Humbert M, Donnai D, Martensson G, Tranebjaerg L, Loyd J E, Trembath R C, Nichols W C. BMPR2 haploinsufficiency as the inherited molecular mechanism for primary pulmonary hypertension. Am J Hum Genet 20016892–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Galvin K M, Donovan M J, Lynch C A, Meyer R I, Paul R J, Lorenz J N, Fairchild‐Huntress V, Dixon K L, Dunmore J H, Gimbrone M A, Falb D, Huszar D. A role for Smad6 in development and homeostasis of the cardiovascular system. Nat Genet 200024171–174. [DOI] [PubMed] [Google Scholar]
- 120.Chaouat A, Coulet F, Favre C, Simonneau G, Weitzenblum E, Soubrier F, Humbert M. Endoglin germline mutation in a patient with hereditary haemorrhagic telangiectasia and dexfenfluramine associated pulmonary arterial hypertension. Thorax 200459446–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Miyaki M, Kuroki T. Role of Smad4 (DPC4) inactivation in human cancer. Biochem Biophys Res Commun 2003306799–804. [DOI] [PubMed] [Google Scholar]
- 123.Marchuk D A, Srinivasan S, Squire T L, Zawistowski J S. Vascular morphogenesis: tales of two syndromes. Hum Mol Genet 200312R97–112. [DOI] [PubMed] [Google Scholar]
- 124.Massague J. TGF‐β signal transduction. Annu Rev Biochem 199867753–791. [DOI] [PubMed] [Google Scholar]