Abstract
During a study to extend our knowledge of the host range and genetic diversity of arenaviruses in Great Britain, 66 of 1,147 rodent blood samples tested for antibody, and 127 of 482 tested by PCR, were found positive. All sequences most closely resembled those of previously identified lymphocytic choriomeningitis virus.
Keywords: LCMV, rodents, disease vectors, serology, prevalence, variation genetics, dispatch
Viruses in the family Arenaviridae are separated into 2 distinct serocomplexes, the New World serocomplex and the Old World serocomplex (1). Several arenavirus species are known to cause human disease, including lymphocytic choriomeningitis virus (LCMV), which causes influenza-like clinical signs, occasionally with neurologic complications. Infection may be asymptomatic in up to one third of patients (2), and serious complications often occur in intrauterine infection (3). Less severe cases of adult human infection are likely underreported and often misdiagnosed (4).
LCMV is found worldwide, probably because of its association with its natural Old World host, the house mouse, Mus musculus (5). Although antibodies have also been detected in other rodent species (6,7), arenaviruses are known to be serologically cross-reactive. Few isolates of LCMV have been obtained from wild rodents so little is known about its genetic diversity. Recent studies on American arenaviruses found that diverse arenaviruses co-evolved with their rodent hosts (8), a finding that suggests that a more thorough study of European rodents might also identify novel arenaviruses. The purpose of this study was therefore to extend our knowledge of LCMV and LCMV-like arenaviruses in rodents in Great Britain.
The Study
In total 1,147 blood samples were collected from rodents: 1,060 were live-trapped, wild animals from <20 sites (Table 1), and 87 blood samples were collected from a captive colony of wild house mice (9) and tested serologically. All animal research was conducted under license, according to UK regulations.
Table 1. Rodent species, numbers tested, seroprevalence, and viral RNA prevalence to LCMV at each UK and Republic of Ireland site*.
| Site code and year | Geographic location | Species tested for antibody | Species tested for viral RNA |
|---|---|---|---|
| PHF 2004 | Cheshire | MMu (0/4), RN (0/2); SP = 0.0 | |
| PHF 2005 | Cheshire | MMu (8/26); SP = 30.8 | |
| BHF 2004 | Cheshire | MMu (4/10); SP = 40.0 | MMu (2/9), RN (0/2) – 2 sequences |
| BHF 2005 | Cheshire | MMu (2/2), RN (0/2); SP = 50.0 | |
| BGF | Cheshire | MMu (0/7), RN (0/2); SP = 0.0 | |
| CLF 2004 | Cheshire | MMu (0/6), RN (0/2); SP = 0.0 | |
| CLF 2005 | Cheshire | MMu (0/12), RN (0/4); SP = 0.0 | |
| MF 2002 | Cheshire | AS (0/10), MG (0/9), MA (0/2), MMu (0/30); SP = 0.0 | AS (1/10), MG (0/1), MA (0/1), MMu (2/4) – 3 sequences |
| MF 2004 | Cheshire | AS (4/10), MG (0/1), MA (0/1), MMu (2/4); SP = 37.5 | |
| CZ 2002 | Cheshire | AS (0/4), MG (0/4), CL (4/61), MMi (0/22), RN (0/2); SP = 4.3 | AS (1/1), CL (0/4), MMi (0/1), MMu (0/1) – 0 sequences |
| CZ spring 2003 | Cheshire | AS (0/9), MG (0/3), MA (0/4), MMi (1/3), SP = 5.3 | |
| CZ autumn 2003 | Cheshire | AS (1/18), MG (0/19), MA (0/4), MMu (0/1), SP = 2.9 | |
| CZ 2004 | Cheshire | MMu (1/19), RN (1/12), SP= 3.2 | |
| DF | Cheshire | MMu (1/69), SP = 1.4 | |
| MW | Cheshire | MG (0/105), AS (0/45), SP = 0.0 | |
| RH | Cheshire | MG (0/19), AS (0/49), SP = 0.0 | |
| LVFS | Cheshire | RN (0/2, SC (0/4), SP = 0.0 | |
| FA | Merseyside | MA (0/2), AS (0/24), SP = 0.0 | |
| KF | Northumberland | MA (2/104), SP = 1.9 | |
| LI | North Devon | RN (0/40), SP = 0.0 | |
| IOW | Isle of Wight | SV (1/18), SP = 5.6 | |
| TF | Thetford | SV (1/21), SP = 4.8 | |
| CF | Cumbria | SC (0/10), SV, (0/4), SP = 0.0 | |
| NI | Northern Ireland | AS (1/149), SP = 0.7 | |
| CA | Republic of Ireland | MG (0/15), SP = 0.0 | |
| CC | Republic of Ireland | AS (0/7), SP = 0.0 | |
| TW | Republic of Ireland | AS (0/10), SP = 0.0 | |
| Other | Various locations | RN (1/6), SC (0/1), SV (2/26), SP = NA | AS (0/1), MA (0/2), MMu (0/31), RN (0/5), SV (1/4) – 0 sequences |
| Captive colony | Captive colony, Cheshire | MMu (30/87), SP = 34.5 | MMu (122/403) – 92 sequences |
*LCMV, lymphocytic choriomeningitis virus; SP, site prevalence (%). Species key: MMu, Mus musculus (house mouse); RN, Rattus norvegicus (brown or Norway rat); AS, Apodemus sylvaticus (wood mouse); MG, Myodes glareolus (bank vole); MA, Microtus agrestis (field vole); MMi, Micromys minutis (harvest mouse); CL, Cynomys ludovicianus (black-tailed prairie dog); SV, Sciurus vulgaris (red squirrel); SC, Sciurus carolensis (gray squirrel); NA, not available. Species containing positive animals are in boldface italics, with number positive and number tested in parentheses.
Serum samples were separated by centrifugation (10,000 rpm, 10 min) and tested for LCMV antibody by using the manufacturer’s protocol for commercial indirect fluorescent antibody assay slides (Charles River Laboratories, Wilmington, MA, USA). A 1:40 dilution of anti-rat or anti-mouse immunoglobulin G fluorescein isothiocyanate (Sigma-Aldrich, Gillingham, UK) or a combination of both were used as secondary antibody. Ninety-three serum samples (from the original serum samples tested for antibody) that were either antibody positive or from sites with high seroprevalence were tested for arenavirus RNA by PCR. Another 379 blood samples from the captive colony of house mice, which had not been previously tested for antibody, were also tested. The PCR targeted a fragment of the glycoprotein precursor gene (GPC) (10). A selection of samples found negative by the GPC PCR were subsequently retested by PCR targeted at a fragment of the nucleoprotein (N) gene (8), by using primers to sequences common to the Old World arenaviruses. Total RNA was extracted by using QIAamp viral RNA mini-kit (QIAGEN, Crawley, UK), converted to cDNA, and amplified by using a single-step kit (Superscript III one-step RT-PCR with Platinum Taq polymerase system; Invitrogen, Paisley, UK) in conjunction with oligonucleotides arena1+ and LCMV322– (10) or 1010C and either OW1696R or NW1696R (8). Products were separated and visualized by agarose gel electrophoresis, and amplicons were purified with the QIAquick PCR purification kit (QIAGEN). Bidirectional sequencing was performed off-site (MWG Biotech AG, Ebersberg, Germany). The 97-nt sequences generated here were deposited with GenBank (accession nos. DQ275199–DQ275295).
The software package MEGA version 4.0 (11) was used to construct an alignment of a 283-nt fragment of the GPC gene nucleotide sequences and predicted amino acid sequences, and for phylogenetic analysis with the neighbor-joining method (p distance model), with bootstrap support based on 1,000 pseudoreplicates. Other GenBank sequences included for comparison are listed in Table 2. Pairwise genetic distances were calculated by using the p distance model; percentage sequence identities were calculated by subtracting the genetic distances from 1.0 and multiplying by 100.
Table 2. Percentage nucleotide identities between the study sample sequences (all and from the captive colony only) and previously isolated LCMV and Lassa virus sequences*.
| Sequence | All study sequences, % | Captive colony sequences only, % |
|---|---|---|
| All study sequences | 93.6–100 | NS |
| Captive colony sequences only | NS | 97.4–100 |
| LCMV CIPV76001 Pasteur (AF095783; France) | 78.7–80.5 | 78.7–80.5 |
| LCMV CIP97001 (AF079517; France) | 79.4– 83.1 | 80.9–83.1 |
| LCMV Marseille (DQ286931; France) | 82.8– 83.9 | 82.8–83.5 |
| LCMV CH5871 (AF325215; Germany) | 81.6–83.1 | 81.6–83.1 |
| LCMV CH5692 (AF325214; Germany) | 81.3–82.8 | 81.3–82.8 |
| LCMV MX (EU195888; Slovakia) | 78.7–80.5 | 78.7–80.5 |
| LCMV Armstrong (M20869; USA) | 82.8– 85.8 | 83.9–85.8 |
| LCMV WE (M22138) | 82.0–84.3 | 82.0–84.3 |
| Lassa LP (AF181853) | 58.1–59.9 | 58.8–59.9 |
*LCMV, lymphocytic choriomeningitis virus; NS, not shown.
Overall, 66 of 1,147 serum samples and 7 of 9 rodent species had antibodies to arenaviruses. Sciurus vulgaris had the highest prevalence, 26%, although only 15 squirrels were tested. M. musculus had the second highest prevalence, 17.5%. Antibodies were also detected in Apodemus sylvaticus, Microtus agrestis, Micromys minutis, captive-housed Cynomys ludovicianus, and Rattus norvegicus. Seroprevalence varied between species (1.4%–26%) and between sites (0%–50%) (Table 1).
GPC PCR amplicons were obtained from 127 of 472 tested samples, and sequences were determined for 97 samples (Table 1). All positive samples were from Mus musculus except 1 from A. sylvaticus. Twenty samples negative in the GPC PCR, but seropositive or from high prevalence sites, were tested by N gene PCR, and 2 were weakly positive: 1 S. vulgaris and 1 A. sylvaticus. In neither case, however, could a sequence be obtained from the amplicon.
Nucleotide and amino acid GPC sequence identities for all the samples in this study ranged from 93.6%–100% (Table 2), and 94.5%–100%, respectively (data not shown). When compared with other arenaviruses, the nucleotide sequences exhibited 78.7%–85.8% identity with LCMV reference sequences and only 58.1%–59.9% identity with Lassa virus (Appendix Figure).
Although antibodies to arenaviruses have been reported in a range of European rodent species, our study provided evidence of arenaviruses infecting red squirrels (S. vulgaris) and European harvest mice (M. minutis). Antibodies to arenaviruses have been reported in introduced S. carolensis in Great Britain (12) but were not detected in this study. We also reported antibodies to arenaviruses in black-tailed prairie dogs (Cynomys ludovicianus): those tested in this study were part of a colony in a zoo, however, and had contact with wild mice, some of which were seropositive. As found in previous studies, Mus musculus was more likely to be infected with LCMV than other rodent species.
The nucleotide sequences of most PCR amplicons clearly identified LCMV as the most frequent cause of the antibody detected. However, the detection of arenaviral RNA in 2 animals by the N gene PCR, but not by the LCMV-specific GPC PCR, may suggest the presence of another species of arenavirus. Further studies are needed to determine if other arenaviruses species are present in European rodent populations (8).
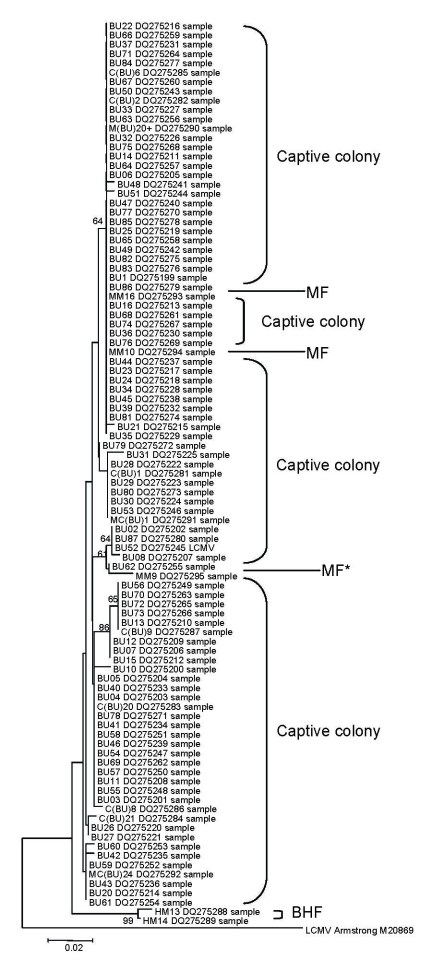
Genetic heterogeneity was present within and between sites (Figure), as seen in previous studies of arenaviruses (13,14). Sequences from animals in the captive colony and a nearby farm (MF) clustered and were different from those from a more distant farm (BHF). Furthermore, all of the British sequences clearly clustered separately from the reference strain sequences (from the United States, France, Germany, or Slovakia). These findings suggest spatial heterogeneity in sequence may be reflected in host range and pathogenicity. Sequencing might be useful in tracing sources of future human outbreaks.
Figure.
Unrooted neighbor-joining tree using the p-distance model (1,000 replicates) for a section of the glycoprotein precursor gene gene, showing bootstrap values of >60 for all sequences identified in this study (283 bp) and indicating site of origin. Captive colony, MF 2004, and BHF 2005 as in Table 1. MF* is from Apodemus sylvaticus, and all other sequences are from Mus musculus. Scale bar indicates number of substitutions per site.
Conclusions
This study has increased the list of European (and North American) rodents that may be infected with LCMV and that might therefore pose a risk to humans. The genetic variation observed and potential variations in pathogenicity may indicate that some wildlife populations pose more of a public health risk than others. Further studies are needed to assess which mutations cause increased pathogenicity and to establish whether or not LCMV represents the only arenavirus present in European rodent populations.
Supplementary Material
Unrooted neighbor-joining radial tree that used the p-distance model (1,000 replicates) for a section of the glycoprotein precursor gene gene, rooted to Lassa virus strain LP. A total of 24 representative sequenced amplicons from wild rodents (283 bp) are shown, with comparisons to previously published lymphocytic choriomeningitis virus (LCMV) sequences and Lassa virus strain LP (GenBank). Scale bar indicates a distance of 0.05 substitutions per site.
Acknowledgments
We thank all those who contributed samples, allowed us access to their animals, and allowed us to trap on their land. Thanks also to Alan Radford for his assistance with sequence analysis.
This research was funded by the European Union grant QLK2-CT-2002-01358, a BBRSC grant to J.H. and studentship to S.D.B.
Biography
Dr Blasdell is based at the Pasteur Institute, Phnom Penh, Cambodia, where she is studying hantaviruses in Southeast Asia. Her research interests are in the ecology and evolution of natural host-virus systems.
Footnotes
Suggested citation for this article: Blasdell KR, Becker SD, Hurst J, Begon M, Bennett M. Host range and genetic diversity of arenaviruses in rodents, United Kingdom. Emerg Infect Dis [serial on the Internet]. 2008 Sep [date cited]. Available from http://www.cdc.gov/EID/content/14/9/1455.htm
References
- 1.Charrel RN, Lemasson JJ, Garbutt M, Khelifa R, De Micco P, Feldmann H, et al. New insights into the evolutionary relationships between arenaviruses provided by comparative analysis of small and large segment sequences. Virology. 2003;317:191–6. 10.1016/j.virol.2003.08.016 [DOI] [PubMed] [Google Scholar]
- 2.Barton LL, Mets MB. Congenital lymphocytic choriomeningitis virus infection: decade of rediscovery. Clin Infect Dis. 2001;33:370–4. 10.1086/321897 [DOI] [PubMed] [Google Scholar]
- 3.Bonthius DJ, Wright R, Tseng B, Barton L, Marco E, Karacay B, et al. Congenital lymphocytic choriomeningitis virus infection: spectrum of disease. Ann Neurol. 2007;62:347–55. 10.1002/ana.21161 [DOI] [PubMed] [Google Scholar]
- 4.Davison KL, Crowcroft NS, Ramsay ME, Brown DW, Andrews NJ. Viral encephalitis in England, 1989–1998: what did we miss? Emerg Infect Dis. 2003;9:234–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salazar-Bravo J, Ruedas LA, Yates TL. Mammalian reservoirs of arenaviruses. Curr Top Microbiol Immunol. 2002;262:25–63. [DOI] [PubMed] [Google Scholar]
- 6.Lledo L, Gegundez MI, Saz JV, Bahamontes N, Beltran M. Lymphocytic choriomeningitis virus infection in a province of Spain: analysis of sera from the general population and wild rodents. J Med Virol. 2003;70:273–5. 10.1002/jmv.10389 [DOI] [PubMed] [Google Scholar]
- 7.El Karamany RM, Imam IZ. Antibodies to lymphocytic choriomeningitis virus in wild rodent sera in Egypt. J Hyg Epidemiol Microbiol Immunol. 1991;35:97–103. [PubMed] [Google Scholar]
- 8.Bowen MD, Peters CJ, Nichol ST. Phylogenetic analysis of the Arenaviridae: patterns of virus evolution and evidence for cospeciation between arenaviruses and their rodent hosts. Mol Phylogenet Evol. 1997;8:301–16. 10.1006/mpev.1997.0436 [DOI] [PubMed] [Google Scholar]
- 9.Becker SD, Bennett M, Stewart JP, Hurst JL. Serological survey of virus infection among wild house mice (Mus domesticus) in the UK. Lab Anim. 2007;41:229–38. 10.1258/002367707780378203 [DOI] [PubMed] [Google Scholar]
- 10.Asper M, Hofmann P, Osmann C, Funk J, Metzger C, Bruns M, et al. First outbreak of callitrichid hepatitis in Germany: genetic characterization of the causative lymphocytic choriomeningitis virus strains. Virology. 2001;284:203–13. 10.1006/viro.2001.0909 [DOI] [PubMed] [Google Scholar]
- 11.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- 12.Greenwood AG, Sanchez S. Serological evidence of murine pathogens in wild grey squirrels (Scuirus carolensis) in North Wales. Vet Rec. 2002;150:543–6. [DOI] [PubMed] [Google Scholar]
- 13.Garcia JB, Morzunov SP, Levis S, Rowe J, Calderon G, Enria D, et al. Genetic diversity of the Junin virus in Argentina: geographic and temporal patterns. Virology. 2000;272:127–36. 10.1006/viro.2000.0345 [DOI] [PubMed] [Google Scholar]
- 14.Bowen MD, Rollin PE, Ksiazek TG, Hustad HL, Bausch DG, Demby AH, et al. Genetic diversity among Lassa virus strains. J Virol. 2000;74:6992–7004. 10.1128/JVI.74.15.6992-7004.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Unrooted neighbor-joining radial tree that used the p-distance model (1,000 replicates) for a section of the glycoprotein precursor gene gene, rooted to Lassa virus strain LP. A total of 24 representative sequenced amplicons from wild rodents (283 bp) are shown, with comparisons to previously published lymphocytic choriomeningitis virus (LCMV) sequences and Lassa virus strain LP (GenBank). Scale bar indicates a distance of 0.05 substitutions per site.