Abstract
Inhibitors of epidermal growth factor receptor (EGFR) tyrosine kinases,such as erlotinib and gefitinib, have not been very effective in the treatment of breast cancer although many breast cancer cells express EGFR. To address this apparent paradox, we examined possible predictors of the sensitivity of 10 breast cancer cell lines to erlotinib in light of cyclin-dependent kinase 2 (CDK2), considered the farthest downstream kinase that controls cell cycling in the EGFR signaling pathway. Expression of EGFR and HER2 were not associated with sensitivity to erlotinib. Expression of phosphorylated (p-)tyrosine, p-Akt, phosphorylated extracellular signal-regulated kinase (p-ERK) 1/ERK2 (p42/p44), and p27 after treatment of erlotinib was not associated with erlotinib sensitivity. However, suppression of CDK2 activity after erlotinib treatment correlated with erlotinib sensitivity (P < 0.0001). Restoration of CDK2 activity partially restored proliferation and induced erlotinib resistance in erlotinib-sensitive cell lines, indicating that sensitivity to erlotinib in these breast cancer cells depends, at least in part, on CDK2 activity. p27, an inhibitor of CDK2, was not translocated into the nucleus in erlotinib-resistant cell lines. Knocking down p27 protein partially blocked erlotinib-induced cell death and cell cycle arrest. These findings indicate that the ability of erlotinib to suppress CDK2 activity is critical for cellular sensitivity to erlotinib, regardless of EGFR expression level, and that the presence of p27 in the cytoplasm also participates in erlotinib resistance.
Introduction
The epidermal growth factor receptor (EGFR) is highly expressed in a variety of solid tumors, including breast cancer. Because activation of EGFR signaling in tumor cells has been linked with decreased apoptosis and increased proliferation, angiogenesis, and metastasis, EGFR is being explored as a potential target for anticancer therapy. Erlotinib HCl (Tarceva; OSI Pharmaceuticals, Inc., and Genentech, Inc.) is an orally available quinazolinamine that competes with ATP for binding with the intracellular catalytic domain of EGFR tyrosine kinase (EGFR-TK) to inhibit the phosphorylation of EGFR-TK. This action blocks downstream signal transduction and inhibits the tumorigenic effects associated with ligand-dependent and ligand-independent EGFR activation (1, 2). In preclinical studies, erlotinib was found to have substantial antitumor activity against various human tumor xenografts (3). However, inhibitors of EGFR-TK have not been particularly effective in women with breast cancer, even if the tumor over-expresses EGFR. Indeed, some reports have indicated that EGFR expression level does not predict sensitivity to EGFR-TK inhibitors (EGFR-TKI; refs. 4-6). Disease in some patients with various types of solid tumors does respond to EGFR-TKIs (6-10); however, factors that might predict responsiveness to EGFR-TKIs have yet to be defined.
Reports that cell lines showing sensitivity to EGFR-TKIs showed G1 arrest after treatment with EGFR-TKIs (11-13) led us to study the potential relationship between cyclin-dependent kinases (CDK), particularly CDK2 (12, 13) and erlotinib sensitivity. CDK2 regulates the G1-S phase transition and, within the EGFR signaling pathway, is the farthest downstream molecule with known kinase activity (13-15). However, whether erlotinib sensitivity is causally linked with CDK2 activity is unknown. Thus, we investigated the involvement of CDK2 in the sensitivity of breast cancer cells to the EGFR-TKI erlotinib. This work provides the first demonstration that sensitivity to erlotinib correlates with and depends on CDK2 activity after erlotinib treatment. We also noted that the presence of p27 in the cytoplasm (as opposed to the nucleus) contributes to erlotinib resistance in breast cancer cells.
Materials and Methods
Cell Lines, Chemicals, and Viruses
We used one human epidermoid carcinoma cell line (A-431) and 10 breast cancer cell lines obtained either from The University of Texas M. D. Anderson Cancer Center Breast Cancer Translational Research Core Laboratory cell line depository (MDA-MB-231, MDA-MB-361, MDA-MB-435, MDA-MB-453, and MDA-MB-468) or from the American Type Culture Collection (A-431, SK-BR-3, BT-20, BT-474, T-47D, and MCF-7). We used A-431 because this cell line is sensitive to EGFR-TKIs through their suppression of EGFR signaling (16). The EGFR-TKI erlotinib was kindly provided by OSI Pharmaceuticals. A stock solution of erlotinib (5 mmol/L) was prepared in DMSO and stored in aliquots at -20°C as described previously (17). The recombinant adenovirus containing cDNA for wild-type CDK2 (Ad.WT-CDK2) was kindly provided by Dr. Joseph R. Nevins (Duke University Medical Center, Durham, NC; ref. 18). Recombinant adenovirus containing cDNA for a dominant-negative (DN) CDK2 construct was created as described elsewhere (19). As a control vector, we used Ad.mock, as described previously (20).
The efficiency of adenoviral gene transfer was evaluated by combining cells with a recombinant adenovirus containing the gene for the expression of green fluorescent protein (Ad.GFP) at multiplicities of infection (MOI) of 1, 3, 10, 30, 100, or 300 and counting the green fluorescent protein-positive cells 48 h later. The optimal MOI was chosen as that which resulted in the highest transfection efficiency with the least adenoviral toxicity.
Western Blotting and Antibodies
Western blotting was done as described previously (17). Immunoblot analyses or immunoprecipitations were done with the following antibodies: mouse anti-EGFR (Ab-12 and Ab-13, LabVision); anti-c-ErbB2/c-Neu antibody (Ab-3, Calbiochem); mouse anti-p-Akt (Ser473), rabbit anti-Akt, and rabbit anti-p-ERK (all from Cell Signaling Technology); mouse anti-phosphotyrosine antibody 4G10 (Upstate Cell Signaling Solutions); mouse anti-CDK2 (sc-6248), rabbit anti-CDK2 (sc-748), rabbit anti-ERK (sc-94), rabbit anti-p27 (sc-776), and rabbit anti-EGFR (sc-03) antibodies (all from Santa Cruz Biotechnology); and mouse anti-β-actin antibody (A-5441; Sigma Chemical Co.).
Immunoprecipitation of EGFR
Immunoprecipitation of EGFR was done as described in our previous report (17). Cells were plated in 100-mm plates, incubated overnight, washed twice with serum-free medium, and then incubated in serum-free medium for 24 h. At that time, the serum-free medium was replaced with medium containing 0.05% fetal bovine serum, and cells were treated (or not treated) with 10 μmol/L erlotinib for 3 h. Cells were then stimulated for 15 min with 100 nmol/L EGF (Upstate Cell Signaling Solutions), washed twice with PBS, collected by scraping, and lysed in lysis buffer [20 mmol/L HEPES (pH 7.9), 10 mmol/L KCl, 2 mmol/L MgCl2, 0.5% NP40, 1 mmol/L sodium fluoride, 1 mmol/L sodium orthovanadate, and 1 μL/mL protease inhibitor cocktail (Sigma)] for 30 min at 4°C. Total protein concentration was determined with a BCA protein assay reagent kit according to the manufacturer’s instructions (Bio-Rad Laboratories). Protein extracts (150 μg per sample for A-431 and MDA-MB-468 cells, 1,000 μg per sample for SK-BR-3 cells) were precleared to decrease nonspecific adsorption to immunoprecipitates by incubating them for 1 h with 15 μL of a protein A-agarose suspension (Calbiochem). The precleared lysates were then subjected to immunoprecipitation with 30 μL of protein A-agarose beads bound to 2 μg of a mouse anti-EGFR antibody (Ab-13). Precipitates were separated by electrophoresis on 6% SDS-polyacrylamide gels, transferred to a polyvinylidene diflouride membrane, and probed with anti-phosphotyrosine antibody (4G10) or anti-EGFR antibody (sc-03).
Cell Viability: 3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide Assay
Assays of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) uptake were done as described previously (21). Briefly, cells were seeded in 96-well plates at a density of 2,000 in 90 μL per well (except for A-431 cells at 200 cells in 90 μL) and cultured overnight, after which 10 μL of erlotinib solution at final concentrations of 0.03, 0.06, 0.1, 0.2, 0.3, 0.6, 1, 2, 3, 6, 10, or 20 μmol/L were added to the individual wells; plates were then incubated for 72 h at 37°C in a humidified atmosphere with 5% CO2. At that time, 10 μL of MTT (Sigma; 5 mg/mL of PBS) was added to each well, and the plates were incubated for 4 h at 37°C. Then, the supernatant was aspirated; 100 μL of DMSO was added into each well; and absorbance was measured at 570 nm. All experiments were done in quadruplicate and SDs were obtained. Results are expressed as percentage of cell viability, calculated as (the absorbance of treated well minus the absorbance of cell-free control) / (absorbance of untreated control minus the absorbance of cell-free control) × 100. IC50 values of erlotinib were determined from the percentage of cell viability measurements.
Cell Viability: Trypan Blue Exclusion Assay
Cells were seeded in 12-well plates at a density of 5,000 per well and cultured overnight. Cells were then incubated for 6, 24, or 72 h of treatment with 0, 1, or 10 μmol/L erlotinib, and total cells and viable cells were counted by trypan blue exclusion as follows. Floating and adherent cells were collected at each time point, sedimented by centrifugation, and resuspended in medium. The cells were then diluted at a 1:9 ratio of 0.4% trypan blue (Sigma) and scored under a light microscope. Viable (unstained) and nonviable (blue-stained) cells were counted, and the total numbers of living and dead cells were calculated.
Clonogenicity Assay
To assess colony-forming ability (an indirect measure of tumorigenicity), cells were seeded in 60-mm culture dishes (Falcon) at a per-dish density of 100 cells for A-431, 200 cells for MDA-MB-231 and MDA-MB-468, or 500 cells for SK-BR-3 and incubated overnight. Cells were then treated with erlotinib at final concentrations of 1 or 10 μmol/L for 72 h. At that time, culture medium (including drug) was removed and the cells were washed twice with PBS and allowed to proliferate in fresh medium for 11 days (for A-431 cells) or for 2 weeks (for the other cell lines). When colonies of ∼50 cells per group had formed, the number of colonies was determined by staining with 1% crystal violet in 20% ethanol. Numbers of colony-forming units in treated cultures were expressed as a percentage of the untreated controls. These experiments were repeated thrice, and SDs were obtained.
Cell Cycle Distribution Analysis
Cell cycle distribution was analyzed by flow cytometry as described in our previous report (17). Briefly, cells were plated in 60-mm dishes, cultured overnight, and then treated or not treated with erlotinib at final concentrations of 1 or 10 μmol/L for 72 h. Floating and adherent cells were then collected by trypsinization, and cells were fixed overnight in 70% ethanol and resuspended in propidium iodide (25 μg/mL) supplemented with 0.1% RNase A and 0.1% Triton X-100. DNA content was measured with a FACScan flow cytometer (Becton Dickinson). These experiments were repeated thrice.
Proliferation Assay
For these experiments, cells were plated in 60-mm dishes, treated or not treated with 1 or 10 μmol/L erlotinib for 72 h, and then incubated with bromodeoxyuridine (BrdUrd; Sigma) for 30 min at 37°C with 5% CO2. Floating and adherent cells were collected by trypsinization, washed with PBS containing 1% bovine serum albumin, fixed overnight in 70% ethanol, stained with anti-BrdUrd-FITC (Becton Dickinson) for 30 min, and then sorted with a FACScan flow cytometer to determine the percentages of proliferating cells.
Immunostaining for p27
To test the subcellular localization of p27, cells were plated on chamber slides and incubated overnight at 37°C, followed by treatment with 0 or 10 μmol/L erlotinib for 72 h, also at 37°C. Cells were then fixed with 100% methanol, blocked in 5% normal goat serum (DakoCytomation), and stained with anti-p27 mouse monoclonal antibody (DakoCytomation) at a 1:200 dilution. Signals were then detected with an FITC anti-mouse antibody (Biosource International) at a 1:250 dilution. Nuclei were stained with ProLong Gold, an antifade reagent with 4′,6-diamidino-2-phenylindole (Invitrogen), and cells were visualized by fluorescence microscopy (ApoTome; Carl Zeiss MicroImaging). The relative intensity of the fluorescence was quantified with the NIH Image program as we described previously (22). Briefly, images are captured with a digital camera and imported into the program, which then converts them to gray scale and binary images to allow the computer to distinguish areas of immunoreactivity from background. The fluorescence intensity of background, total cell area, and nuclear areas is then measured, and background intensity is eliminated through a standardized process. Then, the total cell areas and nuclear areas are selected on an image displayed on the computer monitor, the absolute fluorescence intensity in each cell and nucleus is recorded, and the ratio of nuclear fluorescence intensity to total cell intensity is calculated by Microsoft Excel. After more than 10 cells are examined in this way, the nuclear/total cell intensity ratios for untreated cells are compared with those for cells treated with 10 μmol/L erlotinib. P values of <0.05 were considered statistically significant.
Small Interfering RNA Transfection
To silence p27 gene expression, a single transfection of small interfering RNA (siRNA) duplex was done with Oligofectamine (Invitrogen) according to the manufacturer’s protocol. Functionally validated siRNA/p27 duplex was purchased from Qiagen. For the control experiments, cells were transfected with a siRNA-scrambled duplex (Dharmacon Research). The final siRNA concentration was 5 nmol/L.
CDK2 Assay
CDK2 activity was assayed as described in our previous report (23). Briefly, cells were lysed for 30 min at 4°C in lysis buffer [50 mmol/L Tris-HCl (pH 7.5), 150 mmol/L NaCl, 0.5% NP40, 25 mmol/L sodium fluoride, 200 μmol/L sodium orthovanadate, and 1 μL/mL protease inhibitor cocktail]. Total protein concentrations were determined with a BCA protein assay reagent kit (Bio-Rad Laboratories). Protein extracts (100-250 μg per sample) were precleared by incubation for 1 h with 15 μL of Protein G Plus/Protein A-agarose suspension (Calbiochem). The precleared lysates were then subjected to immunoprecipitation with 30 μL of protein G/protein A-agarose beads bound to 2 μg of anti-CDK2 antibody (Santa Cruz Biotechnology, D-12, sc-6248). After extensive washing, the immunoprecipitates were suspended in 20 μL of kinase buffer [50 mmol/L Tris-HCl (pH 7.5), 10 mmol/L MgCl2, 2.5 mmol/L ethylene glycol bis(b-aminoethylether) tetraacetic acid, 1 mmol/L DTT, 0.1% Triton X-100, 100 μmol/L sodium fluoride, and 100 μmol/L sodium orthovanadate] supplemented with 20 μmol/L unlabeled ATP, 50 μg/mL of histone H1 as substrate, and 2 μCi of [γ-32P]ATP per sample, and incubated for 30 min at 30°C. Reactions were terminated by the addition of 5 μL of 5× sample buffer, and the labeled proteins were resolved by SDS-PAGE and detected by autoradiography. CDK2 activity was determined with the NIH Image program. To measure immunoprecipitated protein levels, the immunoprecipitates were resuspended in 20 μL of sample buffer, separated by SDS-PAGE, and electroblotted onto a polyvinylidene fluoride membrane (Bio-Rad). Protein levels were then analyzed with rabbit polyclonal antibodies to CDK2 (Santa Cruz Biotechnology, H-298). Negative controls for immunoprecipitation were mouse normal IgG (Upstate Cell Signaling Solutions).
Statistical Analysis
Statistical analyses were done with commercially available software (Statview, version 5.0; SAS Institute). We used single regression analysis to assess the ratio of CDK2 activity after treatment with 0 or 10 μmol/L erlotinib, with the percentage cell viability after erlotinib treatment as the dependent variable. Student’s t tests were used to compare the ratio of nuclear intensity/total cell intensity of p27 between cells treated with 0 or 10 μmol/L of erlotinib. Comparisons of Ad.mock versus Ad.WT-CDK2 effects, Ad.mock versus Ad.DN-CDK2 effects, and siRNA/control versus siRNA/p27 were done by two-way ANOVA (when the variant was another cell line) or analysis of covariance (when the covariant was erlotinib dose). Statistical significance was defined as a P value of <0.05.
Results
Sensitivityof Breast Carcinoma Cells to Erlotinib Is Unrelated to EGFR Expression Level
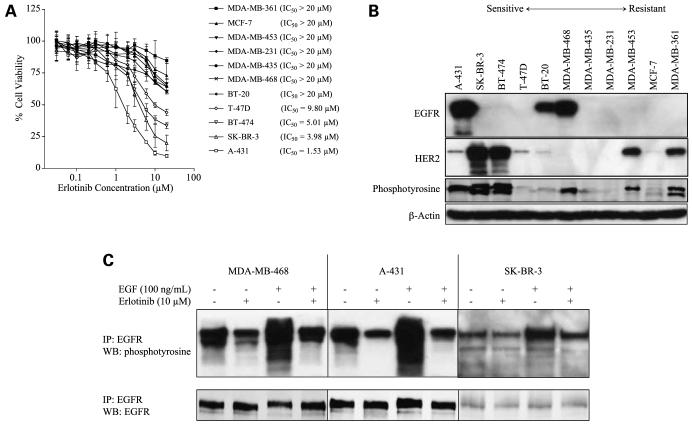
As a first step, 10 breast cancer cell lines expressing various levels of EGFR were tested for their sensitivity to erlotinib in vitro (Fig. 1A; Supplementary Fig. S1A).5 As a positive control, we included the human epidermoid carcinoma cell line A-431, which overexpresses EGFR and is sensitive to erlotinib. MTT assays showed that four cell lines (A-431, SK-BR-3, BT-474, and T-47D) were sensitive to erlotinib (IC50 for A-431 cells, 1.53 μmol/L; for SK-BR-3, 3.98 μmol/L; for BT-474, 5.01 μmol/L; and for T-47D, 9.80 μmol/L). Results of trypan blue exclusion assays after erlotinib treatment were similar to those of the MTT assay (data not shown). We then selected three of the sensitive cell lines (A-431, SK-BR-3, and BT-474) and two of the resistant cell lines (MDA-MB-231, MDA-MB-468) for further experiments. Treatment with erlotinib markedly suppressed clonogenicity in sensitive cell lines but not in resistant cell lines (data not shown). Collectively considering MTT uptake, trypan blue exclusion, and clonogenicity, we defined cellular sensitivity or resistance according to the IC50 from the MTT assay.
Figure 1.
Erlotinib sensitivity is not associated with the expression levels of EGFR and HER2. A, dose-response curves of A-431 cells and 10 breast cancer cell lines after treatment with erlotinib at final concentrations of 0.03, 0.06, 0.1, 0.2, 0.3, 0.6, 1, 2, 3, 6, 10, or 20 μmol/L for 72 h. The surviving fractions were determined by MTT assay. Points, mean percentage cell growth among six wells; bars, SD. B, Western blotting of all 11 cell lines was done by loading equal amounts of protein (50 μg/lane) from untreated cells, followed by SDS-PAGE and immunoblotting. μ-Actin was used as a loading control. C, erlotinib inhibits EGFR phosphorylation in both erlotinib-sensitive and erlotinib-resistant cell lines. Cells were treated with 0 or 10 μmol/L erlotinib and then stimulated (or not) with 100 nmol/L EGF. After cell lysis, the EGFRs were precipitated with specific antibodies and the precipitates were immunoblotted with p-tyrosine antibody. IP, immunoprecipitation; WB, Western blot. Erlotinib blocked EGF signals in each cell line. Precipitation with EGFR antibody followed by detection with EGFR antibody verified that equal amounts of EGFR had been precipitated.
Next, we tested whether erlotinib sensitivity depends on basal expression level of EGFR. With regard to the sensitive cell lines, basal EGFR expression was confirmed in A-431 cells and, when 100 μg (rather than 50 μg) protein was loaded and a different antibody was used, in SK-BR-3 and BT-474 cells (Supplementary Fig. S1B).5 Of the resistant cell lines, BT-20 and MDA-MB-468 cells expressed EGFR (Fig. 1B). Hence, no association was found between EGFR expression levels and erlotinib sensitivity, which suggests that high levels of EGFR are not required to render cells sensitive to erlotinib. Further, neither HER2 nor p-tyrosine expression levels correlated with erlotinib sensitivity (Fig. 1B).
Erlotinib Inhibits the Tyrosine Phosphorylation of EGFR in Both Sensitive and Resistant Cell Lines
Although EGFR was expressed in four of the breast cancer cell lines tested, some of those cell lines were resistant and some were sensitive to erlotinib. Our next step was to confirm that erlotinib was targeting the EGFR by examining whether erlotinib could inhibit the tyrosine phosphorylation of the EGFR. Immunoprecipitation of EGFR followed by Western blotting with anti-phosphotyrosine antibodies revealed EGFR phosphorylation in the A-431, MDA-MB-468, and SK-BR-3 cells (Fig. 1C). In all three of these cell lines, treatment with 10 μmol/L erlotinib blocked EGFR phosphorylation, and stimulation with EGF induced abundant EGFR phosphorylation (Fig. 1C). Furthermore, treatment with 10 μmol/L erlotinib before stimulation with EGF abrogated EGFR phosphorylation in both the erlotinib-sensitive and erlotinib-resistant cell lines (Fig. 1C). These results confirm that erlotinib blocks EGFR signaling in both erlotinib-sensitive and erlotinib-resistant cell lines.
Erlotinib-Sensitive Cell Lines Show G1 Arrest upon Treatment with Erlotinib
We then examined the effect of erlotinib on cell cycle distribution in both erlotinib-sensitive and erlotinib-resistant cell lines. All three erlotinib-sensitive cell lines (A-431, SK-BR-3, and BT-474) exhibited G1 arrest after 24 h of treatment with 10 μmol/L erlotinib (not shown). However, apoptosis (denoted by an increased proportion of cells in sub-G1) was observed in only two of these cell lines after a 72-h treatment with 10 μmol/L erlotinib. Specifically, the percentage of A-431 cells in sub-G1 was 26% and that of SK-BR-3 cells was 10.6%; however, only 1.5% BT-474 cells were in sub-G1. In contrast, the erlotinib-resistant MDA-MB-231 and MDA-MB-468 cells showed no change in cell cycle distribution after erlotinib treatment. We also investigated the effect of erlotinib on S-phase populations, as determined by BrdUrd positivity. The erlotinib-sensitive cell lines all showed decreased proportions of cells in S phase after 72 h of treatment with 10 μmol/L erlotinib, but the erlotinib-resistant cell lines showed no obvious change in the S-phase populations (not shown).
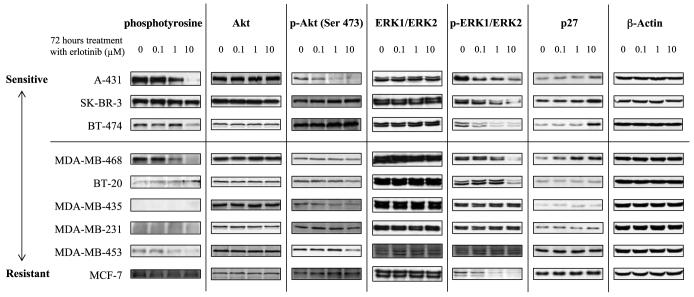
Akt, p-Akt, ERK, p-ERK, and p27 Levels After Erlotinib Treatment Are Not Associated with Sensitivity to Erlotinib
Next, we looked at the effects of erlotinib on the expression of downstream molecules in the EGFR and HER2 signaling pathways (Fig. 2). All three of the erlotinib-sensitive cell lines showed down-regulation of p-ERK1/ERK2 and up-regulation of p27 after a 72-h exposure to erlotinib. However, p-ERK1/ERK2, with p-Akt, was also down-regulated in some resistant cell lines, and p27 was also up-regulated in the resistant MDA-MB-468 cells. Similar results were obtained from time course studies of treatment with 10 μmol/L erlotinib for 6, 12, 24, or 72 h (Supplementary Fig. S2).5 Thus, no association was found between sensitivity to erlotinib and expression levels of Akt/p-Akt, ERK/p-ERK, or p27 in these cells.
Figure 2.
Protein expression levels of molecules downstream of EGFR signaling pathways are not associated with erlotinib sensitivity. Levels of downstream molecules in EGFR signaling pathways were screened by Western blotting after treatment with 0, 0.1, 1, or 10 μmol/L erlotinib. β-Actin was used as a loading control.
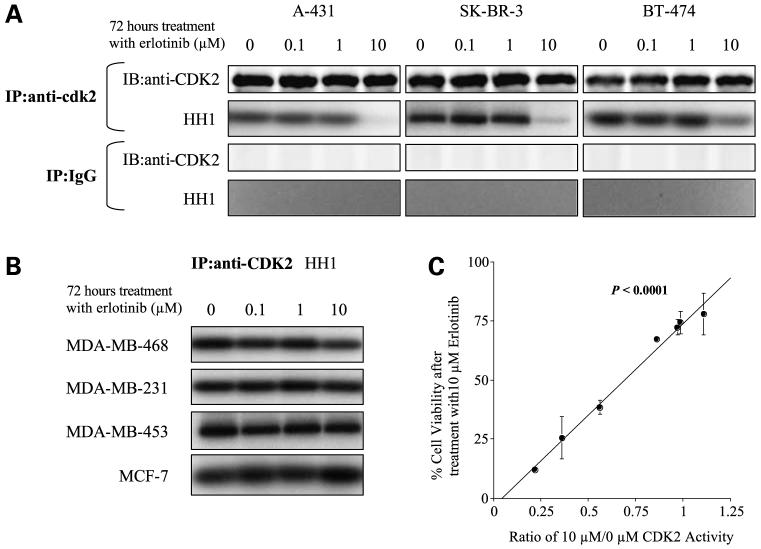
CDK2 ActivityAfter Erlotinib Treatment Correlates with Sensitivity to Erlotinib
Because the erlotinib-sensitive cell lines showed G1 arrest upon treatment with erlotinib, we next examined the activity of CDK1, CDK2, CDK4, CDK6, and some other cell cycle-related proteins, all critical regulators of the cell cycle transition, in response to erlotinib treatment. Upon finding that CDK2 activity, but not that of CDK1, CDK4, CDK6, or other molecules (Supplementary Fig. S3A and SB),5 was associated with erlotinib sensitivity, we narrowed our focus to CDK2 activity in the context of erlotinib sensitivity. Indeed, the three erlotinib-sensitive cell lines showed marked decreases in CDK2 activity after exposure to 10 μmol/L erlotinib (Fig. 3A), whereas CDK2 activity did not change in the erlotinib-resistant cell lines (Fig. 3B). Further, the extent of CDK2 activity after erlotinib treatment (determined by the NIH Image program) correlated strongly with cell viability after erlotinib (determined by MTT assay; P < 0.0001; Fig. 3C).
Figure 3.
CDK2 activity is correlated with erlotinib sensitivity. A, erlotinib-sensitive cells were treated with 0, 0.1, 1, or 10 μmol/L erlotinib for 72 h, followed by measurement of CDK2 activity and immunoprecipitated (IP) CDK2 levels. Mouse normal IgG was used to show the specificity of CDK2 antibody. HH1, histone H1. B, erlotinib-resistant cells were treated with 0, 0.1, 1, or 10 μmol/L erlotinib for 72 h, followed by measurement of CDK2 activity. C, kinase activity (determined with NIH Image), expressed as ratios of CDK2 activity at 10 μmol/L erlotinib versus that at 0 μmol/L erlotinib, is associated with cell viability (determined by MTT assay).
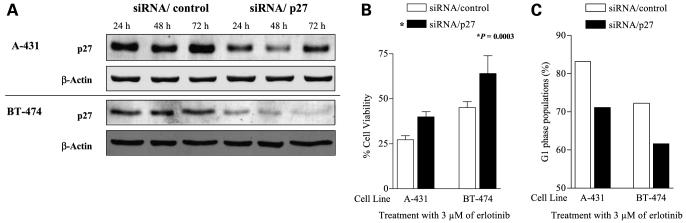
Knocking Down p27 Reduces Sensitivity (Increases Resistance) to Erlotinib
Given that CDK2 correlated with erlotinib sensitivity but expression of p27, an upstream target in the EGFR signaling pathway and a CDK2 inhibitor, did not, we next investigated the effect of knocking down p27 in erlotinib-sensitive cell lines. First, we confirmed that p27 protein expression was knocked down by siRNA/p27 in cells not treated with erlotinib (Fig. 4A) and in cells treated with erlotinib (Supplementary Fig. S4A).5 Then, we checked erlotinib sensitivity and cell cycle distribution after erlotinib treatment in cells with siRNA/p27. Knocking down the p27 protein reduced cell death induced by 3 μmol/L erlotinib [P = 0.003 by MTT assay (Fig. 4B) and P = 0.007 by trypan blue exclusion assay (Supplementary Fig. S4B)].5 Knocking down p27 protein also reduced proportion of G1 arrest induced by erlotinib (Fig. 4C; Supplementary Fig. S4C).5 These results indicate that sensitivity to erlotinib is partially dependent on p27.
Figure 4.
Knocking down p27 protein reduces erlotinib sensitivity (enhances erlotinib resistance) in erlotinib-sensitive cell lines. A, cells (2 × 105) were transiently transfected with siRNA/p27 or siRNA/control for 24, 48, or 72 h, followed by harvest and Western blotting for p27. β-Actin was used as a loading control. B, cells were transfected with siRNA for 24 h, treated with 3 μmol/L erlotinib, then tested for viability with an MTT assay 48 h later. siRNA to p27 significantly increased the viability of the sensitive cells (P = 0.0003). C, cells were transfected with siRNA for 24 h, treated with 3 μmol/L erlotinib, and harvested for cell cycle analysis 48 h later. Columns, percentages of cells in G1.
Cytoplasmic Localization of p27 Is Associated with Erlotinib Resistance
In erlotinib-resistant MDA-MB-468 breast cancer cells, the CDK2 inhibitor p27 was up-regulated in a dose-dependent and time-dependent manner after erlotinib treatment, but no G1 arrest was observed. Thus, we focused on whether the cellular localization of p27 (assessed in terms of nuclear fluorescence intensity/total cell intensity) was related to erlotinib sensitivity. In the three erlotinib-sensitive cell lines, p27 translocated from the cytoplasm into nucleus after treatment with 10 μmol/L of erlotinib (P < 0.0001 for each of the three cell lines; not shown); however, in the two erlotinib-resistant cell lines, p27 did not translocate after erlotinib treatment (MDA-MB-468, P = 0.1305; MDA-MB-231, P = 0.5274; not shown). These results indicate that the presence of p27 in the cytoplasm after treatment with erlotinib may be associated with erlotinib resistance.
Restoring CDK2 Activity by Transfection with Wild-type CDK2 Partially Restores Proliferation After Erlotinib Treatment of Sensitive Cell Lines
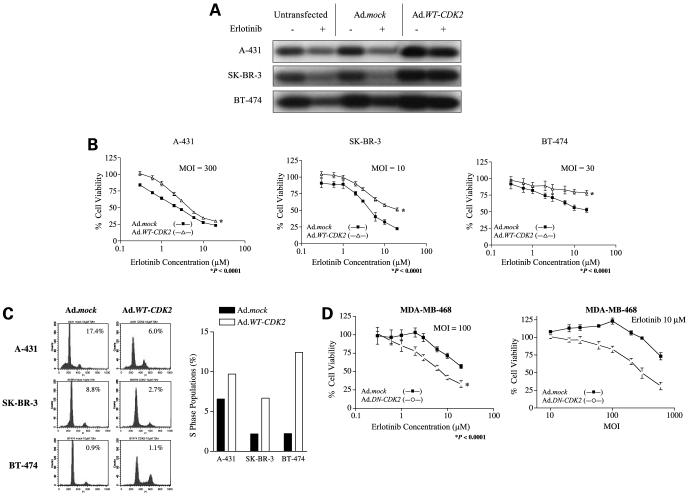
Because p27 is capable to inhibit CDK2 activity, we examined the relationship between CDK2 activity and erlotinib sensitivity by testing the erlotinib sensitivity by restoring CDK2 activity in sensitive and resistant cells by transfecting them with Ad.WT-CDK2, which encodes wild-type CDK2. Transfection of the erlotinib-sensitive A-431, SK-BR-3, and BT-474 cells with Ad.WT-CDK2 at various MOI revealed a dose-dependent effect on CDK2 activity (Supplementary Fig. S5A).5 Next, we tested the effect of Ad.WT-CDK2 in combination with erlotinib in these cells and found that CDK2 activity was restored in all three cell lines (Fig. 5A). Restoration of CDK2 activity was associated with partial restoration of cell viability (Fig. 5B) in the same three erlotinib-sensitive cell lines after treatment with erlotinib. In terms of viability, restoration of CDK2 activity raised the IC50 of A-431 cells to 4.63 μmol/L; the IC50 values for both SK-BR-3 cells and BT-474 cells was >20.00 μmol/L in all three cell lines (P < 0.0001; Fig. 5B; Supplementary Fig. S5B).5 Further, it was dependent on the amount of Ad.WT-CDK2 transfected (Supplementary Fig. S5C).5 Restoration of CDK2 activity also partially restored cell proliferation, as indicated by its blocking erlotinib-induced G1 arrest and apoptosis and its increasing the S-phase population in all three sensitive cell lines (Fig. 5C).
Figure 5.
Sensitivity to erlotinib depends on CDK2 activity. Cells were transfected with Ad.mock, Ad.WT-CDK2, or Ad.DN-CDK2 for 6 h; treated with erlotinib for 72 h; and tested for CDK2 activity (A), viability (B and D), and cell cycle distribution (C). A, CDK2 activity is restored by Ad.WT-CDK2. A-431 cells were transfected with Ad.WT-CDK2 at 300 MOI and treated with 3 μmol/L erlotinib; SK-BR-3 cells were transfected with 10 MOI and treated with 5 μmol/L erlotinib; and BT-474 cells were transfected with 30 MOI and treated with 5 μmol/L erlotinib. Erlotinib concentrations were the IC50 values for those cell lines as determined by MTT assay. B, cell viability (by MTT assay) in A431, SK-BR-3, and BT-474 cells in response to erlotinib is protected dose dependently by Ad.WT-CDK2 transfection. Cells were transfected with the indicated concentrations of Ad.mock or Ad.WT-CDK2 and incubated with the indicated erlotinib concentrations for 72 h. C, Ad.WT-CDK2 blocked G1 arrest and apoptosis and increased the proportions of cells in S phase in response to erlotinib. A-431, SK-BR-3, and BT-474 cells were transfected with adenovirus at the indicated MOI and treated with 10 μmol/L erlotinib for 72 h; cell cycle distributions were analyzed by flow cytometry and presented as percentages of apoptotic cells (as determined by sub-G1 peaks) and proportions of cells in S phase. PI, propidium iodide. D, sensitivity of erlotinib-resistant MDA-MD-468 cells in response to erlotinib is restored in a dose-dependent manner by transfection with a DN form of WT-CDK2.
Suppressing CDK2 Activity by Transfecting Cells with DN-CDK2 Partially Increases Erlotinib Sensitivity
Finally, to confirm whether erlotinib resistance also depends on CDK2 activity, we transfected the erlotinib-resistant cell line MDA-MB-468 with DN-CDK2 (23). Suppression of CDK2 activity through transfection with Ad.DN-CDK2 increased the sensitivity of MDA-MB-468 cells to erlotinib (P < 0.0001), and this effect was dependent on the amount of Ad.DN-CDK2 transfected (Fig. 5D). However, this effect was not observed in MDA-MB-453, MDA-MB-361, and MCF-7 (Supplementary Fig. S5D).5
Discussion
The molecular mechanisms underlying erlotinib resistance in breast cancer have not been well defined. In our screening of breast cancer cell lines, down-regulation of CDK2 after treatment with erlotinib showed association with erlotinib sensitivity. Moreover, our study provides that erlotinib sensitivity is causally linked with CDK2 activity, indicating that erlotinib sensitivity depends, at least in part, on CDK2 activity. Some reports have indicated that the growth-inhibitory effect induced by EGFR-TKIs in sensitive cell lines depends mainly on G1 cell cycle arrest (1, 7, 11, 13); others have shown CDK2 activity to be down-regulated after treatment with an EGFR-TKI (12, 13). However, this is the first report establishing cause and effect relationship between erlotinib sensitivity and CDK2 activity.
The erlotinib sensitivity of breast cancer cell lines has clinical relevance for several reasons. The limited clinical activity of EGFR-TKIs in breast cancer was also echoed by the findings of this in vitro study. Because EGFR-TKIs were designed to block EGFR-TK phosphorylation, EGFR expression was previously thought to be a molecular marker of the effectiveness of EGFR-TKIs. However, in vitro and in vivo preclinical studies (4, 8, 11, 24-26) as well as phase I and II clinical studies (6, 10) suggest that the efficacy of EGFR-TKIs is not directly related to EGFR expression levels. Our own findings indicated a lack of association between the expression of EGFR and p-EGFR and sensitivity to erlotinib, although we did find that erlotinib blocked EGFR phosphorylation even in erlotinib-resistant cell lines.
Other evidence suggests that HER2 overexpression indicates sensitivity to EGFR-TKIs (5, 24, 25). Although one recent article reported that erlotinib directly blocked HER2 kinase and downstream signaling events in cells that did not express EGFR (27), our Western blotting results indicated that HER2 expression level (high in SK-BR-3, BT-474, MDA-MB-453, and MDA-MB-361 cells) was not associated with erlotinib sensitivity. Further, HER2 expression level in SK-BR-3 and BT-474 cells, noted to be sensitive to erlotinib, did not change after erlotinib treatment (data not shown).
Some investigators have investigated the downstream signaling pathways of EGFR and have suggested that down-regulated activity of ERK1/ERK2, mitogen-activated protein kinase, and phosphatidylinositol 3′-kinase/Akt after treatment with EGFR-TKIs might serve as a marker of drug response (6, 9-11, 24, 25, 28). However, firm conclusions about which of these two pathways (if either) is relevant for measuring the efficacy of EGFR-TKIs have not been drawn. Some studies have found down-regulation of the ERK1/ERK2 mitogen-activated protein kinase signal in both EGFR-TKI-sensitive and EGFR-TKI-resistant cancer cell lines, suggesting that ERK1/ERK2 mitogen-activated protein kinase activation is not a reliable marker of EGFR-TKI -induced inhibition of proliferation (5, 8, 11, 29), and some showed that Akt activity could not predict erlotinib sensitivity (5, 10, 29). In our study, no association was found between erlotinib sensitivity and the expression of Akt, p-Akt, ERK1/ERK2, or p-ERK1/ERK2, as detected by Western blotting, after a 72-h exposure to erlotinib. Although molecular defects in the EGFR signaling molecules of the Akt and ERK1/ERK2 mitogen-activated protein kinase pathway cannot be excluded, more complex mechanisms probably underlie the effect of EGFR-TKIs (5).
The CDK inhibitor p27 is a potent negative regulator of the cell cycle involved in both EGFR and non-EGFR signaling pathways, and it is directly upstream of CDK2. p27 inactivation, by down-regulation of its expression or its exclusion from the nucleus, has been implicated in human carcinogenesis (30, 31). Cytoplasmic sequestration of p27 has been reported in breast carcinomas (32-34), and loss of p27 function has been proposed as a marker of malignancy (30, 31). Previous reports have suggested that p27 could be a predictive marker of response to EGFR-TKIs (10, 12, 13). In our study, the erlotinib-sensitive cell lines showed translocation of p27 from the cytoplasm to the nucleus, but the erlotinib-resistant cell lines did not, despite increased p27 expression levels. Our finding that knocking down p27 protein only partially blocked the effects of erlotinib may reflect insufficient transfection efficiency or the participation of other molecules in the erlotinib effect. However, our results do indicate that p27 is an important factor in how erlotinib affects cells, although erlotinib-induced up-regulation of p27 by itself is insufficient to block CDK2 activity.
This is also the first report that p27 cytoplasmic localization is related to resistance to EGFR-TKIs. In other words, “mislocalization” of p27, and the subsequent continuous activation of CDK2, may be an important mechanism of resistance to EGFR-TKIs. Cells in which the Akt pathway was activated subsequent to PTEN deficiency were recently reported to be naturally resistant to the effect of EGFR-TKIs (35-37). In addition, several mechanisms of p27 mislocalization have been proposed, some of which involve its phosphorylation at various sites. For example, phosphorylation of serine 10 by the hKIS kinase promotes the export of p27 from the nucleus (38, 39), whereas phosphorylation of threonine 157 by Akt impairs the import of p27 into the nucleus (32-34). However, future studies will be necessary to identify the phosphorylation site of p27 that is related to its translocation after erlotinib treatment. We can also speculate that phosphorylation status of p27 may have affected the overall efficacy of erlotinib even among the erlotinib-sensitive cell lines.
We found that blocking CDK2 activity with DN-CDK2 led to increased sensitivity to erlotinib in the EGFR-overexpressing cell line MDA-MB-468. This finding supports the concept that CDK2 is a target of erlotinib, at least in some cell lines. However, DN-CDK2 had no effect on other three erlotinib-resistant cell lines (MDA-MB-361, MDA-MB-453, and MCF-7; Supplementary Fig. S5D).5 EGFR expression has been reported to be necessary, if not sufficient, for cell lines to be sensitive to EGFR-TKIs (11). The mechanisms by which DN-CDK2 affected MDA-MB-468 cells but not other cell lines are unclear; however, one possible explanation may be that MDA-MB-468 overexpressed EGFR but MDA-MB-361, MDA-MB-453, and MCF-7 cells did not (as shown by our Western blotting results). Another possibility is that CDK2 down-regulation requires other molecular effects such as p27 up-regulation, p-ERK1/ERK2 down-regulation, as seen in MDA-MB-468 cells, to increase the sensitivity of erlotinib-resistant cell lines. Alternatively, EGFR signaling pathways may be more intact in MDA-MB-468 cells than in other cell lines, as suggested by our findings that p-ERK1/ERK2 were down-regulated and p27 was up-regulated in MDA-MB-468 cells but not in other cell lines. We found that erlotinib blocked EGFR phosphorylation in both EGFR-expressing sensitive cell lines and EGFR-expressing resistant cell lines (Fig. 1A). In the EGFR-expressing erlotinib-sensitive cell lines, CDK2 was suppressed, perhaps subsequent to the movement of p27 into the nucleus; however, CDK2 was not suppressed, and p27 did not move into the nucleus, in EGFR-expressing resistant cell lines. These results indicate that suppression of EGFR, by itself, was not sufficient for cells to be sensitive to erlotinib. In other words, both blockade of EGFR phosphorylation and down-regulation of CDK2 are required for cells to be sensitive to erlotinib. Our findings suggest that targeting CDK2 in EGFR-overexpressing erlotinib-resistant cells could be fruitful. Future studies are necessary to clarify the role of CDK2 and that of molecular effects upstream of CDK2 by erlotinib treatment.
In summary, our results showed that erlotinib did have some effect on breast cancer cells independent of the level of EGFR and HER2 expression; that suppression of CDK2 activity correlated with erlotinib activity regardless of the upstream molecular effects; and that p27 subcellular localization is critical for erlotinib sensitivity.
Supplementary Material
Acknowledgments
We thank Christine F. Wogan, MS, ELS, and Karen F. Phillips, ELS, of the Department of Scientific Publications at M. D. Anderson Cancer Center for their editorial assistance; Tomokazu Yoshida for screening CDK activities; Caimiao Wei and Hiroaki Itamochi for their assistance with statistical analyses; Bill Spohn for his assistance with the Apoptome microscope; Wendy Schober for assistance with flow cytometry; Geoffrey Bartholomeusz for helpful and constructive discussions; and Shalini Das, Mitsuyo Ota, Yoshihiro Kiura, and Yoshinori Kajiwara for their support of this research.
Grant support: Nellie B. Connally Breast Cancer Research Fund and NIH Grant R01 CA123318-01A1.
Footnotes
Supplementary material for this article is available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
References
- 1.Moyer JD, Barbacci EG, Iwata KK, et al. Induction of apoptosis and cell cycle arrest by CP-358,774, an inhibitor of epidermal growth factor receptor tyrosine kinase. Cancer Res. 1997;57:4838–48. [PubMed] [Google Scholar]
- 2.Pollack VA, Savage DM, Baker DA, et al. Inhibition of epidermal growth factor receptor-associated tyrosine phosphorylation in human carcinomas with CP-358,774: dynamics of receptor inhibition in situ and antitumor effects in athymic mice. J Pharmacol Exp Ther. 1999;291:739–48. [PubMed] [Google Scholar]
- 3.Higgins B, Kolinsky K, Smith M, et al. Antitumor activity of erlotinib (OSI-774, Tarceva) alone or in combination in human non-small cell lung cancer tumor xenograft models. Anticancer Drugs. 2004;15:503–12. doi: 10.1097/01.cad.0000127664.66472.60. [DOI] [PubMed] [Google Scholar]
- 4.Sirotnak FM, Zakowski MF, Miller VA, Scher HI, Kris MG. Efficacy of cytotoxic agents against human tumor xenografts is markedly enhanced by coadministration of ZD1839 (Iressa), an inhibitor of EGFR tyrosine kinase. Clin Cancer Res. 2000;6:4885–92. [PubMed] [Google Scholar]
- 5.Campiglio M, Locatelli A, Olgiati C, et al. Inhibition of proliferation and induction of apoptosis in breast cancer cells by the epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor ZD1839 (“Iressa”) is independent of EGFR expression level. J Cell Physiol. 2004;198:259–68. doi: 10.1002/jcp.10411. [DOI] [PubMed] [Google Scholar]
- 6.Baselga J, Albanell J, Ruiz A, et al. Phase II and tumor pharmacodynamic study of gefitinib in patients with advanced breast cancer. J Clin Oncol. 2005;23:5323–33. doi: 10.1200/JCO.2005.08.326. [DOI] [PubMed] [Google Scholar]
- 7.Herbst RS, Fukuoka M, Baselga J. Gefitinib—a novel targeted approach to treating cancer. Nat Rev Cancer. 2004;4:956–65. doi: 10.1038/nrc1506. [DOI] [PubMed] [Google Scholar]
- 8.Brehmer D, Greff Z, Godl K, et al. Cellular targets of gefitinib. Cancer Res. 2005;65:379–82. [PubMed] [Google Scholar]
- 9.Baselga J, Rischin D, Ranson M, et al. Phase I safety, pharmacokinetic, and pharmacodynamic trial of ZD1839, a selective oral epidermal growth factor receptor tyrosine kinase inhibitor, in patients with five selected solid tumor types. J Clin Oncol. 2002;20:4292–302. doi: 10.1200/JCO.2002.03.100. [DOI] [PubMed] [Google Scholar]
- 10.Daneshmand M, Parolin DA, Hirte HW, et al. A pharmacodynamic study of the epidermal growth factor receptor tyrosine kinase inhibitor ZD1839 in metastatic colorectal cancer patients. Clin Cancer Res. 2003;9:2457–64. [PubMed] [Google Scholar]
- 11.Bishop PC, Myers T, Robey R, et al. Differential sensitivity of cancer cells to inhibitors of the epidermal growth factor receptor family. Oncogene. 2002;21:119–27. doi: 10.1038/sj.onc.1205028. [DOI] [PubMed] [Google Scholar]
- 12.Chang GC, Hsu SL, Tsai JR, et al. Molecular mechanisms of ZD1839-induced G1-cell cycle arrest and apoptosis in human lung adenocarcinoma A549 cells. Biochem Pharmacol. 2004;68:1453–64. doi: 10.1016/j.bcp.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Di Gennaro E, Barbarino M, Bruzzese F, et al. Critical role of both p27KIP1 and p21CIP1/WAF1 in the antiproliferative effect of ZD1839 (“Iressa”), an epidermal growth factor receptor tyrosine kinase inhibitor, in head and neck squamous carcinoma cells. J Cell Physiol. 2003;195:139–50. doi: 10.1002/jcp.10239. [DOI] [PubMed] [Google Scholar]
- 14.Wilkinson MG, Millar JB. Control of the eukaryotic cell cycle by MAP kinase signaling pathways. FASEB J. 2000;14:2147–57. doi: 10.1096/fj.00-0102rev. [DOI] [PubMed] [Google Scholar]
- 15.Fan Z, Lu Y, Wu X, DeBlasio A, Koff A, Mendelsohn J. Prolonged induction of p21Cip1/WAF1/CDK2/PCNA complex by epidermal growth factor receptor activation mediates ligand-induced A431 cell growth inhibition. J Cell Biol. 1995;131:235–42. doi: 10.1083/jcb.131.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wakeling AE, Guy SP, Woodburn JR, et al. ZD1839 (Iressa): an orally active inhibitor of epidermal growth factor signaling with potential for cancer therapy. Cancer Res. 2002;62:5749–54. [PubMed] [Google Scholar]
- 17.Yamasaki F, Johansen MJ, Zhang D, et al. Acquired resistance to erlotinib in A-431 epidermoid cancer cells requires downregulation of mutated in multiple advanced cancers 1/phosphatase and tensin homoslogue and up-regulation of phosphorylated Akt. Cancer Res. 2007;67:5779–88. doi: 10.1158/0008-5472.CAN-06-3020. [DOI] [PubMed] [Google Scholar]
- 18.Leone G, DeGregori J, Jakoi L, Cook JG, Nevins JR. Collaborative role of E2F transcriptional activity and G1 cyclin dependent kinase activity in the induction of S phase. Proc Natl Acad Sci U S A. 1999;96:6626–31. doi: 10.1073/pnas.96.12.6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adachi S, Ito H, Tamamori-Adachi M, et al. Cyclin A/cdk2 activation is involved in hypoxia-induced apoptosis in cardiomyocytes. Circ Res. 2001;88:408–14. doi: 10.1161/01.res.88.4.408. [DOI] [PubMed] [Google Scholar]
- 20.Jones N, Shenk T. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell. 1979;17:683–9. doi: 10.1016/0092-8674(79)90275-7. [DOI] [PubMed] [Google Scholar]
- 21.Yamasaki F, Kajiwara Y, Hama S, et al. Retinoblastoma protein prevents staurosporine-induced cell death in a retinoblastoma-defective human glioma cell line. Pathobiology. 2007;74:22–31. doi: 10.1159/000101048. [DOI] [PubMed] [Google Scholar]
- 22.Bartholomeusz C, Itamochi H, Nitta M, Saya H, Ginsberg MH, Ueno NT. Antitumor effect of E1A in ovarian cancer by cytoplasmic sequestration of activated ERK by PEA15. Oncogene. 2006;25:79–90. doi: 10.1038/sj.onc.1209014. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi T, Yamasaki F, Sudo T, et al. Cyclin A-associated kinase activity is needed for paclitaxel sensitivity. Mol Cancer Ther. 2005;4:1039–46. doi: 10.1158/1535-7163.MCT-04-0282. [DOI] [PubMed] [Google Scholar]
- 24.Moasser MM, Basso A, Averbuch SD, Rosen N. The tyrosine kinase inhibitor ZD1839 (“Iressa”) inhibits HER2-driven signaling and suppresses the growth of HER2-overexpressing tumor cells. Cancer Res. 2001;61:7184–8. [PubMed] [Google Scholar]
- 25.Moulder SL, Yakes FM, Muthuswamy SK, Bianco R, Simpson JF, Arteaga CL. Epidermal growth factor receptor (HER1) tyrosine kinase inhibitor ZD1839 (Iressa) inhibits HER2/neu (erbB2)-overexpressing breast cancer cells in vitro and in vivo. Cancer Res. 2001;61:8887–95. [PubMed] [Google Scholar]
- 26.Ciardiello F, Tortora G. A novel approach in the treatment of cancer: targeting the epidermal growth factor receptor. Clin Cancer Res. 2001;7:2958–70. [PubMed] [Google Scholar]
- 27.Schaefer G, Shao L, Totpal K, Akita RW. Erlotinib directly inhibits HER2 kinase activation and downstream signaling events in intact cells lacking epidermal growth factor receptor expression. Cancer Res. 2007;67:1228–38. doi: 10.1158/0008-5472.CAN-06-3493. [DOI] [PubMed] [Google Scholar]
- 28.Albanell J, Rojo F, Averbuch S, et al. Pharmacodynamic studies of the epidermal growth factor receptor inhibitor ZD1839 in skin from cancer patients: histopathologic and molecular consequences of receptor inhibition. J Clin Oncol. 2002;20:110–24. doi: 10.1200/JCO.2002.20.1.110. [DOI] [PubMed] [Google Scholar]
- 29.Tan AR, Yang X, Hewitt SM, et al. Evaluation of biologic end points and pharmacokinetics in patients with metastatic breast cancer after treatment with erlotinib, an epidermal growth factor receptor tyrosine kinase inhibitor. J Clin Oncol. 2004;22:3080–90. doi: 10.1200/JCO.2004.08.189. [DOI] [PubMed] [Google Scholar]
- 30.Slingerland J, Pagano M. Regulation of the cdk inhibitor p27 and its deregulation in cancer. J Cell Physiol. 2000;183:10–7. doi: 10.1002/(SICI)1097-4652(200004)183:1<10::AID-JCP2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 31.Viglietto G, Motti ML, Fusco A. Understanding p27(kip1) deregulation in cancer: down-regulation or mislocalization. Cell Cycle. 2002;1:394–400. doi: 10.4161/cc.1.6.263. [DOI] [PubMed] [Google Scholar]
- 32.Liang J, Zubovitz J, Petrocelli T, et al. PKB/Akt phosphorylates p27, impairs nuclear import of p27 and opposes p27-mediated G1 arrest. Nat Med. 2002;8:1153–60. doi: 10.1038/nm761. [DOI] [PubMed] [Google Scholar]
- 33.Shin I, Yakes FM, Rojo F, et al. PKB/Akt mediates cell-cycle progression by phosphorylation of p27(Kip1) at threonine 157 and modulation of its cellular localization. Nat Med. 2002;8:1145–52. doi: 10.1038/nm759. [DOI] [PubMed] [Google Scholar]
- 34.Viglietto G, Motti ML, Bruni P, et al. Cytoplasmic relocalization and inhibition of the cyclin-dependent kinase inhibitor p27(Kip1) by PKB/Akt-mediated phosphorylation in breast cancer. Nat Med. 2002;8:1136–44. doi: 10.1038/nm762. [DOI] [PubMed] [Google Scholar]
- 35.She QB, Solit D, Basso A, Moasser MM. Resistance to gefitinib in PTEN-null HER-overexpressing tumor cells can be overcome through restoration of PTEN function or pharmacologic modulation of constitutive phosphatidylinositol 3′-kinase/Akt pathway signaling. Clin Cancer Res. 2003;9:4340–6. [PubMed] [Google Scholar]
- 36.Bianco R, Shin I, Ritter CA, et al. Loss of PTEN/MMAC1/TEP in EGF receptor-expressing tumor cells counteracts the antitumor action of EGFR tyrosine kinase inhibitors. Oncogene. 2003;22:2812–22. doi: 10.1038/sj.onc.1206388. [DOI] [PubMed] [Google Scholar]
- 37.Kokubo Y, Gemma A, Noro R, et al. Reduction of PTEN protein and loss of epidermal growth factor receptor gene mutation in lung cancer with natural resistance to gefitinib (IRESSA) Br J Cancer. 2005;92:1711–9. doi: 10.1038/sj.bjc.6602559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodier G, Montagnoli A, Di Marcotullio L, et al. p27 cytoplasmic localization is regulated by phosphorylation on Ser10 and is not a prerequisite for its proteolysis. EMBO J. 2001;20:6672–82. doi: 10.1093/emboj/20.23.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boehm M, Yoshimoto T, Crook MF, et al. A growth factor-dependent nuclear kinase phosphorylates p27(Kip1) and regulates cell cycle progression. EMBO J. 2002;21:3390–401. doi: 10.1093/emboj/cdf343. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.